Abstract
PilC1, a pilus-associated protein in Neisseria menin– gitidis, is a key element in initial meningococcal adhesion to target cells. A promoter element (CREN, contact regulatory element of Neisseria) is responsible for the transient induction of this gene upon cell contact. crgA (contact-regulated gene A) encodes a transcriptional regulator whose expression is also induced upon cell contact from a promoter region similar to the CREN of pilC1. CrgA shows significant sequence homologies to LysR-type transcriptional regulators. Its inactivation in meningococci provokes a dramatic reduction in bacterial adhesion to epithelial cells. Moreover, this mutant is unable to undergo intimate adhesion to epithelial cells or to provoke effacing of microvilli on infected cells. Purified CrgA is able to bind to pilC1 and crgA promoters, and CrgA seems to repress the expression of pilC1 and crgA. Our results support a dynamic model of bacteria–cell interaction involving a network of regulators acting in cascade. CrgA could be an intermediate regulator in such a network.
Keywords: intimate adhesion/LysR-regulator/Neisseria meningitidis
Introduction
Neisseria meningitidis (Nm) is a pathogenic bacterium only encountered in humans, where it provokes septicaemia and/or meningitis. Bacteria first adhere to the epithelium of the nasopharynx and may then be trans– located into the bloodstream. To gain entry into the cerebrospinal fluid (CSF), Nm has to adhere to endothelial cells and cross the blood–CSF barrier. Adhesion, a crucial step in meningococcal pathogenesis, is usually viewed as a two-step process: (i) initial adhesion, which is pilus mediated; and (ii) intimate adhesion, which involves other bacterial and cellular structures. Pili seem to disappear during intimate adhesion (Pujol et al., 1997). They make an essential contribution to Nm pathogenicity by allowing initial localized adhesion to target cells. Meningococcal pili are of type IV and are composed of a major protein subunit called pilin. Pili interact with target cells and trigger a signal transduction response in them. Several cellular components have been proposed to participate in pilus-dependent interaction between bacteria and cells, such as CD46, a complement regulatory glycoprotein. Initial binding causes a transient increase in cytosolic free Ca2+ in target cells (Källström et al., 1998). In a recent study, neisserial type IV pili were reported to be required for cortical plaque formation in epithelial cells (Merz et al., 1999). The Opa family of neisserial proteins may interact with several members of the CD66 family (Virji et al., 1996; Gray-Owen et al., 1997).
Two homologous proteins, PilC1 and PilC2, are also key elements in the structure of pili, since the production of at least one PilC protein is required for pilus assembly. In addition, PilC1 but not PilC2 modulates adhesiveness, most likely by being the adhesin (Jonsson et al., 1991; Nassif et al., 1994; Rudel et al., 1995). The pilC1 and pilC2 genes are controlled by distinct promoters (Taha et al., 1996). Moreover, the expression of pilC1, but not that of pilC2, seems to be induced early and transiently upon contact with viable epithelial cells. A subsequent negative feedback is then observed and the level of expression of pilC1 declines to its basal level at a late (intimate) stage of bacterial adhesion (Taha et al., 1998). This induction depends on the expression of pilC1 from a transcription start point (TSP) that is located in a specific fragment in the promoter region of pilC1 that is absent from pilC2 (Taha et al., 1996, 1998). The inactivation of this TSP abolished the induction of the expression of pilC1 upon contact of the bacteria with the host cells and caused a dramatic reduction in adhesion. As bacterial adhesion to target cells seems to involve several genes, the presence of such a regulatory fragment in the pilC1 promoter region might suggest that other meningococcal genes may possess this fragment and could be controlled coordinately during bacterium–cell interaction. The aim of this report is to identify these genes and to analyse the role of the specific fragment found upstream of pilC1 in the global regulation of the adhesion of Nm to target cells.
Results
The contact regulatory element of Neisseria (CREN) is a conserved regulatory sequence specific for pathogenic Neisseria
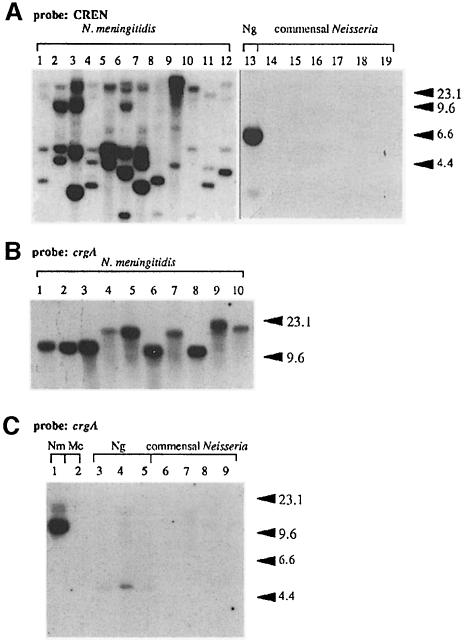
We have previously identified a fragment of DNA (150 bp) upstream of the pilC1 gene in the meningococcal strain 8013 that was shown to be involved in the induction of expression of pilC1 during bacterium–cell interaction (Taha et al., 1998). By Southern hybridization, PCR amplification and DNA sequencing using oligonucleotides hybridizing to this fragment, we first showed that it was conserved upstream of the pilC1 gene in different meningococcal strains belonging to different genetic lineages (Figure 1A and data not shown). We also tested DNA from Neisseria gonorrhoeae strain MS11 and from commensal Neisseria strains. As shown in Figure 1A, only DNA from pathogenic Neisseria (Nm and N.gonorrhoeae) hybridized with the probe under moderately stringent conditions. Each meningococcal strain showed a multiple banding hybridization, indicating that several regions on the meningococcal chromosome harbour homologous sequences to the probe. As this fragment was shown to be involved in the induction of transcription of pilC1 upon contact with target cells, we tentatively named it CREN for contact regulatory element of Neisseria. Data from hybridization experiments suggest that several meningococcal genes could be coordinately regulated by cell contact using CREN.
Fig. 1. Southern blot analysis of ClaI-digested chromosomal DNA from different strains belonging to the genus Neisseria. Hybridization was performed under conditions of moderate stringency (see Materials and methods). The probe used in (A) was a PCR-generated fragment amplified between oligonucleotides C1-152 and C1-8 on the pilC1 promoter and corresponding to the CREN of pilC1. Neisseria meningitidis (Nm) strains were LNP6505 (1), LNP6548 (2), LNP8013 (3), LNP10824 (4), LNP12681 (5), LNP12787 (6), LNP12886 (7), LNP12963 (8), LNP12970 (9), LNP13150 (10), LNP13407 (11), LNP13473 (12). Neisseria gonorrhoeae (Ng) strain was MS11 (13). Commensal Neisseria were: N.mucosa strain LNP405 (14), N.lactamica strain LNP415 (15), N.flavescens strain LNP414 (16), N.cinerea strain LNP415 (17), N.polysaccharia strain LNP462 (18), N.sicca strain LNP3265 (19). The probe used in (B) was an internal fragment of crgA generated by PCR using oligonucleotides 98-4 and 98-7. Nm strains were LNP10824 (1), LNP7381 (2), LNP6505 (3), LNP12873 (4), LNP12792 (5), LNP13146 (6), LNP12870 (7), LNP8013 (8), LNP13083 (9) and LNP13145 (10). These strains belong to different genetic lineages (Guibourdenche et al., 1997). The probe used in (C) was an internal fragment of crgA generated by PCR using oligonucleotides 98-4 and 98-7. Nm strain LNP8013 (1), Moraxella catarrhalis (Mc) LNP417 (2), N.gonorrhoeae strains MS11, LNP403 and LNP6911 (lanes 3–5); commensal Neisseria were: N.lactamica strain LNP415 (6), N.polysaccharia strain LNP462 (7), N.cinerea strain LNP415 (8), N.perflava strain LNP407 (9). All these strains were reported previously (Taha and Marchal, 1990; Guibourdenche et al., 1997).
