Abstract
Two transcription factor families that are activated during multiple conditions of skeletal muscle wasting are nuclear factor κB (NF-κB) and forkhead box O (Foxo). There is clear evidence that both NF-κB and Foxo activation are sufficient to cause muscle fiber atrophy and they are individually required for at least half of the fiber atrophy during muscle disuse, but there is no work determining the combined effect of inhibiting these factors during a physiological condition of muscle atrophy. Here, we determined whether inhibition of Foxo activation plus inhibition of NF-κB activation, the latter by blocking the upstream inhibitor of kappaB kinases (IKKα and IKKβ), would prevent muscle atrophy induced by 7 days of cast immobilization. Results were based on measurements of mean fiber cross-sectional area (CSA) from 72 muscles transfected with 5 different mutant expression plasmids or plasmid combinations. Immobilization caused a 47% decrease in fiber CSA in muscles injected with control plasmids. Fibers from immobilized muscles transfected with dominant negative (d.n.) IKKα-EGFP, d.n. IKKβ-EGFP or d.n. Foxo-DsRed showed a 22%, 57%, and 76% inhibition of atrophy, respectively. Co-expression of d.n. IKKα-EGFP and d.n. Foxo-DsRed significantly inhibited 89% of the immobilization-induced fiber atrophy. Similarly, co-expression of d.n. IKKβ-EGFP and d.n. Foxo-DsRed inhibited the immobilization-induced fiber atrophy by 95%. These findings demonstrate that the combined effects of inhibiting immobilization-induced NF-κB and Foxo transcriptional activity has an additive effect on preventing immobilization-induced atrophy, indicating that NF-κB and Foxo have a cumulative effect on atrophy signaling and/or atrophy gene expression.
Keywords: muscle wasting, NF-κB signaling, Foxo signaling, inhibitor of kappaB kinase
Introduction
Various diseases, such as HIV, cancer, and chronic heart failure, and physiological conditions of muscle disuse, such as unloading, denervation and immobilization result in significant skeletal muscle atrophy. This atrophy leads to muscle weakness, increased fatigability and therefore significantly compromised muscle function. While the triggers that initiate muscle atrophy in each of these conditions vary, signaling pathways that are commonly activated during multiple conditions of muscle wasting induce at least two families of transcription factors: the nuclear factor κB (NF-κB) and forkhead box O (Foxo) [1; 2; 3; 4; 5].
Active forms of the mammalian NF-κB family of transcription factors consist of homo- or heterodimers with various combinations of RelA (p65), RelB, c-Rel, p100/p52 and p105/p50. NF-κB dimers are found in all cell types studied and in unstimulated cells the dimers are mostly retained in the cytosol [6]. A variety of stimuli induce dimer translocation to the nucleus. Binding of dimers to κB sites in target genes often induce, but sometimes repress (p50 or p52 homodimers), gene transcription [7]. This nuclear translocation is commonly regulated by two upstream kinases, inhibitor of kappa B kinase alpha (IKKα) and IKKβ. NF-κB signaling and/or NF-κB transcriptional activity is now known to increase in multiple models of skeletal muscle atrophy, including immobilization [5], unloading [4; 8; 9], cancer cachexia [1], and denervation [1]. This is significant since NF-κB activation alone is sufficient to cause skeletal muscle fiber atrophy [1; 8]. Moreover, inhibition of NF-κB activation during either disuse or cancer cachexia prevents at least 50% of the muscle fiber atrophy, demonstrating that NF-κB activation is necessary for muscle atrophy in these models [1; 8; 10].
In addition to NF-κB, the Foxo family of transcription factors also plays a significant role in regulating muscle atrophy. Although there are four Foxo factors in mammalian cells, only three are present in skeletal muscle: Foxo1, Foxo3a, and Foxo4. The mRNA expression of Foxo1 and Foxo3a is up-regulated in skeletal muscle during conditions of fasting, cancer cachexia, disuse and aging [2; 11; 12; 13], and an increase in either Foxo1 or Foxo3a is alone sufficient to cause atrophy in normal skeletal muscle [3; 14]. Furthermore, we recently demonstrated that inhibition of Foxo transcriptional activity during muscle disuse prevents almost half of the muscle fiber atrophy [12], identifying the requirement of Foxo activation for normal muscle atrophy during a physiological condition.
