Abstract
The mouse mammary tumor virus (MMTV) promoter is induced by glucocorticoid hormone. A robust hormone- and receptor-dependent activation could be reproduced in Xenopus laevis oocytes. The homogeneous response in this system allowed a detailed analysis of the transition in chromatin structure following hormone activation. This revealed two novel findings: hormone activation led to the establishment of specific translational positioning of nucleosomes despite the lack of significant positioning in the inactive state; and, in the active promoter, a subnucleosomal particle encompassing the glucocorticoid receptor (GR)-binding region was detected. The presence of only a single GR-binding site was sufficient for the structural transition to occur. Both basal promoter elements and ongoing transcription were dispensable. These data reveal a stepwise process in the transcriptional activation by glucocorticoid hormone.
Keywords: chromatin structure/glucocorticoid receptor/MMTV promoter/nucleosome positioning/Xenopus oocyte
Introduction
All eukaryotic cells have their DNA packaged into a protein–DNA structure, chromatin. The basic subunit of chromatin is the nucleosome (Luger et al., 1997), which can be located at specific DNA segments in the eukaryotic genome (Simpson, 1991). Translational nucleosome positioning depends on local variations in DNA curvature, helical periodicity and/or boundary effects. When present in gene regulatory regions, nucleosomes can act as a barrier to the process of transcriptional initiation (Han et al., 1988; Perlmann and Wrange, 1991) in a gene-specific manner (Wyrick et al., 1999). This repressive effect of chromatin is modulated at specific loci by the rapid remodeling of the chromatin structure during gene activation. One well studied example of transcription activation-dependent chromatin remodeling is the mouse mammary tumor virus (MMTV) promoter. This promoter is strongly induced by glucocorticoid hormone. Activation is associated with the appearance of a DNase I-hypersensitive site (Zaret and Yamamoto, 1984; Truss et al., 1995) over the glucocorticoid response element (GRE). Micrococcal nuclease (MNase) and methidiumpropyl-EDTA⋅iron(II) complex (MPE) digestion of the MMTV long terminal repeat (LTR) in situ initially revealed that this regulatory DNA segment harbors six translationally positioned nucleosomes and that nucleosome B, positioned over the GRE, undergoes an activation-dependent remodeling (Richard-Foy and Hager, 1987). Further mapping experiments at high resolution showed that nucleosomes, although not precisely positioned, displayed a clustered distribution (Fragoso et al., 1995). Interestingly, despite the drastic increase in transcription activation and the occurrence of a glucocorticoid-dependent DNase I-hypersensitive site (see above), careful in vivo analyses have failed to detect any hormone-induced changes at the level of nucleosomal organization for the MMTV LTR stably incorporated in tissue culture cells (Fragoso et al., 1995; Truss et al., 1995). In these in vivo studies, the positioned nucleosome ladder seemed to remain unchanged independent of the transcriptional status of the promoter.
In vivo footprinting experiments showed that basal transcription factors, such as nuclear factor 1 (NF1) and TATA-box binding factor (TFIID), do not interact with their cognate target sites in the MMTV promoter unless a hormone-induced chromatin remodeling event has taken place (Cordingley et al., 1987; Archer et al., 1992; Truss et al., 1995). This suggested a role for chromatin in keeping the inactive promoter in a closed configuration (Han et al., 1988; Perlmann and Wrange, 1991). As a direct implication, chromatin rearrangements should occur to permit the transition towards an active state. The discovery that transcription coactivators had histone acetyltransferase, histone deacetylase and nucleosome remodeling properties, as reviewed in Kingston and Narlikar (1999), further provided the possible molecular players in this process. However, structural aspects of in vivo chromatin remodeling during gene activation remain obscure.
Xenopus oocytes represent an attractive in vivo system to follow these issues. Estrogen- (Theulaz et al., 1988), glucocorticoid- (Perlmann and Wrange, 1991) and thyroid hormone-dependent (Wong et al., 1998) gene regulation occur in Xenopus oocytes merely by expression of the appropriate receptor protein(s) and by injection of a DNA reporter plasmid. Here, we revealed a robust glucocorticoid-dependent and transcription-coupled chromatin remodeling over the GRE of the MMTV LTR. This remodeling was homogeneous and, in contrast to previous results in tissue culture cells, it involved major rearrangements at the nucleosome level. The hormone-induced chromatin remodeling of the MMTV promoter resulted in induction of translational positioning of initially randomly organized nucleosomes. This was independent of basal promoter elements and of ongoing transcription, but required a high affinity glucocorticoid receptor (GR)-binding site, highlighting the distinct steps involved in hormone activation in vivo. We conclude that nucleosome positioning in the MMTV LTR is not functionally required to achieve hormone-dependent induction but is a consequence of the induction event.
Results
Chromatin assembly and glucocorticoid hormone induction in Xenopus oocytes
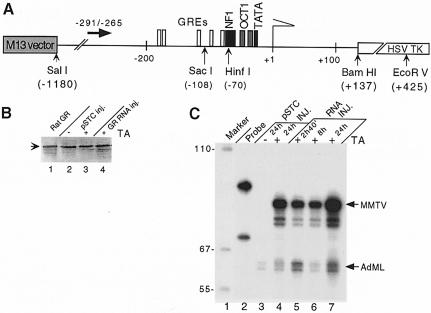
The MMTV LTR was fused to the herpes simplex virus thymidine kinase (HSVTK) gene coding sequence (Buetti and Kuhnel, 1986) and propagated in the M13 filamentous phage (Figure 1A). It was used for intranuclear Xenopus oocyte injections in single-stranded (ss) form. This results in replication-coupled chromatin assembly of the injected ssDNA (Almouzni and Wolffe, 1993), which leads to formation of naturally spaced chromatin. This can be monitored after MNase digestion by the appearance of DNA fragments whose lengths are multiples of the size corresponding to a nucleosome repeat length (Figure 2B).
Fig. 1. Reconstitution of glucocorticoid regulation in Xenopus oocytes. (A) The reporter DNA construct, the pMTV:M13 coding vector with the primer used for primer extension analysis of the SacI in situ accessibility assay (solid black arrow) and the restriction enzyme cleavage sites that are referred to in the text. White boxes, GRE hexanucleotide elements; black box, NF1 site; light gray box, OCT 1 site; and dark gray box, TATA-box sequence. (B) GR expression in oocytes. Western blot of SDS–PAGE: lane 1, GR prepared from rat liver (Perlmann and Wrange, 1988); lanes 2 and 3, one Xenopus oocyte equivalent was analyzed 24 h after injection of 5 ng of pSTC GR 3-795 expression vector; lane 4, one oocyte equivalent injected with 5 ng of in vitro transcribed GR RNA 24 h before analysis. Hormone (TA, 1 μM) was added as indicated. (C) Hormone-dependent MMTV transcription in Xenopus oocytes. Transcription analysis by S1 nuclease protection of MMTV and the AdML promoter. Oocytes in groups of five were injected with 1 ng of pMTV:M13 coding vector ssDNA and 0.25 ng of pAdML reference and either 5 ng of GR expression vector (pSTC GR 3-795) (lanes 3 and 4) or 5 ng of in vitro transcribed GR RNA (lanes 5–7). After 24 h, hormone (TA, 1 μM) was added to oocyte culture media and oocytes were harvested for RNA analysis at the time indicated.
