Abstract
tmRNA, through its tRNA and mRNA properties, adds short peptide tags to abnormal proteins, targeting these proteins for proteolytic degradation. Although the conservation of tmRNA throughout the bacterial kingdom suggests that it must provide a strong selective advantage, it has not been shown to be essential for any bacterium. We report that tmRNA is essential in Neisseria gonorrhoeae. Although tagging per se appears to be required for gonococcal viability, tagging for proteolysis does not. This suggests that the essential roles of tmRNA in N.gonorrhoeae may include resolving stalled translation complexes and/or preventing depletion of free ribosomes. Although derivatives of N.gonorrhoeae expressing Escherichia coli tmRNA as their sole tmRNA were isolated, they appear to form colonies only after acquiring an extragenic suppressor(s).
Keywords: Escherichia coli/10Sa RNA/SsrA RNA/trans-translation
Introduction
The ssrA gene of Escherichia coli encodes a small, abundant and unusual RNA (Ray and Apirion, 1979; Chauhan and Apirion, 1989; Oh et al., 1990), named tmRNA (also known as 10Sa RNA and SsrA RNA) based on its having characteristics of both tRNA and mRNA (Williams and Bartel, 1996). A relationship of tmRNA to tRNA was first suggested by structural models showing that tmRNA could fold into a tRNA-like configuration with a TΨC stem–loop and an acceptor stem with a 3′ end CCA aminoacetylation sequence (Komine et al., 1994). The acceptor stem contains a G:U wobble, which, like the G3:U70 wobble in the acceptor stem of alanyl tRNA (Hou and Schimmel, 1988; McClain et al., 1988), is configured to serve as the signal recognized by alanyl-tRNA synthetase (Komine et al., 1994). In vitro studies showed that E.coli tmRNA could be charged with alanine (Komine et al., 1994). A relationship of tmRNA to mRNA was first suggested by the finding of Tu et al. (1995) that interleukin–6 expressed in E.coli acquired an 11 amino acid C–terminal tag (AANDENYALAA). A search of E.coli genomic sequence revealed the ssrA gene as the probable gene encoding the last 10 amino acids of the tag (Tu et al., 1995). Based on these and their own studies, Keiler et al. (1996) proposed that tmRNA removes prematurely terminated polypeptide from truncated mRNA while adding an 11 amino acid tag that targets the polypeptide for proteolysis. Referred to as trans-translation or co-translation, this model postulates that tmRNA, charged with alanine and functioning as tRNA, enters the acceptor position of the ribosome, which stalls on mRNA lacking an in-frame stop codon. The alanine is added to the C–terminus of an uncompleted polypeptide; then, serving as an mRNA, tmRNA provides the template for the addition of the tag. At least three proteases, Tsp, HflB (FtsH) and Clp, recognize this C–terminal tag (Keiler et al., 1996; Gottesman et al., 1998; Herman et al., 1998). Consistent with the idea that tmRNA is involved in some aspect of translation is the finding that tmRNA is associated with 70S ribosomal particles, but not with the 50S or 30S subunits (Komine et al., 1996; Tadaki et al., 1996).