While it is clear that both Foxo and NF-κB activation are sufficient to cause skeletal muscle fiber atrophy and are individually required for at least 50% of the muscle fiber atrophy during muscle disuse, there has been no work determining the combined effect of inhibiting these two pathways during any physiological condition of skeletal muscle atrophy. Therefore, in the current study we inhibited IKK-mediated activation of NF-κB activity and/or Foxo transcriptional activity during cast immobilization to determine if the inhibition of both pathways blocks muscle fiber atrophy to a greater extent than blocking either pathway alone. To do this we used plasmid DNA electrotransfer of 5 different mutant expression plasmids or plasmid combinations into a total of 72 muscles in vivo. These plasmids encoded d.n. IKKα, d.n. IKKβ and d.n.Foxo proteins each fused to fluorescent proteins to allow for their visualization in muscle fibers. The fusion plasmids were constructed and tested by us, either in the current study or previously [8], to allow for the direct assessment of whether endogenous IKK-mediated NF-κB activation and/or Foxo activation is required for muscle fiber atrophy during skeletal muscle disuse. Thus, the impact and innovation of this work is in: 1) the construction of multiple sophisticated expression plasmids used singly and in combination, and 2) the successful use of these mutant, fluorescent expression proteins in vivo, during a physiological condition. This is a powerful approach to test the difficult question regarding the requirement of these proteins, alone and in combination, for muscle atrophy without the use of transgenic mice.
Materials and Methods
Animals
Male Sprague Dawley rats (200 g), purchased from Charles River Laboratories (Wilmington, MA) were used for all experiments. Animals were housed at the University of Florida and all protocols were approved by the University of Florida Institutional Animal Care and Use Committee.
Plasmids
The d.n.Foxo3a was a gift of Dr. Paul Coffer (University Medical Center, Utrecht, The Netherlands) [15]. The d.n. IKKα-EGFP and d.n. IKKβ-EGFP fusion plasmids were constructed by us and described previously [8]. The DAF16 (FOXO) reporter was obtained from Dr. Alex Toker (Beth Israel Deaconess Medical Center, Boston, MA) [16]. The d.n. Foxo3a consists of only the DNA-binding domain of Foxo3a (amino acids 141–266). However, due to the significant homology between the Foxo-DNA binding domains of the Foxo factors, d.n. Foxo3a likely represses Foxo1 and Foxo4, in addition to Foxo3a [3]. We therefore refer to this construct as d.n. Foxo. In order to visualize the ectopic d.n. Foxo separate from the ectopic IKK-EGFP proteins we constructed a d.n. Foxo-DsRed fusion plasmid by PCR amplifying the d.n. Foxo3a sequence from pSG5 and subcloning it, in frame, into the Hind III and Sal I sites of DsRed2-c1 (Clontech, Palo Alto, CA, USA). Verification of the appropriate fusion sequence was performed by DNA sequencing at the University of Florida DNA Sequencing Core facility. The empty vectors DsRed2-c1, EGFP-c1 and pCMV (Clontech) were used as control plasmids.
Electroporation
Plasmid was injected into the soleus muscle and electrotransfered as previously described [5]. In brief, an incision was made on the lateral side of the lower leg and the soleus muscle exposed. The soleus was injected with a total of 100 µg DNA in 50 µl 1×PBS. Plasmids were electrotransferred using 5 pulses, 20ms duration at 100V/cm. The plasmid quantities used were as follows: 25 µg EGFP-c1, 25 µg DsRed2-c1, 40 µg d.n. Foxo-DsRed, 60 µg d.n. IKKα-EGFP, and 60 µg d.n. IKKβ-EGFP. When the amount of expression plasmid(s) injected did not equal 100 µg, pCMV was used to supplement such that all muscles in all groups received 100 µg total DNA. The rationale for using less Foxo expression plasmid compared to IKK expressing plasmids was to maintain plasmid copy number, and hence the amount of protein overexpressed, to similar molar amounts. When used, 50 µg of DAF16 (FOXO) reporter was co-injected with 40 µg of d.n. Foxo-DsRed.