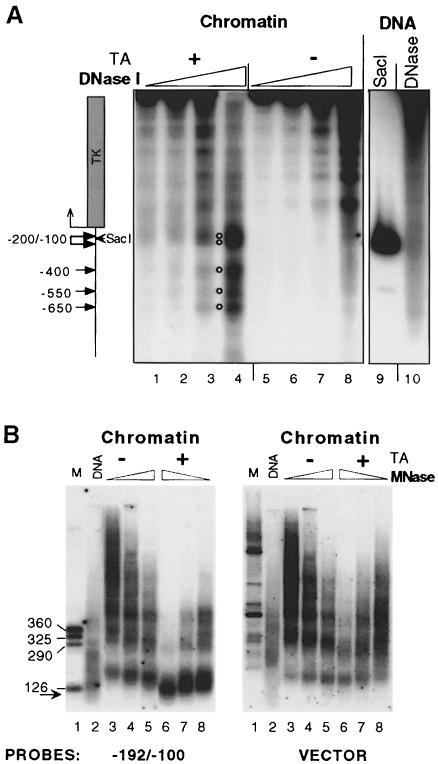
Fig. 2. Chromatin structure of the MMTV promoter. (A) Hormone-dependent DNase I-hypersensitive sites are located in the MMTV LTR. Groups of 12 oocytes were injected with 1 ng of pMTV:M13 coding ssDNA, 5 ng of dsDNA for pSTC GR 3-795 and 0.25 ng for pAdML reference (lanes 1–8). After overnight incubation, hormone was added (TA; 1 μM) (lanes 1–4) or not added (lanes 5–8) and oocytes were harvested after 24 h for the DNase I hypersensitivity assay. Lane 9, internal molecular weight marker showing the position of the SacI restriction enzyme cut. Lane 10, naked dsMMTV promoter DNA digested with DNase I. (B) MNase in situ digestion shows hormone-dependent disruption of the canonical nucleosome structure in the vicinity of GRE elements. Groups of 10 oocytes were injected. The next day, hormone (TA; 1 μM) was added as indicated and oocytes were harvested after 24 h for MNase digestion. DNA was resolved in an agarose gel, transferred and hybridized with a labeled MMTV promoter probe encompassing region –192/–100, and then washed and rehybridized with an M13 vector probe. Lane 1, internal DNA marker; lane 2, naked dsMMTV promoter DNA digested with MNase. The arrow shows a subnucleosomal particle ∼120 bp DNA fragment revealed only after hybridization with specific probe.
GR protein is required to elicit a hormone response in Xenopus oocytes (Perlmann and Wrange, 1991). Full-length GR protein was provided either by nuclear injection of a cytomegalovirus (CMV) promoter-driven rat GR cDNA expression vector (pSTC GR 3-795), or by cytoplasmic injection of in vitro synthesized rat GR RNA. The presence of full-length GR protein was verified by immunoblotting (Figure 1B). The accumulation of the MMTV-specific mRNA was assayed by S1 nuclease protection analysis. The MMTV promoter was virtually silent in the absence of hormone (Figure 1C). In contrast, a strong induction was observed after addition of 1 × 10–6 M synthetic glucocorticoid hormone triamcinolone acetonide (TA) to the oocyte culture medium (Figure 1C). This MMTV promoter-driven transcription was already detectable after 2 h (data not shown). Xenopus oocytes translate injected RNA efficiently (Colman, 1984). The injection of in vitro transcribed GR RNA gave rise to a stronger transcriptional response than that obtained using the GR expression vector strategy (compare lanes 4 and 7 in Figure 1C). As a consequence, GR was usually provided by injection of GR RNA in subsequent experiments.
Glucocorticoid-induced chromatin remodeling in the MMTV LTR
To follow possible chromatin changes upon hormone induction, Xenopus oocytes were injected with ssMMTV reporter DNA and GR expression vector and incubated with TA (1 μM) for 24 h. The oocytes were then homogenized and digested with increasing amounts of DNase I. Digestion products were analyzed by an indirect end-labeling assay (Wu, 1989). These experiments revealed several glucocorticoid-dependent DNase I-hypersensitive segments within the MMTV LTR. The strongest DNase I hypersensitivity was distributed around position –200/–100, which includes the MMTV GRE, –185/–79 (compare Figure 2A, lane 4, labeled with a double circle, with lane 8 non-hormone-treated oocytes, and the location of the SacI restriction site at –108, lane 9). There were additional hormone-dependent DNase I-hypersensitive regions with a lower intensity further upstream within the MMTV LTR. These were distributed around three positions, –400, –550 and –650 (Figure 2A, lane 4, open circles). The DNase I hypersensitivity at the –200/–100 position, however, was always the most prominent, and the extent of DNase I hypersensitivity correlated with the extent of MMTV transcription, as measured by S1 nuclease analysis (data not shown).
MNase digestion was used to examine in more detail the effects of hormone activation on chromatin structure in the vicinity of the GRE (–185/–79) (Payvar et al., 1983; Buetti and Kuhnel, 1986). Injected oocytes were homogenized and digested with MNase. Isolated DNA was resolved on an agarose gel, blotted and probed with a short MMTV promoter fragment –192/–100, covering the strongest DNase I-hypersensitive area. The substantial alteration of the canonical MNase ladder indicated that hormone activation leads to drastically increased MNase cutting of the DNA in the vicinity of the GRE region (Figure 2B left, compare lanes 3–5 with 6–8). Unexpectedly, the mononucleosome fraction at 146–185 bp, which was present in the inactive promoter, was replaced by an unusual subnucleosomal particle protecting a DNA fragment of ∼120 ± 10 bp, in the active promoter (Figure 2B, left, compare lane 5 with lane 6). The relative resistance of this subnucleosomal particle to MNase digestion and its discrete migration on the agarose gel suggest that it represents a defined DNA–protein complex. Importantly, the signal corresponding to this subnucleosomal particle reflects the fact that chromatin remodeling involves the vast majority of the MMTV DNA copies. Reprobing the filter of the MNase in situ digested chromatin with M13 vector DNA as probe (Figure 2B, right) showed that the hormone-induced subnucleosomal particle was not present in the vector DNA (compare lanes 6–8 in the left and right panels). There is, however, a slight but clearly detectable hormone-dependent increase in MNase digestion in the vector DNA (Figure 2B, right, compare lanes 3–5 with lanes 6–8). We attribute this to chromatin ‘domain’ effects of the strong transcriptional response.
We conclude that transcription activation of the MMTV promoter results in (i) increased accessibility of DNA in the vicinity of the GRE segment; (ii) reorganization of the DNA harboring the GRE into a subnucleosomal protein–DNA complex that protects ∼120 ± 10 bp of DNA; and (iii) virtually all copies of the MMTV template undergoing a similar nucleosome remodeling event in the vicinity of the GRE region.
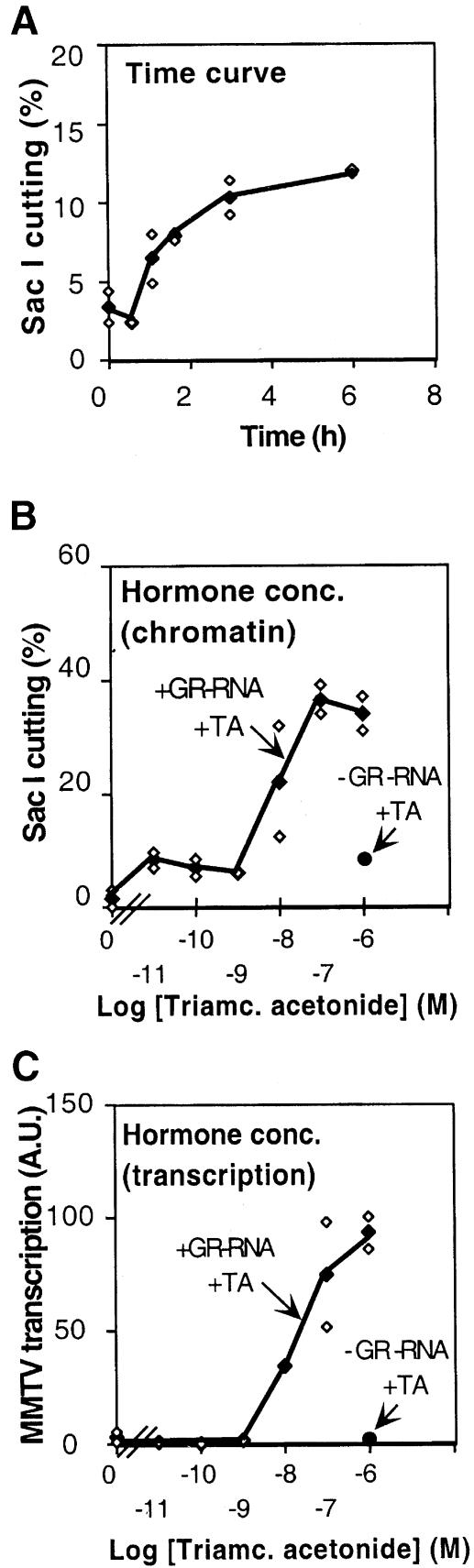
Chromatin remodeling can be followed by in situ digestion of DNA with an appropriate restriction enzyme that cuts within the remodeled chromatin region (Archer et al., 1992; Truss et al., 1995; Fragoso et al., 1998). We used the SacI restriction enzyme, which cuts the MMTV promoter at position –108 within the GRE segment. For quantitation, we carried out a primer extension analysis with a primer annealing to the DNA strand that is synthesized in the oocyte after ssDNA injection. In this way, molecules assembled into chromatin are specifically revealed. A distinct increase in SacI cutting 1 h after hormone addition was observed and it reached a plateau after 3–6 h (Figure 3A). In addition, a significant stimulation of both chromatin remodeling (Figure 3B) and MMTV transcription (Figure 3C) was detected at a hormone concentration of 10 nM. This illustrates the parallel between chromatin remodeling and accumulation of MMTV RNA. Both hormone-dependent SacI cutting and MMTV transcription were dependent on the presence of GR, which was provided here by GR RNA injection (see inset right part of Figure 3B and C for oocytes not injected with GR RNA). We conclude that glucocorticoid hormone-induced chromatin remodeling (quantified by a SacI in situ cutting assay) and transcription are closely correlated in the MMTV promoter.
