Abstract
Advances in cell biology and biophysics revealed that cellular membranes consist of multiple microdomains with specific sets of components such as lipid rafts and tetraspanin-enriched microdomains. An increasing number of enveloped viruses have been shown to utilize these microdomains during their assembly. Among them, association of HIV-1 and other retroviruses with lipid rafts and tetraspanin-enriched microdomains within the plasma membrane is well documented. In this review, I will describe our current knowledge on interrelationships between plasma membrane microdomain organization and the HIV-1 particle assembly process. Microdomain association during virus particle assembly may also modulate subsequent virus spread. Potential roles played by microdomains will be discussed with regard to two post-assembly events, i.e., inhibition of virus release by a raft-associated protein BST-2/tetherin and cell-to-cell HIV-1 transmission at virological synapses.
Keywords: lipid rafts; tetraspanin; Gag; virus assembly; plasma membrane; tetherin; virological synapse; phosphatidylinositol-(4,5)-bisphosphate
Introduction
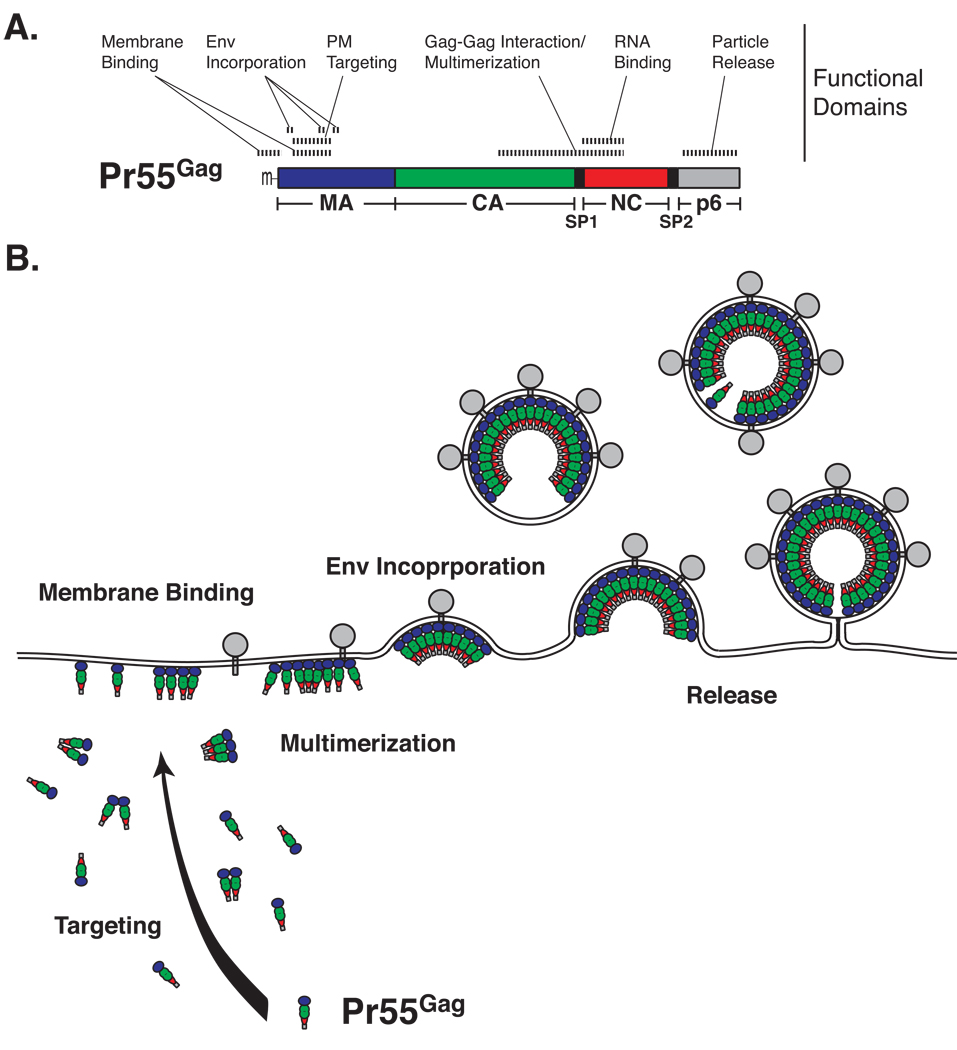
In most cell types including natural hosts such as T cells, HIV-1 assembles at the plasma membrane (PM). HIV-1 particle formation is a multi-step process driven by the viral structural protein Gag (Adamson and Freed, 2007) (Fig. 1). This process includes: 1) targeting of Gag to the PM, 2) Gag binding to membrane, 3) Gag multimerization, 4) encapsidation of viral genomic RNA, 5) incorporation of the viral Env glycoprotein, and 6) budding and release of virus particles. Although the order of some of these steps remains to be determined, regions in Gag involved in each step are well defined. Gag is a multidomain protein consisting of four major domains, matrix (MA), capsid (CA), nucleocapsid (NC), and p6 as well as spacer peptides, SP1 and SP2. MA mediates Gag targeting and binding to the PM and Env incorporation, whereas the CA C-terminal domain (CTD) and NC promote Gag multimerization. The CA-CTD contains a dimerization interface, while NC, an RNA binding domain, is thought to promote higher-order multimerization as scaffolding through NC-RNA-NC binding. NC also contains zinc finger motifs that mediate specific recognition of viral RNA for packaging of the genome. p6 recruits cellular ESCRT complexes that facilitate fission of virions from the PM.
Fig. 1.
HIV-1 Gag and virus particle assembly. A. Structural and functional domains are shown. MA, matrix; CA, capsid; NC, nucleocapsid; SP, spacer peptide. N-terminal myristylation is shown as (m−). B. A general outline of virus assembly process is shown. For clarity, RNA molecules associated with NC are not depicted.
Advances in cell biology and biophysics have revealed that the PM is heterogeneous, consisting of multiple microdomains that contain specific sets of lipids and proteins. These microdomains may have various lifetimes, sizes, and dynamics and may coalesce or dissociate from each other, thereby modulating cellular functions. Among them, lipid rafts and tetraspanin-enriched microdomains (TEMs) have been implicated in various aspects of the HIV-1 life cycle. In this review, I will focus on interrelationships between these two specific types of microdomains and HIV-1 assembly. I will also discuss potential roles of these microdomains in two post-assembly events currently under intense scrutiny, i.e., BST-2/tetherin-mediated virion release inhibition and cell-to-cell HIV-1 transmission at virological synapses.
PM microdomains associated with the late phase of HIV-1 replication cycle
Lipid rafts
Lipid rafts, also known as membrane rafts, are microdomains enriched with cholesterol, glycosphingolipids, and other saturated lipids, as well as specific types of proteins. In the original concept of lipid rafts, formation of these microdomains was thought to rely on the propensity of participating lipids to form a liquid-ordered state through lipid-lipid interactions (Simons and Ikonen, 1997; London and Brown, 2000). According to this model, this cholesterol-dependent liquid-ordered structure coexists with liquid-disordered domains in cell membranes, and membrane-associated proteins partition to either of these domains depending on their membrane binding modes. Partitioning of molecules with lipid rafts in cells has primarily been assessed based on their association with detergent resistant membrane (DRM) fractions (Brown and Rose, 1992). However, it is of note that DRM association reflects only a preference for rafts by proteins or lipids of interest and does not prove that raft association precedes experimental manipulation (Lichtenberg et al., 2005). Using this and other methods, proteins anchored to the extracellular leaflet of the PM by a glycosylphosphatidylinositol (GPI) moiety are identified as raft-associated proteins (Brown and London, 2000; Simons and Toomre, 2000). Cytoplasmic proteins modified with saturated acyl chains such as palmitoyl moieties constitute another representative class of raft proteins. Unsaturated lipid modifications such as prenylation are less favored in rafts (Melkonian et al., 1999; Zacharias et al., 2002). Some transmembrane proteins such as transferrin receptor (TfR) and CD45 are also known to be excluded from rafts.
Lipid rafts are thought to serve as delivery or concentration platforms in various cellular functions including signaling, protein sorting, and cell polarity. However, the involvement of rafts in these cell functions, and even the very existence of rafts in intact cells, have been matters of debate because of their submicroscopic size, and because of concerns over methods for studying rafts, in particular, the use of detergent-resistancebased isolation methods (Edidin, 2003; Munro, 2003; Hancock, 2006). Nonetheless, advanced biophysical and cell biological techniques have provided strong support to the presence of cholesterol-dependent microdomains in the PM (Kusumi et al., 2004; Mayor and Rao, 2004; Hancock, 2006; Jacobson et al., 2007; Day and Kenworthy, 2009). The definition of rafts put forth in the 2006 Keystone Symposium of Lipid Rafts and Cell Function summarizes the current consensus as following: “Lipid rafts are small, heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions.” (Pike, 2006).