Isolation of recombinant plasmids harbouring CREN-homologous fragments
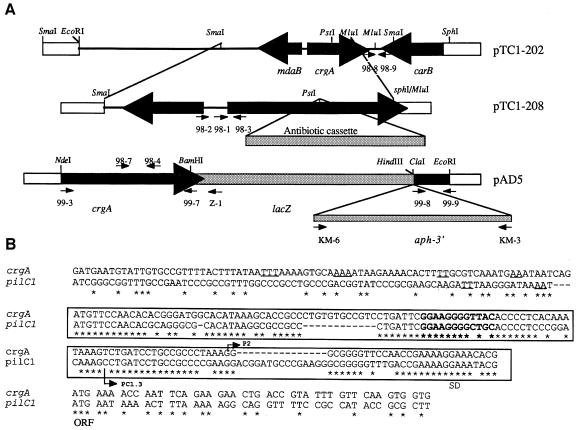
Next we tried to isolate a gene(s) that could be controlled by CREN in Nm. A cosmid-born gene bank from Nm strain 8013 was maintained in Escherichia coli. By colony blotting using the CREN of pilC1 as a probe, we isolated several positive E.coli clones. One of them (harbouring cosmid pNM5-266) was chosen for further study. Using Southern blot analysis, the ClaI fragment (9.5 kb) on pNM5-266 was shown to hybridize with the probe; it was subcloned into pUC18 vector to obtain the recombinant plasmid pTC1-202 (Figure 2) SmaI fragment on pTC1-202, which hybridized with the probe, was separated into several subclones and the nucleotide sequence was determined. An open reading frame (ORF), encoding a 299 aa protein, was found in this SmaI fragment. Upstream of this ORF is a sequence homologous to the CREN of pilC1 (Figure 2B). The CREN of pilC1 and crgA shared 65% homology. Like pilC1, crgA has in its CREN the unusual motif GG-N8-AC with a single mismatch (A instead of G) (Figure 2B). Indeed, sequences from other CRENs showed GG-N8-(A/G)C (data not shown). We have reported previously that this motif, which is close but not identical to the –24/–12 consensus sequence, is important for the expression of pilC1 upon contact with target cells (Taha et al., 1998).
Fig. 2. (A) Schematic representation of the crgA locus in Nm (Clone 12). Recombinant plasmid pTC1-202 was constructed from the original cosmid pNM5-266 as described in the text. Thick black arrows indicate the organization of ORFs. Recombinant plasmid pTC1-208 was constructed by subcloning the fragment SmaI–MluI of pTC1-202 into pUC18 vector. crgA was then disrupted by the insertion of a cassette encoding an antibiotic resistance (kanamycin or spectinomycin) into the PstI site. Recombinant plasmid pAD5 contains the crgA–lacZ–aph-3′ transcriptional fusion (see Materials and methods for detailed description). Small arrows indicate oligonucleotides used in this study, white boxes correspond to DNA of vectors. The figure is not drawn to scale. (B) Alignment of the nucleotide sequence of the CREN of pilC1 and crgA. The TSPs (P2 for crgA and PC1.3 for pilC1), the beginnings of the ORFs and the Shine–Dalgarno sequence (SD) are indicated. The CREN of both genes is in a box. The GG-N8-(A/G)C motif is in bold. The sequences of pilC1 and crgA that bind CrgA are upstream of the boxed region and the T-N11-A motifs are underlined.
The sequence of the ORF was submitted to DDBJ/EMBL/GenBank under accession No. AF190471. This ORF was tentatively named crgA for contact-regulated gene A. Upstream of crgA is another divergent ORF that encodes a potential protein homologous to the mdaB gene of E.coli. mdaB-encoded protein is involved in the modulation of the sensitivity of bacteria to tumoricidal agents (Chatterjee and Sternberg, 1995). Downstream of crgA is the 3′ end of an ORF homologous to the carB gene of N.gonorrhoea, which encodes one of the subunits of the carbamoyl-phosphate synthase (Lawson et al., 1995).
Next we performed Southern blotting experiments under moderate stringent conditions using as a probe an internal fragment of crgA generated by PCR using oligonucleotides 98-4 and 98-7. All Nm strains tested harboured a copy of the crgA gene (Figure 1B). DNA from strains of N.gonorrhoeae hybridized with the probe while no hybridization was observed with DNA from commensal Neisseria (Figure 1C).
crgA is induced upon contact with target cells
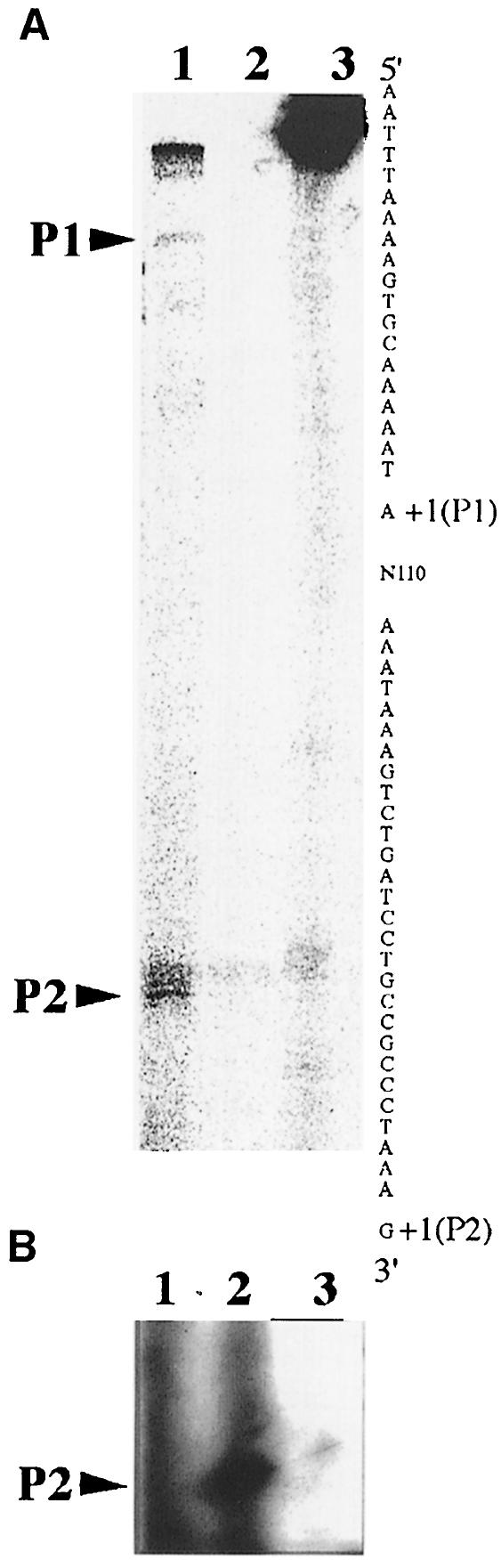
As crgA, like pilC1, is preceded by a CREN, we first analysed the expression of crgA. Total RNA was extracted from Clone 12 (wild-type meningococcal strain); an S1 mapping experiment was performed using a PCR-amplified fragment generated from oligonucleotides 98-2 and 98-3 (Figure 3A). TSPs were detected and were named P1 and P2. Similar results were also obtained by primer extension experiments using the oligonucleotide 98-3 (data not shown). Like pilC1, crgA has a TSP within the CREN (P2), which is preceded by the motif GG-N8-AC (Figures 2B and 3A). crgA is hence likely to be upregulated by target cell contact. To test this hypothesis, a primer extension analysis was performed with cell-associated bacteria harvested from a 1 h infected monolayer of Hec-1-B epithelial cells. In this analysis, we compared identical amounts of cell-associated bacteria with bacteria grown alone in cell culture media. As shown in Figure 3B, crgA seemed to be expressed at a higher level from P2 when bacteria interacted with the Hec-1-B epithelial cells. As an alternative test for the induction of crgA transcription upon cell contact, a promoterless lacZ gene was inserted downstream of the crgA stop codon (Figure 2A). crgA–lacZ was then introduced into Clone 12 (a wild-type meningococcal strain for crgA) to give strain NM99-3. In the absence of cells, crgA seemed to be expressed at a very low level (Table I) when Hec-1-B epithelial cells were infected (see Materials and methods), the level of β-galactosidase activity was found to be increased 3–6 times in cell-associated bacteria compared with bacteria grown alone in cell culture medium in the case of strain NM99-3 (crgA–lacZ) and strain KZ1 (pilC1–lacZ) (Table I). No induction was observed for strain EZ1 (pilE–lacZ), which showed a reduction in the level of β–galactosidase activity in cell-associated bacteria after 9 h of infection (Table II). Like pilC1, crgA seems to be induced upon cell contact.