Fig. 3. Evaluation of the effect of time and hormone concentration on hormone-induced chromatin remodeling. (A) Oocytes were injected with 10 ng of GR RNA and 1 ng of ssDNA pMTV:M13 coding strand, and 0.25 ng of pAdML for reference. After overnight incubation, oocytes were divided into 12 groups of five oocytes each; 1 μM TA was added at various times. Oocytes were homogenized and two-thirds taken for SacI in situ accessibility assay and one-third for RNA analysis (not shown). White diamonds signify each individual analysis as quantified by PhosophorImager, and black diamonds the mean value for each double sample. (B) Oocytes were injected with DNA and GR RNA (+GR-RNA) or with DNA only (–GR-RNA) and the next day divided into 16 groups with six oocytes in each and treated with the indicated concentrations of hormone (TA) for 9 h and then homogenized. Two-thirds was taken for SacI and one-third for RNA analysis. Symbols as in (A). Log [TA] is given on the abscissa. (C) Quantitaion of MMTV RNA relative to AdML RNA of the experiment described in (B) using S1 nuclease protection assay and PhosphorImager analysis, arbitrary units (A.U.). Symbols as in (A).
Hormone activation induces translational nucleosome positioning in the MMTV LTR
The presence of many sequence-specific MNase cut sites in the MMTV LTR (Richard-Foy and Hager, 1987) made the use of this enzyme inappropriate to determine nucleosome positioning over these sequences. Thus, we used the chemical nuclease MPE, which has a strong preference for internucleosomal regions and that, due to its small size, shows almost no sequence specificity in DNA cleavage (Richard-Foy and Hager, 1987; Truss et al., 1995).
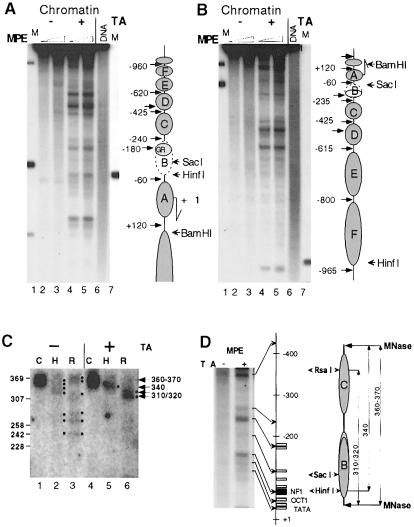
Figure 4A and B shows the MPE cleavage pattern of the MMTV promoter. Surprisingly, we observed no obvious nucleosomal pattern in the inactive MMTV promoter (Figure 4A and B, lanes 2 and 3). After hormone-induced transcription activation, however, a distinct cleavage pattern was seen, suggesting a strong nucleosome positioning over the entire MMTV LTR (compare lanes 2 and 3 with lanes 4 and 5 in Figure 4A and B). The nucleosome positioning revealed here by MPE digestion experiments in hormone-treated Xenopus oocytes coincides with that shown previously for the MMTV promoter stably incorporated into bovine papilloma virus (BPV)-based episomal vector constructs in tissue culture cells (Richard-Foy and Hager, 1987; Truss et al., 1995). In agreement with these results, our MPE cleavage pattern suggests that at least six nucleosomes are positioned over the MMTV LTR. Our interpretation of this MPE-induced pattern with respect to nucleosome positioning is shown (Figure 4A and B, diagram on the right). We have termed these positioned nucleosomes A–F following the previously used nomenclature (Richard-Foy and Hager, 1987). Contrary to previous findings, however, we did not observe any significant translational nucleosome positioning in the inactive MMTV promoter.
Fig. 4. Hormone-induced nucleosome positioning analyzed by MPE and MNase digestion in situ. (A and B) Transcriptional activation leads to establishment of nucleosome positioning along the MMTV promoter. Injected oocytes were analyzed after 24 h of hormone treatment. MPE digestion was performed for 3 min (lanes 2 and 4) and 10 min (lanes 3 and 5). Isolated DNA was digested with SalI and EcoRV, resolved on agarose, blotted and hybridized first with a random-primed labeled fragment adjacent to the EcoRV site (EcoRV–SacI fragment in A) and then stripped and reprobed with SalI–RsaI (B). Lanes 1 and 7, internal molecular weight markers (see map to the right); lanes 2–5, MPE digestion of injected oocytes, treated (lanes 4 and 5) and not treated (lanes 2 and 3) with hormone; lane 6, naked dsMMTV promoter DNA digested with MPE. To the right in (A) and (B) is a schematic summary of MPE cuts along the MMTV LTR with putative nucleosome positions. (C) Mapping of dinucleosome borders suggests that nucleosomes are translationally positioned along the MMTV promoter only after activation of transcription. Groups of 10 oocytes were hormone treated as in (A) and (B) and MNase digested as in Figure 2B (lanes 3 and 8). DNA was isolated and resolved in a 4% NuSieve GTG agarose gel together with size markers. The band corresponding to dinucleosome DNA (360–370 bp in length) was excised from the gel, DNA was eluted and analyzed as a control (lanes 1 and 4) or digested either with HinfI (lanes 2 and 5) or RsaI (lanes 3 and 6). DNA was resolved in a 1% SeaKem GTG + 2.5% NuSieve GTG agarose gel, blotted and hybridized with a random-primed probe encompassing region –415/–100. Black dots outline the DNA bands revealed by hybridization. (D) Chromatin organization of the MMTV promoter as revealed by MNase and MPE mapping. A magnified section of lanes 3 and 4 in (A) is shown together with a schematic presentation of the MMTV LTR and the restriction enzyme cleavage sites. All symbols are as in Figure 1A. The positions of the nucleosomes (on the right) are based on the results in (A–C). The co-localizations of MPE cuts with internucleosome linkers and/or factor-binding sites are indicated by arrows.