Immobilization
To achieve disuse muscle atrophy, the hind limbs of rats were immobilized in plaster casts in a plantar flexed position as previously described [5]. For atrophy experiments, limbs were immobilized for seven days; for the FOXO reporter activity experiment, limbs were immobilized for three days.
Reporter Plasmid Activity
To test the function of the d.n. Foxo-DsRed fusion protein on Foxo reporter activity, muscles were homogenized in passive lysis buffer (Promega, Madison, WI, USA) and luciferase activity measured according to standard protocol, as previously described [5].
Histology
Muscles were sectioned through the mid-belly (10 µm) with a Microm HM 550 Cryostat (Microm International, Walldorf, Germany). Sections were fixed for 20 minutes in 4% paraformaldehyde followed by extensive washing in phosphate buffered saline (PBS) and then incubated with Alexa Fluor 350 conjugated wheat germ agglutinin (Invitrogen, Carlsbad, CA, USA) for two hours in the dark. Hoescht 33342 dye was used in additional sections to visualize nuclei. Images were captured using an Olympus IX50 camera. To measure cross sectional area (CSA), transfected fibers were traced and measured using Image Pro Discovery software (Media Cybernetics, Bethesda, MD, USA).
Western Blotting
Frozen muscles were homogenized in passive lysis buffer (Promega, Madison, WI, USA) and centrifuged for 10 minutes at 4°C, as previously described [8]. Protein concentrations were determined using a detergent compatible assay from Bio-Rad (Hercules, CA, USA). 100 micrograms of protein were separated on SDS-polyacrylamide gels and transferred to Immobilon-FL polyvinylidene fluoride membrane (Millipore, Bedford, PA, USA) as previously described [8]. Membranes were blocked in Odyssey blocking buffer (Li-Cor Biosciences, Lincoln, NE, USA), and incubated with primary antibody (IKKα antibody sc-7182, Santa Cruz Biotechnology, Santa Cruz, CA; IKKβ Antibody c.s. 2684, Cell Signaling Technology, Danvers, MA; or DsRed antibody sc-33354 Santa Cruz Biotechnology, Santa Cruz, CA) according to the manufacturer’s instructions. Anti-GAPDH antibody (S9545, Sigma-Aldrich, St. Louis, MO) was used as a loading control. Alexa Fluor 680 fluorescent dye-conjugated secondary antibodies (Invitrogen, Carlsbad, CA) were used for visualization with the Li-Cor Odyssey fluorescent detection system as described previously [8].
Statistics
Data were analyzed using a 1- or 2-way ANOVA followed by Bonferroni corrections when necessary (GraphPad). Data are presented as means ± standard error of the mean. Significance was set at p<0.05.
Results