Fig. 3. Nuclease S1 and reverse transcriptase promoter mapping of the TSPs of crgA. (A) S1 mapping was performed using an end-labelled fragment obtained by PCR between oligonucleotides 98-2 and 98-3. Total RNA was extracted from Clone 12 grown on GCB medium. (1) Total RNA from Clone 12 was incubated with the PCR fragment in the presence of S1 enzyme; (2) the PCR fragment was incubated in the presence of S1 enzyme but no RNA was added; (3) total RNA from Clone 12 was incubated with the PCR fragment but no S1 enzyme was added. Arrowheads indicate the TSPs P1 and P2. DNA sequence of the coding strand is shown with P1 and P2 indicated. (B) Reverse transcriptase mapping experiments were performed using total RNA extracted (1) from Hec-1-B epithelial cells, (2) from Hec-1-B cells infected with Clone 12 for 1 h, and (3) from Clone 12 grown alone in cell culture medium. The arrowhead indicates the TSP P2.
Table I. β-galactosidase activity during bacterium–cell interaction.
| Strain | crgA | Gene fusion | 1 h of infection |
4 h of infection |
9 h of infection |
|||
|---|---|---|---|---|---|---|---|---|
| bacteria alone | cell-associated | bacteria alone | cell-associated | bacteria alone | cell-associated | |||
| Clone 12 | wild-type crgA | no fusion | no activity | no activity | no activity | no activity | no activity | no activity |
| NM98-3 | inactivated | no fusion | no activity | no activty | no activity | no activity | no activity | no activity |
| KZ1 | wild-type crgA | pilC1–lacZ | 204 ± 53 | 950 ± 429 | 248 ± 52 | 418 ± 308 | 258 ± 82 | 249 ± 69 |
| NM98-5 | inactivated | pilC1–lacZ | 185 ± 45 | 2374 ± 1558 | 120 ± 27 | 764 ± 47 | 148 ± 54 | 772 ± 158 |
| EZ1 | wild-type crgA | pilE–lacZ | 422 ± 61 | 280 ± 27 | 373 ± 37 | 284 ± 25 | 367 ± 105 | 108 ± 1 |
| NM99-3 | wild-type crgA | crgA–lacZ | 14 ± 8 | 40 ± 4 | 12 ± 7 | 75 ± 10 | 14 ± 5 | 60 ± 24 |
Table II. Oligonucleotides used in this study.
| Oligonucleotide | Sequence | Relevant characteristic |
|---|---|---|
| C1-8 | 5′-ctgcctttttaaagttttattcatcgt-3′ | 3′ of the non-coding strand of pilC1 |
| C1-354 | 5′-ggataacagtaatattcaaagattat-3′ | 5′ of the coding strand of pilC1 |
| C1-152 | 5′-cctgcccgacggtatcccgcgaagcaagat-3′ | 5′ of the coding strand of pilC1 |
| C1-128 | 5′-cacgcagggcgcacataaggc-3′ | 5′ of the coding strand of pilC1 |
| Z-1 | 5′-cccgtaatcttacgtcagtaactt-3′ | 3′ of the non-coding strand of lacZ |
| KM6 | 5′-cccagcgaaccatttgagg-3′ | 5′ of the coding strand of aph-3′ gene |
| KM3 | 5′-gcggaagcttgccgtctgaatgctttttagacatctaaatctagg-3′ | 5′ of the non-coding strand of aph-3′ |
| 98-1 | 5′-gcgtcaaatgaaataatcagatgt-3′ | Figure 2 |
| 98-2 | 5′-gatgaatgtattgtgccgttttac-3′ | Figure 2 |
| 98-3 | 5′-caccacttgaacaaatacggtcagttcttc-3′ | Figure 2 |
| 98-4 | 5′-cgttcagccgtgcgcgagagcttggcatgg-3′ | Figure 2 |
| 98-7 | 5′-gcttcggaagaaacgagcgaaagtc-3′ | Figure 2 |
| 99-3 | 5′-cgtcatatgaaaaccaattcagaagaactgacc-3′ | 5′ of the coding strand of crgA |
| 99-5 | 5′-gctggtctcccatgaaaaccaattcagaagaactgacc-3′ | 5′ of the coding strand of crgA |
| 99-6 | 5′-cacctcgagtccacagagattgtttcccagttc-3′ | 5′ of the non-coding strand of crgA |
| 99-7 | 5′-gccggatccttatccacagagattgtttcccagtcc-3′ | 5′ of the non-coding strand of crgA |
| 99-8 | 5′-gctatcgatttctataaaaacctgtcataaaattg-3′ | Figure 2 |
| 99-9 | 5′-gctgaattcaaaaagccgagaccatctggg-3′ | Figure 2 |
| 99-17 | 5′-aatacacacctgcaaaggcgg-3′ | 5′ of the coding strand of pilT |
| 99-18 | 5′-tttagcgccgaaggcgagtaagt-3′ | 3′ of the coding strand of pilT |
| porA0 | 5′-gatgtcagcctatacggcgaaatcaaa-3′ | 5′ of the coding strand of porA |
| porA101 | 5′-gccgataaacgagccgaaatc-3′ | 3′ of the non-coding strand of porA |
Characterization of crgA as a new regulatory gene in Nm that is involved in bacterial adhesion to epithelial cells
Searches in non-redundant DDBJ/EMBL/GenBank CDS translations + PDB + SwissProt + Spupdate + PIR indicated a strong homology (ranging between 47 and 77%) of CrgA to LysR-type transcriptional regulators (LTTRs), possibly the most common type of transcriptional regulators in prokaryotes. LTTRs are involved in very diverse biological functions such as amino acid biosynthesis and regulation of virulence factors (Schell, 1993).
To study the possible regulatory role of crgA, we inactivated it by inserting a kanamycin- or spectinomycin-resistance gene (strains NM98-1 and NM98-3, respectively) (Figure 2A). When grown in GCB medium (Difco), these mutants produced amounts of pilin similar to the wild-type strain as judged by Western blotting (data not shown). Thus, crgA does not affect pilE expression under these conditions (absence of target cells).