As noted in this consensus definition of lipid rafts, in addition to lipid-lipid interactions, protein-protein and protein-lipid interactions have been gaining recognition as important factors in dynamics of these microdomains (Douglass and Vale, 2005; Gaus et al., 2005; Larson et al., 2005; Hancock, 2006). In particular, oligomeric proteins that bind rafts (e.g., caveolins and flotillins) modulate stability, size, and/or structure of membrane domains, creating specific subsets of microdomains (Langhorst et al., 2005; Morrow and Parton, 2005; Parton and Simons, 2007).
Tetraspanin-enriched microdomains (TEMs)
TEMs are another type of membrane microdomains primarily organized by tetraspanins, a family of proteins with four transmembrane domains (Hemler, 2005; Levy and Shoham, 2005; Berditchevski and Odintsova, 2007; Charrin et al., 2009; Yanez-Mo et al., 2009). The importance of protein-protein interactions has been well recognized for formation of TEMs. These interactions include homo- and hetero-oligomerization of tetraspanins and interactions of tetraspanins with other proteins (e.g., integrins) (Hemler, 2005; Levy and Shoham, 2005; Berditchevski and Odintsova, 2007; Charrin et al., 2009; Yanez-Mo et al., 2009). Because of these interactions with other proteins, TEMs are involved in a wide variety of cellular functions including intra- and inter-cellular signaling and cell-cell adhesion (Hemler, 2005; Levy and Shoham, 2005; Berditchevski and Odintsova, 2007; Charrin et al., 2009; Yanez-Mo et al., 2009).
TEMs are detected as microscopically visible patches using antibodies recognizing specific tetraspanins (Claas et al., 2001; Nydegger et al., 2006; Espenel et al., 2008). Live cell microscopy showed that these patches are stable in shape and localization (Espenel et al., 2008). In addition to these patches, single-molecule tracking of the tetraspanin CD9 demonstrated that there is another population of tetraspanins not associated with TEMs and that this population of molecules is in a dynamic exchange with those associated with TEMs (Espenel et al., 2008). Somewhat analogous to lipid rafts, at least a portion of the non-TEM-associated CD9 appears to form small, dynamic clusters (Espenel et al., 2008). Notably, formation of these clusters is dependent on PM cholesterol and CD9 palmitoylation (Espenel et al., 2008). Furthermore, under some experimental conditions, tetraspanins were observed to associate with DRM fractions (Claas et al., 2001; Charrin et al., 2002; Charrin et al., 2003). However, lipid rafts and TEMs (or related clusters) are generally regarded as distinct types of microdomains because detergent resistance and sensitivity to cholesterol depletion displayed by tetraspanins are qualitatively different from those of raft proteins (Claas et al., 2001; Charrin et al., 2002; Hemler, 2005; Le Naour et al., 2006; Charrin et al., 2009; Yanez-Mo et al., 2009). As described later, this view is also supported by recent microscopy-based experiments that demonstrated clear segregation of tetraspanins from raft-associated proteins (Nydegger et al., 2006; Barreiro et al., 2008; Espenel et al., 2008).
Relationships between microdomain organization and HIV-1 assembly
Roles played by lipid rafts during HIV-1 assembly
During virus particle assembly, Gag and Env associate with DRM that presumably originates from rafts (Nguyen and Hildreth, 2000; Lindwasser and Resh, 2001; Ono and Freed, 2001; Lindwasser and Resh, 2002; Ding et al., 2003; Halwani et al., 2003; Holm et al., 2003; Ono et al., 2005; Bhattacharya et al., 2006; Dou et al., 2009). Immunofluorescence microscopy studies showed that Gag colocalizes with lipid raft markers in cells (Nguyen and Hildreth, 2000; Holm et al., 2003; Ono and Freed, 2005). The lipid bilayer of the viral envelope is enriched in raft-associated lipids and proteins (Aloia et al., 1993; Saifuddin et al., 1995; Graham et al., 2003; Brugger et al., 2006; Chertova et al., 2006; Chan et al., 2008; Ott, 2008) and adopts cholesterol-dependent liquid-ordered structure (Lorizate et al., 2009). Cellular cholesterol depletion, which disrupts rafts, inhibits virus particle production (Ono and Freed, 2001; Pickl et al., 2001) by impairing Gag membrane binding, multimerization, and/or mobility at the PM (Gomez and Hope, 2006; Ono et al., 2007). Substitution of myristate at the Gag N-terminus with an unsaturated analogue blocks Gag-DRM association and impairs virus particle production (Lindwasser and Resh, 2002). Due to technical limitations inherent to the methodologies applied in these studies, some of these data can be explained without implicating rafts in HIV-1 assembly. Collectively, however, these reports are consistent with the notion that lipid rafts or cholesterol-dependent microdomains play an important role in HIV-1 particle assembly.
Not only other retroviruses [e.g., HTLV-1 (Pickl et al., 2001; Feng et al., 2003) and murine leukemia virus (Pickl et al., 2001; Nitta et al., 2009)] but also other families of enveloped and non-enveloped viruses are thought to utilize lipid rafts during the late phase of their life cycles (Suomalainen, 2002; Briggs et al., 2003; Ono and Freed, 2005; Metzner et al., 2008). These viruses include paramyxoviruses (e.g., measles virus, Sendai virus, and respiratory syncytial virus) (Sanderson et al., 1995; Ali and Nayak, 2000; Manie et al., 2000; Vincent et al., 2000; Henderson et al., 2002; McCurdy and Graham, 2003), orthomyxoviruses (e.g., influenza A virus) (Scheiffele et al., 1999; Ali et al., 2000; Barman and Nayak, 2000; Zhang et al., 2000; Leser and Lamb, 2005), filoviruses (e.g., Ebola virus) (Bavari et al., 2002; Panchal et al., 2003), herpes viruses (e.g., herpes simplex virus and pseudorabies virus) (Lee et al., 2003; Favoreel et al., 2004; Lyman et al., 2008), and reoviruses (e.g., Bluetongue virus) (Bhattacharya and Roy, 2008). In most cases, lipid rafts are postulated to serve as delivery vehicles or concentration platforms for viral structural elements, thereby facilitating assembly and release of infectious virions. However, the exact nature of association between lipid rafts and structural proteins of these viruses (including HIV-1), in particular, the impact of viral proteins on lipid raft organization, is less well understood.
MA as the interface between Gag and lipid rafts
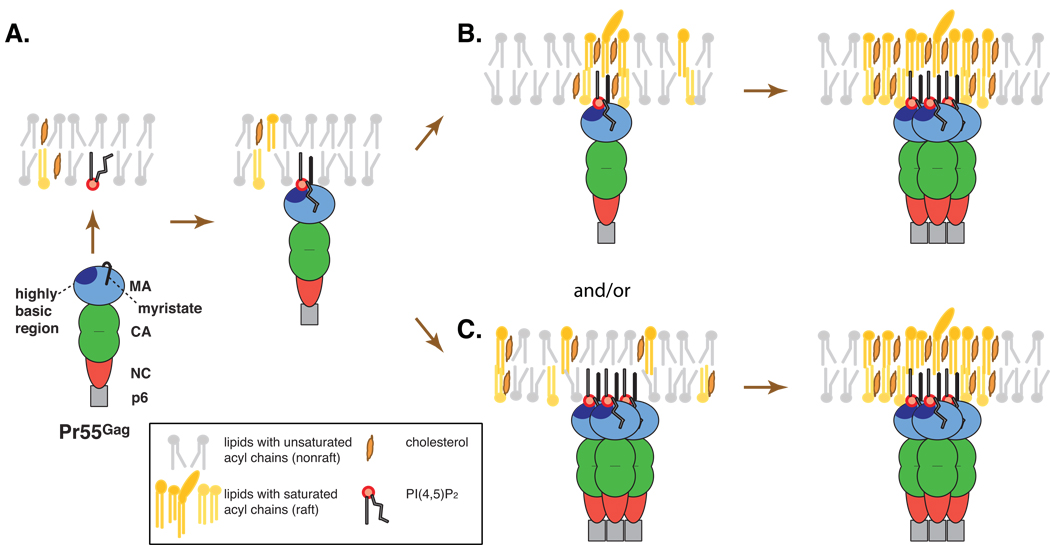
In the case of HIV-1 Gag, both the N-terminal myristate and a highly basic region of MA, which are essential for binding of MA to the PM bilayer (Gottlinger et al., 1989; Bryant and Ratner, 1990; Zhou et al., 1994; Hill et al., 1996), are implicated in lipid raft association. The MA highly basic domain has been shown to interact with a PM-specific phospholipids, PI(4,5)P2 (Saad et al., 2006; Shkriabai et al., 2006; Chan et al., 2008; Chukkapalli et al., 2008; Alfadhli et al., 2009a; Alfadhli et al., 2009b; Chukkapalli et al., 2010); reviewed in (Ono, 2009)]. This interaction promotes Gag-membrane binding and Gag localization to the PM, thereby facilitating HIV-1 particle assembly and release (Ono et al., 2004; Chan et al., 2008; Chukkapalli et al., 2008). Gag proteins of other retroviruses including murine leukemia virus, Mason-Pfizer monkey virus, and HIV-2 also bind PI(4,5)P2 (Hamard-Peron et al.; Stansell et al., 2007; Chan et al., 2008; Saad et al., 2008). Notably, NMR studies showed that exposure of the myristate moiety, which is otherwise sequestered inside MA, is induced by binding of PI(4,5)P2 to the MA highly basic domain (Saad et al., 2006). The NMR studies further suggest that when HIV-1 MA binds PI(4,5)P2, it sequesters the highly unsaturated 2’ acyl chain of PI(4,5)P2, leaving the saturated 1’ chain available for association with membrane (Saad et al., 2006) (Fig. 2). This mode of MA-PI(4,5)P2 binding is predicted to promote Gag interaction with lipid rafts (Saad et al., 2006). In addition, as described earlier, substitution of myristate with unsaturated analogues blocks Gag association with DRM, suggesting that the N-terminal myristate moiety plays a key role in Gag association with lipid rafts (Lindwasser and Resh, 2002). Therefore, it is likely that MA does not simply function as a membrane-binding domain but also serves as a specific interface between Gag and membrane microdomains.