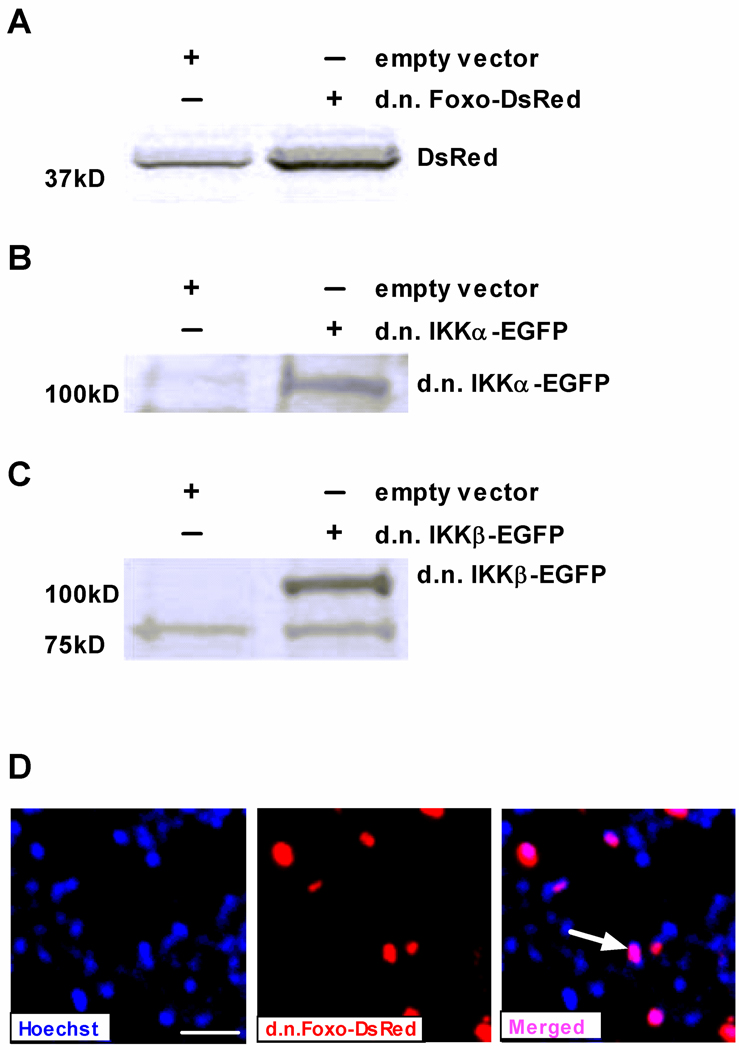
Two plasmids of a similar size co-transduce the same fibers almost 100% of the time [17]. Thus, in previous work we co-injected d.n. Foxo (5 kb) with EGFP (4.7 kb) to visualize d.n. Foxo-transfected fibers. However in the current study we needed to co-express d.n. Foxo with either d.n. IKKα-EGFP or d.n. IKKβ-EGFP, so we created a d.n. Foxo-DsRed fusion plasmid by subcloning d.n. Foxo in-frame into a DsRed expressing plasmid. To ensure that function of the d.n. Foxo protein was not altered by fusion to the DsRed protein (27 kDa) and to confirm expression of d.n. Foxo-DsRed we co-injected a DAF-16 (FOXO) reporter plasmid with an empty vector or d.n. Foxo-DsRed into the solei of rats that were subsequently assigned to weight bearing or 3 day cast immobilization groups. The increase in Foxo reporter activity following immobilization was completely abolished by expression of d.n. Foxo-DsRed as we have previously found with d.n. Foxo [5], thereby confirming that the d.n. Foxo-DsRed fusion protein functions similarly as d.n. Foxo (data not shown). Verification that d.n. IKKα-EGFP and d.n. IKKβ-EGFP function as well as their respective non-fusion protein forms has previously been confirmed [8]. Expression of full-length d.n. Foxo-DsRed was confirmed using anti-DsRed, as no antibody is currently available to the truncated form of Foxo (Fig. 1A). Expression of full-length d.n. IKKα-EGFP and d.n. IKKβ-EGFP were confirmed in whole muscle cell lysates probed with anti-IKKα and anti-IKKβ (Fig. 1B, C).
Figure 1.
Expression of electrotransfered plasmids in skeletal muscle. Immunoblots were performed on lysates from whole soleus muscles injected with (A) d.n. Foxo-DsRed, (B) d.n. IKKα-EGFP or (C) d.n. IKKβ-EGFP. Blots were incubated with anti-DsRed, anti-IKKα or anti-IKKβ antibodies, respectively. The predicted molecular weight of d.n. Foxo is ~14 kDa and DsRed is 27 kDa. Therefore the molecular weight of d.n. Foxo-DsRed is ~41 kDa. A non-specific band is also seen at this molecular weight (empty vector lane, Fig 1A) but is substantially less intense than the d.n. Foxo-DsRed band. (D) Nuclear localization of d.n. Foxo-DsRed was identified in a representative cross section by merging a d.n. Foxo-DsRed image with a Hoechst 33342 stained image to visualize nuclei (blue). Pink nuclei in the merged image (shown by arrow) are nuclei showing d.n.Foxo-DsRed localization. There are more blue nuclei than red fluorescence because Hoechst stains all DNA in the cross section, while the red fluorescence is only visualized in fibers that have nuclei expressing d.n.Foxo-DsRed. Scale bar: 50 µm.