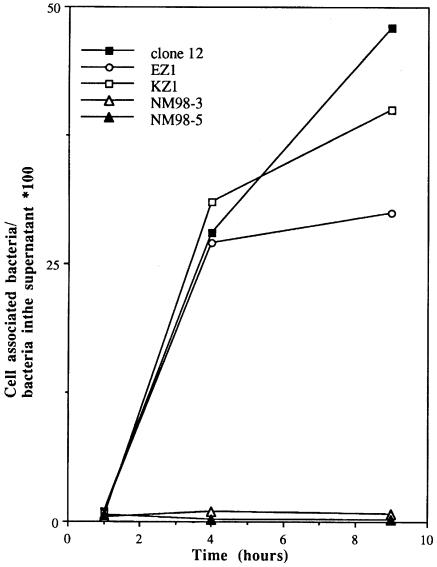
We next analysed the effect of the inactivation of crgA on the adhesion of Nm to human epithelial Hec-1-B cells. Monolayers of these cells were infected by Clone 12 (wild-type strain) or its crgA mutants and, after 1, 4 and 9 h the adhesion level was scored as described in Materials and methods. After 1 h of infection, the crgA mutants and wild-type strain showed comparable levels of adhesion to epithelial cells (Figure 4 and data not shown). However, adhesion of both mutant strains was dramatically reduced (20–50 times) after 4 and 9 h of infection (Figure 4). Moreover, the strain that had a kanamycin-resistance gene inserted downstream of the stop codon of crgA behaved like the wild-type strain (data not shown), ruling out the possibility that the phenotype observed in crgA mutants might be provoked by a polar effect on a downstream gene. These results indicate that the crgA gene is involved in the regulation of the adhesion of Nm to target cells and in particular, in a late step most likely corresponding to intimate adhesion.
Fig. 4. Adhesion level of Clone 12 and its derivatives on Hec-1-B epithelial cells. Infection was performed as described in Materials and methods. At 1, 4 and 9 h post-infection, cells were lifted off the plates and the adhesion level was measured.
crgA is involved in intimate adhesion of Nm and in effacing of microvilli on the surface of target cells
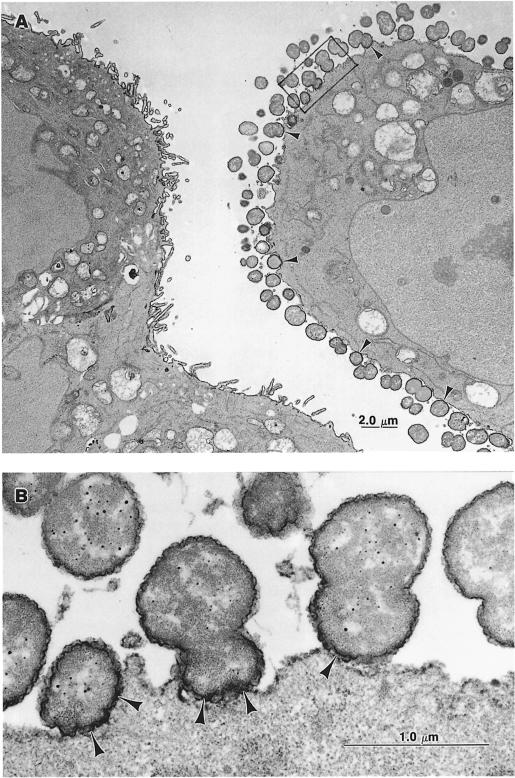
We next analysed the Nm–cell interactions at the ultrastructural level. Morphological examinations and incubation with concanavalin A (conA), which binds preferentially to α-d-mannose and α-d-glucose residues at the cellular surface, revealed selective association of the wild-type strain of Nm (Clone 12) with non-villous cell surfaces. These surfaces also showed weak conA binding (Figure 5A, compare infected with non-infected cells). Clone 12 was able to establish intimate adhesion with target cells (Figure 5B). It is noteworthy that the conA reaction product preferentially stained microvilli at the surface of bacteria-free cells.
Fig. 5. EM micrographs after conA/peroxidase staining of Hec-1-B epithelial cells infected for 9 h by Clone 12 (wild-type). (A) Numerous bacteria are in close contact with the cell on the right (arrowheads). The cell on the left is devoid of bacteria. Note that the conA/peroxidase reaction preferentially delimited microvilli on this cell. (B) A higher magnification from (A) (framed). Arrowheads indicate bacteria intimately adhered to the cell surface.
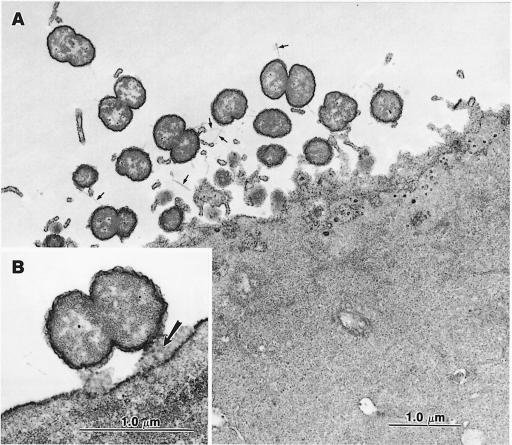
When a crgA mutant was tested, it showed a quite different interaction with target cells. Indeed, the crgA mutant was unable to adhere intimately to cells. Bacteria were always distant from the cell surface; bacterial and cellular membranes were never observed to come into direct contact (Figure 6). Moreover, microvilli were still present at the surface of the cells. Taken together, these data strongly suggest that the crgA mutant is impaired in adhesion to target cells and that the crgA gene is involved in the establishment of an intimate adhesion between Nm and target cells.
Fig. 6. EM micrographs after conA/peroxidase staining Hec-1-B epithelial cells infected for 9 h by strain NM98-3 (crgA mutant). (A) Bacteria are spread on the cell surface, but are not involved in intimate adhesion. Few pili (small arrows) are present. (B) One diplococcus is seen in close proxmity to the cell membrane but is still separated from the cell surface by a space in which the product of the conA/peroxidase reaction is present (arrow).