To determine to what extent the observed MPE cleavage pattern was caused by nucleosome positioning, we used an alternative strategy to map the borders of the putative dinucleosome that covers the –425/–60 segment of the MMTV promoter. This was achieved by isolating dinucleosomal DNA (360–370 bp) from an agarose gel after MNase digestion and determining its borders by restriction enzyme cutting. We reasoned that the agarose gel size selection would reduce the influence of any local sequence specificity of MNase cutting. Oocytes were processed for MNase digestion. Dinucleosomal DNA was isolated and cleaved with either HinfI or RsaI restriction enzymes (Figure 4D; the diagram on the right side displays the locations of restriction sites). The resulting DNA samples were resolved in a 3.5% agarose gel, blotted and hybridized with an α–32P-labeled MMTV promoter probe that encompassed the –415/–100 segment of MMTV DNA. The hormone-activated MMTV promoter cleavage with HinfI resulted in conversion of the 360–370 bp dinucleosome DNA fragment into a discrete 340 bp fragment, while cleavage with RsaI generated distinct bands of 310 and 320 bp (Figure 4C, lanes 5 and 6). In contrast, digestion of the dinucleosomal DNA from the inactive MMTV promoter (not treated with hormone) with either HinfI or RsaI resulted in several bands ranging from 240 to 370 bp in size (Figure 4C, lanes 2 and 3). This result is in agreement with the MPE data, and the results taken together strongly suggest that the hormone-dependent activation of the MMTV promoter induces a precise nucleosome positioning of initially randomly organized nucleosomes. The additional MPE cut sites, which do not coincide with the location of the nucleosome linkers, could possibly reflect transactivating factors binding to DNA (Figure 4D, e.g. the –180 cut site, which coincides with a strong GR-binding site). These cut sites are clustered within and proximal to the nucleosome B segment where most transactive factors bind. Overall, the nucleosome B segment displays hormone-dependent hypercutting. This is in contrast to the nucleosome C segment, which shows hormone-induced MPE protection, probably reflecting translational positioning and the relative lack of factor binding in this region (Figure 4A, B and D).
Hormone-induced nucleosome positioning depends on the presence of GR-binding site(s) but not on OCT1, NF1 or TATA-box elements
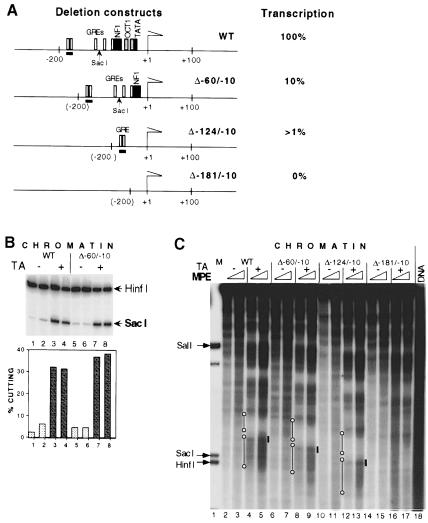
To evaluate the influence of different promoter elements and their cognate transactivating factors on hormone-dependent chromatin remodeling, we created three MMTV LTR deletion mutants. These mutants and the wild type are represented in Figure 5A. Their hormone-dependent transcriptional activity was 10%, 1% and non-detectable for the Δ–60/–10, Δ–124/–10 and Δ–181/–10 mutants, respectively, relative to the wild type (data not shown). None of the mutants displayed constitutive MMTV expression.
Fig. 5. Nucleosome remodeling and establishment of nucleosome positioning are dependent on GR binding but not on other basal promotor elements. (A) Maps of MMTV deletion mutants. Names of mutants signify the base pairs that were deleted relative to the transcription initiation start (+1). The strong GRE site at position –185/–171 in the wild type and the corresponding site in Δ–60/–10 and Δ–124/–10 mutants are underlined. Hormone-dependent transcriptional efficiency relative to wild type, as measured by S1 nuclease protection, is given on the right. (B) Nucleosome remodeling in the vicinity of the GRE elements in wild type (lanes 1–4) and Δ–60/–10 mutant (lanes 5–8) as revealed by SacI restriction enzyme accessibility assay. Groups of five oocytes were subjected to the SacI restriction enzyme accessibility assay. Arrows show specific bands generated by SacI and HinfI. The diagram below shows SacI cutting as a percentage of total DNA. (C) MPE analysis. See Figure 4A legend for details. Lane 1, internal molecular weight marker, showing the positions of HinfI and SacI restriction enzyme cuts. Lanes 2–17, wild-type or mutant ssDNA injected as indicated. Lane 18, naked dsMMTV promoter DNA digested with MPE. Solid black lines mark the position of the strong GRE elements at –185/–171 in the wild type and in Δ–60/–10 and Δ–124/–10 mutants. Open circles connected with a black line mark the hormone-induced positioning of nucleosomes C and B (from top to bottom).
We analyzed the MMTV chromatin structure of the Δ–60/–10 mutant using two assays: MNase digestion and SacI in situ cutting. In the first assay, a similar hormone-dependent increase in MNase cutting was revealed in both the mutant and wild-type promoter (not shown). The second assay further confirmed that these two promoters could respond to the hormone in a similar way at the structural level. The SacI cutting profiles were superimposable (Figure 5B). MPE digestion experiments showed strong hormone-dependent nucleosome positioning along the MMTV LTR, and hormone-dependent hypercutting, indicating chromatin remodeling, around the GRE segment within the nucleosome B region in the wild-type MMTV promoter as well as in the Δ–60/–10 and Δ–124/–10 mutants (Figure 5C). As observed earlier (Figure 4A and B), the nucleosome C region displayed as a hormone-dependent, MPE-protected region in the wild type and in these two deletion mutants (Figure 5C). Conversely, the nucleosome C region was not protected in the Δ–181/–10 mutant where all GR-binding sites had been deleted. In this mutant, 67% of the wild-type –240/–60 nucleosome B segment has been deleted; however, the –425/–240 nucleosome C region as defined above remains intact. This demonstrates that positioning of nucleosome C depends on the more proximal region of the promoter, which is deleted in the Δ–181/–10 mutant and that contains a strong GR-binding site(s) (Payvar et al., 1983).
Therefore, a high affinity GR-binding site(s) seems to be necessary and sufficient for the establishment of chromatin remodeling and nucleosome positioning in the proximal part of the MMTV LTR. Since only 1% of transcriptional activity remained in the Δ–124/–10 mutant, while a significant level of chromatin remodeling and distinct nucleosome positioning was detected, these experiments further suggest that chromatin remodeling can be uncoupled from transcription.
Chromatin remodeling does not depend on ongoing transcription
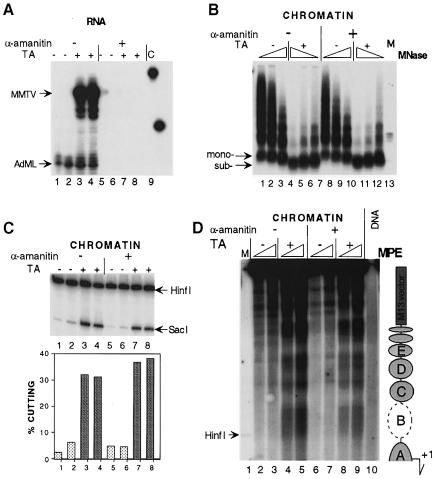
To achieve a complete transcriptional arrest, we injected α–amanitin, a toxin known for its ability to arrest RNA polymerase II-driven transcription. The toxin was co-injected with ssMMTV DNA and AdML reference DNA. S1 nuclease protection showed that transcription from both the MMTV and the AdML promoter was abolished by α–amanitin at a final intracellular concentration of ∼0.5 μg/ml (Figure 6A, lanes 5–8). However, hormone-dependent chromatin remodeling of the MMTV promoter occurred independently of transcription as revealed by (i) MNase (Figure 6B, compare lanes 1–3 and 4–6 with lanes 7–9 and 10–12), (ii) SacI restriction enzyme accessibility assay (Figure 6C, compare lanes 1, 2 and 3, 4 with lanes 5, 6 and 7, 8), and (iii) MPE footprinting (Figure 6D, compare lanes 2, 3 and 4, 5 with lanes 6, 7 and 8, 9). Hormone-activated chromatin remodeling was thus demonstrated to be independent of the transcriptional activity of the MMTV promoter. This was confirmed in a separate experiment in which α–amanitin was injected into the cytoplasm of the oocyte prior to hormone induction (data not shown). Therefore, ongoing transcription is not required for the establishment of chromatin remodeling and nucleosome positioning. This is in agreement with previous results looking at hormone-induced chromatin remodeling of the MMTV promoter in tissue culture cells (Truss et al., 1995) and for the thyroid hormone-induced remodeling of the TRβA promoter in Xenopus oocytes (Wong et al., 1995).
