Abstract
The 9–cis retinoic acid receptor (retinoid X receptor, RXR) forms heterodimers with the all-trans retinoic acid receptor (RAR) and other nuclear receptors on DNA regulatory sites composed of tandem binding elements. We describe the 1.70 Å resolution structure of the ternary complex of RXR and RAR DNA-binding regions in complex with the retinoic acid response element DR1. The receptors recognize identical half-sites through extensive base-specific contacts; however, RXR binds exclusively to the 3′ site to form an asymmetric complex with the reverse polarity of other RXR heterodimers. The subunits associate in a strictly DNA-dependent manner using the T–box of RXR and the Zn–II region of RAR, both of which are reshaped in forming the complex. The protein–DNA contacts, the dimerization interface and the DNA curvature in the RXR–RAR complex are distinct from those of the RXR homodimer, which also binds DR1. Together, these structures illustrate how the nuclear receptor superfamily exploits conformational flexibility and locally induced structures to generate combinatorial transcription factors.
Keywords: nuclear receptor/RAR/RXR/structure/transcription factor
Introduction
The 9-cis retinoic acid receptor (retinoid X receptor, RXR) is an important member of the nuclear receptor family because it forms heterodimers with many other receptors, including the all-trans retinoic acid receptor (RAR), vitamin D3 receptor (VDR), thyroid hormone receptor (TR), the peroxisome proliferator activated receptor (PPAR) and the nerve growth factor induced–B (NGFI–B) receptor (Leblanc and Stunnenberg, 1995; Mangelsdorf and Evans, 1995; Mangelsdorf et al., 1995). RAR and RXR, each of which have α, β and γ isotypes, transduce the retinoid signal into a variety of genetic responses, and their functions underlie the essential role played by retinoids in the development and homeostasis of vertebrates (Chambon, 1996). A consensus sequence 5′–AGGTCA–3′ is the core recognition element for the entire family of nearly 150 non-steroid nuclear receptors to which RXR and RAR belong (Mangelsdorf and Evans, 1995). However, RXR and RAR alone display negligible binding to this consensus site as monomers (Leid et al., 1992). Heterodimerization increases their joint affinity and selectivity for retinoic acid response elements (RAREs), which are composed of both binding sites arranged in tandem. The RXR–RAR heterodimer binds more efficiently to RAREs than either of the homodimeric forms of RXR or RAR (Leid et al., 1992).
The core sequence of the DNA-binding domain (DBD) is highly conserved in the nuclear receptor superfamily, with >40% amino acid identity over a 67–residue region. This sequence is comprised of two zinc-nucleated modules and two α–helices that fold into a single globular domain. The use of a highly conserved receptor DBD together with a single major core binding sequence raises an important question as to how diverse signaling pathways are created in this transcription factor superfamily (Gronemeyer and Moras, 1995). Formation of combinatorial transcription factors involving RXR is one important mechanism by which complex DNA sites can be employed with greater selectivity (Yu et al., 1991; Bugge et al., 1992; Kliewer et al., 1992b; Marks et al., 1992). The functional consequence of forming receptor heterodimers is that in some cases two distinct ligands can co-regulate transcription from a single gene-regulatory element (Forman et al., 1995).
Importantly, the DBDs of the nuclear receptors can generate the same pattern of DNA selectivity and dimerization as the full-length receptors (Mader et al., 1993; Perlmann et al., 1993; Zechel et al., 1994). A second dimerization function in the ligand-binding domains of some receptors has no selective role for response element recognition, but can further stabilize some dimeric assemblies (Perlmann et al., 1996). All RXR heterodimers preferentially bind response elements composed of two AGGTCA sites arranged in a direct repeat configuration with characteristic inter-half-site spacings of 1–5 bp (these are known as DR1–DR5, respectively; Umesono et al., 1991; Mangelsdorf and Evans, 1995). Interestingly, the sequence of the spacer is less critical than its size for discrimination. The selective recognition of DRs based on inter-half-site spacings has been formalized in a 1–5 rule that defines the highest affinity binding sites for each RXR heterodimer (Umesono et al., 1991; Mangelsdorf and Evans, 1995). However, the binding repertoire for other members of the nuclear superfamily extends beyond the usage of direct repeat geometry, as some factors bind efficiently to inverted and everted repeats of core hexamer (Mangelsdorf and Evans, 1995). The multiple arrangements with which the same consensus AGGTCA sequence is utilized in this family suggest that combinatorial nuclear receptors possess a remarkable readout mechanism that allows them to decipher the geometry of a binding site in addition to its sequence.
An associated feature of any asymmetric response element is the polarity adopted by the subunits of the bound receptor heterodimer. The non-symmetric RXR–RAR complexes allow the position of each receptor to be distinguished as upstream or downstream, even though the two hexameric binding sites may have an identical sequence. The RXR–RAR complex, therefore, can adopt different polarities when bound to its various response elements, and these alternative arrangements have been shown to confer different functions on the heterodimer in terms of hormone and co-repressor binding (Kurokawa et al., 1994, 1995). A previous structural analysis of the RXR–TR DBD complex bound to a thyroid response element DR4 allowed the direct visualization of the polarity associated with that complex (Rastinejad et al., 1995). The asymmetric assembly was established through the cooperation between DBD subunits, which occurs only when the inter-half-site spacing is 4 bp in length and RXR is positioned upstream of TR (Rastinejad et al., 1995).
The response element repertoire of the RXR–RAR complex is considerably less selective than that of the RXR–TR complex, which was restricted to a single direct repeat element (DR4). In fact, the RXR–RAR heterodimer mediates transcriptional control through DR1, DR2 and DR5 sequences found in naturally occurring RAREs (Mangelsdorf et al., 1991; Predki et al., 1994; Chambon, 1996). The multiple high-affinity DNA-binding targets suggest a large degree of versatility on the part of their DBDs in forming favorable interaction surfaces, each of which has a distinct effect on target gene regulation (Kurokawa et al., 1995; Chambon, 1996). To explore how the joint assembly of RXR and RAR on DNA is mediated, we determined the 1.7 Å crystal structure of the ternary complex containing RXR and RAR DBDs bound to the DR1 element, the sequence first identified in the promoter of the cytosolic retinol-binding protein II gene (Mangelsdorf et al., 1991). In vivo, RXR–RAR heterodimers on DR1 repress transcription in both the absence and presence of their hormones (Kurokawa et al., 1995). In examining the structure of the complex, we asked how the DR1 specifies the polar assembly of the RXR and RAR DBDs, how cooperativity and target selectivity are achieved, and how DNA-induced changes in the secondary and tertiary structures of RXR and RAR promote the assembly of their complex.