The fact that ssrA is highly conserved in the eubacteria, homologs being identified in all but one of the nearly 70 bacterial species examined (Williams, 1999), suggests that tmRNA provides an important activity to bacteria. It is surprising, then, that in the three bacterial species previously examined, E.coli (Oh and Apirion, 1991), Vibrio cholerae (C.Huang, V.DiRita and D.I.Friedman, unpublished data) and Bacillus subtilis (Muto et al., 1998), tmRNA was shown not to be essential. Escherichia coli mutants that do not express tmRNA, however, exhibit some phenotypic characteristics, such as slower growth rates and altered motility at high temperature, and slower recovery from carbon starvation (Oh and Apirion, 1991; Komine et al., 1994). Also, E.coli mutants lacking a functional tmRNA fail to support growth of certain hybrid phages (Strauch et al., 1986; Retallack et al., 1994; Withey and Friedman, 1999). These phages, as a group called λimmP22, have varying amounts of genetic material from colipage λ and Salmonella phage P22 (Gemski et al., 1972; Botstein and Herskowitz, 1974). As recently reported, the parental P22 phage fails to grow in Salmonella typhimurium derivatives lacking tmRNA activity (Karzai et al., 1999). Although growth of some λimmP22 hybrids is supported effectively by mutant tmRNAs which add tags that do not contain the signal for protein degradation, growth of these hybrids is not supported by a mutant tmRNA that cannot be charged with alanine (Withey and Friedman, 1999). These observations led to the proposal that tagging for proteolysis might not be the primary role for tmRNA; rather, in some cases, tmRNA may only be required to remove the stalled ribosome from intact mRNA, with degradation of polypeptides serving only an ancillary role (Withey and Friedman, 1999). Recent studies have shown that a protein encoded by the smpB gene, located in E.coli immediately upstream of the ssrA gene (ssrAEc), is also required for tmRNA activity (Karzai et al., 1999; J.Withey, C.Huang and D.I.Friedman, in preparation).
Although studies of tmRNA have focused on E.coli ssrA variants where tmRNA is not essential, it is plausible to consider that studies in other bacteria might uncover more information on the physiological role of this unique and ubiquitous RNA. Because Neisseria gonorrhoeae (gonococcus) differs from E.coli in having an extremely limited ecological niche and a relatively small genome size, we chose to determine the role of tmRNA in the growth of N.gonorrhoeae. We identified and cloned the ssrA homolog from N.gonorrhoeae (ssrANg) and found that tmRNA is essential for growth of N.gonorrhoeae. Consistent with the results of studies with λimmP22 phages, our studies show that while charging of tmRNA, and presumably tagging per se, are required, degradation of the tagged polypeptides is not required for the growth of N.gonorrhoeae. Further, these studies suggest that E.coli tmRNA supports the growth of N.gonorrhoeae only when the bacterium has acquired an as yet unidentified extragenic suppressor mutation(s).
Results
Identification and cloning of the ssrA gene of N.gonorrhoeae
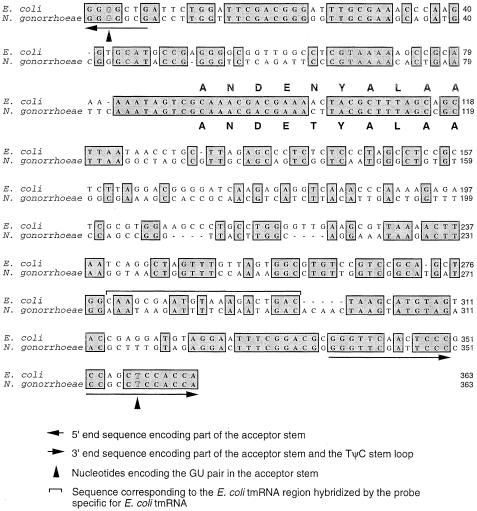
We identified a candidate for the ssrA gene of N.gonorrhoeae strain N400 (Freitag et al., 1995) based on the results of a homology search of the genome sequence of N.gonorrhoeae strain FA1090 (Roe et al.) using the sequence of the ssrAEc gene as the search template. The putative ssrANg gene shares 59% nucleotide identity with the ssrAEc gene. Based on the defined 5′ and 3′ ends of the E.coli tmRNA (Chauhan and Apirion, 1989; Komine et al., 1994), we have determined that the ssrANg gene encodes an RNA of 363 nucleotides, equal in length to that of the mature E.coli tmRNA (Figure 1).
Fig. 1. Sequence alignment of E.coli and N.gonorrhoeae (strain N400) ssrA genes. Boxes indicate positions of sequence identity. Letters denoting amino acid tag sequences are placed either above (for E.coli) or below (for N.gonorrhoeae) the codons of the tagging sequences. The tag sequences of the two ssrA genes differ at only one codon; codon 5 encodes asparagine in ssrAEc and threonine in ssrANg.
