Abstract
Dysregulated Protein Arginine Deiminase (PAD) activity, particularly PAD4, has been suggested to play a role in the onset and progression of numerous human diseases, including Rheumatoid Arthritis (RA). Given the potential role of PAD4 in RA, we set out to develop inhibitors/inactivators that could be used to modulate PAD activity and disease progression. This effort led to the discovery of two mechanism-based inactivators, denoted F- and Cl-amidine, that inactivate PAD4 via the covalent modification of an active site cysteine that is critical for catalysis. To gain further insights into the mechanism of inactivation by these compounds, the effect of pH on the rates of inactivation were determined. These results, combined with the results of solvent isotope effect and proton inventory studies, strongly suggest that the inactivation of PAD4 by F- and Cl-amidine proceeds via a multi-step mechanism that involves the protonation and stabilization of the tetrahedral intermediate formed upon nucleophilic attack by the active site cysteine, i.e. Cys645. Stabilization of this intermediate would help to drive the halide-displacement reaction, which results in the formation of a three-membered sulfonium ring that ultimately collapses to form the inactivated enzyme. This finding - that protonation of the tetrahedral intermediate is important for enzyme inactivation - may also suggest that during catalysis, protonation of the analogous intermediate is required for efficient substrate turnover.
Keywords: Deiminase, Citrulline, Cl-amidine, rheumatoid arthritis, inactivator
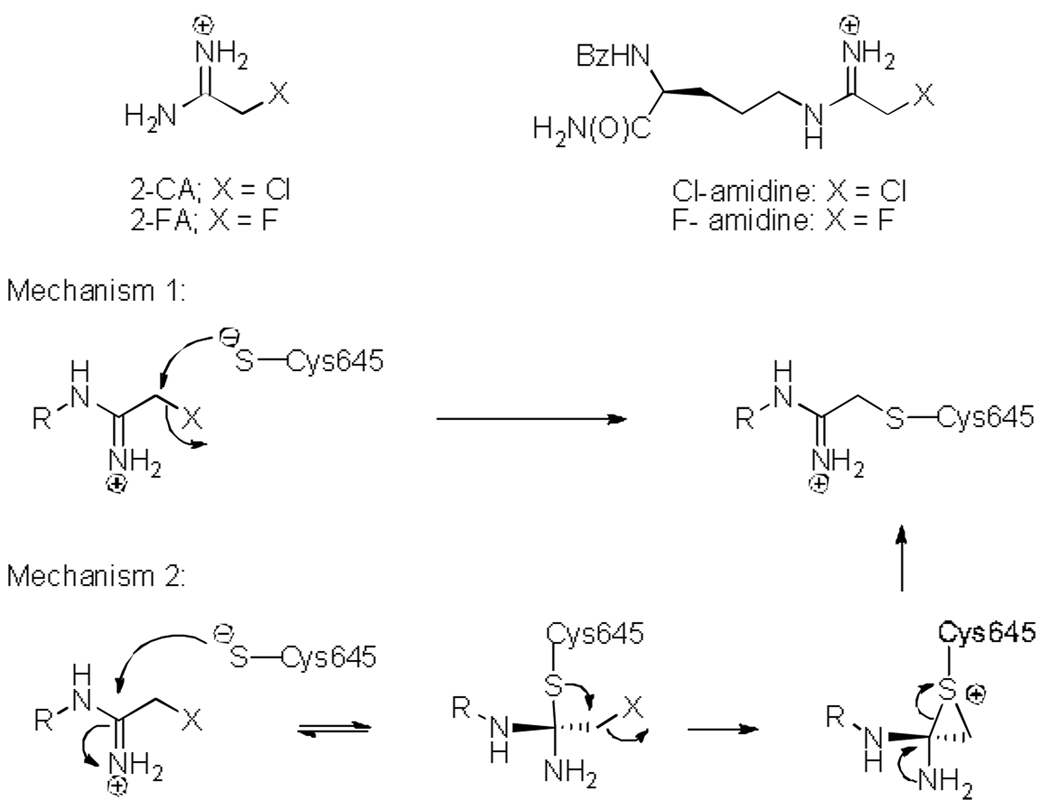
In nature, a myriad of posttranslational modifications are found in proteins. These modifications, and the requisite modifying enzymes, can have far-reaching effects on living systems. Within the family of protein modifying enzymes are the Protein Arginine Deiminases (PADs). These enzymes catalyze the hydrolysis of arginine residues to form citrulline.[1–3] Much effort from our lab has been focused on gaining insight into the mechanism of the PADs, and, in particular, PAD4. [3–5] Our interest in PAD4, and the PADs in general, was piqued as evidence linking dysregulated PAD activity to the increased incidence and severity of Rheumatoid Arthritis (RA) emerged.[2, 3, 6] This disease, which afflicts nearly 1% of the population, is an autoimmune disorder that appears to be triggered, at least in part, in response to aberrant citrullination, a result of dysregulated PAD activity. Based on this apparent causal relationship, we set out to develop inhibitors/inactivators that could be used to modulate PAD activity and disease progression. This effort led to the discovery of two mechanism-based inactivators, denoted F- and Cl-amidine (Figure 1), which are the most potent PAD inhibitors described to date.[7, 8] At the same time, Fast and colleagues reported that 2-chloroacetamidine, i.e. the warhead in Cl-amidine, also inactivates PAD4, and other members of the guanidinium modifying family of enzymes.[9]
Figure 1.