Results
Overview of the structure
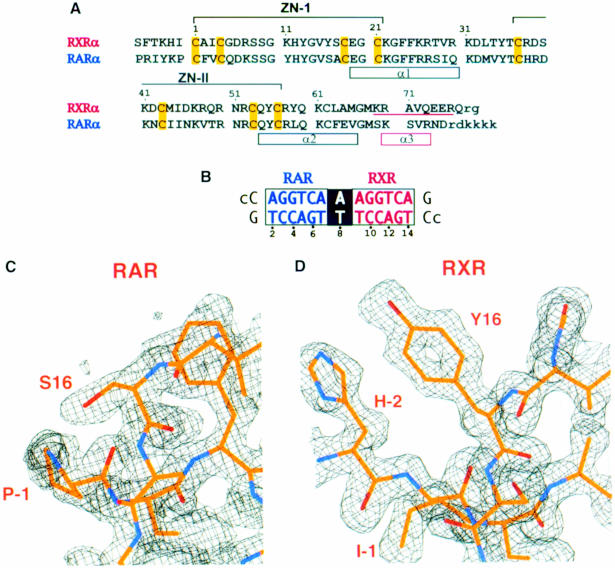
The protein and DNA constructs used in co-crystallization are shown in Figure 1A and B. The polypeptides were designed to support the ability of RXR and RAR to form cooperative interactions on DR1. Using these constructs, the heterodimer of RXR and RAR DBDs gives half-maximal DR1 binding when each protein is at a concentration of ∼300–350 nM (data not shown). In contrast, the DBD of RXR alone shows a >10–fold reduction in binding to DNA sites containing a single AGGTCA sequence (Leid et al., 1992; Lee et al., 1993; Zhao et al., 2000). The T–box sequence of RXR, which has the sequence KREAVQEER at the C–terminal extension of the DBD, is required for the formation of the RXR homodimer DR1 complex, as well as the formation of other heterodimeric complexes when RXR occupies the site downstream of DR1 (Wilson et al., 1992; Predki et al., 1994; Ijpenberg et al., 1997).
Fig. 1. The protein and DNA constructs used in structure determination. (A) The DNA-binding regions of RXR (residues 129–212) and RAR (residues 82–167) used for co-crystallization. The polypeptides contain two zinc-nucleated modules whose coordinating cysteines are indicated in yellow. Each protein contains two α–helices (α1, α2) in its core domain. A third α–helix, α3, in the T–box of RXR (underlined) is seen in the absence of DNA, but is disrupted upon DNA binding (Holmbeck et al., 1998a,b). Disordered residues not found in the electron density maps are indicated by the small letters. The numbering of the amino acids is referenced to the first Zn-coordinating cysteine (1) for simplicity. (B) The 15 bp DR1 sequence with the consensus recognition half-sites and spacer sequence shown in white and black boxes, respectively. The 5′ overhangs, indicated by the small letters, are not seen in the electron density maps. (C) 2Fo – Fc map revealing the identity of the RAR subunit. RAR is distinguished by clear densities for sites such as Ser16 and Pro(–1). (D) 2Fo – Fc map showing how RXR can be distinguished by unambiguous density at sites such as Tyr16, His(–2) and Ile(–1)
There is a good structural overlap between the backbone atoms of RXR and RAR DBDs in their homologous 67–residue core sequences, and this fold is shared with the corresponding DBD structures of TR, RevErb, estrogen receptor (ER), glucocorticoid receptor (GR), NGFI–B and by extension the entire nuclear receptor family (Luisi et al., 1991; Schwabe et al., 1993; Rastinejad, et al., 1995; Zhao et al., 1998; Meinke and Sigler, 1999). To identify the polarity associated with the complex, specific and unambiguous assignments of RXR and RAR in the electron density maps were required. The polypeptides differ in 34 amino acid positions within the region –2 to 75, but most of these residues are conservative substitutions or solvent-exposed positions that are not fully ordered. The high-resolution X–ray data proved helpful in this regard. Figure 1C and D shows that the electron densities belonging to Y16 and its counterpart S16 are unambiguous, allowing the protein electron density containing the former to be assigned to RAR and the latter to RXR. Similarly, the identification of non-conserved residues that precede the core DBD, such as His(–2), Ile(–1) and Pro(–1), also proved helpful. These and other observations are consistent with the binding of RAR and RXR to the upstream and downstream half-sites, respectively.