Fig. 2.
Association of Gag with lipid raft microdomains. NMR studies suggest that binding of PI(4,5)P2 to the MA highly basic region induces exposure of the N-terminal myristate moiety as well as sequestration of the highly unsaturated 2’-acyl chain of PI(4,5)P2 (A). Two exposed saturated acyl chains, i.e., the N-terminal myristate and the 1’ acyl chain of PI(4,5)P2, are postulated to promote partitioning of the Gag molecule to a raft domain, which may become stabilized or coalesce with other rafts upon Gag multimerization (B). Multiple saturated acyl chains associated with a Gag cluster may also induce formation of a stable raft-like domain (C). Possibilities shown in B and C are not mutually exclusive.
Potential effects of HIV-1 assembly on the organization of lipid rafts
Besides MA, NC may affect Gag interaction with lipid rafts. Although NC-mediated higher-order multimerization is not essential for initial DRM association of Gag (Ono et al., 2005), NC increases density of the DRM with which Gag associates (Lindwasser and Resh, 2001). Therefore, as postulated previously (Lindwasser and Resh, 2001; Ono and Freed, 2001; Dalton et al., 2007), Gag membrane binding and multimerization may alter the size and/or structure of lipid rafts (Fig. 2). In this regard, it is important to note that, except for caveolae, which constitute a unique subset of rafts organized by caveolin, lipid rafts are generally considered to be small, with the estimated diameter of 5–50 nm (Pralle et al., 2000; Prior et al., 2003; Sharma et al., 2004; Eggeling et al., 2009). In contrast, the diameter of an HIV-1 particle is approximately 100–150 nm. Therefore, it is unlikely that a virus particle assembles within and buds from a single lipid raft. Rather, it is more likely that virus particle assembly involves recruitment and coalescence of small rafts into large stable rafts at assembly sites. When Gag forms multimers at the PM, saturated acyl chains associated with MA likely create a lipid environment suitable for recruiting raft-associated molecules in the PM cytoplasmic leaflet (Fig. 2). Therefore, during virus assembly, Gag may induce formation of lipid-raft-like microdomains de novo. As mentioned earlier, oligomeric membrane-bound proteins such as flotillin can stabilize lipid rafts to form microscopically visible structures (Langhorst et al., 2005; Morrow and Parton, 2005; Parton and Simons, 2007). It is tempting to speculate that membrane-bound Gag multimers play an analogous role during association with lipid rafts.
Roles played by TEMs during HIV-1 assembly
TEMs have also been shown to colocalize with assembling Gag in T cells as well as other cell types (Mazurov et al., 2006; Nydegger et al., 2006; Jolly and Sattentau, 2007; Grigorov et al., 2009; Hogue et al., 2009). Biochemical studies showed that HIV-1 Gag can be coimmunoprecipitated with some tetraspanins from cell lysates (Grigorov et al., 2009). In some cases, somewhat larger membrane domains enriched with tetraspanins, termed endosome-like domain (ELD), are observed to be sites of virus assembly (Booth et al., 2006). Because of their similarity, these two domains will be hereafter referred to as TEM/ELDs in this review. In macrophages, HIV-1 Gag has been observed to assemble virus particles within apparently intracellular, tetraspanin-enriched compartments (Raposo et al., 2002; Pelchen-Matthews et al., 2003). At least subpopulations of these compartments were later shown to be convoluted invaginations of the PM (Deneka et al., 2007; Jouve et al., 2007; Welsch et al., 2007; Bennett et al., 2009). Therefore, these compartments may be derived from TEM/ELDs.
Like raft components, tetraspanins are incorporated into retrovirus particles (Nguyen et al., 2003; Pelchen-Matthews et al., 2003; Chertova et al., 2006; Jolly and Sattentau, 2007; Khurana et al., 2007; Medina et al., 2008; Sato et al., 2008). Notably, these virion-associated tetraspanins impair virus infectivity by inhibiting Env-mediated fusion (Sato et al., 2008; Grigorov et al., 2009; Krementsov et al., 2009; Weng et al., 2009). Consistent with the effect of tetraspanins on virus-cell fusion, tetraspanins also inhibit cell-cell fusion mediated by cell-associated Env (Sato et al., 2008; Grigorov et al., 2009; Krementsov et al., 2009; Weng et al., 2009). It is of note that this inhibitory effect seems to require Gag-dependent clustering of Env and resulting localization of Env to TEMs (Weng et al., 2009). Therefore, HIV-1 assembly in TEM/ELDs likely prevents virus-expressing cells in contact with uninfected cells from fusing prematurely and forming syncytia (Krementsov et al., 2009; Weng et al., 2009). In contrast to the inhibitory effect on Env-mediated fusion, a consensus has not yet emerged on whether TEM/ELDs or particular tetraspanins actively regulate the HIV-1 particle assembly process. RNAi-mediated knock-down of tetraspanins have produced contradictory data thus far (Chen et al., 2008; Ruiz-Mateos et al., 2008; Grigorov et al., 2009; Krementsov et al., 2009). Careful comparison of cell types, expression levels of each of the major tetraspanins in these cells, and timing after expression of Gag and siRNA molecules are likely needed for determining the effect of TEM/ELDs on virus assembly and release.
Interestingly, it has been shown that association of proteins with TEM/ELDs requires membrane binding and higher-order multimerization of the proteins (Fang et al., 2007). Therefore, considering the dynamic nature of tetraspanin partitioning to membrane domains described above (Barreiro et al., 2008; Espenel et al., 2008), it is possible that, like lipid rafts, TEM/ELDs are also recruited to or stabilized at virus assembly sites due to Gag multimerization at the PM.
Relationships between lipid rafts and TEM/ELDs at HIV-1 assembly sites
As described earlier, biochemical data suggest that lipid rafts and TEM/ELDs are distinct microdomains. Microscopy studies that examined copatching of raft proteins and tetraspanins also support this view (Nydegger et al., 2006; Espenel et al., 2008). Copatching has been frequently used for studies of membrane microdomains that are submicroscopic in size (Harder et al., 1998; Janes et al., 1999; Shvartsman et al., 2003; Gri et al., 2004; Meder et al., 2006; Lingwood et al., 2008). In a copatching assay, membrane proteins are homotypically crosslinked with specific divalent antibodies before fixation. This induces formation of microscopically visible patches. If two proteins share affinity to the same microdomain, and crosslinked proteins retain association with the microdomain stably enough, they colocalize in the same patch, or copatch. For example, a transmembrane raft marker, influenza HA, and another raft marker, PLAP (a GPI-anchored protein), colocalize at discrete patches when each are crosslinked by specific antibodies before fixation. In contrast, when stained after fixation, both show diffuse localization over the cell surface (Harder et al., 1998). Such copatching is not observed between PLAP and a nonraft marker, TfR (Harder et al., 1998). Using this method, tetraspanins were observed to segregate from HA and CD55, a GPI-anchored raft protein (Nydegger et al., 2006; Espenel et al., 2008) (Hogue and AO, unpublished data). Consistent with copatching experiments, live cell microscopy coupled with advanced techniques, including fluorescent recovery after photobleaching and single-molecule tracking, further showed that dynamic behaviors of tetraspanins and GPI-anchored proteins are clearly distinct (Barreiro et al., 2008; Espenel et al., 2008).