The putative ssrANg gene, like other identified ssrA homologs, possesses conserved sequences in regions thought to be essential for tmRNA function, including those at the 5′ terminus, in the sequence encoding the peptide tag and at the 3′ terminus (Figure 1). The putative tag sequence encoded by ssrANg differs by only one codon from that encoded by ssrAEc, having a threonine at position 5 instead of an asparagine (Figure 1). We cloned a 1.8 kb PCR product (GCtm) containing the ssrANg gene from strain N400 into pUP6, a plasmid with two gonococcal uptake sequences (GCU) in the polylinker (Wolfgang et al., 1999), yielding plasmid pGCtm.
Functional analysis of N.gonorrhoeae tmRNA in E.coli
We tested whether gonococcal tmRNA could complement an E.coli strain lacking tmRNA for growth of λimmP22hy25 (Hilliker and Botstein, 1976), a phage requiring tmRNA for growth (Retallack et al., 1994; Withey and Friedman, 1999). Plasmid pGCtm was introduced into an E.coli derivative, K8619 (Withey and Friedman, 1999), that carries an ssrAEc allele inactivated with a chloramphenicol acetyltransferase (cat) gene insertion (ssrAEc::cat) (Kirby et al., 1994), creating strain K8745. Northern blot analysis using the coding sequence of ssrAEc as the probe showed that pGCtm in K8745 expressed an RNA having the size of and homology with E.coli tmRNA (Figure 2).
Fig. 2. Northern blot analysis identifying tmRNAs of E.coli and N.gonorrhoeae. The probe was synthesized using ssrAEc as template. Strains examined were: K37, wild-type for ssrAEc (lane 1), K8619, isogenic with K37 except that it has the ssrAEc::cat allele (lane 3), and variants of K8619 carrying pGCtm (lane 2) or derivatives of pGCtm with ssrANg mutant alleles (lanes 4–6).
Phage growth was measured as efficiency of plating (EOP). When the EOP of λimmP22hy25 on the bacterial lawns formed from K37 (wild type for ssrAEc) was taken as 1, the EOP of λimmP22hy25 on the bacterial lawns formed from the ssrAEc::cat strain K8619 was 3.8 × 10–5, while the EOP on lawns formed from the strain, K8745, which only expresses tmRNA from the plasmid-encoded ssrANg gene, was 0.5 (Table I). Thus, based on its capacity to support growth of λimmP22hy25, the gonococcal tmRNA expressed from pGCtm is functional.
Table I. Ability of ssrA alleles to support the growth of λimmP22hy25.
| Strain | ssrA alleles | EOPa |
|---|---|---|
| K37 | ssrAEc | 1 |
| K8619 | ssrAEc::cat | 3.8 × 10–5 |
| K8745 | ssrAEc::cat /ssrANg | 0.5 |
| K9335 | ssrAEc::cat/ssrANg/ochre | 5.4 × 10–5 |
| K9336 | ssrAEc::cat/ssrANg/DD | 4.0 × 10–5 |
| K9337 | ssrAEc::cat/ssrANg/HindIII | 0.4 |
| K9101 | ssrAEc::cat/ssrANg/–StyI | 0.5 |
| K9102 | ssrAEc::cat/ssrANg/UG | 5.3 × 10–5 |
aEOP represents the titer of the phage on a bacterial lawn formed from the indicated strain divided by the titer on a lawn formed on K37, our standard E.coli strain with a wild-type ssrAEc allele.