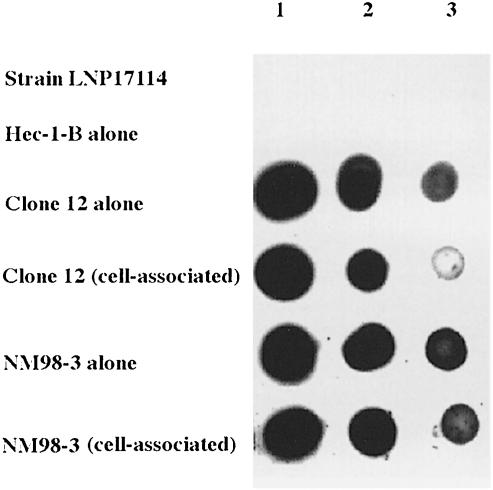
The expression of pilC1 is modulated through Nm–cell interaction (Table I; Taha et al., 1998). One possible explanation of the phenotype of the crgA mutant could be the absence of pilC1 modulation. We therefore tested the effect of crgA inactivation on the expression of pilC1. The insertionally inactivated crgA gene was introduced by transformation and allelic replacement into the previously described strain KZ1 to obtain the strain NM98-5. Strain KZ1 harbours the pilC1–lacZ transcriptional fusion (Taha et al., 1998). In adhesion assays, strains KZ1 and NM98–5 behaved like wild-type strain and CrgA– mutant, respectively (Figure 4). Both KZ1 and NM98-5 showed an increase in β-galactosidase activity in cell-associated bacteria, reflecting the induction of pilC1. In contrast to the wild-type strain, this induction was maintained throughout the adhesion in the strain NM98-5 (crgA mutant) (Table I). These results suggest that at least part of the effect of crgA inactivation (diminution of adhesion and absence of intimate adhesion) might be due to the absence of negative feedback regulation on pilC1 during adhesion to target cells. Therefore, CrgA could be a negative regulator of pilC1 during the intimate phase of bacterial adhesion. As adhesion assays have been performed using encapsulated strains, and as capsule is known to hinder bacterium–cell contact, we tested whether capsule level was modified during Nm–cell interaction. Colony blotting analysis was used to monitor capsule level in cell-associated bacteria and in bacteria grown alone in cell culture medium. As shown in Figure 7, when Clone 12 (wild-type strain) was tested, cell-associated bacteria had a lower capsule level than bacteria grown alone in cell culture medium. No difference was observed when strain NM98-3 (crgA mutant) was tested. Similar results were also obtained from an agglutination test (data not shown). These results indicate that capsule synthesis is also downregulated during intimate Nm–cell interaction in the wild-type strain but not in the crgA mutant.
Fig. 7. Colony blotting to monitor capsule level. Adhesion of Clone 12 (wild-type) and strain NM98-3 was performed as described in Materials and methods. After bacterial counting, serial dilutions of whole bacteria were spotted onto a nitrocellulose membrane. Equal amounts of bacteria were spotted, 2 × 107 (1), 4 × 106 (2) and 8 × 105 (3). The membrane was immunoblotted using a rabbit anti-serogroup C serum and revealed using the ECL-kit (Amersham). A strain of serogroup B (LNP17114) and epithelial cells alone were also tested as controls.
Purified CrgA binds pilC1 and crgA promoters
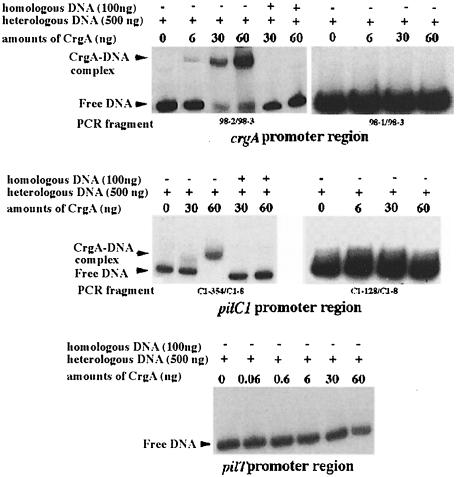
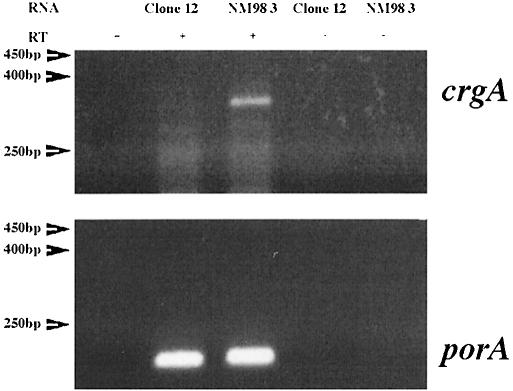
To test the hypothesis that CrgA regulates the expression of pilC1 by binding directly to its promoter region, crgA was cloned in pET28b (Novagen), creating a gene coding for CrgA with six C-terminal histidine residues. This gene fusion was expressed under the control of T7 bacteriophage promoter (see Materials and methods). His-tagged CrgA was overproduced and purified to >95% purity (data not shown). Purified CrgA was used in gel mobility shift assays. As LTTRs are known to bind to their own promoters and to produce in general a negative autoregulatory effect (Schell, 1993), we first used a PCR-generated DNA fragment of crgA (259 bp, oligonucleotides 98-2/98-3) corresponding to positions –187 to +72 with respect to P2, the major TSP of crgA (Figures 2B and 3A). As shown in Figure 8 CrgA bound specifically to its own promoter in a dose-dependent manner. However, no gel shift was observed using another PCR-generated DNA fragment of crgA (197 bp, oligonucleotides 98-1/98-3) corresponding to positions –125 to +72 with respect to P2 (Figure 8). These results suggest that CrgA binds to its own promoter between the positions –125 and –187. Interestingly, this region contains several copies of the T-N11-A motif, which has been suggested to be the recognition site for LTTRs (Figure 2B) (Schell, 1993). To study the effect of CrgA on its own expression, RT–PCR analysis of the transcription of crgA in Clone 12 (wild-type) and strain NM98-3 (crgA mutant) was performed. As shown in Figure 9, strain NM98-3 showed a higher transcription of crgA than the wild-type strain. As a control, no difference was observed in the transcription of the porA gene, which does not harbour a CREN (Arhin et al., 1998), and which encodes one of the major outer membrane porins of Nm (Figure 9). These results suggest that CrgA has a negative autoregulatory effect.
Fig. 8. Gel retardation analysis with purified CrgA. The promoter regions used were those of crgA, pilC1 and pilT. These regions were amplified by PCR using oligonucleotides 98-2/98-3 and 98-1/98-3 (crgA), C1-354/C1-8 and C1-128/C1-8 (pilC1) and 99-17 and 99-18 (pilT). PCR fragments were end labelled using T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mmol; Amersham). For each sample, 25 ng of labelled fragment were incubated with increasing amounts of purified CrgA (top) as described in Materials and methods. Heterologous DNA (500 ng) was present in all cases. Competition experiments with unlabelled homologous DNA (100 ng) were also performed for crgA and pilC1.
Fig. 9. RT–PCR analysis of the expression of crgA and porA genes. Oligonucleotides used were 98-4 and 98-7 (crgA) and porA0 and porA101 (porA). The bacteria tested are indicated above each lane. Size markers are indicated on the left. RT, reverse transcriptase.
We next tested a PCR-generated fragment (408 bp, oligonucleotides C1-354/C1-8) of the pilC1 gene corresponding to positions –319 to +89 with respect to the major TSP (PC1.3) (Taha et al., 1996). This fragment harbours the major TSP involved in the induction of expression of pilC1 upon cell contact (Taha et al., 1998). CrgA bound specifically to the pilC1 promoter region (Figure 8). Another PCR-generated fragment (205 bp, oligonucleotides C1-152/C1-8, positions –116 and +89) of pilC1 also showed a gel shift in the presence of CrgA (data not shown). However, no gel shift was observed using a PCR-generated DNA fragment of pilC1 of 162 bp between oligonucleotides C1-128/C1-8, and corresponding to positions –63 to +89 with respect to PC1.3 (Figures 2B and 8). These results suggest that CrgA binds specifically to pilC1 promoter between the positions –63 and –116. Interestingly, this region also contains the motif T-N11-A (Figure 2B).
For both crgA and pilC1, the addition of an excess of unlabelled heterologous DNA did not prevent DNA binding, while the addition of an excess of unlabelled homologous DNA effectively prevented DNA binding (Figure 8). These results demonstrate the specificity of CrgA binding to crgA and pilC1 promoters.