The findings described above indicate that, in normal cells, TEM/ELDs and lipid rafts are distinct microdomains. As described earlier, however, HIV-1 particle assembly associates with both lipid rafts and TEM/ELDs. This raises a question whether HIV-1 induces coalescence of these two types of microdomains at virus assembly sites. One study compared distributions of HA and tetraspanins in the presence of other influenza proteins and HIV-1 Gag (Khurana et al., 2007). In this study, Gag copatched well with TEM/ELD markers but HA remained segregated (Khurana et al., 2007). However, because other influenza proteins such as M1 likely restricted free movement of HA or HA-containing lipid rafts, it remains unknown whether lipid rafts and TEM/ELDs can be co-recruited to the HIV-1 assembly sites, or whether they form separate assembly sites.
In this regard, it is notable that a recent report suggests that two viral glycoproteins (HIV-1 Env and Ebola GP) are incorporated into distinct populations of progeny virions even when both glycoproteins are expressed in the same cells (Leung et al., 2008). Incorporation of viral glycoproteins into retrovirus particles has been suggested to involve lipid rafts or other membrane microdomains (Pickl et al., 2001; Briggs et al., 2003; Bhattacharya et al., 2004; Bhattacharya et al., 2006; Metzner et al., 2008; Jorgenson et al., 2009)(but see (Yang et al., 2009)). Therefore, the observation of bimodal incorporation of HIV-1 Env and Ebola GP was interpreted as indicating that a single Gag particle quantally associates with a microdomain containing single species of viral glycoprotein (Leung et al., 2008). Thus, it is possible that qualitatively different microdomains undergo homotypic but not heterotypic coalescence and independently constitute or become recruited to different assembly sites. Clearly, further investigation is needed for better understanding of dynamic reorganization of PM microdomains during HIV-1 assembly.
Potential roles played by microdomains in steps following virus particle assembly
Involvement of PM microdomains in a cellular defense mechanism
PM microdomains may be involved not only in particle assembly and viral glycoprotein incorporation but also in a recently discovered cellular defense mechanism that inhibits a post-assembly process. The HIV-1 protein Vpu has been known to enhance virus release in a cell-type-specific manner (Klimkait et al., 1990; Varthakavi et al., 2003; Neil et al., 2006). In the absence of Vpu, virus particles are tethered to the surface of non-permissive virus-producing cells and are eventually endocytosed (Klimkait et al., 1990; Neil et al., 2006). These tethered virions can be released after treatment with a protease, subtilisin (Neil et al., 2006). Recent studies demonstrated that a cellular protein known as HM1.24, CD317, BST-2, or tetherin (hereafter referred to as BST-2/tetherin) is a subtilisin-sensitive, Vpu-responsive inhibitor of virion release (Neil et al., 2008; Van Damme et al., 2008). Expression of this protein, which can be augmented by interferon (Neil et al., 2007), renders permissive cells non-permissive to HIV-1 lacking Vpu (Neil et al., 2008; Van Damme et al., 2008). Depletion of BST-2/tetherin rescues virion release from non-permissive cells in the absence of Vpu (Neil et al., 2008; Van Damme et al., 2008). Vpu antagonizes this protein at least partly through downregulation from the cell surface by ubiquitin-dependent mechanisms and/or through retention in the trans-Golgi network (Bartee et al., 2006; Van Damme et al., 2008; Douglas et al., 2009; Dube et al., 2009; Goffinet et al., 2009; Gupta et al., 2009; McNatt et al., 2009; Mitchell et al., 2009; Miyagi et al., 2009; Rong et al., 2009; Sato et al., 2009).
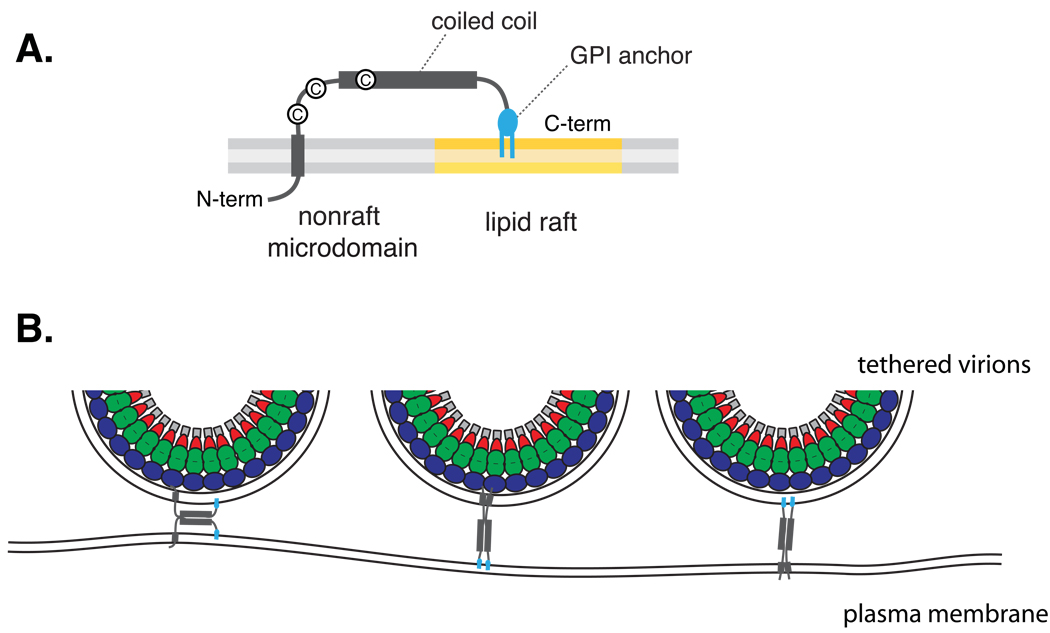
BST-2/tetherin is a type II transmembrane protein that consists of a short N-terminal cytoplasmic domain, a transmembrane domain, a dimerizing extracellular region containing a coiled-coil structure, and a C-terminal GPI anchor (Kupzig et al., 2003)(Fig. 3). Based on detergent resistance, this protein was identified as a raft-associated protein (Kupzig et al., 2003; Rollason et al., 2007). The GPI anchor but not the transmembrane domain mediates this raft association (Kupzig et al., 2003). Based on these findings, this protein was speculated to reside at the boundary between raft and nonraft regions (Kupzig et al., 2003). Because of its topology and dimerization ability, BST-2/tetherin is proposed to tether virus particles to the surface of producer cells by physically linking viral and plasma membranes (Neil et al., 2008) (Fig. 3), although an alternative or additional mechanisms may be possible (Goffinet et al., 2009; Miyagi et al., 2009). Supporting the physical tethering model, BST-2/tetherin has been observed to colocalize with Gag puncta on the cell surface (Jouvenet et al., 2008; Neil et al., 2008; Van Damme et al., 2008; Goffinet et al., 2009; Mitchell et al., 2009) and associate with budding virus particles (Perez-Caballero et al., 2009). Interestingly, BST-2/tetherin can inhibit the release of a wide variety of enveloped viruses including retroviruses, filoviruses, and arenaviruses (Jouvenet et al., 2008; Kaletsky et al., 2009; Sakuma et al., 2009) and inhibit HIV-1 release in cells derived from various species (Sato et al., 2009). Notably, a completely artificial protein designed to contain key structural features of BST-2/tetherin inhibits virus release like native BST-2/tetherin, despite the lack of sequence homology (Perez-Caballero et al., 2009). Therefore, if physical association between BST-2/tetherin and assembling virus particles is needed for its antiviral activity, such association is likely dependent on a common cellular structure rather than specific viral or cellular cofactor proteins. Together with the notion that many enveloped viruses associate with lipid rafts or other microdomains (Suomalainen, 2002; Briggs et al., 2003; Ono and Freed, 2005; Metzner et al., 2008), it was suggested that microdomain association of BST-2/tetherin may promote incorporation of this protein into assembling particles (Jouvenet et al., 2009). Consistent with a potential role for microdomains in BST-2/tetherin function, a cholesterol-binding compound amphotericin B methylester blocks the antagonistic activity of Vpu against BST-2/tetherin (Waheed et al., 2008). Furthermore, a BST-2/tetherin derivative lacking the GPI-anchor, which is unable to associate with DRMs (Kupzig et al., 2003), fails to inhibit virus release despite proper transport to the PM (Neil et al., 2008; Perez-Caballero et al., 2009). However, further investigation is needed to determine whether association of BST-2/tetherin with microdomains is essential for inhibition of virus release by this protein.