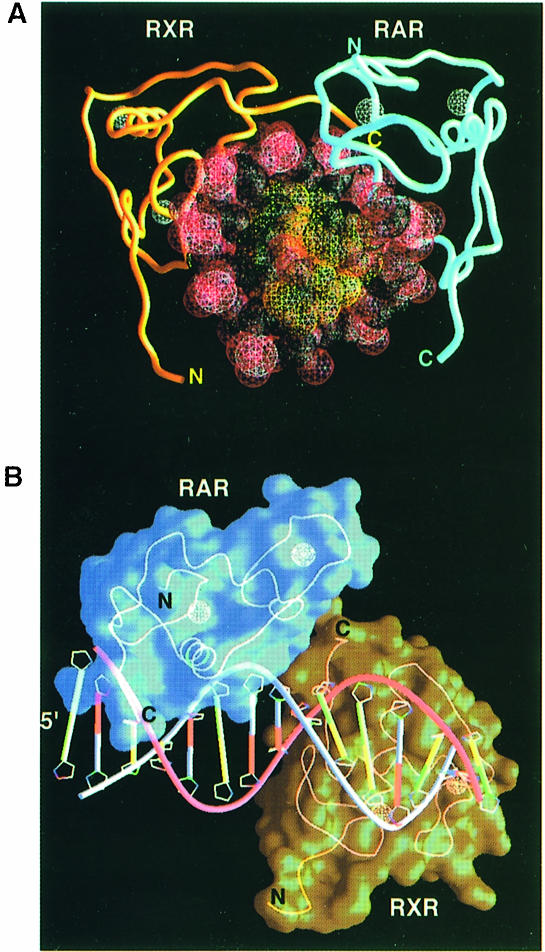
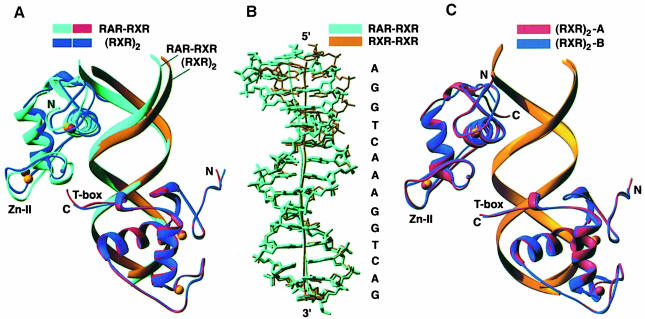
Figure 2 shows the overall structure of the complex. The observed mode of polarity agrees with a previous biochemical study that examined DR1 binding by full-length RAR and RXR polypeptides (Kurokawa et al., 1994). The binding of RXR at the downstream site is uniquely associated with DR1 elements, since most RXR heterodimers studied to date, including the two other RXR–RAR heterodimers on DR2 and DR5, the RXR–VDR heterodimer on DR3, the RXR–TR and RXR–Liver X receptor (LXR) heterodimers on DR4, and the RXR–NGFI–B interactions on DR5, use the opposite polarity, in which RXR binds exclusively at the upstream position. However, the DR1 sequence also supports the cooperative binding of RXR–PPAR with 3′ RXR binding as observed in the current structure (Ijpenberg et al., 1997). The unique asymmetry is a consequence of the subunit's ability to form productive protein–protein contacts in one orientation and not the other.
Fig. 2. Overall structure of the ternary complex. (A) Top view looking down the 5′ end of the DNA. The DNA phosphates and the zinc atoms are shown in red and white, respectively. The DNA recognition helices (α1) of RXR and RAR are partly invisible in this orientation as they insert into the major grooves of DR1. The C–terminally positioned T–box of RXR points towards the RAR Zn–II module to mediate the subunit interactions. (B) Side view showing the complementary surfaces formed between RAR and RXR and the polarity associated with the complex. Figures are made with the program GRASP (Nicholls et al., 1991).
The characteristic spacing of the DR1 response element sets the rotation and translation geometry by which the RXR and RAR subunits must interact while maintaining a favorable registration with respect to the hexameric half-site. The minor groove of the DR1 reaches its narrowest point (5.6 Å) at the spacer base pair 8, precisely where the two proteins meet. The DNA helical axis deviates a total of 6° from that of standard B–DNA. The two AGGTCA sequences are contacted in the major groove by the recognition helix of each DBD (α1 in Figure 1A). These contacts account for all of the major groove interactions of the RXR–RAR complex. In their complex, ∼3500 Å2 of water-accessible surface area is buried, 1900 Å2 of which derives from the RXR–DNA interactions and 1600 Å2 of which is from the RAR–DNA interface. These values are in line with other protein–DNA interactions that involve similar sized recognition modules and DNA-binding sites (Nadassy et al., 1999).
The heterodimerization interface and its DNA dependence
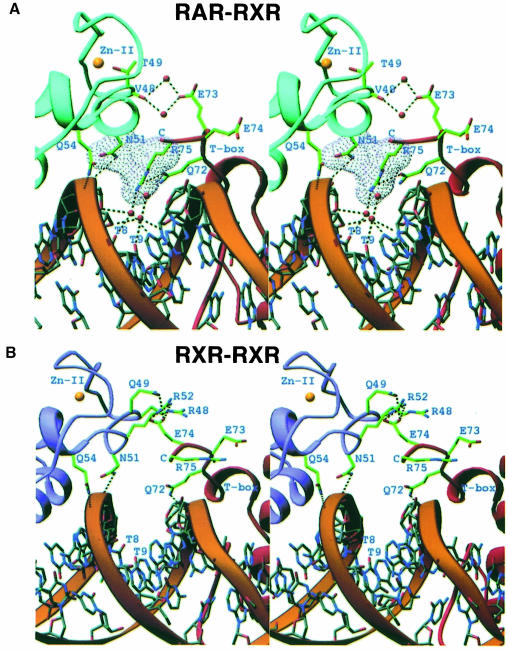
RXR and RAR DBDs do not form homo- or heterodimeric associations in the absence of specific response elements. Figure 3A shows that the contacts between RXR and RAR in their complex take advantage of the polar functional groups in the minor grooves of base pairs 8 and 9, as well as the well ordered hydration spine associated with DNA structure (Shui et al., 1998). The intimate role of the DNA in aligning the subunits explains why stable interactions between RXR and RAR DNA-binding regions are not possible off DNA. The minor groove compression at the protein–protein junction (base pair 8) is electrostatically favorable, bringing the negatively charged DNA phosphates into closer proximity with the guanidino group of Arg75. This compression, as well as hydrogen-bonding with Gln72 and the ordered water molecules in the minor groove, reduces the conformational flexibility of Arg75, precisely orienting its side-chain to make van der Waals contact with Asn51 of RAR. The orientation of Asn51 with respect to the dimer interface is also restricted through hydrogen-bonding with DNA. An interaction between Glu73 of RXR, Val48 of RAR and bridging water gives rise to a secondary stabilizing interface between subunits.
Fig. 3. Stereo diagrams showing the contacts between the protein subunits. (A) The RXR–RAR interface involves the DNA minor groove and well ordered water molecules (red spheres). Dotted lines indicate hydrogen-bonding between atoms. The yellow spheres indicate the positions of the Zn–II atoms. Arg75 and Asn51 form complementary van der Waals interactions. (B) The corresponding region of the RXR–DBD homodimer interface on DR1 (Zhao et al., 2000). These and the following molecular display graphics were made using the program Ribbons (Carson and Bugg, 1986).