Structures of compounds described in this communication and proposed mechanisms of PAD4 inactivation by Cl- or F-amidine.
Much work has been done to characterize the mechanism of inactivation, including dialysis experiments verifying irreversible inhibition. Additionally, analysis of the kinetics of inactivation demonstrated that F- and Cl-amidine possess kinact/KI values of 3,000 M−1min−1 and 13,000 M−1min−1, respectively.[7, 8] Subsequent crystallographic data confirmed that inactivation was due to the covalent modification of an active site cysteine (Cys645) that is critical for catalysis – this residue promotes arginine deimination by covalent catalysis using a mechanism that is analogous to the Cys proteases.[5] While this finding conclusively demonstrated the mode of inactivation – alkylation of the thiolate – the precise mechanism of inactivation has yet to be established.
While unclear, the mechanism of inactivation presumably proceeds through one of at least two routes: direct displacement of the halogen via an SN2 mechanism or initial attack on the imminium carbon, followed by displacement of the halide to form a sulfonium ring, and ending with concomitant reformation of the imine and opening of the sulfonium ring (Figure 1). While the former possibility is the most intuitive, the latter is analogous to the mechanism by which the fluoromethylketones inactivate the cysteine proteases[10–12] and therefore warranted further investigation.
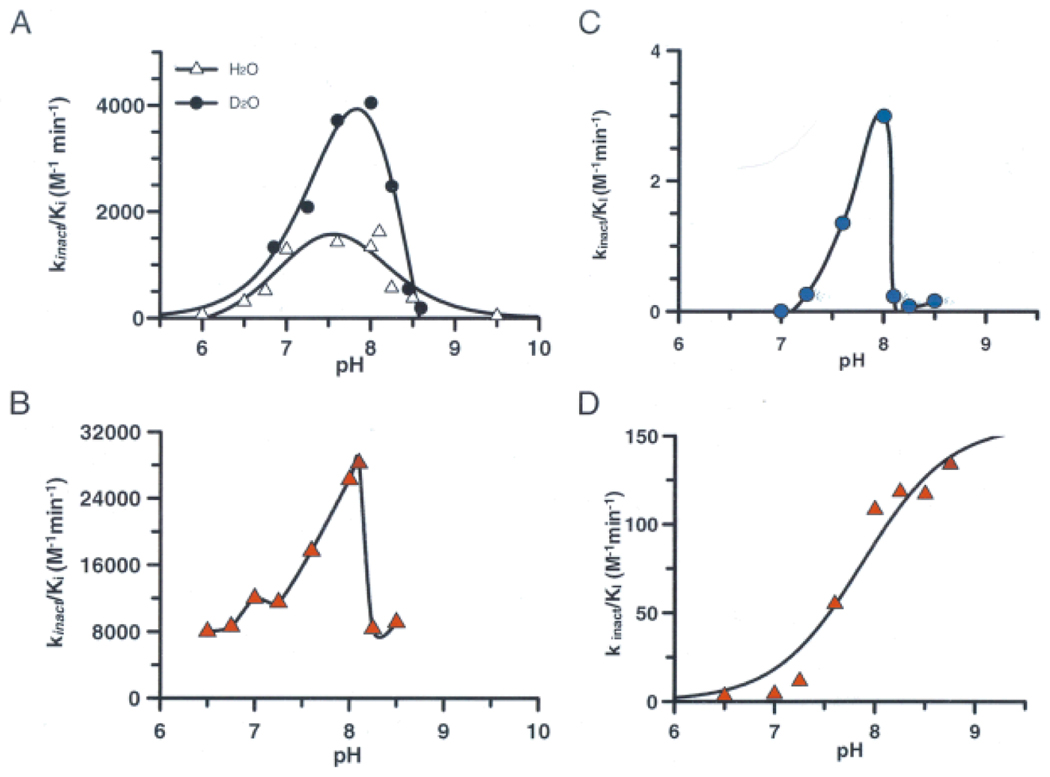
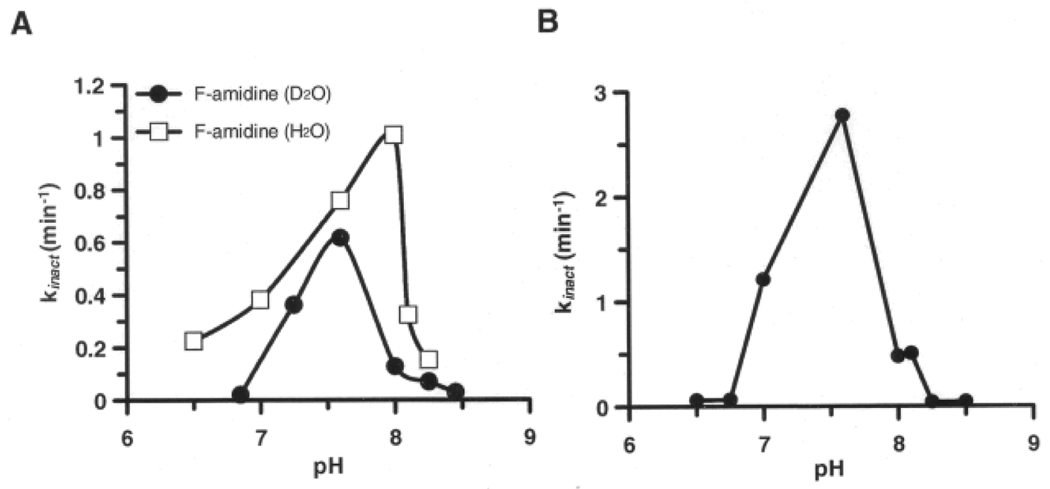