In previous work when we co-injected d.n. Foxo with EGFP [12] it was not possible to detect the cellular localization of d.n. Foxo. However, in the current experiments d.n. Foxo-DsRed shows distinct localization at the fiber periphery at what appears to be muscle nuclei. To confirm this, we stained a representative cross section with Hoescht 33342 dye and, indeed, d.n. Foxo-DsRed appears to be localized to the nucleus as demonstrated by the pink (vs. red or blue) colored nuclei in the muscle cross section (Fig. 1D). Importantly, this is distinct from the localization of DsRed, which is primarily cytosolic (Fig. 2A). As previously shown, d.n. IKKα-EGFP and d.n. IKKβ-EGFP show cytosolic distribution (Fig. 2A) [8].
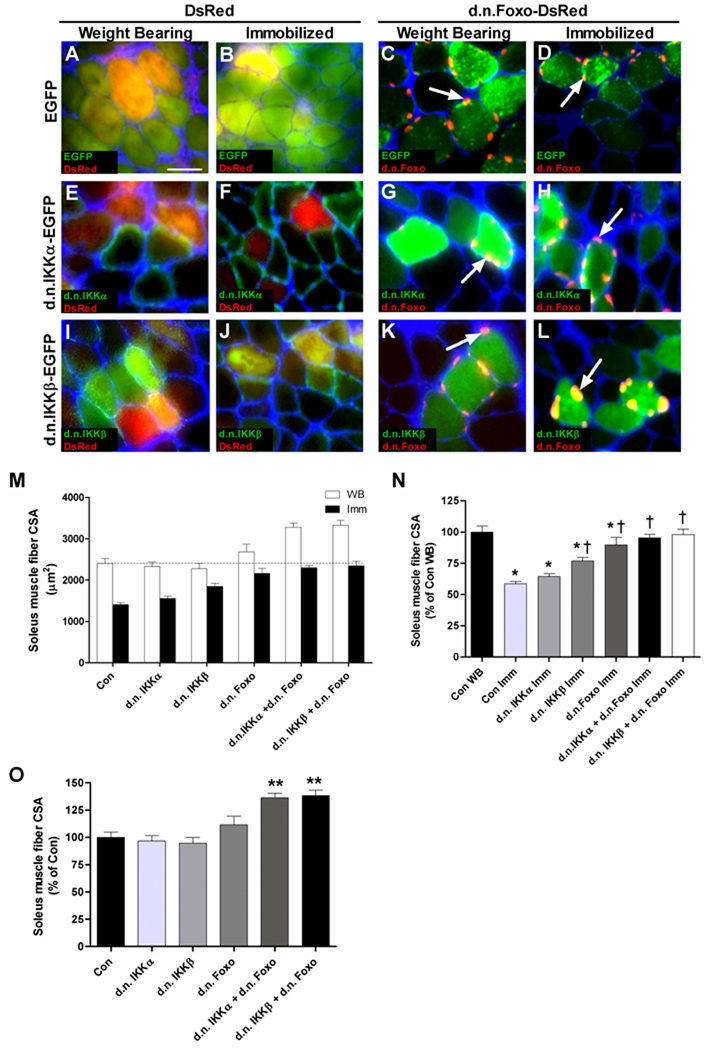
Figure 2.
Inhibition of IKK and/or Foxo blocks disuse muscle atrophy. Representative cross sections (A–L) of muscles from weight bearing or cast immobilized groups injected and electroporated with various plasmid combinations (see Results section for details). When merging images of cytoplasmic localized green proteins (EGFP, d.n.IKKα-EGFP and d.n.IKKβ-EGFP) and cytoplasmic localized red proteins (DsRed), yellow fibers showing co-expression can be visualized. Co-expression can also be visualized when merging images of cytoplasmic localized green proteins with the nuclear localized red protein, d.n.Foxo-DsRed. Arrows in C, D, G, H, K and L point to d.n.Foxo-DsRed. Sections were incubated with wheat germ agglutinin Alexa Fluor 350 conjugate (blue) that binds to glycoproteins predominant in the sarcolemma-extracellular matrix and allows for visualization of muscle fibers. (M) Mean fiber CSA of weight bearing (WB) and immobilized (Imm) muscles injected with the various plasmid combinations. (N) Mean CSA of immobilized muscles injected with the various plasmid combinations expressed as a percentage of weight bearing control (data normalized from M). (O) Mean CSA of weight bearing muscles injected with the various plasmid combinations expressed as a percentage of control (data normalized from M). Data are presented as mean + SEM. N=6 muscles per group. *p<0.05 compared to control weight bearing muscles. †p<0.05 compared to control immobilized muscles. Scale bar in (A): 50 µm.