In all, the subunit interactions lead to a total loss of less than ∼400 Å2 of solvent-accessible surface from both proteins. This suggests that the observed dimer interface is inferior to bona fide protein–protein interfaces, as these typically bury >700 Å2 of solvent-exposed area (Janin et al., 1998). However, the extent of the solvent-excluded surface is comparable to and in some cases even favorable relative to other transcription factor interfaces that form exclusively on DNA (Klemm and Pabo, 1996; Zhao et al., 1998, 2000; Fuji et al., 1999; Zheng et al., 1999). The interactions in the RXR–RAR complex are also distinct from those of the homo-cooperative complex of RXR, which forms on the same DR1 response element (see Figure 3B; Zhao et al., 2000). But in both cases, the subunits utilize the DNA infrastructure to stabilize their interactions. This observation suggests that these and other combinatorial RXR complexes do not pre-assemble their DNA-recognition surfaces in solution, but rather assemble only when an appropriate gene-regulatory site is available.
Relation to other nuclear receptor assemblies on DR1
In both the homo- and heterodimeric complexes shown in Figure 3, the downstream RXR uses exclusively its T–box to mediate protein–protein interactions with the upstream Zn–II region. However, there are differences in terms of the induced T–box conformation, the protein–protein and the protein–DNA interactions in these assemblies. For example, the subunit interactions in the homodimer rely to some extent on hydrogen-bonding interactions between Glu74 from the T–box of the downstream subunit, and Gln49 and Arg52 from the Zn–II region of the upstream subunit (see Figure 3B). This is not possible in the RXR–RAR interaction, since RAR has a T49 in place of Q49, disabling the salt-bridge at the dimerization surface (Figure 3).
The DR1 configuration is also used by the RXR–PPAR complex (Kliewer et al., 1992a; Gearing et al., 1993; Palmer et al., 1995; Ijpenberg et al., 1997). Interestingly, PPARα, PPARβ and PPARγ all contain the same Asn51 and Glu54 residues that provide the upstream contact surfaces shown in Figure 3A. In addition, an A⋅T base pair is also highly preferred in the DR1 spacer of PPAR response elements, suggesting the possibility of a similar role for the DNA minor groove in aligning subunit interactions (Ijpenberg et al., 1997). However, the binding site for RXR–PPAR is extended relative to that of the RXR–RXR and RXR–RAR, since PPAR also recognizes the minor grooves of 5′ flanking sequences using a C–terminal extension of its core DBD known as the GRIP-box (Ijpenberg et al., 1997; Zhao et al., 1998). These distinctions suggest that a graded affinity is associated with each of these RXR complexes, depending on the sequence of the spacer, the induced curvature in the DNA and the unique sequences flanking the DR1.
Factors responsible for the cooperative assembly
Figure 3A suggests that protein occupancy at one site can lead to facilitated binding at the second site via protein–protein interactions. An extended DNA-binding surface formed through tight protein–protein interactions can result in a considerable gain in DNA-binding cooperativity. However, given the limited extent of the subunit contacts, it is likely that additional indirect effects must also contribute to cooperation between RXR and RAR. One such mechanism is adjacent DNA site stabilization, when binding of one subunit reduces the conformational flexibility at the adjacent site, thus pre-paying some of the entropic costs associated with its tight binding. It has been shown that the POU domains of Oct–1 use partially overlapping DNA contacts at the center of their octamer site to achieve cooperative binding, even though there is a total absence of protein–protein interactions (Klemm and Pabo, 1996). Similarly, Ubx–Exd subunits cooperate to a significant extent through their partially overlapping DNA contacts at the center of their binding site, which is enough to produce a cooperative enhancement even if their primary protein–protein interactions are removed (Chan et al., 1994; Passner et al., 1999; Piper et al., 1999). Tandem site stabilization may also enhance the assembly of RXR and RAR on DR1, given that the half-sites are separated by only 1 bp and that the subunits form overlapping contacts at the spacer (see section on DNA sequence recognition and Figure 5).
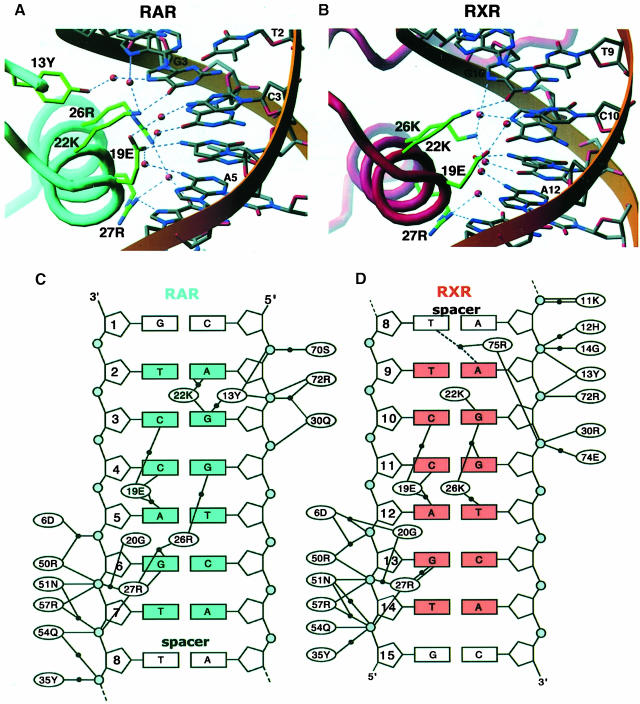
Fig. 5. DNA sequence recognition by RXR and RAR. (A) A view along the DNA-recognition helix (α1) of RAR showing residues Tyr13, Arg26, Lys22, Glu19 and Arg27 and their direct and water-mediated base contacts. Hydrogen-bonds and water molecules are shown as dotted blue lines and red spheres, respectively. The DNA sequence is numbered as in Figure 1B. (B) The corresponding view of the RXR interface. (C) Summary of the protein–DNA contacts of RAR. Bridging water molecules are shown as black circles. The base pairs in blue form the consensus AGGTCA recognition element. The gray circles indicate the DNA phosphates. (D) Summary of RXR–DNA interactions. The base pairs in orange form the consensus element.