The binding of CrgA to pilT promoter was also tested. Indeed, pilT, one of the pilus-related genes, has recently been reported to be required for the induction of intimate adhesion of Nm to epithelial cells (Pujol et al., 1999). A PCR-generated fragment (530 bp) containing the upstream region of the pilT gene (between –494 and +36 with respect to the start codon) was used to test DNA binding. No CrgA–DNA complex of reduced mobility was observed under our experimental conditions with this fragment (Figure 8). These results indicate that CrgA regulates the expression of pilC1 by binding specifically to its promoter region but does not bind to the pilT promoter.
Discussion
Initial interactions between pathogenic bacteria and target cells are crucial events in cell infection. The results reported here confirm the two-step model of Nm–cell interaction. Bacteria first attach to epithelial cells in a pilus-dependent manner (initial adhesion). We have previously reported cross-talk between Nm and target cells that induces pilC1 gene expression and that is necessary for an optimal bacterial adhesion. This induction required the CREN element in the pilC1 promoter and was transient. The expression of pilE does not seem to be induced upon contact with cells, and the pilE promoter region does not harbour a CREN-like element (Taha et al., 1996).
During the second step, intimate adhesion, the expression of pilC1 decreased to its basal level after 9 h of adhesion (Taha et al., 1998). It is tempting to speculate that a negative feedback mechanism represses pilC1 expression during intimate adhesion of the bacteria to the cells. Pili also seem to disappear during intimate adhesion.The level of expression of the pilin structure gene, pilE, declines as the adhesion progresses (Table II).
Trans-acting regulators of the LTTR family that are able to modulate the expression of virulence factors have been reported in several bacteria. One example is Salmonella typhimurium SpvR, which regulates the expression of spv virulence genes involved in spleen invasion (Caldwell and Gulig, 1991; Coynault et al., 1992). LTTRs are thought to act in a coinducer-responsive manner in order to permit optimal bacterial survival and adaptation to environmental factors (Schell, 1993). In Nm, crgA, a new member of the LTTR family, seems to be expressed at a very low level in the absence of contact with target cells. However, it is noteworthy that the expression of crgA, like that of pilC1, is induced upon Nm–cell contact. The nature of the signal responsible for the induction of expression of crgA and pilC1, as well as the regulatory protein(s) required, remains to be determined. However, this induction depends on the presence of CREN in the promoter region. Once crgA is induced it seems to repress the expression of pilC1 after the initial phase of induction. The fact that CrgA is able to bind to pilC1 promoter immediately upstream of the CREN is in favour of this hypothesis. Indeed, the CrgA binding site is distinct from the sequence necessary for pilC1 transcriptional stimulation by Nm–cell contact (Figure 2; Taha et al., 1998). CrgA may modulate the function of CREN by binding DNA in the near vicinity of CREN.
Repression of pilC1 might be necessary for bacterial adhesion to progress further into intimate adhesion. The latter may occur by the unmasking of structures involved in intimate adhesion. In our study, it is unlikely that Opa proteins or Opc protein plays a major role in this process as Clone 12 and its derivatives, which are used in this study, are Opa– and Opc–. Moreover, the capsule does not prevent intimate interaction as it seems to be downregulated upon intimate adhesion. The nature of this regulation remains to be elucidated. The fact that the capsule level is not changed in the crgA mutant suggests that CrgA may directly repress capsule synthesis. However, capsule diminution may occur after intimate adhesion rather than prior to it. Alternatively, CrgA, once induced, might stimulate the expression of the gene(s) involved in intimate adhesion. These genes remain to be identified, and the purification of CrgA reported here should facilitate this identification. According to the second hypothesis, CrgA might act as both a positive and negative regulator of the transcription of target genes. Regulators of the LTTR family are very diverse. In general, they are positive transcriptional regulators with a negative autoregulatory effect (Schell, 1993). However, several LTTRs can act as repressors or even as repressor–activators, such as TfdS protein encoded by pJP4 in Alcaligenes eutrophus (Kaphammer and Olsen, 1990).
Adhesion of Nm to target cells provokes bacterium–cell cross-talk. The target cells seem to undergo several modifications such as loss of microvilli and carbohydrate-carrying surface structures, possibly as a result of signal transduction in the cell. Whether these modifications are induced by Nm or provoked by different physiological states of target cells remains to be analysed. Neisseria gonorrhoeae, which is closely related to Nm, was shown to induce the production of inflammatory cytokines by epithelial cells. This induction is dependent on the activation of the transcriptional factor NF-κB and requires the adhesion of N.gonorrhoeae to epithelial cells (Naumann et al., 1997). Moreover, we have recently shown that piliated (adhesive) meningococci but not non-piliated (not adhesive) meningococci are capable of inducing expression of the TNF-α encoding gene in target cells (Taha, 2000). This induction could be provoked by a pilus-activated signal transduction pathway in the cells. Interestingly, cortical plaque formation in Nm-infected epithelial cells also required pilus-mediated adhesion (Merz et al., 1999).
A new aspect in this Nm–cell interaction is the fact that the bacterium also undergoes an adaptive response through a signal transduction involving a network of regulators acting in cascade. CrgA could be a member of this network and could be involved in a coordinate regulation of bacterial genes such as pilC1 and other genes involved in the intimate adhesion. Other bacterial regulators in this pathway remain to be identified.
Epithelial cells are located at the interface between external environment and the host. Pilus-mediated adhesion could be responsible for targeting the interaction of bacteria to these cells (Abraham et al., 1998). This interaction would be expected to facilitate the passage of bacteria to internal compartments of the host. The activation through signal transduction pathways of bacteria and these strategically located cells is therefore a key element in neisserial pathogenesis.
Materials and methods
Bacterial strains and media
Clone 12 is a derivative of 8013, a serogroup C, class 1 strain of Nm. The relevant phenotype of this strain is P+, Opa–, Opc–, PilC1+/PilC2+. Nm was grown on GCB medium (Difco) containing the supplements described by Kellogg et al. (1963). Escherichia coli strains were DH5 (Hanahan, 1983) for plasmid preparation and BL21(DE3) pLysS, which was employed for overproduction of His6-tagged CrgA (Studier et al., 1990). Kanamycin and ampicillin were used at final concentrations of 100 μg/ml. Spectinomycin was used at a final concentration of 75 μg/ml. Other strains of Neisseria used in this study (see the legend to Figure 1) were described previously (Taha and Marchal, 1990; Guibourdenche et al., 1997). Transformation of Nm was performed as described previously (Taha et al., 1998). Strains EZ1 and KZ1 were described previously (Taha et al., 1998).
DNA techniques and immunoblotting
All recombinant DNA protocols, RNA extractions, hybridizations and primer extensions were performed as described elsewhere (Taha et al., 1988; Sambrook et al., 1989). Oligonucleotides used in this study are listed in Table II. Primer extension for crgA was performed using oligonucleotide 98-3.
Immunoblotting for pilin was performed as described previously (Dupuy et al., 1991). Colony blotting to monitor capsule level was performed by spotting a serial dilution of whole bacteria onto a nitrocellulose membrane, which was imunoblotted using a rabbit anti-serogroup C polyclonal serum (Institut Pasteur). This serum was also used to perform agglutination, which was estimated by microscopic examination using a Petroff–Hausser counting chamber (Touzart & Matignon).