Evidence that tmRNA is essential for the growth of N.gonorrhoeae
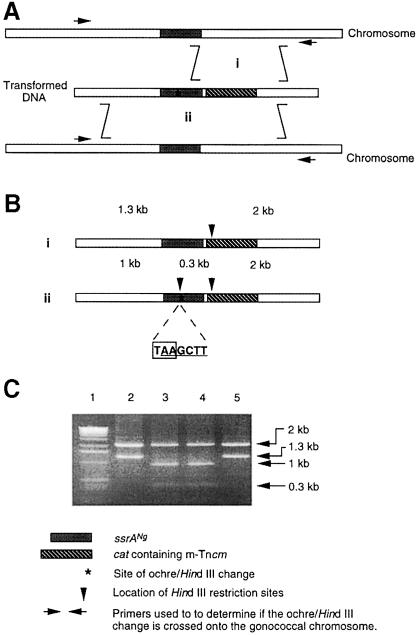
To determine if tmRNA is essential in N.gonorrhoeae, we attempted to construct a derivative with an insertionally inactivated ssrA gene. pGCtm/ssrANg::cat, a derivative of pGCtm with a cat gene inserted into the ssrANg gene, was used in these experiments. It would be expected that when N.gonorrhoeae is transformed with the linear GCtm/ssrANg::cat fragment, chloramphenicol-resistant (Cmr) transformants (i.e. recombinants) will form by a double crossover event that results in an allelic replacement of the wild-type ssrANg gene with the ssrANg::cat allele. We were unable to obtain stable Cmr recombinants following transformation with the GCtm/ssrANg::cat fragment when the recipient N.gonorrhoeae was haploid for ssrANg (strain N400). However, we were able to obtain stable Cmr recombinants when the recipient N.gonorrhoeae was diploid for ssrANg (strain C101). The latter observation ruled out a number of possible technical reasons for our failure to obtain allelic replacement with ssrANg::cat in the haploid strain, including problems in the DNA preparation, defects in transformation or failure of recombination in the region of the genome containing the ssrANg gene. Thus, the generation of stable recombinants in the diploid strain suggested that the failure to obtain allelic replacements in the haploid strain with ssrANg::cat is due to the essential nature of the product of the ssrA gene in N.gonorrhoeae.
We next determined whether insertion mutations adjacent to but not in ssrANg could be crossed into the haploid strain. A library of mini-transposon insertions in the GCtm insert in pGCtm was constructed by shuttle mutagenesis (Seifert et al., 1986) using the m-Tncm transposon, a derivative of Tn3 with a cat gene (Drake et al, 1997). Insertion mutations were located upstream of the ssrANg gene (group A), in the ssrANg gene (group B) or downstream of the ssrANg gene (group C) (Figure 3). Thirteen representative m-Tncm insertion mutations were tested for their ability to complement the ssrAEc::cat mutation for λimmP22hy25 phage growth in E.coli as well as the formation of Cmr colonies when transformed into N.gonorrhoeae either haploid or diploid for the ssrANg gene. As shown in Figure 3, for all insertion mutations having m-Tncm in the ssrANg gene (group B), λimmP22hy25 growth in E.coli was not supported and Cmr recombinants could not be isolated from attempted transformations of an N.gonorrhoeae strain haploid for the ssrANg. However, Cmr colonies could be isolated from transformations of a strain diploid for ssrANg. For all variants having m-Tncm insertions outside the ssrANg gene (groups A and C), λimmP22hy25 growth in E.coli was supported and Cmr recombinants were obtained with N.gonorrhoeae haploid or diploid for the ssrANg gene. These results argue that m-Tncm insertions in ssrANg affect gonococcal viability directly, not by interfering in a polar manner with expression of downstream genes.
Fig. 3. Functional analysis of ssrANg alleles with transposon insertions in and surrounding ssrANg. A library of 13 m-Tncm insertions was created in plasmid pGCtm. The positions (shown by downward pointing arrows) in the GCtm fragment of these insertions were located by DNA sequencing. Insertions were located in three regions: A, upstream of ssrA; B, within ssrA; and C, downstream of ssrA. The effect on tmRNA activity of each insertion was determined by assessing growth of λimmP22hy25 in derivatives of the ssrAEc::cat E.coli strain K8619 carrying variants of pGCtm with the respective 13 insertions (row 1); (+) support of phage growth and (–) no support of phage growth. Formation of stable recombinants with mini-transposon inserted ssrANg alleles in N.gonorrhoeae either haploid (row 2) or diploid (row 3) for ssrANg was determined by selecting Cmr colonies; (+) Cmr colonies were isolated and (–) Cmr colonies were not isolated.