The RXR and RAR proteins also cooperate through DNA- and dimerization-induced structural transitions in RXR that overcome an inhibition in its DNA binding. We have shown previously that the RXR T–box, in its α–helical structure off DNA (Holmbeck et al., 1998a), poses unfavorable contacts with the DNA via Glu74 and Glu75 and also blocks the DNA-binding surface in α1. A reconfiguration of the T–box structure overcomes both inhibitory effects in the RXR homodimer (Holmbeck et al., 1998b; Zhao et al., 2000). Interestingly, either of the two extended structures seen in downstream subunits (Figure 3) leads to an effective dimerization surface (one in the homodimer, the other in the heterodimer), and relieves the inhibitory effects on DNA binding. A similar mechanism by which the DNA element and a heterodimeric partner can help overcome an inhibitory DNA function associated with a transcription factor has also been described for the Hox–Exd complex (Chan et al., 1996).
The role of DNA curvature
Further cooperation in a ternary transcriptional complex is achieved when protein occupancy at the one DNA site induces structural distortions at the second site to accommodate protein binding better. The importance of even small DNA deformations, especially bends in the DNA helix, for fine-tuning protein–DNA interactions has been pointed out previously (Gewirth and Sigler, 1995; Dickerson, 1998; Dickerson and Chiu, 1998). The bending of the DNA has also been suggested to be responsible for the cooperative binding of the interferon regulatory factor 1 (IRF–1)–DNA complex, which does not use any contacts between DBDs in its cooperative assembly (Fuji et al., 1999). The utilization of this mechanism in achieving cooperativity in the RXR–RAR–DR1 is illustrated in Figure 4A and B, which shows that the DNA helical axis bends, especially at the 5′ binding site. This distortion appears to facilitate RAR and RXR interactions with their half-sites while being engaged as a heterodimer.
Fig. 4. Topological comparison of the RXR–RAR–DR1 complex with the (RXR)2–DR1 complex. (A) The superposition of the two complexes shows that the DNA structure and the subunit position at the 5′ (top) half-sites are different. (B) Superposition of the DNA molecules shows a divergence in the helical axes associated with DR1 in its complexes with the RXR homodimer and the RXR–RAR heterodimer. The divergence is most severe at the 5′ half-sites, which are occupied by different proteins in these complexes. (C) For comparison, the superposition of two crystallographically distinct RXR DBD homodimer–DR1 complexes, showing very close overlap throughout, despite different crystal packing environments (Zhao et al., 2000).
The comparison in Figure 4B demonstrates how structural versatility in DNA structure is exploited in accommodating two distinct types of receptor interactions with RXR. The RXR homodimer induces a 12° kink in DR1 compared with the RXR–RAR complex, which bends the DNA by 6°. These induced curvatures are a direct result of the distinct subunit associations on DR1, since the DNA sequences in the two complexes are otherwise identical along 14 out of 15 bp. The kinks in both cases are mainly localized to the central spacer, which adopts a different combination of tilt and roll angles to give rise to the 12 and 6° bends, respectively. Because the spacer is also the convergence point for the protein–protein interactions in each complex, the different helical distortions are a direct consequence of the dimerization arrangements. Figure 4C shows that two RXR homodimer–DR1 complexes from two different crystallographic environments (these being two distinct complexes in the asymmetric unit of the RXR–RXR–DNA crystals) are nearly superimposable with respect to their DNA curvatures (Zhao et al., 2000). This observation suggests that the distinct distortions observed in the helical axes of the RXR–RAR and RXR–RXR binding sites are not caused merely by their different crystallographic packing environments.
Recognition of DNA sequence
The DNA contacts for both proteins are shown in Figure 5. The base-specific contacts of RXR and RAR occur along 6 and 5 bp, respectively. In the RXR–DNA interface, two of the six base contacts occur along the minor grooves of base pairs 8 and 9. The pattern of major groove contacts for these proteins is similar, as shown in Figure 5A and B. The recognition helix of each receptor forms direct and water-mediated base contacts using Arg27, Lys22, Glu19 and Lys26/Arg26. RXR and RAR both also use a number of highly ordered water molecules to bridge their base-specific and phosphate interactions. In addition, RXR and RAR each form extensive backbone contacts along 7 bp of DNA (Figure 5C and D).
We compared the DNA contacts with those seen in the RXR homodimer–DR1 complex (Zhao et al., 2000). In terms of the number of specific protein–DNA interactions, the RXR–RAR complex has a clear advantage over the homodimeric RXR complex. In the homodimer, the subunits together form specific contacts with only six positions along the entire DR1, compared with 11 contacts formed by the heterodimer. Moreover, the three base-specific contacts made by each RXR in the homodimer vary in each independent crystallographic representation of RXR (Zhao et al., 2000). The weak, and by comparison relaxed, binding of the homodimer to DR1 has also been demonstrated by biochemical studies (Dowhan et al., 1994; Yang et al., 1995; Castelein et al., 1996).
Protein structural transitions in the complex
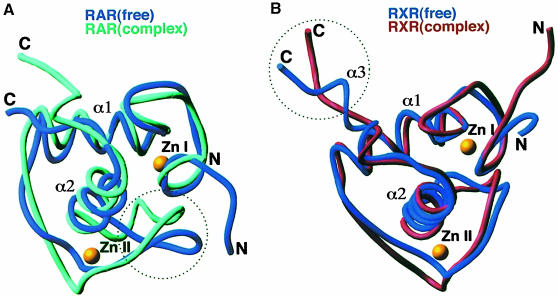
The structures of the isolated RXR and RAR DBD polypeptides have previously been studied by NMR (Knegtel et al., 1993; Holmbeck et al., 1998a; van Tilborg et al., 1999). In comparing these protein structures with their counterparts bound to DR1, we asked what secondary and tertiary protein structural changes are associated with the formation of the complex. Figure 6 indicates that the two regions undergoing the most dramatic structural rearrangement in each subunit are the T–box and the Zn–II module, both of which interact to stabilize the RXR–RAR–DR1 complex and can adopt other conformations in different contexts (Holmbeck et al., 1998b; van Tilborg et al., 1999). The ability of the highly dynamic Zn–II region of RXR to facilitate the assembly of the RXR–RAR–DR5 complex was suggested in a recent dynamics study of these proteins (van Tilborg et al., 1999). Furthermore, the Zn–II regions of nuclear receptors can in some cases undergo transitions between helix and loop in binding and dissociating from DNA (Schwabe et al., 1993; Holmbeck et al., 1998a,b). Similarly, the T–box sequence of RXR has an inherently large degree of structural freedom, and this property is suggestive of its ability to form cooperative protein–DNA complexes (Lee et al., 1993; Holmbeck, 1998a,b).