Fig. 3.
Structure and function of BST-2/tetherin. A. Key structural features and a possible mode of microdomain partitioning are shown. Cysteine residues that form disulfide bonds in BST-2/tetherin dimers are depicted. Glycosylation sites are not shown. B. Proposed modes for BST-2/tetherin-mediated physical linkage between virions and the plasma membrane are illustrated.
Involvement of PM microdomains in cell-to-cell virus transmission
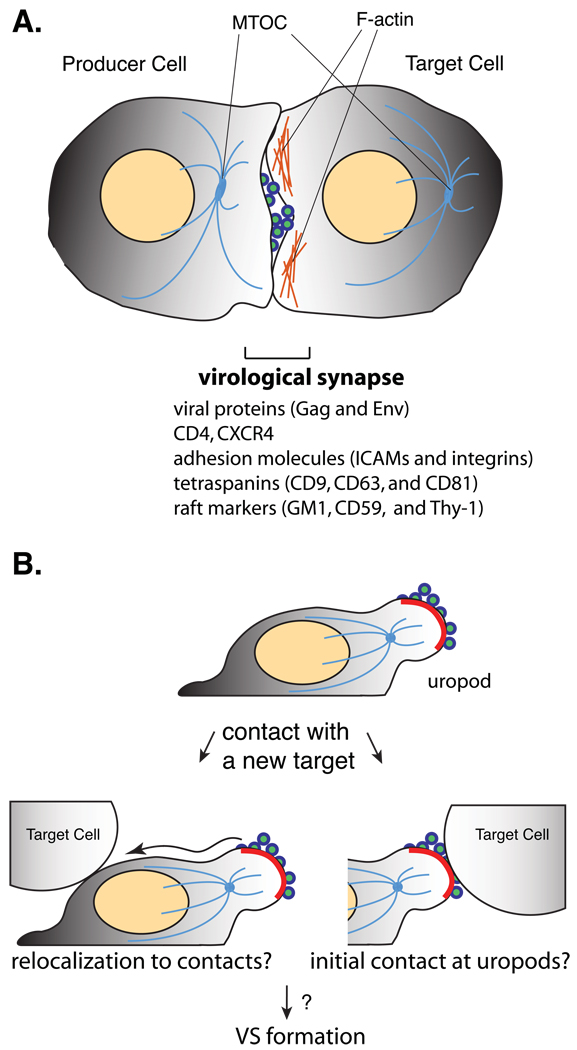
Another post-assembly process that may involve PM microdomains is cell-to-cell virus transmission. Virus transmission from infected cells to adjacent uninfected cells at cell-cell contacts has been shown to allow more efficient virus spread than infection by cell-free virions (Sato et al., 1992; Chen et al., 2007; Sourisseau et al., 2007; Sattentau, 2008; Sherer and Mothes, 2008). Recent microscopy-based studies revealed that retrovirus-producing cells form several different types of contact structures with target cells through which nascent virions can be transferred (Haller and Fackler, 2008; Sattentau, 2008; Sherer and Mothes, 2008). These structures include filopodial bridges, tunneling or membrane nanotubes, and virological synapses (VS) (Haller and Fackler, 2008; Sattentau, 2008; Sherer and Mothes, 2008). Attachment of tips of filopodia extended from one cell to another initiates filopodial bridges (Sherer et al., 2007), whereas nanotubes are likely formed after two conjugated cells detach from each other, at least in the case of T-cell nanotubes (Sowinski et al., 2008). Live cell imaging demonstrated that nascent virus particles move along the surface of these thin membranous structures from infected cells to uninfected cells (Sherer et al., 2007; Sowinski et al., 2008; Jin et al., 2009; Rudnicka et al., 2009). In contrast to filopodial bridges and nanotubes, the VS facilitates massive virus transfer at a short distance. The VS is a zone of contact formed between HIV- or HTLV-infected T cells and target T cells. This contact zone is enriched in viral Env and Gag proteins, cell adhesion and signaling molecules, and cytoskeletal proteins (Igakura et al., 2003; Jolly et al., 2004; Jolly et al., 2007a; Jolly et al., 2007b; Sol-Foulon et al., 2007; Arthos et al., 2008; Vasiliver-Shamis et al., 2009). A large number of assembled or assembling virus particles are observed in this junction (Fig. 4) (Jolly et al., 2004; Hubner et al., 2009; Rudnicka et al., 2009). Notably, markers for lipid rafts and TEM/ELDs are also enriched at the VS (Jolly and Sattentau, 2005; Jolly and Sattentau, 2007; Rudnicka et al., 2009). Furthermore, cholesterol depletion and treatment with anti-tetraspanin antibodies reduce formation of the VS (Jolly and Sattentau, 2005; Jolly and Sattentau, 2007). However, it is currently unknown what roles lipid rafts and/or TEM/ELDs play in VS formation.
Fig. 4.
Virological synapse and potential involvement of uropods in its formation. A. Key features of the virological synapse formed between a virus-producing T cell and a target T cell are shown. Major components of the virological synapse are listed. B. Accumulation of assembling and assembled viruses to uropods enriched in cell adhesion molecules may facilitate formation of virological synapses upon contact of uropods with a new target cell. Alternatively, virus-laden platforms may be formed at uropods and laterally move over the plasma membrane to cell-cell contacts at other area of virus-producing cells.
Although intact cytoskeletons (microtubules and actin filaments) (Jolly et al., 2007b; Sol-Foulon et al., 2007) and cell adhesion molecules (Jolly et al., 2007a; Arthos et al., 2008) (but see also (Puigdomenech et al., 2008)) have been shown to facilitate VS formation, little is understood about cellular events leading to establishment of the VS. In particular, whether precursors of the VS exist or whether the VS is formed de novo upon cell-cell contact remain to be determined. In recent live cell microscopy experiments, preexisting Gag-containing patches were observed to laterally move toward contact sites (Hubner et al., 2009; Rudnicka et al., 2009). Therefore, it is possible that Gag multimers and assembling particles first accumulate at a specific membrane region and form Gag-laden patches, which eventually constitute the VS upon cell-cell contact. In this regard, it is of note that in T cells, viral proteins often accumulate at one cellular pole (Fais et al., 1995; Deschambeault et al., 1999; Nguyen and Hildreth, 2000; Chen et al., 2007; Fang et al., 2007; Grigorov et al., 2009). In some cases, Gag was observed to localize to a protrusion resembling a uropod (Nguyen and Hildreth, 2000; Chen et al., 2007), a rear-end structure formed in migrating T cells (Sanchez-Madrid and Serrador, 2009). Indeed, we have observed that Gag highly colocalizes with several uropod markers including PSGL-1 and CD43 in polarized primary CD4+ T cells (Llewellyn and AO, unpublished data). Such specific localization of Gag to uropods may be particularly relevant for cell-to-cell spread because T cells are known to adopt a polarized morphology while migrating within lymphoid organs where cell-to-cell HIV-1 transmission likely occurs frequently (Germain et al., 2006; Krummel and Macara, 2006; Cahalan and Parker, 2008; Sanchez-Madrid and Serrador, 2009). The uropod is enriched in adhesion molecules and observed to mediate T cell-T cell contacts (Sanchez-Madrid and Serrador, 2009). Thus, polarized localization of viral components to a uropod may form a putative precursor for the VS. Notably, markers for lipid rafts and TEM/ELDs are known to accumulate to uropods in polarized T cells (Gomez-Mouton et al., 2001; Sala-Valdes et al., 2006). Lipid rafts containing flotillin are implicated in trafficking of PSGL-1 to uropods (Rossy et al., 2009). We observed that the tetraspanin CD81 copatches substantially with Gag at uropods (Llewellyn and AO, unpublished data). Notably, siRNA-mediated depletion of CD81, but not other tetraspanins, was observed to disperse polarized localization of Gag in T cells (Grigorov et al., 2009). Therefore, it is conceivable that PM microdomains associated with Gag multimers mediate polarized localization and assembly of Gag at a uropod, which eventually serves as a preformed platform for the VS. Filopodial contacts were found to induce polarized virus assembly in infected cells during cell-to-cell virus transmission between adherent cells (Jin et al., 2009). In the case of VS formation between migrating T cells, however, such polarity may already be established before cells initiate contact. In future studies on the cell-to-cell HIV-1 transmission, elucidating mechanisms promoting polarized localization of Gag and associated microdomains will likely be an important part of efforts toward better understanding of this mode of virus spread.