Fig. 6. Nucleosome remodeling and establishment of translational nucleosome positioning are not dependent on ongoing transcription. (A) Transcription analysis by S1 nuclease protection of MMTV and AdML RNA. In half of the oocytes, α–amanitin was co-injected together with the DNA (lanes 5–8). After 24 h, 1 μM TA was added (lanes 3, 4, 7 and 8) or not added (lanes 1, 2, 5 and 6) and oocytes were harvested another 24 h later for RNA analysis. Lane 9, undigested S1 probe. (B) MNase analysis. DNA was resolved in agarose, transferred and hybridized with a probe encompassing region –192/–100 of MMTV. Arrows show positions for mononucleosomal (mono-) and subnucleosomal (sub-) particles. (C) SacI accessibility assay. Oocytes in groups of six for each analysis. Symbols as in Figure 5B. (D) MPE footprinting. Oocytes in groups of seven were analyzed by MPE digestion. Isolated DNA was assayed according to the indirect end-labeling protocol as in Figure 4A except that digested DNA was only cleaved with EcoRV (+425). Lane 1, internal molecular weight marker, showing the positions of HinfI cleavage. Lane 10, naked dsMMTV promoter DNA digested with MPE. To the right is a schematic summary of MPE cuts along the MMTV LTR with putative nucleosome positions.
Discussion
The Xenopus oocyte has a potential for use as a ‘biological test tube’ where heterologous DNA and protein can be highly expressed in vivo (Colman, 1984). Studies of thyroid hormone-dependent gene expression have exploited Xenopus oocytes to study hormone-induced chromatin remodeling in vivo in the TRβA promoter (Wong et al., 1998). The optimization of this system by use of ssDNA injection (Almouzni and Wolffe, 1993) and the GR mRNA injection strategy allowed us to demonstrate, for the first time, a hormone-induced nucleosome positioning on the MMTV promoter. In contrast to previous studies of chromatin structure in the MMTV promoter in vivo in tissue culture cells (Richard-Foy and Hager, 1987; Fragoso et al., 1995; Truss et al., 1995) or in vitro reconstituted MMTV promoter in Drosophila embryo extracts (Venditti et al., 1998; Di Croce et al., 1999), the Xenopus oocytes do not harbor positioned nucleosomes in this promoter prior to hormone activation. This has led to the discovery that GR-mediated chromatin remodeling is able to induce translational nucleosome positioning over this promoter. Furthermore, it shows that a pre-set nucleosome positioning is not required for glucocorticoid-mediated transcription activation but is rather a consequence of the induction event. The nucleosome positioning and additional structural changes in the MMTV promoter depend on the presence of a GRE but are independent of basal promoter elements and of ongoing transcription.
Comparison with previous chromatin studies of MMTV LTR in tissue culture cells
Previous studies of the chromatin structure in the MMTV LTR using tissue culture cells have shown clear-cut hormone-induced effects at the chromatin level. These effects could be monitored as the appearance of a DNase I-hypersensitive site (Zaret and Yamamoto, 1984; Richard-Foy and Hager, 1987; Truss et al., 1995), increased restriction enzyme cutting (Archer et al., 1992; Truss et al., 1995; Fragoso et al., 1998) and increased MPE digestion over nucleosome B (Richard-Foy and Hager, 1987). In these studies, nucleosomes were demonstrated to be translationally positioned already in the inactive MMTV LTR (see, however, Fragoso et al., 1995), and several careful in vivo studies have not detected any effect on nucleosome positioning by hormone activation (Richard-Foy and Hager, 1987; Fragoso et al., 1995; Truss et al., 1995). The same is true for in vitro reconstituted chromatin on subclones of MMTV LTR-derived DNA. Again there is a clear preference in translational positioning of nucleosome B, similar to that found in vivo. This is the case both in pure in vitro reconstitution systems (Perlmann and Wrange, 1988; Pina et al., 1990; Archer et al., 1991; Flaus and Richmond, 1998) and in Drosophila embryo extracts using plasmid DNA (Venditti et al., 1998; Di Croce et al., 1999). These in vitro studies have demonstrated that MMTV LTR DNA harbors nucleosome positioning elements.
Why is translational nucleosome positioning not observed in the inactive MMTV promoter in Xenopus oocytes? Previous in vivo studies of MMTV LTR nucleosome positioning in tissue culture cells (Richard-Foy and Hager, 1987; Truss et al., 1995) were often based on the use of MMTV reporter constructs propagated in an episomal multicopy BPV vector. Under these conditions, hormone induction engages a minority of the gene copies, 15–20% (Bresnick et al., 1992; Fragoso et al., 1995). This inevitably results in heterogeneous chromatin patterns. However, studies of the nucleosome structure of the MMTV promoter have also involved cell lines harboring a single copy of stably integrated MMTV LTR-driven reporters (see, for example, Truss et al., 1995). These cells also showed a low but significant transcription activity in the absence of exogenously added hormone and again showed no difference in nucleosome positioning in the presence or absence of hormone induction (Truss et al., 1995). We speculate that the lack of nucleosome positioning in the Xenopus oocytes is due to the complete silence of the MMTV promoter in the absence of added hormone (cf. Figure 1C). A low frequency of transcription may be required and sufficient for nucleosomes to move to their preferred positions along the MMTV LTR, perhaps then retained in these translational positions by DNA sequence-directed positioning elements. This might explain the pre-positioned nucleosomes in MMTV LTR in the tissue culture cell lines. We do not know whether this basal transcription is due to traces of glucocorticoid hormone or to the status of GR or chromatin in these cells. Anyway, this suggests that the Xenopus oocyte system offers an unusual opportunity to follow the activation-induced chromatin reorganization of a previously inactive and newly replicated promoter.
Additional differences of possible relevance are the following: in Xenopus oocytes, the injected ssDNA is assembled in a process coordinated to the second strand synthesis of our M13 MMTV derivative, presumably without any sequence-specific initiation. In contrast, the BPV vectors used in the previous studies have a defined origin of replication. Furthermore, tissue culture cells are kept at 37°C while Xenopus oocytes are kept at 18–19°C. In vitro studies show that nucleosome sliding is increased by an increase in temperature (Flaus and Richmond, 1998). Along these lines, we obtained a weak but significant rearrangement of chromatin in the MMTV LTR by incubating the oocytes for 30 min at 37°C prior to homogenization and MPE footprinting (S.Belikov and Ö.Wrange, unpublished observation). In addition, the inactive MMTV LTR in Xenopus oocytes does show some weak and variable multiframe nucleosome positioning (see Figure 5C). In none of these cases does the nucleosome pattern become ordered into the strict translational positioning that we obtained in the activated MMTV LTR (Figure 4).
Importantly, our results demonstrate that in the MMTV LTR pre-positioning of nucleosomes is not required to elicit a strong GR-induced response involving chromatin remodeling of virtually all MMTV templates. Rather, the positioning is an integrated part of the chromatin remodeling event. This suggests that GR is able to bind its nucleosomal targets in MMTV LTR irrespective of the translational nucleosome frame in each individual template. This agrees with the results of in vitro GR binding (Perlmann and Wrange, 1988) and in vivo progesterone receptor binding (Truss et al., 1995) studies showing that all five GREs are occupied in spite of their different rotational positioning on the positioned MMTV B nucleosome. Nucleosomes have in several cases been shown to contribute to promoter architecture (Schild et al., 1993; Lu et al., 1995; Sewack and Hansen, 1997). In these studies, it has been suggested that the main function of a positioned nucleosome is to bring various factor-binding sites into close proximity. Although our results show that pre-positioning of nucleosomes is not functionally required, they do not exclude the possibility that a stronger and/or a more reactive hormone response may be elicited if nucleosomes are pre-positioned. Such induction kinetics will be difficult to address in Xenopus oocytes where lipophilic steroid hormones will be retained for a long time after hormone withdrawal.