Fig. 6. Protein structural transitions induced in the RXR–RAR–DR1 complex. (A) The superposition of the free RAR DBD structure derived from NMR studies with its structure in the RXR–RAR–DR1 complex. The dotted circle indicates that a major structural rearrangement induced on assembly occurs along the Zn–II loop. (B) The superposition of the free RXR DBD structure from NMR studies with its structure in the RXR–RAR–DR1 complex. The dotted circle indicates that a major structural reorganization occurs along the α3 helix of the T–box, which is disrupted in forming the complex.
Discussion
The current structure illustrates the ability of a DNA regulatory element to assume the role of a classic allosteric ligand, inducing new conformations and/or interactions that ultimately enhance its own binding. Conformational changes that occur with DNA binding have not often been studied in detail, as relatively few proteins have their high-resolution, three-dimensional structures known for both DNA-bound and free states. Our analysis suggests that two distinct types of allosteric changes could be conferred on nuclear receptors by their DNA response element. First, there are conformational changes that can occur within a DBD, such as the deformation of the T–box α–helix, which leads to the enhanced binding of RXR to DNA. A second type of allosteric influence involves reshaping DNA structures to facilitate protein–protein contacts. Favorable interactions between RXR and RAR give rise to a substantial enhancement in their DNA affinity. In the context of the full-length nuclear receptors, the response elements are also known to have important effects on the interactions of these factors with their ligands, co-repressors, coactivators, AP–1 and other factors that can influence gene expression (Kurokawa et al., 1993, 1994, 1995; Glass et al., 1997; Lefstin and Yamamoto, 1998).
The current study also provides important lessons about the usefulness of flexible protein and DNA surfaces that can be precisely altered on a DNA site. The inter-half-site spacing of the retinoid response elements specifies a fixed geometry through which a pair of nuclear receptors must interact, since a change of one nucleotide in the spacer causes RXR and its partners to rotate ∼35° around the double helix and be displaced from each other by 3.4 Å. Therefore, RXR DBD must either possess a number of fixed and distinct surfaces to accommodate its many combinatorial interactions, or more efficiently make use of a few adaptable protein elements that can adjust to the rotations, displacements and polarity of these binding sites with one or more receptor partners. The essential role played by the DNA in conferring the protein structures required for dimerization also means that the various combinatorial complexes of RXR need not be preformed until they are required at particular control sites. In this way, there is substantial economy gained given the large number of pairwise interactions that RXR can form with its partners.
Materials and methods
Crystallization
The DBDs were expressed in Escherichia coli as fusions with glutathione S–Sepharose, using the pGEX-4T vector (Pharmacia). The proteins and the DNA oligonucleotides were purified as described previously (Rastinejad et al., 1995; Zhao et al., 1998). The crystals used for data collection grew at 8°C in hanging drops made up of 2 μl of protein/DNA solution in 25 mM Tris buffer pH 7.5 and 2 μl of the reservoir solution that contained 18–23% PEG3350, 25 mM Tris pH 7.5, 5 mM MgCl2 and 0.4 M NH4Cl. The final protein and DNA concentrations were 0.90 and 0.45 mM, respectively.
Data collection
X–ray data were collected at –160°C on beamline X25 at the Brookhaven National Laboratory on an MAR image plate detector (λ = 0.98 Å). The data were processed and scaled to 1.70 Å using HKL (Otwinowski and Minor, 1997). The crystals belong to space group P212121 with cell parameters a = 80.66, b = 30.90 and c = 101.86 Å, and one complex per asymmetric unit. The detailed data parameters are summarized in Table I. The Rmerge value was calculated as in Zhao et al. (2000).
Table I. Summary of data collection and refinement.
| Diffraction data | |
| Resolution (Å) | 1.70 |
| Unique reflections | 29 834 |
| Completeness (%) | 94.8 |
| last shell (1.76–1.70 Å) | 98.0 |
| Average I/σ(I) | 19.9 |
| last shell | 8.8 |
| Rmerge (%) | 7.1 |
| last shell | 17.0 |
| Redundancy | 4.1 |
| Crystallographic refinement | |
| Resolution range (Å) | 50–1.70 |
| Reflections used | 28 322 |
| Total number of non-hydrogen atoms [<B> (Å2)] | |
| protein | 1289 (21.5) |
| DNA | 609 (19.4) |
| zinc | 4 (16.1) |
| solvent | 342 (30.5) |
| R (%) | 19.8 |
| Rfree (%) | 26.7 |
| R.m.s. deviation from ideal values | |
| bond lengths (Å) | 0.009 |
| angles (°) | 2.1 |
Structure solution and refinement
The structure was solved by molecular replacement using the program AMORE (Navaza, 1994). The search model was based on a polyalanine ‘mutant’ made from the refined coordinates of the RXR homodimer on its response element (Zhao et al., 2000). All zinc ions and terminal residues (preceding residue 1 or following residue 67; see Figure 1A) were omitted from the search. The rotation and translation function searches each gave distinct solutions. Rigid body refinement resulted in an R–factor of 47.7%, which dropped to 43.8% after the first cycle of simulated annealing with X–PLOR (Brünger, 1993). At this point, metal atoms, conserved side-chains and C–terminal residues were built into their Fo – Fc and 2Fo – Fc electron density maps with the program O (Jones et al., 1991). Only after almost all the identical RXR and RAR residues had been modeled were the resulting difference maps inspected for electron density that could be assigned uniquely to RXR or RAR. Many RXR- and RAR-specific side-chains were clearly identifiable in all subsequent difference maps. Iterative steps of map inspection, model building and simulated annealing cycles resulted in an R–factor of 25.4% for 2σ reflections between 6.0 and 1.7 Å (Rfree for 5% of the data was 29.9%). The refinement procedure was then changed to the conjugate gradient least-squares method employed in SHELXL–97. Restraints for the DNA base pairs were included according to geometric parameters described by Parkinson et al. (1996), with bond angles addressed by their 1,3–distances. Purine and pyrimidine atoms were restrained to the plane of the aromatic ring system. We did not impose any torsion restraints on the nucleotides. The stereochemistry at the chiral carbon atoms, however, was fixed by restraining their chiral volume. Zinc and sulfur atoms were refined anisotropically to account for disorder visible in the final difference maps. The final model included 342 defined water molecules, refined to the values given in Table I. The R and Rfree were calculated as in Zhao et al. (2000).