Atrophy
To determine the effects of inhibiting Foxo and/or IKK activation in skeletal muscle during disuse the CSA area of fibers expressing dominant negative forms of one or both of these proteins was measured. Fiber area was measured in weight bearing and cast immobilized soleus muscles expressing EGFP and DsRed (Fig. 2A,B), d.n. Foxo-DsRed + EGFP (Fig. 2C,D), d.n. IKKα-EGFP + DsRed (Fig. 2E,F), d.n. IKKα-EGFP + d.n. Foxo-DsRed (Fig. 2G,H), d.n. IKKβ-EGFP + DsRed (Fig. 2I,J), or d.n. IKKβ-EGFP + d.n. Foxo-DsRed (Fig. 2K,L). These 6 plasmid groups expressed in weight bearing and in cast immobilized muscles produced 12 different groups. With n=6 muscles per group, 72 total muscles were analyzed for muscle fiber CSA. As expected, 7 days of cast immobilization significantly decreased muscle fiber CSA (47%, p<0.001) in control injected muscles (Fig. 2M,N). Although muscle fibers transfected with d.n. IKKα-EGFP showed a 22% inhibition of disuse atrophy, this did not reach statistical significance (Fig. 2M,N). However, muscle fiber atrophy was significantly inhibited (57%) in fibers transfected with d.n. IKKβ-EGFP (Fig. 2M,N). There was no significant difference in fiber CSA of weight bearing groups transfected with control, d.n. IKKα or d.n. IKKβ plasmids (Fig. 2M,O). Interestingly, overexpression of d.n. Foxo-DsRed in weight bearing muscles increased muscle fiber size by 11%, although this did not reach statistical significance. Furthermore, overexpression of d.n. Foxo-DsRed alone in immobilized muscles prevented disuse fiber atrophy by 76% (Fig. 2M–O). Overexpression of d.n. IKKα-EGFP and d.n. Foxo-DsRed significantly increased muscle fiber CSA by 36% in weight bearing muscles and inhibited immobilization-induced fiber atrophy by 89% (Fig. 2M–O). Similarly, overexpression of d.n. IKKβ-EGFP and d.n. Foxo-DsRed in weight bearing muscles significantly increased muscle fiber CSA by 38% and inhibited the immobilization-induced fiber atrophy by 95% (Fig. 2M–O). Since EGFP + DsRed plasmid-injected muscles in the weight bearing group reflect the baseline muscle fiber CSA, all data are presented as a percent of this group. Figure 2C shows the percent decrease in fiber CSA due to immobilization in all groups, while Figure 2D depicts the percent increase in fiber CSA in all groups during weight bearing conditions.
Discussion
Signaling pathways that are activated in multiple models of muscle atrophy lead to the activation of NF-κB and Foxo mediated transcription. Recent work demonstrated that individual expression of d.n. IKKα or d.n. IKKβ completely inhibits hind limb unloading-induced NF-κB activation and inhibits the associated muscle fiber atrophy by 50% [8]. Moreover, co-expression of d.n. IKKα and d.n. IKKβ showed a 70% inhibition of unloading-induced muscle fiber atrophy, demonstrating a partial additive effect. In other work, expression of d.n. Foxo inhibited cast immobilization-induced muscle fiber atrophy by 43% [12]. However, to our knowledge the present study is the first to inhibit both NF-κB and Foxo mediated transcription during any atrophy condition to determine if there is an additive effect on disuse fiber atrophy.
We found that expression of d.n. IKKβ attenuated 57% of disuse muscle fiber atrophy, which is in close agreement with recent work demonstrating a 50% attenuation of atrophy in fibers expressing d.n. IKKβ during hind limb suspension [8]. However in contrast to this recent work, which also found a 50% inhibition of fiber atrophy in fibers expressing d.n. IKKα during hind limb suspension, expression of d.n. IKKα in the current study only inhibited 22% of the muscle fiber atrophy, which was not statistically significant. This difference is presumably a function of the different models used (hind limb unloading versus cast immobilization). Indeed, Bodine et al. found that most genes altered during immobilization are similarly regulated during denervation but unaltered during unloading, despite comparable rates of atrophy in each of the three disuse conditions [18]. There may be, then, some potential differences in the signaling pathways that are activated during unloading and immobilization. During hind limb unloading the limbs can move freely through a normal range of motion but load bearing is completely eliminated and therefore any muscle contraction is characterized by low force. However during cast immobilization the limbs cannot move through any range of motion, but isometric contractions with high force can occur. These differences have been used to explain some of the different functional adaptations that occur during unloading versus immobilization [19].