Chromatin structure of the activated MMTV promoter
Although hormone activation results in a translationally positioned array of at least six nucleosomes, our results show that each individual nucleosome in this array has a different structure (compare nucleosome B and C in Figures 4A and B, 5C and 6D). Nucleosome B shows hormone-dependent MPE hypercutting, while its upstream neighbor, nucleosome C, shows hormone-dependent protection. These MPE data are consistent with the MNase and DNase I analyses. Nucleosome B is positioned over the GRE, which harbors binding sites for 4–5 GR homodimers (Payvar et al., 1983; Truss et al., 1995) and includes the NF1 site (Cordingley et al., 1987). It is not surprising that the binding of these transactivating factors results in significant remodeling, as revealed by MPE hypercutting, either due to steric effects arising from the factor binding, or due to targeting of chromatin remodeling complex(es) (Fryer and Archer, 1998; Di Croce et al., 1999; Rachez et al., 1999; Xu et al., 1999). The strong and hormone-dependent increase in protection of the neighboring nucleosome C, despite MPE hypercutting in nucleosome B in the active promoter, shows that transactive factor(s)-mediated chromatin disruption is locally restricted. The distinct hormone-dependent remodeling of nucleosome D and to some extent nucleosome E (Figures 4A and B, 6C and 6D) has not been analyzed further but is likely to reflect transactivating factor binding. Several upstream enhancer segments have been found in the MMTV LTR (Gouilleux et al., 1991; Lefebvre et al., 1991; Kusk et al., 1996). We cannot exclude the possibility that the strong upstream MPE-cut sites may represent alternative translational nucleosome frames, but our in vivo nucleosome mapping experiments suggest a single frame of nucleosome positioning in the active MMTV promoter.
This study is the first report on the appearance of a subnucleosomal 120 bp particle in vivo as well as the hypercutting of MNase in the active MMTV promoter. MNase-induced subnucleosomal particles have been observed previously in other promoters both in vivo (Wong et al., 1998) and in vitro (Liu et al., 1999) and represent hallmarks of remodeled promoters during transcription activation. This supports the notion that the subnucleosome observed here does indeed reflect the active structural state of the MMTV promoter. We note that a histone H3–H4 tetramer can form an MNase-protected DNA fragment of 120 bp (Hayes et al., 1991) and that partially purified SWI/SNF complex promotes ATP-dependent remodeling of the MMTV B nucleosome octamer DNase I pattern into a tetramer-like pattern in vitro (Spangenberg et al., 1998). Since the subnucleosome complex is formed following glucocorticoid activation and encompasses a cluster of 4–5 GR-binding sites, it is likely to contain GR. Our efforts are now directed towards the characterization of the protein and DNA components in this subnucleosomal complex.
The mechanism of establishment of hormone-induced translational nucleosome positioning in the MMTV promoter
In vitro nucleosome reconstitution studies suggest that positioning over the nucleosome A and B region in the MMTV LTR is directed to a large extent by the DNA sequence itself (Perlmann and Wrange, 1988; Pina et al., 1990; Archer et al., 1991; Flaus and Richmond, 1998). We show here, however, that these sequence determinants are overcome in the in vivo reconstituted inactive MMTV LTR in Xenopus oocytes. On the other hand, during hormone activation, one high affinity GR-binding site is able to mediate nucleosome positioning in vivo (Figure 5C). The GR protein has been shown to interact with both the SWI/SNF complex (Yoshinaga et al., 1992; Muchardt and Yaniv, 1993; Östlund Farrants et al., 1997; Fryer and Archer, 1998) and the histone acetyltransferase-containing coactivator complexes such as GRIP1 (Xu et al., 1999). Furthermore, the homologous progesterone receptor was recently shown to target ISWI and NURF38 to the MMTV promoter (Di Croce et al., 1999). The ability to reposition nucleosomes along DNA was shown recently for several nucleosome remodeling complexes such as NURF (Hamiche et al., 1999), CHRAC (Längst et al., 1999) and SWI/SNF complex (Whitehouse et al., 1999). For example, in the presence of NURF, nucleosomes in the reconstituted hsp70 promoter adopt one predominant position from a variety of possible locations (Hamiche et al., 1999). However, NURF had little effect on 5S nucleosomes, arguing for a definite role of DNA sequence determinants, such as anisotropic flexibility of nucleosomal core DNA and/or rigidity of internucleosomal linker, in establishing nucleosome positioning. It is possible that chromatin remodeling complexes, most probably SWI/SNF (Yoshinaga et al., 1992; Östlund Farrants et al., 1997; Fryer and Archer, 1998) and/or NURF (Di Croce et al., 1999), recruited to the MMTV LTR by GR, participate in chromatin opening. This opening leads to increased nucleosome mobility and allows nucleosomes to slide along the DNA. The sliding then focuses the nucleosomes into a preferred translational frame due to DNA sequence-directed nucleosome positioning and/or sequence-specific DNA-binding transcription factors. These factors may serve as steric barriers, or as nucleosome positioning proteins, as has been shown for HNF3. This factor directs nucleosome positioning in the albumin enhancer (Shim et al., 1998). We hypothesize that nucleosome positioning in the MMTV LTR may thus be driven by the cooperative forces of a GR-mediated chromatin remodeling, DNA sequence-directed bending and DNA-binding factors.
Materials and methods
Plasmids, DNA and RNA preparation
The 3 kb SalI–PvuII fragment containing the MMTV LTR/TK gene sequence from pLSwt (Buetti and Kuhnel, 1986) was inserted into M13 mp9 SalI–Sma to generate the pMTV:M13 coding strand. The pβGR/RN3P construct designated for in vitro transcription of GR RNA was generated by cloning of the BamHI–NotI fragment obtained by PCR using specific primers and pBal 117 plasmid (Miesfeld et al., 1986) containing the complete cDNA for rat GR as a template into the vector of BamHI–NotI-cleaved plasmid pβGFP/RN3P (Zernika-Goetz et al., 1996). 5′–capped GR RNA was prepared by in vitro transcription using the Message Machine™ kit (Ambion). Deletion pMTV:M13 coding strand mutants were created by ‘long’ PCR using primers containing a unique XhoI recognition site and positioned such that the indicated DNA segments were deleted in the final constructs (see Figure 5A) with subsequent cleavage with XhoI and ligation. The success of cloning was confirmed by sequencing.
Oocyte microinjection and maintenance
Defolliculated stage VI Xenopus laevis oocytes were prepared by collagenase treatment (Almouzni and Wolffe, 1993). The oocytes were incubated overnight at 18–19°C in OR2 medium containing 1 mM CaCl2 (Colman, 1984). The next day, healthy oocytes were injected with DNA into nuclei and/or RNA into the cytoplasm (20 nl). In a typical experiment, 10 ng of in vitro transcribed GR RNA were injected and 2–6 h later 1 ng of ssDNA of pMTV:M13 coding strand and 0.25 ng of pAdML dsDNA (Ohlsson and Edlund, 1986) were injected, the latter as a transcription reference (Perlmann and Wrange, 1991). In the experiments indicated, GR was introduced as a GR expression vector (pSTC GR 3–795) (Wieland et al., 1988), which was co-injected with the other DNAs as 5 ng of dsDNA. At 16–24 h after injections, the synthetic glucocorticoid hormone TA (Sigma-Aldrich), routinely at 1 μM, was added to the oocyte culture medium. Oocytes were harvested for analysis at the time indicated. For RNA polymerase II transcription inhibition, 20 nl of α–amanitin was co-injected into oocyte nuclei at a concentration of 30 μg/ml in the course of the DNA injection.
RNA analysis and immunoblotting
RNA analysis and immunoblotting using a polyclonal rabbit antiserum against rat GR were performed as previously described (Gelius et al., 1999).