We began our investigations by examining the influence of pH on kincat/KI, i.e. the second order rate constant of enzyme inactivation. These studies were pursued because pH rate profiles can suggest the identity of catalytically important functional groups, in the inactivator or free enzyme, up to and including the first irreversible step of the reaction. For these studies, kincat/KI values were determined for both Cl- and F-amidine over a pH range of 6.0–8.5. Interestingly, the rates of inactivation increased until ~ pH 8.1, after which point further increases in alkalinity resulted in a dramatic drop in the rates of inactivation (Figure 2A and 2C).
Figure 2.
Inactivation pH profiles of Cys645 using A) F-amidine B) Cl-amidine C) 2-fluoroacetamidine and D) 2-chloroacetamidine.
Fits of the F-amidine versus pH rate data give estimates of the pKa values for the ascending and descending limbs of 7.05 and 8.05, respectively, with a pH independent value of 1425 M−1min−1. Interestingly, these values are in reasonable agreement with those obtained for substrate turnover (i.e., 7.3 and 8.2), and, as such, they likely correspond to the pKas of His471 and Cys645, respectively. Given that His471 is thought to act as a general acid in the acylation step of the reaction, and that this drop in reactivity mimics the pH profile for substrate turnover,[5] this data suggests that general acid catalysis also contributes to enzyme inactivation by F-amidine. Although the data for Cl-amidine fit poorly to canonical pH versus rate equations, the trends of the pH rate profiles are similar (i.e., decreased inactivation at pH values >8), thereby suggesting that general acid catalysis also plays a role in the inactivation of PAD4 by this compound.