Acknowledgments
Acknowledgements
The authors wish to thank Scott A.Chasse and Srikripa Devarakonda for assistance with biochemical studies, Christine S.Wright and Michael L.Sierk for computational advice, and the staff of beamline X25 at Brookhaven National Laboratory for assistance with data collection. The coordinates can be obtained from the corresponding authors, or from the Protein Data Bank. T.W. was supported in part by the Alexander von Humboldt Foundation. This study was made possible by grant support from the Leukemia Society of America (S.K.) and the National Institutes of Health GM55217 (F.R.).
References
- Brünger A.T. (1993) X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT. [Google Scholar]
- Bugge T.H., Pohl, J., Lonnoy, O. and Stunnenberg, H.G. (1992) RXR α, a promiscuous partner of retinoic acid and thyroid hormone receptors. EMBO J., 11, 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. and Bugg, C.E. (1986) Algorithm for ribbon models of proteins. J. Mol. Graph., 4, 121–122. [Google Scholar]
- Castelein H., Janssen, A., Declercq, P.E. and Baes, M. (1996) Sequence requirements for high affinity retinoid X receptor-α homodimer binding. Mol. Cell. Endocrinol., 119, 11–20. [DOI] [PubMed] [Google Scholar]
- Chambon P. (1996) A decade of molecular biology of retinoic acid. FASEB J., 10, 940–954. [PubMed] [Google Scholar]
- Chan S.-K., Jaffe, L., Copovilla, M., Botas, J. and Mann, R.S. (1994) The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeodomain. Cell, 78, 603–615. [DOI] [PubMed] [Google Scholar]
- Chan S.-K., Pöpperl, H., Krumlauf, R. and Mann, R.S. (1996) An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. EMBO J., 15, 2476–2487. [PMC free article] [PubMed] [Google Scholar]
- Dickerson R.E. (1998) DNA bending–the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res., 26, 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R.E. and Chiu, T.K. (1998) Helix bending as a factor in protein/DNA recogntion. Biopolymers, 44, 361–403. [DOI] [PubMed] [Google Scholar]
- Dowhan D.H., Downes, M., Sturm, R.A. and Muscat, G.E. (1994) Identification of deoxyribonucleic acid sequences that bind retinoid-X receptor-γ with high affinity. Endocrinology, 135, 2595–2607. [DOI] [PubMed] [Google Scholar]
- Forman B.M., Umesono, K., Chen, J. and Evans, R.M. (1995) Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell, 81, 541–550. [DOI] [PubMed] [Google Scholar]
- Fuji Y., Shimizu, T., Kusumoto, M., Kyogoku, Y., Tadatsugu, T. and Hakoshima, T. (1999) Crystal structure of an IRF–DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J., 18, 5028–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing K.L., Gottlicher, M., Teboul, M., Widmark, E. and Gustafsson, J.A. (1993) Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc. Natl Acad. Sci. USA, 90, 1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirth D.T. and Sigler, P.B. (1995) The basis for half-site specificity explored through a non-cognate steroid receptor–DNA complex. Nature Struct. Biol., 2, 386–394. [DOI] [PubMed] [Google Scholar]
- Glass C.K., Rose, D.W. and Rosenfeld, M.G. (1997) Nuclear receptor coactivators. Curr. Opin. Cell Biol., 9, 222–232. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H. and Moras, D. (1995) How to finger DNA. Nature, 375, 190–191. [DOI] [PubMed] [Google Scholar]
- Holmbeck S.M.A., Foster, M.P., Casimoro, D.R., Sem, D.S., Dyson, H.J. and Wright, P.E. (1998a) High-resolution solution structure of the retinoid X receptor DNA-binding domain. J. Mol. Biol., 281, 271–284. [DOI] [PubMed] [Google Scholar]
- Holmbeck S.M.A., Dyson, H.J. and Wright, P.E. (1998b) DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retionoid X receptor. J. Mol. Biol., 284, 533–539. [DOI] [PubMed] [Google Scholar]
- Ijpenberg A., Jeannin, E., Wahli, W. and Desvergne, B. (1997) Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. J. Biol. Chem., 272, 20108–20117. [DOI] [PubMed] [Google Scholar]
- Janin J., Miller, S. and Chothia, C. (1998) Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol., 204, 155–164. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W. and Kjeldgaard, M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Klemm J.D. and Pabo, C.O. (1996) Oct-1 POU domain–DNA interactions: cooperative binding of isolated subdomains and effects of covalent linkage. Genes Dev., 10, 27–36. [DOI] [PubMed] [Google Scholar]
- Kliewer S.A., Umesono, K., Noonan, D.J., Heyman, R.A. and Evans, R.M. (1992a) Convergence of 9–cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature, 358, 771–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer S.A., Umesono, K., Mangelsdorf, D.J. and Evans, R.M. (1992b) Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature, 355, 446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knegtel R.M.A., Katahira, M., Schilthuis, J.G., Bonvin, A.M., Boelens, R., Eib, D., van der Saag, P.T. and Kaptein, R. (1993) The solution structure of the human retinoic acid receptor-β DNA-binding domain. J. Biomol. NMR, 3, 1–17. [DOI] [PubMed] [Google Scholar]
- Kurokawa R., Yu, V.C., Näär, A., Kyakumoto, S., Han, Z., Silverman, S., Rosenfeld, M.G. and Glass, C.K. (1993) Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev., 7, 1423–1435. [DOI] [PubMed] [Google Scholar]
- Kurokawa R., DiRenzo, J., Boehm, M., Sugarman, J., Gloss, B., Rosenfeld, M.G., Heyman, R.A. and Glass, C.K. (1994) Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature, 371, 528–531. [DOI] [PubMed] [Google Scholar]
- Kurokawa R., Soderstrom, M., Horlein, A., Halachmi, S., Brown, M., Rosenfeld, M.G. and Glass, C.K. (1995) Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature, 377, 451–454. [DOI] [PubMed] [Google Scholar]
- Leblanc B.P. and Stunnenberg, H.G. (1995) 9–cis retinoic acid signaling: changing partners causes some excitement. Genes Dev., 9, 1811–1816. [DOI] [PubMed] [Google Scholar]