Cloning of meningococcal crgA and construction of crgA–lacZ transcriptional fusions and His6-tagged CrgA
A cosmid library of Nm strain 8013 (Taha et al., 1996) was screened in a colony hybridization experiment using a PCR-generated DNA fragment corresponding to the specific promoter region of pilC1 (Taha et al., 1998). The probe was labelled using T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mmol; Amersham). For moderate stringent hybridization, the nitrocellulose filters were pre-hybridized for 2 h at 55°C and then hybridized overnight at 40°C. After several washings at 55°C, the filters were autoradiographed. The positive clones were further analysed by Southern blotting to define the minimal region hybridizing with the probe, and to determine the DNA sequence. A 9 kb fragment on the original plasmid (pNM5-266) was shown to hybridize with the probe. This fragment was subcloned into the AccI site of pUC18 vector (Pharmacia) to obtain the recombinant plasmid pTC1-202. The fragment MluI–SmaI was subsequently blunt-ended and subcloned between the SphI and SmaI sites of pUC18 to obtain the recombinant plasmid pTC1-208. crgA was then inactivated by the insertion of a blunt-ended DNA fragment carrying a cassette conferring resistance to kanamycin or spectinomycin into the blunt-ended PstI site of crgA on the recombinant plasmid pTC1-208 (Figure 2). The resulting recombinant plasmids were used to transform Clone 12 as previously described (Taha et al., 1998).
Olignucleotides 99-5 and 99-6 (Table II) were used to amplify the entire ORF of crgA. These oligonucleotides harbour BsaI and XhoI sites at their 5′ ends, respectively. This fragment was digested by BsaI and XhoI restriction enzymes, cloned into pET28b (Novagen) that was digested with NcoI (compatible ends with BsaI) and XhoI. The recombinant plasmid was named pAD2 and was used to overproduce CrgA.
Oligonucleotides 99-7 (with a 5′ BamHI adaptor) and 99-3 (with a 5′ NdeI adaptor) were used to amplify the entire ORF of crgA. The amplicon was cloned between the NdeI and BamHI sites in the pET16b vector (Novagen) to obtain the recombinant plasmid pAD1. A PCR-generated fragment of 250 bp (olignucleotides 99-8 and 99-9) located immediately downstream of crgA on the meningococcal chromosome was subsequently cloned between the ClaI and EcoRI sites that are located downstream of crgA in pAD1. The new recombinant plasmid was named pAD3. A promoterless lacZ gene (Perkins and Youngman, 1986) was then introduced between the BamHI and HindIII sites on pAD3 to obtain the recombinant plasmid pAD4. Finally, the aph-3′ gene (Taha et al., 1998), which confers kanamycin resistance, was then inserted into the blunt-ended HindIII site at the end of the lacZ fragment to obtain the recombinant plasmid pAD5. The recombinant plasmid pAD5 harbours the crgA–lacZ–aph-3′ operon, where lacZ is expressed under the control of the crgA promoter. Plasmid pAD5 was then used to transform Clone 12. Transformants were selected on standard GCB medium in the presence of 100 μg/ml kanamycin. Integration by homologous recombination into crgA on the Nm chromosome was verified by PCR analysis using oligonucleotides Z-1 and 98-1 (Table II and Figure 2). One transformant, designated NM99-3, was selected for further analysis.
Purification of CrgA
The recombinant plasmid pAD2 was used to transform strain BL21 (DE3) pLysS. This strain harbours the T7 RNA polymerase gene under the control of the inducible lacUV5 promoter. Moreover, this strain is deficient in Lon and OmpT proteases (Studier et al., 1990). The resulting strain was grown in LB medium at 37°C until early-exponential phase (absorbance at 600 nm of 0.2), isopropyl-β-d-thiogalactopyranoside (IPTG) was added (1 mM) and the incubation was continued for 2 h at 37°C. The cells were harvested by centrifugation at 10 000 g for 30 min and CrgA was purified on 3.5 ml of Ni–NTA agarose (Qiagen) as described previously (Derré et al., 1999).
Cell culture, adherence assays and β-galactosidase assays
Adhesion assays were performed using Hec-1-B cells as described previously (Taha et al., 1998), except that in β-galactosidase assay experiments, bacteria were centrifuged on the monolayer at 1000 g for 3 min. Adhesion was calculated as the ratio of cell-associated c.f.u. to c.f.u. present in the supernatant × 100. The production of β-galactosidase was performed with bacteria obtained from different fractions, i.e. cell-associated Nm, Nm obtained from the supernatant of infected monolayers or Nm grown in cell culture medium. At appropriate time points, Hec-1-B cells and cell-associated Nm were lifted off the plates. The number of c.f.u. in each fraction under study, i.e. cell-associated Nm, non-adherent Nm obtained from the supernatant of an infected Hec-1-B monolayer or Nm grown in cell culture medium, was determined by plating serial dilutions on GCB plates supplemented with the appropriate antibiotics. The number of c.f.u. in each fraction was used to standardize the β-galactosidase assays. The number of c.f.u. was converted to optical density OD600 using the equation: 1 OD600 = 9 × 108 bacteria/ml. β–galactosidase assays were performed using o-nitrophenol-β-galactoside (ONPG). Results are expressed as ‘Miller’ units, which are proportional to the increase in the absorbance of free o-nitrophenol per minute per constant cell density (Miller, 1972).
Gel retardation experiments
Fragments encompassing the crgA, pilC1 and pilT promoters were obtained by PCR amplification. The oligonucleotides used were 98-1, 98-2 and 98-3 (crgA), C1-128, C1-152, C1-354 and C1-8 (pilC1) and 99-17 and 99-18 (pilT) (Table II). These fragments were labelled using T4 polynucleotide kinase as recommended by the manufacturer (USB) with [γ-32P]ATP (3000 Ci/mmol; Amersham). For each sample, 25 ng of labelled DNA fragment were incubated with increasing amounts of CrgA (0.03 to 60 ng), and 500 ng of unlabelled heterologous DNA (pUC19 vector) were also added to eliminate non-specific binding. Experiments using an excess (100 ng) of the homologous unlabelled fragment were also performed. Loading buffer was added to the reaction mixture and samples were loaded onto a 6% non-denaturing polyacrylamide gel. After electrophoresis, the gel was dried and autoradiographed.
Reverse transcriptase–polymerase chain reaction (RT–PCR)
Total RNA was prepared using the SV Total RNA Isolation System (Promega). Expression of the crgA gene was monitored by RT–PCR. Five nanograms of total RNA were reverse transcribed into a single-strand cDNA using the oligonucleotide 98-7 (Table II) and reverse transcriptase (RT) (Promega). Amplification was then performed using the two oligonucleotides 98-4 and 98-7 (Table II) to obtain a 320 bp fragment. As a control, RT–PCR analysis was similarly applied to analyse expression of the porA gene. Five nanograms of total RNA were reverse transcribed into a single-strand cDNA using the oligonucleotide porA101 (Table II). Amplification was then performed using the two oligonucleotides porA0 and porA101 (Table II) to obtain a 170 bp fragment. Reverse transcriptase and PCR protocols were as described previously (Sambrook et al., 1989). PCR products were electrophoresed on 2% agarose gel and stained with ethidium bromide.
Electron microscopy
For electron microscopy (EM), Hec-1-B cells were infected by Clone 12 (wild type) or by strain NM98-3 (crgA mutant). After 9 h of infection, monolayers were washed with Sörensen's, 0.1 M phosphate buffer pH 7.3, and prefixed in 1.6% gluteraldehyde in the same buffer at 4°C for 1 h (morphological examination) or for 30 min (cytochemical reaction). For conventional analysis, monolayers were postfixed in buffered 1% OsO4, dehydrated in ethanol and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate.