To gain further insight into this process, the effect of pH on PAD4 inactivation by 2-fluoroacetamidine (FA) – the fluoroacetamidine warhead alone – was examined. As with Cl- and F-amidine, the rates of inactivation are roughly bell-shaped (Figure 2C). In contrast, the rate at which 2-chloroacetamidine (CA) inactivates PAD4 does not decrease with increasing alkalinity (Figure 2D).[5] This break from the trend suggests that CA inactivates PAD4 via a mechanism that is different from the mechanism by which F-amidine, Cl-amidine, and FA inactivate the enzyme. Given that chloride is an inherently better leaving group than fluoride, the most reasonable assumption is that inactivation by CA can proceed through the direct SN2 displacement of chloride, i.e. mechanism 1 in Figure 1. Based on the fact that general acid catalysis is unlikely to enhance the rate of this reaction, this assumption is not only plausible, but likely.
The break in the trend for CA also argues, albeit indirectly, that PAD4 inactivation by Cl-amidine, F-amidine, and FA proceeds through the proposed multi-step mechanism, i.e. mechanism 2. This argument, however, does not explain why the chloride of Cl-amidine is not directly displaced. The simplest and most reasonable hypothesis to explain this discrepancy is that CA, lacking any peripheral structural elements, has far fewer limitations on its orientation within the active site. As such, this molecule can inactivate PAD4 via a direct SN2 displacement that is solely dependent on the concentration of the thiolate. By contrast, the “backbone” amide groups of Cl-amidine are instrumental in binding,[4, 7, 8, 13] and therefore restrict conformational freedom, and thus lead to a mode of binding that is not conducive to an SN2 displacement.
While this hypothesis rationalizes the observed differences in the kinetics of inactivation, it does not directly explain the observed decrease in inactivation rate at higher pH. Although we have suggested that this drop in reactivity suggests a role for general acid catalysis, alternative explanations for the observed drop in reactivity include deprotonation of the warhead and or decomposition of the inhibitors.
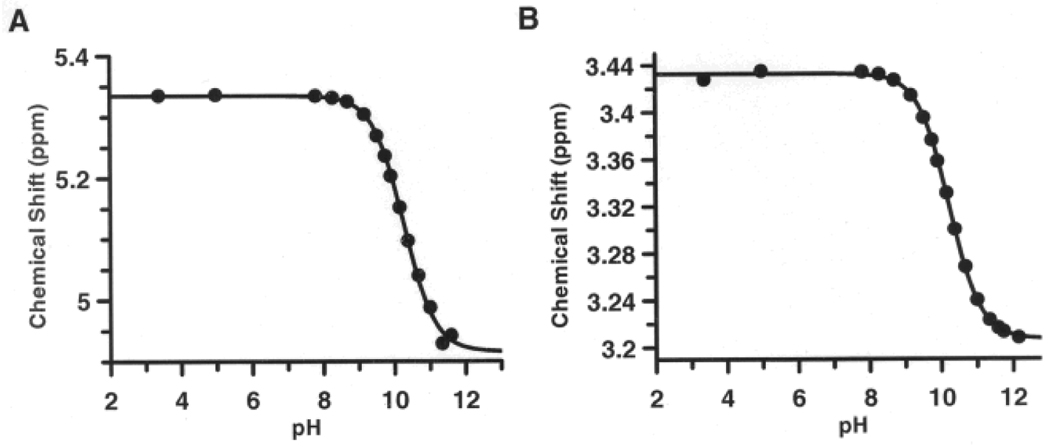
Warhead deprotonation would be expected to decrease the rate of inactivation because the positive charge of the protonated arginine side chain is crucial for substrate binding and catalysis.[4] To investigate this possibility, the pKa of the fluoroacetamidine warhead in F-amidine was determined by NMR. By monitoring the chemical shifts of the protons that are in closest proximity to the amidine, i.e. the protons adjacent to the fluoride and the protons on the δ-carbon, the pKa of the warhead was found to be 10.2 (Figure 3). Virtually identical results were obtained by fluorine NMR (Figure S1). While this pKa is significantly lower than that of arginine (pKa = 12.5), it is high enough to ensure complete protonation in the pH regime of our experiments.
Figure 3.
NMR titration curves: (A) protons adjacent to the fluoride (B) protons on the δ-carbon.