- Lee M.S., Kliewer, S.A., Provencal, J., Wright, P.E. and Evans, R.M. (1993) Structure of the retinoid X receptor α DNA binding domain: a helix required for homodimeric DNA binding. Science, 260, 1117–1121. [DOI] [PubMed] [Google Scholar]
- Lefstin J.A. and Yamamoto, K.R. (1998) Allosteric effects of DNA on transcriptional regulators. Nature, 392, 885–888. [DOI] [PubMed] [Google Scholar]
- Leid M., et al. (1992) Purification, cloning and RXR identity of the HeLa cell facor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell, 68, 377–395. [DOI] [PubMed] [Google Scholar]
- Luisi B.F., Xu, W.X., Otwinowski, Z., Freedman, L.P., Yamamoto, K.R. and Sigler, P.B. (1991) Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature, 352, 497–505. [DOI] [PubMed] [Google Scholar]
- Mader S., Chen, J.Y., Chen, Z., White, J., Chambon, P. and Gronemeyer, H. (1993) The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificites of the DNA binding domains. EMBO J., 12, 5029–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D.J. and Evans, R.M. (1995) The RXR heterodimers and orphan receptors. Cell, 83, 841–850. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Umesono, K., Kliewer, S.A., Borgmeyer, U., Ong, E.S. and Evans, R.M. (1991) A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell, 66, 555–561. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M.S., Hallenbeck, P.L., Nagata, T., Segars, J.H., Appella, E., Nikodem, V.M. and Ozato, K. (1992) H-2RIIBP (RXR β) heterodimerization provides a mechanism for combinatorial diversity in the regulation of retinoic acid and thyroid hormone responsive genes. EMBO J., 11, 1419–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke G. and Sigler, P.B. (1999) DNA-binding mechanism of the monomeric orphan receptor NGFI–B. Nature Struct. Biol., 6, 471–477. [DOI] [PubMed] [Google Scholar]
- Nadassy K., Wodak, S.J. and Janin, J. (1999) Structural features of protein–nucleic acid recognition sites. Biochemistry, 38, 1999–2017. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Palmer C.N.A., Hsu, M.-H., Griffin, K.J. and Johnson, E.F. (1995) Novel sequence determinants in peroxisome proliferator signaling. J. Biol. Chem., 270, 16114–16121. [DOI] [PubMed] [Google Scholar]
- Parkinson G., Vojtechovsky, J., Clowney, L., Brünger, A.T. and Berman, H.M. (1996) New parameters for the refinement of nucleic acid containing structures. Acta Crystallogr. D, 52, 57–64. [DOI] [PubMed] [Google Scholar]
- Passner J.M., Ryoo, H.D., Shen, L., Mann, R.S. and Aggarwal, A.K. (1999) Structure of a DNA-bound Ultrabithorax–Extradenticle homeodomain complex. Nature, 397, 714–719. [DOI] [PubMed] [Google Scholar]
- Perlmann T., Rangarajan, P.N., Umesono, K. and Evans, R.M. (1993) Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev., 7, 1411–1422. [DOI] [PubMed] [Google Scholar]
- Perlmann T., Umesono, K., Rangarajan, P.N., Forman, B.M. and Evans, R.M. (1996) Two distinct dimerization interfaces differentially modulate target gene specificity of nuclear hormone receptors. Mol. Endocrinol., 10, 958–966. [DOI] [PubMed] [Google Scholar]
- Piper D.E., Batchelor, A.H., Chang, C.-P., Cleary, M.L. and Wolberger, C. (1999) Structure of a HoxB1–Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell, 96, 587–597. [DOI] [PubMed] [Google Scholar]
- Predki P.F., Zamble, D., Sarkar, B. and Giguere, V. (1994) Ordered binding of retinoic acid and retinoid-X receptor to asymmetric response elements involves determinants adjacent to the DNA-binding domain. Mol. Endocrinol., 8, 31–39. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Perlmann, T., Evans, R.M. and Sigler, P.B. (1995) Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature, 375, 203–211. [DOI] [PubMed] [Google Scholar]
- Schwabe J.W., Chapman, L., Finch, J.T. and Rhodes, D. (1993) The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell, 75, 567–578. [DOI] [PubMed] [Google Scholar]
- Shui X., McFail-Isom, L., Hu, G.G. and Williams, L.D. (1998) The B–DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry, 37, 8341–8355. [DOI] [PubMed] [Google Scholar]
- Umesono K., Murakami, K.K., Thompson, C.C. and Evans, R.M. (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid and vitamin D3 receptors. Cell, 65, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tilborg P.J.A., et al. (1999) Millisecond to microsecond time scale dynamics of the retinoid X and retinoic acid receptor DNA-binding domains and dimeric complex formation. Biochemistry, 38, 1951–1956. [DOI] [PubMed] [Google Scholar]
- Wilson T.E., Paulsen, R.E., Padgett, K.A. and Milbrandt, J. (1992) Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science, 256, 107–110. [DOI] [PubMed] [Google Scholar]
- Yang Y.Z., Subauste, J.S. and Koenig, R.J. (1995) Retinoid X receptor α binds with the highest affinity to an imperfect direct repeat response element. Endocrinology, 136, 2896–2903. [DOI] [PubMed] [Google Scholar]
- Yu V.C., et al. (1991) RXR β: a coregulator that enhances binding of retinoic acid, thyroid hormone and vitamin D receptors to their cognate response elements. Cell, 67, 1251–1266. [DOI] [PubMed] [Google Scholar]
- Zechel C., Shen, X.-Q., Chen, J.-Y., Chen, Z.-P., Chambon, P. and Gronemeyer, H. (1994) The dimerization interfaces formed between the DNA binding domains of RXR, RAR and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. EMBO J., 13, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Khorasanizadeh, S., Miyoshi, Y., Lazar, M.A. and Rastinejad, F. (1998) Structural elements of an orphan nuclear receptor–DNA complex. Mol. Cell, 1, 849–861. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Chasse,S.A., Devarakonda,S., Sierk,M.L., Ahvazi,B. and Rastinejad,F. (2000) Structural basis of RXR–DNA interactions. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Zheng N., Fraenkel, E., Pabo, C.O. and Pavletich, N.P. (1999) Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev., 13, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]