Concluding remarks
Biochemical and microscopy-based studies showed that two types of PM microdomains, lipid rafts and TEMs, are associated with the process of HIV-1 particle assembly. This association may depend on intrinsic affinity of HIV-1 Gag for preexisting microdomains, but it is also likely that Gag multimerization on the cytoplasmic leaflet creates a membrane environment suitable for recruiting microdomain components. Interactions of Gag with these microdomains likely facilitate particle assembly, Env incorporation into nascent virions, and/or efficient virus spread through cell-cell contacts. On the other hand, these interactions may also set the stage for host cells to impose restrictions on virus particle release by BST-2/tetherin or restrictions on Env-mediated virus-cell fusion by tetraspanins. With advances in our understanding of dynamic aspects of PM organization, future studies will no doubt elucidate molecular mechanisms underlying relationships between microdomains and assembling particles.
Acknowledgments
I thank Dr. Eric Freed and members of the Ono laboratory for helpful discussions and critical review of this manuscript. Work in my laboratory is supported by National Institute of Allergy and Infectious Diseases (R01 AI071727), American Heart Association (0850133Z), and amfAR (107449-45-RGHF).
Abbreviations list
- CA
capsid
- CTD
C-terminal domain
- DRM
detergent-resistant membrane
- ELD
endosome-like domain
- HIV-1
human immunodeficiency virus type 1
- MA
matrix
- NC
nucleocapsid
- PI(4,5)P2
phosphatidylinositol-(4,5)-bisphosphate
- PM
plasma membrane
- TEM
tetraspanin-enriched microdomain
- VS
virological synapse
References
- Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- Alfadhli A, Barklis RL, Barklis E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology. 2009a;387:466–472. doi: 10.1016/j.virol.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfadhli A, Still A, Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol. 2009b;83:12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Nayak DP. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology. 2000;276:289–303. doi: 10.1006/viro.2000.0556. [DOI] [PubMed] [Google Scholar]
- Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci U S A. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- Barman S, Nayak DP. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J Virol. 2000;74:6538–6545. doi: 10.1128/jvi.74.14.6538-6545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro O, Zamai M, Yanez-Mo M, Tejera E, Lopez-Romero P, Monk PN, Gratton E, Caiolfa VR, Sanchez-Madrid F. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol. 2008;183:527–542. doi: 10.1083/jcb.200805076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee E, McCormack A, Fruh K. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2006;2:e107. doi: 10.1371/journal.ppat.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavari S, Bosio CM, Wiegand E, Ruthel G, Will AB, Geisbert TW, Hevey M, Schmaljohn C, Schmaljohn A, Aman MJ. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exp Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AE, Narayan K, Shi D, Hartnell LM, Gousset K, He H, Lowekamp BC, Yoo TS, Bliss D, Freed EO, Subramaniam S. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 2009;5:e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8:89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya B, Roy P. Bluetongue virus outer capsid protein VP5 interacts with membrane lipid rafts via a SNARE domain. J Virol. 2008;82:10600–10612. doi: 10.1128/JVI.01274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya J, Peters PJ, Clapham PR. Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J Virol. 2004;78:5500–5506. doi: 10.1128/JVI.78.10.5500-5506.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya J, Repik A, Clapham PR. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J Virol. 2006;80:5292–5300. doi: 10.1128/JVI.01469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Wilk T, Fuller SD. Do lipid rafts mediate virus assembly and pseudotyping? J Gen Virol. 2003;84:757–768. doi: 10.1099/vir.0.18779-0. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci U S A. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahalan MD, Parker I. Choreography of cell motility and interaction dynamics imaged by two-photon microscopy in lymphoid organs. Annu Rev Immunol. 2008;26:585–626. doi: 10.1146/annurev.immunol.24.021605.090620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R, Uchil PD, Jin J, Shui G, Ott DE, Mothes W, Wenk MR. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420:133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- Charrin S, Manie S, Billard M, Ashman L, Gerlier D, Boucheix C, Rubinstein E. Multiple levels of interactions within the tetraspanin web. Biochem Biophys Res Commun. 2003;304:107–112. doi: 10.1016/s0006-291x(03)00545-x. [DOI] [PubMed] [Google Scholar]
- Charrin S, Manie S, Oualid M, Billard M, Boucheix C, Rubinstein E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 2002;516:139–144. doi: 10.1016/s0014-5793(02)02522-x. [DOI] [PubMed] [Google Scholar]
- Chen H, Dziuba N, Friedrich B, von Lindern J, Murray JL, Rojo DR, Hodge TW, O'Brien WA, Ferguson MR. A critical role for CD63 in HIV replication and infection of macrophages and cell lines. Virology. 2008;379:191–196. doi: 10.1016/j.virol.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Hubner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0908661107. Published online before print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claas C, Stipp CS, Hemler ME. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J Biol Chem. 2001;276:7974–7984. doi: 10.1074/jbc.M008650200. [DOI] [PubMed] [Google Scholar]
- Dalton AK, Ako-Adjei D, Murray PS, Murray D, Vogt VM. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J Virol. 2007;81:6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CA, Kenworthy AK. Tracking microdomain dynamics in cell membranes. Biochim Biophys Acta. 2009;1788:245–253. doi: 10.1016/j.bbamem.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol. 2007;177:329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschambeault J, Lalonde JP, Cervantes-Acosta G, Lodge R, Cohen EA, Lemay G. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol. 1999;73:5010–5017. doi: 10.1128/jvi.73.6.5010-5017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Derdowski A, Wang JJ, Spearman P. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J Virol. 2003;77:1916–1926. doi: 10.1128/JVI.77.3.1916-1926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou J, Wang JJ, Chen X, Li H, Ding L, Spearman P. Characterization of a myristoylated, monomeric HIV Gag protein. Virology. 2009;387:341–352. doi: 10.1016/j.virol.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Fruh K, Moses AV. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J Virol. 2009;83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M, Roy BB, Guiot-Guillain P, Mercier J, Binette J, Leung G, Cohen EA. Suppression of Tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J Virol. 2009;83:4574–4590. doi: 10.1128/JVI.01800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edidin M. The state of lipid rafts: from model membranes to cells. Annu Rev Biophys Biomol Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- Espenel C, Margeat E, Dosset P, Arduise C, Le Grimellec C, Royer CA, Boucheix C, Rubinstein E, Milhiet PE. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J Cell Biol. 2008;182:765–776. doi: 10.1083/jcb.200803010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais S, Capobianchi MR, Abbate I, Castilletti C, Gentile M, Cordiali Fei P, Ameglio F, Dianzani F. Unidirectional budding of HIV-1 at the site of cell-to-cell contact is associated with co-polarization of intercellular adhesion molecules and HIV-1 viral matrix protein. Aids. 1995;9:329–335. [PubMed] [Google Scholar]
- Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5:e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favoreel HW, Mettenleiter TC, Nauwynck HJ. Copatching and lipid raft association of different viral glycoproteins expressed on the surfaces of pseudorabies virus-infected cells. J Virol. 2004;78:5279–5287. doi: 10.1128/JVI.78.10.5279-5287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Heyden NV, Ratner L. Alpha interferon inhibits human T-cell leukemia virus type 1 assembly by preventing Gag interaction with rafts. J Virol. 2003;77:13389–13395. doi: 10.1128/JVI.77.24.13389-13395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus K, Chklovskaia E, Fazekas de St Groth B, Jessup W, Harder T. Condensation of the plasma membrane at the site of T lymphocyte activation. J Cell Biol. 2005;171:121–131. doi: 10.1083/jcb.200505047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain RN, Miller MJ, Dustin ML, Nussenzweig MC. Dynamic imaging of the immune system: progress, pitfalls and promise. Nat Rev Immunol. 2006;6:497–507. doi: 10.1038/nri1884. [DOI] [PubMed] [Google Scholar]
- Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, Oberbremer L, Kern C, Tibroni N, Welsch S, Krijnse-Locker J, Banting G, Krausslich HG, Fackler OT, Keppler OT. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Gomez CY, Hope TJ. Mobility of human immunodeficiency virus type 1 Pr55Gag in living cells. J Virol. 2006;80:8796–8806. doi: 10.1128/JVI.02159-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mouton C, Abad JL, Mira E, Lacalle RA, Gallardo E, Jimenez-Baranda S, Illa I, Bernad A, Manes S, Martinez AC. Segregation of leading-edge and uropod components into specific lipid rafts during T cell polarization. Proc Natl Acad Sci U S A. 2001;98:9642–9647. doi: 10.1073/pnas.171160298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DR, Chertova E, Hilburn JM, Arthur LO, Hildreth JE. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J Virol. 2003;77:8237–8248. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gri G, Molon B, Manes S, Pozzan T, Viola A. The inner side of T cell lipid rafts. Immunol Lett. 2004;94:247–252. doi: 10.1016/j.imlet.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Grigorov B, Attuil-Audenis V, Perugi F, Nedelec M, Watson S, Pique C, Darlix JL, Conjeaud H, Muriaux D. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology. 2009;6:28. doi: 10.1186/1742-4690-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Hue S, Schaller T, Verschoor E, Pillay D, Towers GJ. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 2009;5:e1000443. doi: 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller C, Fackler OT. HIV-1 at the immunological and T-lymphocytic virological synapse. Biol Chem. 2008;389:1253–1260. doi: 10.1515/BC.2008.143. [DOI] [PubMed] [Google Scholar]
- Halwani R, Khorchid A, Cen S, Kleiman L. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1. J Virol. 2003;77:3973–3984. doi: 10.1128/JVI.77.7.3973-3984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard-Peron E, Juillard F, Saad JS, Roy C, Roingeard P, Summers MF, Darlix JL, Picart C, Muriaux D. Targeting of murine leukemia virus gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the matrix. J Virol. 84:503–515. doi: 10.1128/JVI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- Henderson G, Murray J, Yeo RP. Sorting of the respiratory syncytial virus matrix protein into detergent-resistant structures is dependent on cell-surface expression of the glycoproteins. Virology. 2002;300:244–254. doi: 10.1006/viro.2002.1540. [DOI] [PubMed] [Google Scholar]
- Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci U S A. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue IB, Hoppe A, Ono A. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding. J Virol. 2009;83:7322–7336. doi: 10.1128/JVI.02545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm K, Weclewicz K, Hewson R, Suomalainen M. Human Immunodeficiency Virus Type 1 Assembly and Lipid Rafts: Pr55(gag) Associates with Membrane Domains That Are Largely Resistant to Brij98 but Sensitive to Triton X-100. J Virol. 2003;77:4805–4817. doi: 10.1128/JVI.77.8.4805-4817.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, Li XD, Asmuth DM, Huser T, Chen BK. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igakura T, Stinchcombe JC, Goon PK, Taylor GP, Weber JN, Griffiths GM, Tanaka Y, Osame M, Bangham CR. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science. 2003;299:1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- Jacobson K, Mouritsen OG, Anderson RG. Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- Janes PW, Ley SC, Magee AI. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Sherer NM, Heidecker G, Derse D, Mothes W. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 2009;7:e1000163. doi: 10.1371/journal.pbio.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. HIV-1 Cell to Cell Transfer across an Env-induced, Actin-dependent Synapse. J Exp Med. 2004;199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Mitar I, Sattentau QJ. Adhesion molecule interactions facilitate human immunodeficiency virus type 1-induced virological synapse formation between T cells. J Virol. 2007a;81:13916–13921. doi: 10.1128/JVI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Mitar I, Sattentau QJ. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J Virol. 2007b;81:5547–5560. doi: 10.1128/JVI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 virological synapse formation in T cells requires lipid raft integrity. J Virol. 2005;79:12088–12094. doi: 10.1128/JVI.79.18.12088-12094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81:7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson RL, Vogt VM, Johnson MC. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J Virol. 2009;83:4060–4067. doi: 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouve M, Sol-Foulon N, Watson S, Schwartz O, Benaroch P. HIV-1 buds and accumulates in "nonacidic" endosomes of macrophages. Cell Host Microbe. 2007;2:85–95. doi: 10.1016/j.chom.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2008 doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Krementsov DN, de Parseval A, Elder JH, Foti M, Thali M. Human immunodeficiency virus type 1 and influenza virus exit via different membrane microdomains. J Virol. 2007;81:12630–12640. doi: 10.1128/JVI.01255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krementsov DN, Weng J, Lambele M, Roy NH, Thali M. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology. 2009;6:64. doi: 10.1186/1742-4690-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel MF, Macara I. Maintenance and modulation of T cell polarity. Nat Immunol. 2006;7:1143–1149. doi: 10.1038/ni1404. [DOI] [PubMed] [Google Scholar]
- Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Koyama-Honda I, Suzuki K. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic. 2004;5:213–230. doi: 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- Langhorst MF, Reuter A, Stuermer CA. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol Life Sci. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Gosse JA, Holowka DA, Baird BA, Webb WW. Temporally resolved interactions between antigen-stimulated IgE receptors and Lyn kinase on living cells. J Cell Biol. 2005;171:527–536. doi: 10.1083/jcb.200503110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Naour F, Andre M, Boucheix C, Rubinstein E. Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics. 2006;6:6447–6454. doi: 10.1002/pmic.200600282. [DOI] [PubMed] [Google Scholar]
- Lee GE, Church GA, Wilson DW. A subpopulation of tegument protein vhs localizes to detergent-insoluble lipid rafts in herpes simplex virus-infected cells. J Virol. 2003;77:2038–2045. doi: 10.1128/JVI.77.3.2038-2045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- Leung K, Kim JO, Ganesh L, Kabat J, Schwartz O, Nabel GJ. HIV-1 assembly: viral glycoproteins segregate quantally to lipid rafts that associate individually with HIV-1 capsids and virions. Cell Host Microbe. 2008;3:285–292. doi: 10.1016/j.chom.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- Lichtenberg D, Goni FM, Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci. 2005;30:430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Lindwasser OW, Resh MD. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J Virol. 2001;75:7913–7924. doi: 10.1128/JVI.75.17.7913-7924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindwasser OW, Resh MD. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc Natl Acad Sci U S A. 2002;99:13037–13042. doi: 10.1073/pnas.212409999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Ries J, Schwille P, Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc Natl Acad Sci U S A. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London E, Brown DA. Insolubility of lipids in triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts) Biochim Biophys Acta. 2000;1508:182–195. doi: 10.1016/s0304-4157(00)00007-1. [DOI] [PubMed] [Google Scholar]
- Lorizate M, Brugger B, Akiyama H, Glass B, Muller B, Anderluh G, Wieland FT, Krausslich HG. Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. J Biol Chem. 2009;284:22238–22247. doi: 10.1074/jbc.M109.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman MG, Curanovic D, Enquist LW. Targeting of pseudorabies virus structural proteins to axons requires association of the viral Us9 protein with lipid rafts. PLoS Pathog. 2008;4:e1000065. doi: 10.1371/journal.ppat.1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manie SN, Debreyne S, Vincent S, Gerlier D. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J Virol. 2000;74:305–311. doi: 10.1128/jvi.74.1.305-311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S, Rao M. Rafts: scale-dependent, active lipid organization at the cell surface. Traffic. 2004;5:231–240. doi: 10.1111/j.1600-0854.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- Mazurov D, Heidecker G, Derse D. HTLV-1 Gag protein associates with CD82 tetraspanin microdomains at the plasma membrane. Virology. 2006;346:194–204. doi: 10.1016/j.virol.2005.10.033. [DOI] [PubMed] [Google Scholar]