As described above, the dramatic loss in inactivator potency could be due to inhibitor decomposition at higher pHs. To test this possibility, samples of the inhibitors were subjected to reaction conditions, in the absence of enzyme, and subsequently analyzed by analytical HPLC. Essentially no decomposition was observed (Figure S2). This possibility was further ruled out by a slight modification to our assay procedure. Typically, the inactivator is incubated in assay buffer for 10 min prior to the addition of PAD4 to initiate the inactivation reaction. Decreasing this time to 2 min did not affect the rate of inactivation (data not shown). These results clearly rule out decomposition as the reason for the diminished potency.
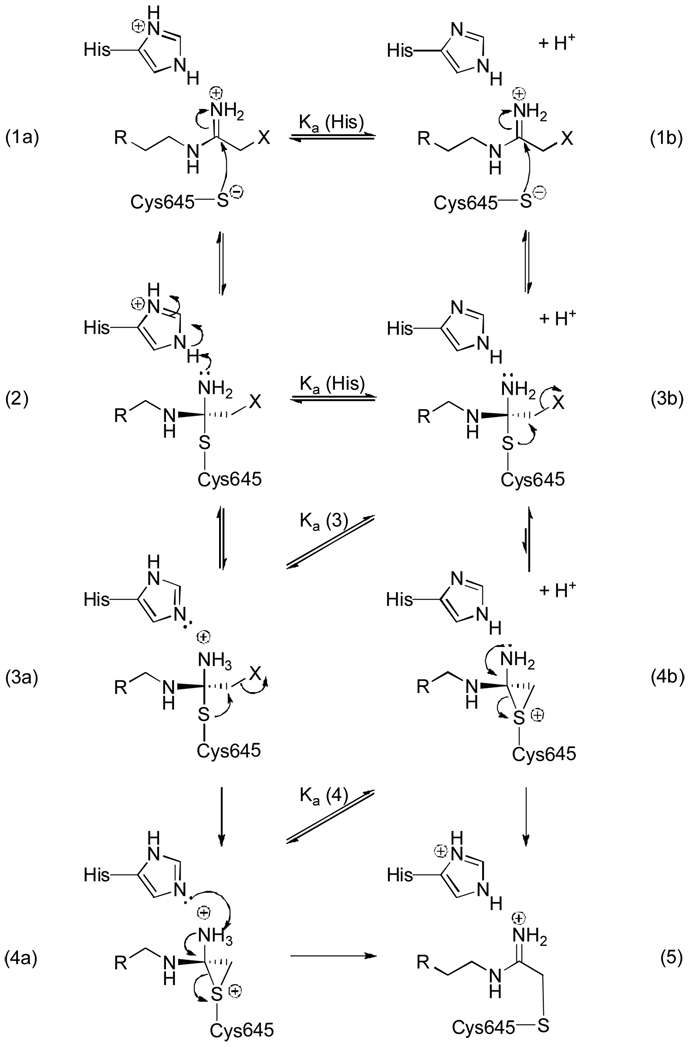
Having ruled out inactivator deprotonation and decomposition as reasons for the sharp decline in inactivation rates above pH 8.0, the most reasonable explanation for the observed effect on kinact/KI, which reports on ionizable groups in the free enzyme, is the presence of a protonated active site residue that is important for binding and orienting the warhead for nucleophilic attack by the active site thiol. If, as we expect (see below), this group represents the imidazolium form of His471, it could polarize the acetamidine, fix charge on one of the two NH2 groups, and thereby promote reaction with the active site thiolate.
To gain further insights into this process, kinact values were determined for F- and Cl-amidine as a function of pH. These experiments were performed to identify catalytically important functional groups in the enzyme●inactivator complex. Additionally, we reasoned that for mechanism 1, the simple SN2 mechanism, the plots of kinact versus pH should rise with increasing pH plateauing at a limiting value that is dependent on the concentration of the active site thiolate. In contrast, for the more elaborate mechanism depicted in Figure 4, we reasoned that if general acid catalysis plays a role in promoting enzyme inactivation after the inactivator has bound the enzyme, then kinact would rise to a limiting value and then fall as the pH is increased. Note that kinact values were not determined for FA and CA because the high KI values for these compounds made the determination of kinact values impractical. Although these data fit poorly to canonical pH versus rate equations, the results of these studies demonstrated that the plots of kinact versus pH plots are also bell-shaped (Figure 5), with the groups contributing to the overall shape of the curve possessing an average pKa of 7.5. As described above, these results further discount the simple SN2 mechanism. Additionally, the drop in reactivity at the higher pH values indicates that general acid catalysis promotes enzyme inactivation after the formation of the initial enzyme●inactivator complex. These results also indicate that the decrease in kinact is the major contributor to the observed effect on kincat/KI, indirectly arguing against a pH effect on inactivator binding.