DNase I hypersensitivity assay
Twelve injected oocytes were collected and homogenized in 10 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM dithiothreitol (DTT), 5 mM MgCl2 and 5% glycerol (buffer A) (three oocytes in 100 μl) by pipeting up and down at 0°C until an even homogenate is obtained (∼15–20 times). The homogenate was divided into four tubes and DNase I (Boehringer Mannheim) was added (6, 9, 13.5 and 20.25 U per tube). Following incubation at 20°C for 3 min, the reaction was stopped by addition of SDS to 0.5% and EDTA to 10 mM. DNA was purified by proteinase K treatment, phenol/chloroform extractions and ethanol precipitations. The samples were RNase A treated (30 μg/ml) for 1 h at 37°C and purified DNA cleaved with SalI, resolved in a 1% agarose gel (Sambrook et al., 1989), vacuum transferred and hybridized with the 32P-labeled single-stranded SalI–SacI fragment of pLSwt (Buetti and Kuhnel, 1986), obtained by synthesis of cDNA (Church and Gilbert, 1984).
MNase digestion
Injected oocytes in groups of 10 were homogenized in buffer A with 3 mM CaCl2 (three oocytes in 100 μl). The homogenate was divided into three parts and MNase (Amersham Pharmacia Biotech) was added (15, 30 and 60 U per tube). After 5 min incubation (20°C), the reaction mixture was adjusted to 0.5% with respect to SDS and 10 mM EDTA. DNA was purified as above, resolved in a 1.6% agarose gel in 0.5× TBE (Sambrook et al., 1989), blotted and hybridized in succession with different random-primed 32P-labeled probes as indicated.
SacI in situ accessibility assay
Oocytes in groups of 5–10 were rinsed twice in SacI in situ buffer [20 mM Tris–HCl pH 8.3, 40 mM NaCl, 1 mM MgCl2, 0.5 mM spermine, 0.15 mM spermidine, 5% glycerol (v/v), 1 mM DTT] and then homogenized in 10 μl per oocyte of the same buffer. We found that the pH of the oocyte homogenization buffer used in the SacI in situ cutting experiments had a surprisingly dramatic effect selectively on the hormone-dependent SacI cutting: the hormone-dependent stimulation of SacI in situ cutting increased from nearly undetectable levels at pH 7.0–7.4 to 5- to 10–fold at pH 8.0–8.6 (data not shown); SacI cutting of naked MMTV DNA in Xenopus oocyte homogenates did not display this profile.
Eight units of SacI restriction endonuclease (NE Biolabs) were added per oocyte and incubated for 10 min at 18°C. The reaction was stopped by addition of SDS, EDTA and proteinase K to a final concentration of 1%, 30 mM and 325 μg/ml, respectively. DNA was then extracted as above. The purified DNA was cleaved to completion by 2 U of HinfI and 50 μg/ml RNase A in 100 μl of 1× HinfI buffer. The reaction was stopped by adding an equal volume of phenol/chloroform (2:1), and DNA was purified as above, dissolved in 10 μl of 10 mM Tris–HCl pH 7.5 and 0.1 mM EDTA. One half to one oocyte equivalent was then analyzed by primer extension using a 32P-labeled primer (–291/–265, coding strand), which was labeled by use of T4 polynucleotide kinase (Sambrook et al., 1989). Primer extension generated a 183 and a 221 nucleotide product when cleaved at SacI or HinfI, respectively (Figure 1A). Primer extension reaction was carried out in a 10 μl volume containing the sample DNA, 0.5 pmol of 5′-end-labeled primer, 0.2 mM each of the four dNTPs and 0.2 μl (0.4 U) of Vent polymerase exo™ (NE Biolabs) in 1× Vent polymerase buffer. Primer extension was performed in a thermal cycler under standard conditions (95°C/1 min; 55°C/2 min; 72°C/3 min) for 20 cycles followed by analysis on a 6% polyacrylamide sequencing gel. ‘SacI cutting’ was always calculated as SacI-cut/total DNA (i.e. the sum of HinfI- and SacI-cut DNA) as quantified by PhosphorImager analysis (Molecular Dynamics)
In situ cleavage by MPE
Seven injected oocytes were homogenized in 350 μl of MPE digestion buffer (10 mM Tris–HCl pH 7.5, 60 mM KCl, 15 mM NaCl, 0.15 mM spermidine, 0.5 mM spermine, 0.3 M sucrose) and hydrogen peroxide was added to oocyte homogenate at a final concentration of 1–2 mM. MPE⋅iron(II) (Sigma-Aldrich) complex was prepared by mixing 18.8 μl of 625 μM MPE solution (from the 5 mM aqueous stock) with 18.8 μl of 625 μM ferrous ammonium sulfate solution (from freshly prepared 5 mM aqueous stock). Immediately before use, DTT was added to the solution of MPE⋅Fe(II) to achieve a final concentration of 10 mM (from a freshly prepared 1 M stock). The cleavage reaction was started by mixing oocyte homogenate with MPE⋅Fe(II) complex and incubation was at 25°C for the indicated time. The reaction was stopped by addition of 1/10 volume of 50 mM bathophenanthroline disulfonate (Merck). DNA was isolated as described above and cleaved by restriction enzyme for indirect end-labeling assay (Wu, 1989).
Acknowledgments
Acknowledgements
We gratefully acknowledge Ulla Björk for skillful technical assistance, Peter Becker, for constructive critique and Drs T.Klenka and P.Ridgway for correction of the English. Ö.W. especially thanks Danièle Roche, J.–P.Quivy and P.-H.Gaillard for helping to handle Xenopus oocytes and for related techniques. We also thank Dr S.Rusconi for providing the rat glucocorticoid expression vector pSTC GR 3-795, Dr E.Buetti for the pLSwt MMTV plasmid, Drs R.Miesfeldt and K.R.Yamamto for pRBal 117 (rat GR cDNA), and Dr J.B.Gurdon for pβGFP/RN3P. This work was supported by the Swedish Cancer Foundation (project 2222-B98-14XAC) and a guest research fellowship to S.B. (3745-B97-02VAA), by the Royal Swedish Academy of Sciences (12682) and by the European Commission, TMR, to G.A. and Ö.W. (Network Contract ERBFMRXCT-98-0191).