- McCurdy LH, Graham BS. Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J Virol. 2003;77:1747–1756. doi: 10.1128/JVI.77.3.1747-1756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder D, Moreno MJ, Verkade P, Vaz WL, Simons K. Phase coexistence and connectivity in the apical membrane of polarized epithelial cells. Proc Natl Acad Sci U S A. 2006;103:329–334. doi: 10.1073/pnas.0509885103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina G, Pincetic A, Ehrlich LS, Zhang Y, Tang Y, Leis J, Carter CA. Tsg101 can replace Nedd4 function in ASV Gag release but not membrane targeting. Virology. 2008;377:30–38. doi: 10.1016/j.virol.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian KA, Ostermeyer AG, Chen JZ, Roth MG, Brown DA. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J Biol Chem. 1999;274:3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- Metzner C, Salmons B, Gunzburg WH, Dangerfield JA. Rafts, anchors and viruses--a role for glycosylphosphatidylinositol anchored proteins in the modification of enveloped viruses and viral vectors. Virology. 2008;382:125–131. doi: 10.1016/j.virol.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 2009;5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Andrew AJ, Kao S, Strebel K. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc Natl Acad Sci U S A. 2009;106:2868–2873. doi: 10.1073/pnas.0813223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow IC, Parton RG. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- Neil SJ, Eastman SW, Jouvenet N, Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta T, Kuznetsov Y, McPherson A, Fan H. Murine leukemia virus glycosylated Gag (gPr80gag) facilitates interferon-sensitive virus release through lipid rafts. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0908660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol. 2006;173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A. HIV-1 Assembly at the Plasma Membrane: Gag Trafficking and Localization. Future Virol. 2009;4:241–257. doi: 10.2217/fvl.09.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. The role of lipid rafts in virus replication. Elsevier. 2005:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- Ono A, Waheed AA, Freed EO. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology. 2007;360:27–35. doi: 10.1016/j.virol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Waheed AA, Joshi A, Freed EO. Association of human immunodeficiency virus type 1 gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J Virol. 2005;79:14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE. Cellular proteins detected in HIV-1. Rev Med Virol. 2008;18:159–175. doi: 10.1002/rmv.570. [DOI] [PubMed] [Google Scholar]
- Panchal RG, Ruthel G, Kenny TA, Kallstrom GH, Lane D, Badie SS, Li L, Bavari S, Aman MJ. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc Natl Acad Sci U S A. 2003;100:15936–15941. doi: 10.1073/pnas.2533915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews A, Kramer B, Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol. 2003;162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickl WF, Pimentel-Muinos FX, Seed B. Lipid rafts and pseudotyping. J Virol. 2001;75:7175–7183. doi: 10.1128/JVI.75.15.7175-7183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- Pralle A, Keller P, Florin EL, Simons K, Horber JK. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J Cell Biol. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigdomenech I, Massanella M, Izquierdo-Useros N, Ruiz-Hernandez R, Curriu M, Bofill M, Martinez-Picado J, Juan M, Clotet B, Blanco J. HIV transfer between CD4 T cells does not require LFA-1 binding to ICAM-1 and is governed by the interaction of HIV envelope glycoprotein with CD4. Retrovirology. 2008;5:32. doi: 10.1186/1742-4690-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 2002;3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- Rollason R, Korolchuk V, Hamilton C, Schu P, Banting G. Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J Cell Sci. 2007;120:3850–3858. doi: 10.1242/jcs.003343. [DOI] [PubMed] [Google Scholar]
- Rong L, Zhang J, Lu J, Pan Q, Lorgeoux RP, Aloysius C, Guo F, Liu SL, Wainberg MA, Liang C. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J Virol. 2009;83:7536–7546. doi: 10.1128/JVI.00620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossy J, Schlicht D, Engelhardt B, Niggli V. Flotillins interact with PSGL-1 in neutrophils and, upon stimulation, rapidly organize into membrane domains subsequently accumulating in the uropod. PLoS One. 2009;4:e5403. doi: 10.1371/journal.pone.0005403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicka D, Feldmann J, Porrot F, Wietgrefe S, Guadagnini S, Prevost MC, Estaquier J, Haase AT, Sol-Foulon N, Schwartz O. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J Virol. 2009;83:6234–6246. doi: 10.1128/JVI.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Mateos E, Pelchen-Matthews A, Deneka M, Marsh M. CD63 is not required for production of infectious human immunodeficiency virus type 1 in human macrophages. J Virol. 2008;82:4751–4761. doi: 10.1128/JVI.02320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, Soheilian F, Freed EO, Summers MF. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382:434–447. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifuddin M, Parker CJ, Peeples ME, Gorny MK, Zolla-Pazner S, Ghassemi M, Rooney IA, Atkinson JP, Spear GT. Role of virion-associated glycosylphosphatidylinositol-linked proteins CD55 and CD59 in complement resistance of cell line-derived and primary isolates of HIV-1. J Exp Med. 1995;182:501–509. doi: 10.1084/jem.182.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Noda T, Urata S, Kawaoka Y, Yasuda J. Inhibition of Lassa and Marburg virus production by tetherin. J Virol. 2009;83:2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sanchez-Madrid F, Yanez-Mo M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem. 2006;281:19665–19675. doi: 10.1074/jbc.M602116200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Madrid F, Serrador JM. Bringing up the rear: defining the roles of the uropod. Nat Rev Mol Cell Biol. 2009;10:353–359. doi: 10.1038/nrm2680. [DOI] [PubMed] [Google Scholar]
- Sanderson CM, Avalos R, Kundu A, Nayak DP. Interaction of Sendai viral F, HN, and M proteins with host cytoskeletal and lipid components in Sendai virus-infected BHK cells. Virology. 1995;209:701–707. doi: 10.1006/viro.1995.1308. [DOI] [PubMed] [Google Scholar]
- Sato H, Orenstein J, Dimitrov D, Martin M. Cell-to-cell spread of HIV-1 occurs within minutes and may not involve the participation of virus particles. Virology. 1992;186:712–724. doi: 10.1016/0042-6822(92)90038-q. [DOI] [PubMed] [Google Scholar]
- Sato K, Aoki J, Misawa N, Daikoku E, Sano K, Tanaka Y, Koyanagi Y. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. J Virol. 2008;82:1021–1033. doi: 10.1128/JVI.01044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Yamamoto SP, Misawa N, Yoshida T, Miyazawa T, Koyanagi Y. Comparative study on the effect of human BST-2/Tetherin on HIV-1 release in cells of various species. Retrovirology. 2009;6:53. doi: 10.1186/1742-4690-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat Rev Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- Sharma P, Varma R, Sarasij RC, Ira, Gousset K, Krishnamoorthy G, Rao M, Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, Mothes W. Cytonemes and tunneling nanotubules in cell-cell communication and viral pathogenesis. Trends Cell Biol. 2008;18:414–420. doi: 10.1016/j.tcb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkriabai N, Datta SA, Zhao Z, Hess S, Rein A, Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- Shvartsman DE, Kotler M, Tall RD, Roth MG, Henis YI. Differently anchored influenza hemagglutinin mutants display distinct interaction dynamics with mutual rafts. J Cell Biol. 2003;163:879–888. doi: 10.1083/jcb.200308142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. Lipid Rafts and Signal Transduction. Nature Reviews. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- Sol-Foulon N, Sourisseau M, Porrot F, Thoulouze MI, Trouillet C, Nobile C, Blanchet F, di Bartolo V, Noraz N, Taylor N, Alcover A, Hivroz C, Schwartz O. ZAP-70 kinase regulates HIV cell-to-cell spread and virological synapse formation. Embo J. 2007;26:516–526. doi: 10.1038/sj.emboj.7601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J Virol. 2007;81:1000–1012. doi: 10.1128/JVI.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Kohler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, Onfelt B, Sattentau Q, Davis DM. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol. 2008;10:211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- Stansell E, Apkarian R, Haubova S, Diehl WE, Tytler EM, Hunter E. Basic residues in the Mason-Pfizer monkey virus gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J Virol. 2007;81:8977–8988. doi: 10.1128/JVI.00657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M. Lipid rafts and assembly of enveloped viruses. Traffic. 2002;3:705–709. doi: 10.1034/j.1600-0854.2002.31002.x. [DOI] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varthakavi V, Smith RM, Bour SP, Strebel K, Spearman P. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc Natl Acad Sci U S A. 2003;100:15154–15159. doi: 10.1073/pnas.2433165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliver-Shamis G, Cho MW, Hioe CE, Dustin ML. Human immunodeficiency virus type 1 envelope gp120-induced partial T-cell receptor signaling creates an F-actin-depleted zone in the virological synapse. J Virol. 2009;83:11341–11355. doi: 10.1128/JVI.01440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S, Gerlier D, Manie SN. Measles virus assembly within membrane rafts. J Virol. 2000;74:9911–9915. doi: 10.1128/jvi.74.21.9911-9915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed AA, Ablan SD, Soheilian F, Nagashima K, Ono A, Schaffner CP, Freed EO. Inhibition of human immunodeficiency virus type 1 assembly and release by the cholesterol-binding compound amphotericin B methyl ester: evidence for Vpu dependence. J Virol. 2008;82:9776–9781. doi: 10.1128/JVI.00917-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Krausslich HG. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 2007;3:e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Krementsov DN, Khurana S, Roy NH, Thali M. Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. J Virol. 2009;83:7467–7474. doi: 10.1128/JVI.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19:434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Yang P, Ai LS, Huang SC, Li HF, Chan WE, Chang CW, Ko CY, Chen SS. The Cytoplasmic Domain of Human Immunodeficiency Virus Type 1 Transmembrane Protein Gp41 Harbors Lipid Raft-Association Determinants. J Virol. 2009 doi: 10.1128/JVI.00899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zhang J, Pekosz A, Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J Virol. 2000;74:4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]