Figure 4.
Proposed general acid catalyzed mechanism of inactivation.
Figure 5.
Rates of PAD4 inactivation by A) F-amidine in H2O and D2O and B) Cl-amidine in H2O.
The observed effect on kinact is again consistent with a role for general acid catalysis in enzyme inactivation, specifically in the enzyme●inactivator complex. Based on the proposed mechanism of catalysis (Figure S3), the most likely residue is His471, which serves to protonate the departing ammonia through the course of catalysis. Although we and others have suggested that, for substrate turnover, proton donation occurs during the collapse of the tetrahedral intermediate,[1, 14] it is equally plausible that protonation of the intermediate occurs during the formation of this intermediate in either in a stepwise fashion or concomitant with the change in hybridization that occurs upon thiolate attack on the guanidinium carbon. If either of the latter possibilities are correct, the above described inactivation data would imply a role for this residue in stabilizing the tetrahedral intermediate formed during the inactivation reaction (Figure 4).
The pKa of His471 has been assigned to be 7.3;[5] thus at higher pH, this residue is unprotonated and cannot donate a proton to stabilize the tetrahedral intermediate formed upon thiolate attack on the iminium carbon (3b; Figure 4). Without this stabilization, which is found when the histidine is protonated, the equilibrium is greatly shifted towards form 1b. In contrast, by stabilizing the tetrahedral intermediate, the displacement of the halide (3a) would be favored. The protonation state of the intermediate (3) is also likely influenced by the increasing pH. Based on model compounds, the pKa of this intermediate is expected to be 2 units lower than the parent compound. Thus, at pH > 8, a similar decline in reactivity would be expected. These overlapping effects, i.e. loss of stabilization from the protonated histidine (2) and deprotonation of intermediate (3), may account for the sharp declines in the rates of inactivation that are observed above pH 8.
Further evidence to support the proposed mechanism of inactivation comes from measurements of the deuterium solvent isotope effect (SIE). For these studies, we focused our efforts on F-amidine, as opposed to CA or Cl-amidine, because this compound can presumably only inactivate the enzyme via the second mechanism, and the KI values are low enough to facilitate the measurement of kinact values.
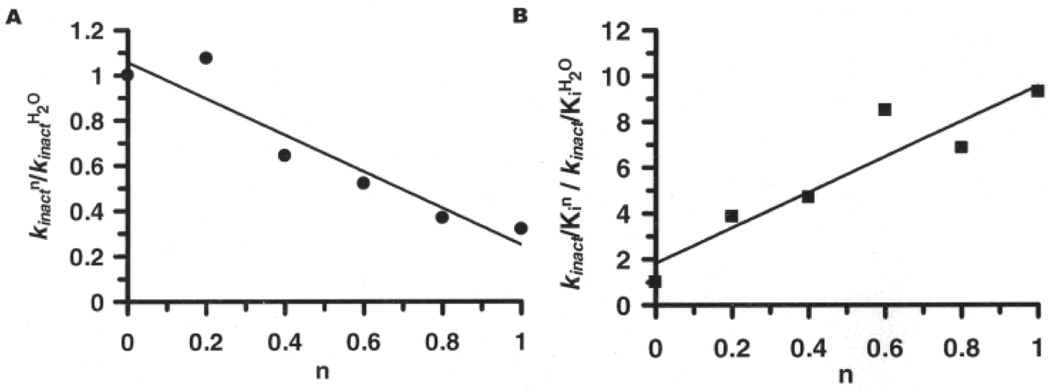
The average of two independent experiments indicate that the SIE on kinact is normal (kinactH/kinactD = 2.17±0.94; Figures 5 and 6), whereas for kinact/KI, the SIE was inverse over the entire pL range studied (kinact/KIH/kinact/KID = 0.24±0.14; at the pL optimum (Figures 2 and 6)). The normal SIE on kinact is suggestive of a proton in flight in the transition state. Such an effect is consistent with the proposed mechanism of inactivation, wherein proton donation to the tetrahedral intermediate would be expected to yield a normal isotope effect. Consistent with this possibility is the fact that proton inventory analyses performed at the pL optimum are linear (r2 = 0.947), which is consistent with a single hydrogenic site in the isotope sensitive step of enzyme inactivation (Figure 6).