Use of mutant ssrANg alleles to determine activities of tmRNA required by N.gonorrhoeae
To explore a possible role(s) of tmRNA in growth of N.gonorrhoeae, we constructed mutant ssrANg alleles based on the set of ssrAEc mutants used in the studies examining the requirement for λimmP22 growth in E.coli (Withey and Friedman, 1999). The DNA sequences encoding the acceptor stem and the tag in N.gonorrhoeae tmRNA are nearly identical to those encoding E.coli tmRNA. We assumed that changes affecting E.coli tmRNA action should, when duplicated in N.gonorrhoeae tmRNA, similarly affect the action of that tmRNA. The construction of ssrANg mutant alleles, and the subsequent selection of N.gonorrhoeae recombinants carrying those mutant alleles, was facilitated by the use of the m-Tncm transposon insertion C7, located 16 nucleotides downstream of the 3′ end of the ssrANg gene (see Figure 3). This insertion was tightly linked to ssrANg, but did not interfere with expression of functional tmRNA (see previous section). Thus, we could use chloramphenicol resistance as a linked selectable marker for allelic replacement of the chromosomal ssrANg gene with the desired ssrANg mutant alleles.
We looked first at the effect of ssrANg mutations designed to allow tagging, but not subsequent degradation of tagged proteins. The ssrANg/ochre allele has a glutamic acid codon, GAA, at position 4 of the tagging sequence changed to the ochre translation termination codon, TAA (Figure 4). The ochre mutation should result in premature termination of translation, producing a tag lacking the terminal ALAA required for proteolytic degradation (Keiler et al., 1996). The ssrANg/DD allele has the two alanine codons at positions 9 and 10 changed to two aspartic acid codons (Figure 4). This change, referred to as DD, results in a tag in which the critical terminal alanines are replaced with negatively charged amino acids shown to interfere with degradation (Parsell et al., 1990; Keiler et al., 1996). Thus, the presence of either of these mutant tags should not signal proteolysis.
Fig. 4. Variants used in determining the properties of gonococcal tmRNA required for N.gonorrhoeae growth. The DD mutation was generated by replacing the two terminal alanine codons with two aspartic acid codons, while the ochre mutation was generated by changing the fourth codon from GAA to the sequence encoding the ochre translation terminator UAA. Both mutants also carry a single base substitution (from AAACTT to AAGCTT) that provides an identifying HindIII site, as indicated by the underlined sequence above the tag sequence. Two changes were made in the sequences encoding the acceptor stem: these result in the change of the G:U base pair required for recognition of alanyl-tRNA synthetase to a U:G base pair. This mutant also carries a single base substitution (from CCTTGG to CGTTGG) that provides an identifying marker by eliminating a StyI site that is only five nucleotides away from the G3 position, indicated by the asterisk.
It has been shown in a previous study from our laboratory (Withey and Friedman, 1999) that E.coli having ssrAEc alleles with either the ochre or DD changes (ssrA° or ssrADD, respectively) as their only functional ssrAEc gene support the growth of λimmP22hy25 nearly as well as an isogenic strain with a wild-type ssrAEc. Therefore, we assumed that the ssrANg ochre and DD alleles (ssrANg/ochre and ssrANg/DD, respectively) would also function in E.coli to support the growth of λimmP22hy25, but this was not the case. As shown in Table I, neither variant ssrANg allele complemented the E.coli strain K8619 for the growth of λimmP22hy25. Nevertheless, E.coli carrying either of these ssrANg mutant alleles expressed stable tmRNA comparable in amount with that expressed by the wild-type ssrANg allele as detected by Northern blotting (Figure 2). Therefore, it is unlikely that instability of these tmRNA molecules can explain the failure to support phage growth.
Derivatives of N.gonorrhoeae with either the ssrANg/ochre or the ssrANg/DD allele were obtained by replacement of the wild-type ssrANg with the appropriate mutant allele (Table II and Figure 5, see also Materials and methods). These results suggest that directing proteins toward degradation is unlikely to be an essential activity for tmRNA in N.gonorrhoeae.