References
- Almouzni G. and Wolffe, A.P. (1993) Replication-coupled chromatin assembly is required for repression of basal transcription in vivo. Genes Dev., 7, 2033–2047. [DOI] [PubMed] [Google Scholar]
- Archer T.K., Cordingley, M.G., Wolford, R.G. and Hager, G.L. (1991) Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol. Cell. Biol., 11, 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer T.K., Lefebvre, P., Wolford, R.G. and Hager, G.L. (1992) Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science, 255, 1573–1576. [DOI] [PubMed] [Google Scholar]
- Bresnick E.H., Bustin, M., Marsaud, V., Richard-Foy, H. and Hager, G.L. (1992) The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res., 20, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetti E. and Kuhnel, B. (1986) Distinct sequence elements involved in the glucocorticoid regulation of the mouse mammary tumor virus promoter identified by linker scanning mutagenesis. J. Mol. Biol., 190, 379–389. [DOI] [PubMed] [Google Scholar]
- Church G.M. and Gilbert, W. (1984) Genomic sequencing. Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman A. (1984) Expression of exogenous DNA in Xenopus oocytes. In Hames,B.D. and Higgins,S.J. (eds), Transcription and Translation–A Practical Approach. IRL Press, Oxford, UK, pp. 49–69. [Google Scholar]
- Cordingley M.G., Riegel, A.T. and Hager, G.L. (1987) Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell, 48, 261–270. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Koop, R., Vendetti, P., Westphal, H.M., Nightingale, K.P., Corona, D.F.V., Pecker, P.B. and Beato, M. (1999) Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol. Cell, 4, 45–54. [DOI] [PubMed] [Google Scholar]
- Flaus A. and Richmond, T.J. (1998) Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol., 275, 427–441. [DOI] [PubMed] [Google Scholar]
- Fragoso G., John, S., Roberts, M.S. and Hager, G.L. (1995) Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev., 9, 1933–1947. [DOI] [PubMed] [Google Scholar]
- Fragoso G., Pennie, W., John, S. and Hager, G.L. (1998) The position and length of the steroid dependent hypersensitive region in the MMTV LTR is invariant despite multiple nucleosome B frames. Mol. Cell. Biol., 18, 3633–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C.J. and Archer, T.K. (1998) Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature, 393, 88–91. [DOI] [PubMed] [Google Scholar]
- Gelius B., Wade, P., Wolffe, A.P., Wrange, Ö. and Östlund Farrants, A.-K. (1999) Characterization of a chromatin remodeling activity in Xenopus oocytes. Eur. J. Biochem., 262, 426–434. [DOI] [PubMed] [Google Scholar]
- Gouilleux F., Sola, B., Cuette, B. and Richard-Foy, H. (1991) Cooperation between structural elements in hormone-regulated transcription from the mouse mammary tumor virus promoter. Nucleic Acids Res., 19, 1563–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos, R., Gdula, A.A. and Wu, C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodelling complex NURF. Cell, 97, 833–842. [DOI] [PubMed] [Google Scholar]
- Han M., Kim, U.-J., Kayne, P. and Grunstein, M. (1988) Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J., 7, 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J.J., Clark, D.J. and Wolffe, A.P. (1991) Histone contributions to the structure of DNA in the nucleosome. Proc. Natl Acad. Sci. USA, 88, 6829–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar, G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Kusk P., John, S., Fragoso, G., Michelotti, J. and Hager, G.L. (1996) Characterization of an NF-1/CTF family member as a functional activator of the mouse mammary tumor virus long terminal repeat 5′ enhancer. J. Biol. Chem., 271, 31269–31276. [DOI] [PubMed] [Google Scholar]
- Längst G., Bonte, E.J., Corona, D.F.V. and Becker, P. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Lefebvre P., Berard, D.S., Cordingley, M.G. and Hager, G.L. (1991) Two regions of the mouse mammary long terminal repeat regulate the activity of its promoter in mammary cell lines. Mol. Cell. Biol., 11, 2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wong, J., Tsai, M.J. and O'Malley, B.W. (1999) Steroid receptor coactivator-1 (SRC-1) enhances ligand-dependent and receptor-dependent cell-free transcription in chromatin. Proc. Natl Acad. Sci. USA, 96, 9485–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Wallrath, L.L. and Elgin, S.C. (1995) The role of a positioned nucleosome in the Drosophila melanogaster hsp26 promoter. EMBO J., 14, 4738–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder, A.W., Richmond, R.K., Sargent, D.F. and Richmond, T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Rusconi, S., Godowski, P.J., Maler, B.A., Okret, S., Wikström, A.-C., Gustafsson, J.-Å. and Yamamoto, K.R. (1986) Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell, 46, 389–399. [DOI] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv, M. (1993) A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J., 12, 4279–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson H. and Edlund, T. (1986) Sequence-specific interactions of nuclear factors with insulin gene enhancer. Cell, 45, 35–44. [DOI] [PubMed] [Google Scholar]
- Östlund Farrants A.-K., Blomquist, P., Kwon, H. and Wrange, Ö. (1997) Glucocorticoid receptor–glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol. Cell. Biol., 17, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payvar F., DeFranco, D., Firestone, G.L., Edgar, B., Wrange, Ö., Okret, S., Gustafsson, J.-Å. and Yamamoto, K.R. (1983) Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell, 35, 381–392. [DOI] [PubMed] [Google Scholar]
- Perlmann T. and Wrange, Ö. (1988) Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J., 7, 3073–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann T. and Wrange, Ö. (1991) Inhibition of chromatin assembly in Xenopus oocytes correlates with derepression of the mouse mammary tumor virus promoter. Mol. Cell. Biol., 11, 5259–5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B., Bruggemeier, U. and Beato, M. (1990) Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell, 60, 719–731. [DOI] [PubMed] [Google Scholar]
- Rachez C., Lemon, B.D., Suldan, Z., Bromleight, V., Gamble, M., Näär, A.M., Erdjument-Bromage, H., Tempst, P. and Freedman, L.P. (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature, 398, 824–828. [DOI] [PubMed] [Google Scholar]
- Richard-Foy H. and Hager, G.L. (1987) Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J., 6, 2321–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schild C., Claret, F.-X., Wahli, W. and Wolffe, A.P. (1993) A nucleosome dependent static loop potentiates estrogen-regulated transcription from Xenopus vitellogenin B1 promoter in vitro. EMBO J., 12, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewack G.F. and Hansen, U. (1997) Nucleosome positioning and transcription-associated chromatin alterations on the human estrogen-responsive pS2 promoter. J. Biol. Chem., 272, 31118–31129. [DOI] [PubMed] [Google Scholar]
- Shim E.Y., Woodcock, C. and Zaret, K.S. (1998) Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev., 12, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.T. (1991) Nucleosome positioning: occurrence, mechanisms and functional consequences. Prog. Nucleic Acid Res. Mol. Biol., 40, 143–184. [DOI] [PubMed] [Google Scholar]
- Spangenberg C., Eisfeld, K., Stünkel, W., Luger, K., Richmond, T.J., Truss, M. and Beato, M. (1998) The mouse mammary tumor virus promoter positioned on a tetramer of histones H3 and H4 binds nuclear factor 1 and OTF1. J. Mol. Biol., 278, 725–739. [DOI] [PubMed] [Google Scholar]
- Theulaz I., Hipskind, R., Heggeler-Bordier, B., Green, S., Kumar, V., Chambon, P. and Wahli, W. (1988) Expression of human estrogen receptor mutants in Xenopus oocytes: correlation between transcriptional activity and ability to form protein–DNA complexes. EMBO J., 7, 1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truss M., Bartsch, J., Schulbert, A., Hache, R.J.G. and Beato, M. (1995) Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J., 14, 1737–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti P., Di Croce, L., Kauer, M., Blank, T., Becker, P. and Beato, M. (1998) Assembly of MMTV minichromosomes with positioned nucleosomes precludes NF1 access but not restriction enzyme cleavage. Nucleic Acids Res., 26, 3657–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I., Flaus, A., Cairns, B.R., White, M.F., Workman, J.L. and Owen-Hughes, T. (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature, 400, 784–787. [DOI] [PubMed] [Google Scholar]
- Wieland S.Y., Schaffner, W. and Rusconi, S. (1988) Metal binding finger structures in the glucocorticoid receptor defined by site-directed mutagenesis. EMBO J., 7, 2503–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Shi, Y.-B. and Wolffe, A.P. (1995) A role for nucleosome assembly in both silencing and activation of the Xenopus TRβ A gene by the thyroid hormone receptor. Genes Dev., 9, 2696–2711. [DOI] [PubMed] [Google Scholar]
- Wong J., Patterton, D., Imhof, A., Gushin, D., Shi, Y.-B. and Wolffe, A.P. (1998) Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J., 17, 520–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. (1989) Analysis of hypersensitive sites in chromatin. Methods Enzymol., 170, 269–289. [DOI] [PubMed] [Google Scholar]
- Wyrick J.J., Holstege, F.C., Jennings, E.G., Causton, H.C., Shore, D., Grunstein, M., Lander, E.S. and Young, R.A. (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature, 402, 418–421. [DOI] [PubMed] [Google Scholar]
- Xu L., Glass, C.K. and Rosenfeld, M.G. (1999) Coactivators and corepressors in nuclear receptor function. Curr. Opin. Genet. Dev., 9, 140–147. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S.K., Peterson, C.l., Herskowitz, I. and Yamamoto, K.R. (1992) Roles of SWI1, SWI2 and SWI3 proteins for transcriptional enhancement by steroid receptors. Science, 258, 1598–1604. [DOI] [PubMed] [Google Scholar]
- Zaret K.S. and Yamamoto, K.R. (1984) Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell, 38, 29–38. [DOI] [PubMed] [Google Scholar]
- Zernika-Goetz M., Pines, J., Ryan, K., Siemering, K.R., Haseloff, J., Evans, M.J. and Gurdon, J.B. (1996) An indelible lineage marker for Xenopus using a mutated green fluorescence protein. Development, 122, 3719–3724. [DOI] [PubMed] [Google Scholar]