Figure 6.
Protonic Inventories obtained for the F-amidine induced inactivation of PAD4 A) kinact and B) kinact/Ki. The term n refers to the fraction of D2O in the reaction. Data was obtained at pL = 7.6.
The large and inverse SIE observed for kinact/KI is consistent with the inverse SIE obtained for substrate turnover,[5] further suggesting that similarities exist between the mechanism of substrate turnover and enzyme inactivation. As previously described for substrate turnover,[5] this large inverse SIE could be explained by pre-equilibrium ionization of the thiol to form the thiolate. Consistent with this possibility is the fact that while the displacement of the pKa is expected to be ~0.4–0.6 pH units higher in D2O compared to H2O, the observed difference (~0.2) is smaller; such smaller differences are highly suggestive of ionization of a thiol to the thiolate. Also consistent with a single ionization of an active site residue is the fact that the proton inventory for kinact/KI is linear (Figure 6B; r2 = 0.931).
In conclusion, the data strongly suggest that the inactivation of PAD4 by F- and Cl-amidine proceeds via the multi-step mechanism, i.e. mechanism 2. Our data further suggests that protonation of the tetrahedral intermediate (3; Figure 4) helps to drive the halide-displacement to form the three-membered sulfonium ring (4), which ultimately collapses to form the inactivated enzyme (5). This finding - that protonation of the tetrahedral intermediate is important for enzyme inactivation - may also suggest that, during catalysis, protonation of the analogous intermediate is required for efficient substrate turnover.
Experimental Section
Chemicals: Dithiothreitol (DTT), benzoyl L-arginine ethyl ester (BAEE), Tris- (2-carboxyethyl)phosphine hydrochloride (TCEP) were purchased from Sigma-Aldrich (St. Louis, MO). F-amidine and Cl-amidine were synthesized as previously described.[7, 8] 2-chloroacetamidine was obtained from Oakwood Products (Columbia, SC). Protein Arginine Deiminase 4 (PAD4) was purified as previously described.[5]
Inactivation Kinetics: Inactivation experiments with F-amidine, Cl-amidine, and 2-fluoroacetamidine were performed as previously described.[5] Data were fit to equations described previously.[5] The inactivation of PAD4 by 2-chloroacetamidine was previously described.[5] The inactivation rates obtained for F-amidine, Cl-amidine, and 2-fluoroacetamidine from this analysis were plotted against pH. Given that the dramatic decrease in activity at pH values > 8 precluded fitting of the data to standard equations, the curves of 2-fluoroacetamidine and Cl-amidine were simply generated using the line function in GraFit™ (version 5.0.11) [15]. The fit for the F-amidine data was determined graphically.
Solvent Isotope Effects: These experiments were performed as previously described,[5] except that the reaction was carried out in > 95% D2O. Data was fit to equations described previously.[5]
Proton Inventory Studies: Briefly, the experiments were performed as described previously for the inactivation kinetic experiments.[5] The only difference was that inactivation reactions were performed in the presence of various D2O concentrations (0–95%). The data collected was processed using the methods described above. kinact and kinact/KI values were obtained and plotted as a ratio of D2O to H2O as a function of D2O concentration (n). The resulting plot was fit using the line function in Grafit™ (version 5.0.1.1).[15]
Synthesis of 2-fluoroacetamidine: To a solution of 2-fluoro-ethylthioimidate (250 mg, 1.6 mmol) in dry ethanol (10 mL) at 0 °C, was added ammonia (1.6 mL, 2M in MeOH). The reaction was allowed to warm to rt. After stirring for 16 h, the volatiles were removed under reduced pressure to afford a white powder that contained both the title compound and ammonium chloride. The final composition of this mixture was determined by NMR using an internal standard. 1H NMR (300 MHz, D2O): δ 5.19 (d, JH-F = 44.7 Hz, 2H). 13C NMR (100 MHz, D2O): δ 166.12 (d, JC-F = 19.9 Hz), 77.49 (d, JC-F = 178.3 Hz). 19F NMR (379 MHz, D2O): δ −233.67 (t, JH-F = 44.8 Hz). HRMS-ESI (C2H6FN2+): calcd 77.0515, observed 77.0526.