In our previous study we co-injected d.n. Foxo with EGFP and found a 43% inhibition of immobilization induced muscle fiber atrophy [12]. In the present study we found a 76% inhibition of fiber atrophy due to expression of d.n. Foxo-DsRed. One explanation for the differing levels of atrophy inhibition could be that in the former study, the proteins were not expressed in equal amounts in each transfected fiber. The consequence of this possibility is a diminution of the effect of overexpressed d.n. Foxo. Another explanation could be related to the amounts of plasmid DNA electrotransfered in each study. In the present study we used 40 µg of d.n. Foxo-DsRed, whereas in the previous study we used 10 µg of d.n. Foxo. Therefore the greater attenuation of disuse fiber atrophy found in the current study could be related to the higher plasmid amount used, and thus the amount of protein made. However, the findings of both studies do agree in clearly demonstrating that Foxo activation is necessary for disuse muscle fiber atrophy.
Since expression of d.n. Foxo alone has only a modest effect on fiber CSA in weight bearing muscles, and expression of d.n. IKKα or d.n. IKKβ have no effect on fiber CSA in weight bearing muscles, the 36% and 38% increase in CSA in fibers co-expressing d.n. Foxo plus d.n. IKKα or d.n. Foxo plus d.n. IKKβ, respectively, compared to control fibers indicates a synergistic effect of inhibiting NF-κB activity and Foxo activity on muscle fiber size. The increase in fiber CSA in these groups suggests that basal levels of NF-κB and Foxo activity may repress protein synthesis. In support of this idea, there are data to suggest that both IKK and Foxo regulate signaling that affects protein synthesis. Knockout mice with muscle specific depletion of IKKβ show a marked increase in phospho(active)-Akt, a potent activator of protein synthesis. However, muscle fiber CSA remained unaltered in these mice, demonstrating that the increase in phospho-Akt was not sufficient to increase muscle fiber size [20]. Foxo may be a negative regulator of protein synthesis by inducing the degradation of components of mTOR signaling. Indeed protein levels of raptor, mTOR, S6 protein kinase 1 (S6K1), and TSC2 are eliminated in C2C12 cells stably expressing c.a. Foxo1 [21]. It is unknown whether d.n. Foxo alters the expression of these regulators of protein synthesis in the present study, or whether d.n. IKKβ (or d.n. IKKα) alters phospho-Akt levels. The possibility exists that co-inhibition of IKK and Foxo in the current study altered muscle fiber size in weight bearing muscle through the combined regulation of these variables. This warrants further investigation.
When IKK-mediated NF-κB and Foxo transcriptional activity are inhibited by co-expression of either d.n. IKKα plus d.n. Foxo, or d.n. IKKβ plus d.n. Foxo, there is an additive effect of complete blockage of immobilization-induced muscle atrophy. There are several potential explanations for these findings. It is possible that NF-κB and Foxo have distinct atrophy target genes and therefore inhibition of both pathways represses the transcription of a broader number of atrophy genes. Alternatively, some atrophy genes may contain both κB sites and Foxo binding elements in their regulatory regions, and inhibition of both NF-κB and Foxo may cause a greater repression of gene transcription than inhibition of either pathway alone. In fact, analysis of the 5’ flanking region of several atrophy-related genes reveals both κB sites and Foxo binding elements in their promoter regions, including MuRF1, atrogin-1, cathepsin L, and 4E-BP1. The effect of inhibiting both NF-κB and Foxo on the transcription of these genes during a physiological condition of muscle wasting, however, has yet to be determined.