For cytochemistry (ultrastructure visualization of carbohydrate components), we used the technique described by Bernhard and Avrameas (1971). Cells were prefixed for 30 min in glutaraldehyde and then rinsed in phosphate-buffered saline (PBS) and incubated in PBS containing conA (0.05 mg/ml) (Sigma) at room temperature for 15 min with gentle agitation. Cells were rinsed in PBS and treated with horseradish peroxidase (0.05 mg/ml in PBS) (HRP, grade VI; Sigma) at room temperature for 15 min. The peroxidase activity was visualized by means of diaminobenzidine (DAM) reaction. Postfixation was in 2% OSO4–1.5% potassium ferrocyanide solution for 1 h. After dehydration in ethanol, cells were embedded in epoxy resin in situ in culture wells. Ultrathin sections were cut tangentially or vertically to the basal surface of the plastic wells and examined unstained or slightly counter-stained with lead citrate. Examinations were performed in a JEOL-JEM 1010 electron microscope.
Acknowledgments
Acknowledgements
We thank Jean-Michel Alonso for his generous support, Tarek Msadek for his valuable help in protein purification, Bruno Dupuy for the generous gift of vectors, Magaly Ducos for the anti-serogroup C serum, and Tony Pugsley for careful and critical reading of this manuscript. This work was supported by the Institut Pasteur.
References
- Abraham S.N., Jonsson, A.B. and Normark, S. (1998) Fimbriae-mediated host–pathogen cross-talk. Curr. Opin. Microbiol., 1, 75–81. [DOI] [PubMed] [Google Scholar]
- Arhin F.F., Moreau, F., Coulton, J.W. and Mills, E.L. (1998) Sequencing of porA from clinical isolates of Neisseria meningitidis defines a subtyping scheme and its genetic regulation. Can. J. Microbiol., 44, 56–63. [PubMed] [Google Scholar]
- Bernhard W. and Avrameas, S. (1971) Ultrastructural visualization of cellular carbohydrate components by means of concanavalin A. Exp. Cell Res., 64, 232–236. [DOI] [PubMed] [Google Scholar]
- Caldwell A.L. and Gulig, P.A. (1991) The Salmonella typhimurium virulence plasmid encodes a positive regulator of a plasmid-encoded virulence gene. J. Bacteriol., 173, 7176–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P.K. and Sternberg, N.L. (1995) A general genetic approach in Escherichia coli for determining the mechanism(s) of action of tumoricidal agents: application to DMP 840, a tumoricidal agent. Proc. Natl Acad. Sci. USA, 92, 8950–8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coynault C., Robbe-Saule, V., Popoff, M.Y. and Norel, F. (1992) Growth phase and SpvR regulation of transcription of Salmonella typhimurium spvABC virulence genes. Microb. Pathog., 13, 133–143. [DOI] [PubMed] [Google Scholar]
- Derré I., Rapoport, G. and Msadek, T. (1999) CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol. Microbiol., 31, 117–131. [DOI] [PubMed] [Google Scholar]
- Dupuy B., Taha, M.-K., Pugsley, A.P. and Marchal, C. (1991) Neisseria gonorrhoeae prepilin export studied in Escherichia coli. J. Bacteriol., 173, 7589–7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Owen S.D., Dehio, C., Haude, A., Grunert, F. and Meyer, T.F. (1997) CD66 carcinoembryonic antigens mediate interactions between Opa-expressing Neisseria gonorrhoeae and human polymorphonuclear phagocytes. EMBO J., 16, 3435–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibourdenche M., Giorgini, D., Guèye, A., Larribe, M., Riou, J.-Y. and Taha, M.-K. (1997) Genetic analysis of a meningococcal population based on the polymorphism of pilA-pilB locus: a molecular approach for meningococcal epidemiology. J. Clin. Microbiol., 35, 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol., 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Jonsson A.-B., Nyberg, G. and Normark, S. (1991) Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J., 10, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källström H., Islam, M.S., Berggren, P.O. and Jonsson, A.B. (1998) Cell signaling by the type IV pili of pathogenic Neisseria. J. Biol. Chem., 273, 21777–21782. [DOI] [PubMed] [Google Scholar]
- Kaphammer B. and Olsen, R.H. (1990) Cloning and characterization of tfdS, the repressor–activator gene of tfdB, from the 2,4-dichlorophenoxyacetic acid catabolic plasmid pJP4. J. Bacteriol., 172, 5856–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.S., Peacock, W.L., Deacon, W.E., Brown, L. and Pirkle, C.I. (1963) Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol., 85, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson F.S., Billowes, F.M. and Dillon, J.A. (1995) Organization of carbamoyl-phosphate synthase genes in Neisseria gonorrhoeae includes a large, variable intergenic sequence which is also present in other Neisseria species. Microbiology, 141, 1183–1191. [DOI] [PubMed] [Google Scholar]
- Merz A.J., Enns, C.A. and So, M. (1999) Type IV pili of pathogenic Neisseriae elicit cortical plaque formation in epithelial cells. Mol. Microbiol., 32, 1316–1332. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Nassif X., Beretti, J.-C., Lowy, J., Stenberg, P., O'Gaora, P., Pefifer, J., Normark, S. and So, M. (1994) Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. Proc. Natl Acad. Sci. USA, 91, 3769–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M., Webler, S., Bartsch, C., Wieland, B. and Meyer, T.F. (1997) Neisseria gonorrhoeae epithelial cell interaction leads to the activation of the transcription factor nuclear factor κB and activator protein 1 and the induction of inflammatory cytokines. J. Exp. Med., 186, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J.B. and Youngman, P.J. (1986) Construction and properties of Tn917-lac, a transposon derivative that mediates transcriptional gene fusions in Bacillus subtilis. Proc. Natl Acad. Sci. USA, 83, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C., Eugène, E., de Saint Martin, L. and Nassif, X. (1997) Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect. Immun., 65, 4836–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C., Eugène, E., Marceau, M. and Nassif, X. (1999) The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl Acad. Sci. USA, 96, 4017–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel T., Scheuerpflug, I. and Meyer, T.F. (1995) Neisseria PilC protein identified as type-4 pilus tip-located adhesion. Nature, 373, 357–359. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schell M.A. (1993) Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol., 47, 597–626. [DOI] [PubMed] [Google Scholar]
- Studier F.W., Rosenberg, A.H., Dunn, J.J. and Dubendorff, J.W. (1990) Use of T7 polymerase to direct expression of cloned genes. Methods Enzymol., 185, 60–89. [DOI] [PubMed] [Google Scholar]
- Taha M.-K. (2000) Neisseria meningitidis induces the expression of the TNF-α gene in endothelial cells. Cytokine, 12, 21–25. [DOI] [PubMed] [Google Scholar]
- Taha M.-K. and Marchal, C. (1990) Conservation of Neisseria gonorrhoeae pilus expression pilA and pilB regulatory genes in the Neisseria genus. Infect. Immun., 58, 4145–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha M.-K., Giorgini, D. and Nassif, X. (1996) The pilA regulatory gene modulates the pilus-mediated adhesion of Neisseria meningitidis by controlling the transcription of pilC1. Mol. Microbiol., 19, 1073–1084. [DOI] [PubMed] [Google Scholar]
- Taha M.-K., Morand, P.C., Pereira, Y., Eugène, E., Giorgini, D., Larribe, M. and Nassif, X. (1998) Pilus-mediated adhesion of Neisseria meningitidis: the essential role of cell contact-dependent transcriptional upregulation of the PilC1 protein. Mol. Microbiol., 28, 1153–1163. [DOI] [PubMed] [Google Scholar]
- Virji M., Makepeace, K., Ferguson, D.J. and Watt, S.M. (1996) Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol., 22, 941–950. [DOI] [PubMed] [Google Scholar]