NMR titration: NMR titrations were conducted using a 400 MHz NMR in H2O (5% D2O). Proton chemical shifts were relative to DSS and fluoride chemical shifts were relative to TFA (internal tube). Upon dissolution of F-amidine, the pH was measured using a micro-pH meter and the first spectra were taken. For subsequent measurements, the sample was transferred to a clean vial where the pH was adjusted with NaOH (1M) before being transferred back to the NMR tube. Changes in chemical shift values were plotted against pH. pKa values were determined by fitting the data to equation 1,
| (1) |
using GraFit™ (version 5.0.11).[15] Lim1 is the minimum rate and Lim2 is the maximum rate.
Decomposition experiments: To investigate the effect of the reactions conditions on Cl-amidine and F-amidine, these compounds were subjected to the assay conditions in the absence of enzyme. Briefly, each inhibitor was dissolved in assay buffers (pH 6.5, 7.0, 7.5, 8.0, and 8.5) and incubated at 37 °C. After 30 min, the samples were flash frozen in liquid nitrogen. Each sample was analyzed by reverse phase (C18) analytical HPLC. The C18 column was pre-equilibrated with ddH2O/0.05%TFA and the reaction components were separated using a linear gradient of acetonitrile/0.05%TFA.
Footnotes
This work was supported by NIH grant GM079357 to P.R.T. and by the Grayce B. Kerr Endowment to the University of Oklahoma and to P.F.C.. The authors thank Philip Cole for critical reading of the manuscript.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Paul F. Cook, Email: pcook@ou.edu.
Paul R. Thompson, Email: Thompson@mail.chem.sc.edu.
References
- 1.Thompson PR, Fast W. ACS Chem Biol. 2006;1:433. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 2.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. Bioessays. 2003;25:1106. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 3.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Curr Opin Drug Discov Devel. 2009;12:616. [PMC free article] [PubMed] [Google Scholar]
- 4.Kearney PL, Bhatia M, Jones NG, Luo Y, Glascock MC, Catchings KL, Yamada M, Thompson PR. Biochemistry. 2005;44:10570. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 5.Knuckley B, Bhatia M, Thompson PR. Biochemistry. 2007;46:6578. doi: 10.1021/bi700095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vossenaar ER, Van Venrooij WJ. Arthritis Res. Ther. 2004;6:107. doi: 10.1186/ar1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo Y, Arita K, Bhatia M, Knuckley B, Lee YH, Stallcup MR, Thompson PR. Biochemistry. 2006;45:11727. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Y, Knuckley B, Lee YH, Stallcup MR, Thompson PR. J Am Chem Soc. 2006;128:1092. doi: 10.1021/ja0576233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stone EM, Schaller TH, Bianchi H, Person MD, Fast W. Biochemistry. 2005;44:13744. doi: 10.1021/bi051341y. [DOI] [PubMed] [Google Scholar]
- 10.Powers JC, Asgian JL, Ekici OD, James KE. Chem. Rev. 2002;102:4639. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 11.Kreutter K, Steinmetz AC, Liang TC, Prorok M, Abeles RH, Ringe D. Biochemistry. 1994;33:13792. doi: 10.1021/bi00250a033. [DOI] [PubMed] [Google Scholar]
- 12.Drenth J, Kalk KH, Swen HM. Biochemistry. 1976;15:3731. doi: 10.1021/bi00662a014. [DOI] [PubMed] [Google Scholar]
- 13.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Nat. Struct. Mol. Biol. 2004;11:777. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 14.Stone EM, Costello AL, Tierney DL, Fast W. Biochemistry. 2006;45:5618. doi: 10.1021/bi052595m. [DOI] [PubMed] [Google Scholar]
- 15.Leatherbarrow RJ. Erathicus Software. Staines, UK: 2004. [Google Scholar]