In summary, both NF-κB and Foxo activity are increased during multiple models of muscle atrophy, and their increases are separately responsible for at least 50% of the muscle fiber atrophy during disuse. Findings in the present study show that blocking both IKK-mediated NF-κB activity and Foxo transcriptional activity during disuse has a cumulative effect that completely abolishes disuse-induced muscle fiber atrophy.
Acknowledgements
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) Grant R03AR056418 (to A. R. Judge) and NIAMS Grant R01AR41705 (to S.C. Kandarian).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cai D, Frantz JD, Tawa NE, Jr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 2.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. Faseb J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 3.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter RB, Stevenson E, Koncarevic A, Mitchell-Felton H, Essig DA, Kandarian SC. Activation of an alternative NF-kappaB pathway in skeletal muscle during disuse atrophy. Faseb J. 2002;16:529–538. doi: 10.1096/fj.01-0866com. [DOI] [PubMed] [Google Scholar]
- 5.Senf SM, Dodd SL, McClung JM, Judge AR. Hsp70 overexpression inhibits NF-kappaB and Foxo3a transcriptional activities and prevents skeletal muscle atrophy. Faseb J. 2008;22:3836–3845. doi: 10.1096/fj.08-110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 7.Pereira SG, Oakley F. Nuclear factor-kappaB1: regulation and function. Int J Biochem Cell Biol. 2008;40:1425–1430. doi: 10.1016/j.biocel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Van Gammeren D, Damrauer JS, Jackman RW, Kandarian SC. The IkappaB kinases IKKalpha and IKKbeta are necessary and sufficient for skeletal muscle atrophy. Faseb J. 2009;23:362–370. doi: 10.1096/fj.08-114249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter RB, Kandarian SC. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J Clin Invest. 2004;114:1504–1511. doi: 10.1172/JCI21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Judge AR, Koncarevic A, Hunter RB, Liou HC, Jackman RW, Kandarian SC. Role for IkappaBalpha, but not c-Rel, in skeletal muscle atrophy. Am J Physiol Cell Physiol. 2007;292:C372–C382. doi: 10.1152/ajpcell.00293.2006. [DOI] [PubMed] [Google Scholar]
- 11.Kamei Y, Mizukami J, Miura S, Suzuki M, Takahashi N, Kawada T, Taniguchi T, Ezaki O. A forkhead transcription factor FKHR up-regulates lipoprotein lipase expression in skeletal muscle. FEBS Lett. 2003;536:232–236. doi: 10.1016/s0014-5793(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 12.Sent SM, Dodd SL, Judge AR. FOXO signaling is required for disuse muscle atrophy and is directly regulated by Hsp70. Am J Physiol Cell Physiol. 2010;298:C38–C45. doi: 10.1152/ajpcell.00315.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, Fielding RA, Kandarian SC. Identification of a molecular signature of sarcopenia. Physiol Genomics. 2005;21:253–263. doi: 10.1152/physiolgenomics.00249.2004. [DOI] [PubMed] [Google Scholar]
- 14.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 15.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kipl and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 16.Storz P, Doppler H, Toker A. Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol. 2005;25:8520–8530. doi: 10.1128/MCB.25.19.8520-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rana ZA, Ekmark M, Gundersen K. Coexpression after electroporation of plasmid mixtures into muscle in vivo. Acta Physiol Scand. 2004;181:233–238. doi: 10.1111/j.1365-201X.2004.01282.x. [DOI] [PubMed] [Google Scholar]
- 18.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 19.Fitts RH, Metzger JM, Riley DA, Unsworth BR. Models of disuse: a comparison of hindlimb suspension and immobilization. J Appl Physiol. 1986;60:1946–1953. doi: 10.1152/jappl.1986.60.6.1946. [DOI] [PubMed] [Google Scholar]
- 20.Mourkioti F, Kratsios P, Luedde T, Song YH, Delafontaine P, Adami R, Parente V, Bottinelli R, Pasparakis M, Rosenthal N. Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J Clin Invest. 2006;116:2945–2954. doi: 10.1172/JCI28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu AL, Kim JH, Zhang C, Unterman TG, Chen J. Forkhead box protein O1 negatively regulates skeletal myocyte differentiation through degradation of mammalian target of rapamycin pathway components. Endocrinology. 2008;149:1407–1414. doi: 10.1210/en.2007-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]