Table II. ssrA allele and N.gonorrhoeae viability.
| ssrA allele linked to Cmr markerb | Allelic replacementa (No. of Cmr colonies) |
|
|---|---|---|
| N400 (ssrANg haploid) | C101 (ssrANg diploid) | |
| ssrANg | 118 | 120 |
| ssrANg/HindIII | 124 | 122 |
| ssrANg/DD | 130 | 140 |
| ssrANg/ochre | 41 | 28 |
| ssrANg/–StyI | 58 | 84 |
| ssrANg/UG | 0 | 38 |
aOne of three experiments that had essentially the same results. Approximately 3 × 106 bacteria transformed with ∼3 μg of DNA were plated for each transformation.
bThe Cmr marker is located in the linked transposon 16 nucleotides downstream of the ssrANg gene.
Fig. 5. Details of the allelic replacement that placed ssrANg mutant alleles on the gonococcal chromosome. Construction of a recombinant chromosomal ssrANg gene with the ochre mutation is used as an example of the procedure. (A) The double crossover recombination events resulting in allelic replacement. Selection is for Cmr carried by the linked m-Tncm. The m-Tncm will be crossed onto the chromosome without the ochre mutation if the recombination occurs as shown in (i). The ochre mutation (indicated by the asterisk) will be crossed with the m-Tncm onto the chromosome if the recombination occurs as shown in (ii). Arrows indicate the positions of the primers used in (B) and (C). (B) Schematic representation of the PCR products of the allelic replacements diagrammed in (A). The downward pointing arrows indicate the locations of the HindIII sites used in the analysis of the PCR products shown in (C). The location of the ochre codon is shown in (ii). (C) Cleavage of PCR products confirming proper allelic replacement. Cmr transformant colonies were used as the source of template DNA for the PCRs. PCR products, generated using the primers indicated in (A), were cleaved with HindIII and the products analyzed by electrophoresis in an agarose gel [source of DNA: lane 1, DNA 1 kb ladder (Gibco); lane 2, wild-type allele; lane 3, plasmid with cloned ssrANg/ochre allele; lanes 4 and 5, transformants]. Because an engineered HindIII site was positioned adjacent to the ochre mutation, PCR products from transformants that have an extra HindIII site, as shown in lane 4, are likely to carry the ochre mutation. Therefore, these transformants were chosen for sequence analysis to determine definitively if the transformant had the desired mutation.
We next examined a mutation altering the G:U base pair in the acceptor stem that blocks the alanyl aminoacylation of tmRNA and, in turn, blocks trans-translation at the earliest step. Like the G:U in the acceptor stem of alanyl tRNA, this G:U base pair is essential for charging the E.coli tmRNA with alanine (Komine et al., 1994). A mutant allele of the ssrANg gene, ssrANg/UG, was constructed so that the G:U base pair in the acceptor stem of the tmRNA (G3 and U357) is changed to U:G (U3, G357, respectively) (Figure 4). We made changes as conservative as possible to minimize any alteration in the tmRNA structure (Komine et al., 1994; Williams and Bartel, 1996; Felden et al., 1997; Zwieb et al., 1999) that might result from the mutations. An ssrAEc gene similarly mutated to alter the U:G charging signal encodes a tmRNA that fails to support growth of λimmP22 phages (Withey and Friedman, 1999). Not surprisingly, we find that when placed in E.coli, the ssrANg/UG mutant allele of N.gonorrhoeae also fails to support growth of λimmP22hy25 (Table I). We next determined whether N.gonorrhoeae with ssrANg/UG as the sole ssrA gene is viable by attempting to construct a strain with that allele as the only ssrA gene. To this end, a DNA fragment carrying ssrANg/UG with the linked Cmr marker was transformed into N.gonorrhoeae either haploid or diploid for ssrANg, and Cmr colonies were selected. We failed to obtain Cmr recombinants following transformation of ssrANg haploid strain N400. However, we did obtain viable Cmr colonies following transformation of the ssrANg diploid strain C101 (Table II). These results indicate that alanyl aminoacylation of tmRNA is required for the essential role(s) that tmRNA plays in supporting growth of N.gonorrhoeae.
Action of E.coli tmRNA in N.gonorrhoeae
To determine whether E.coli tmRNA supports growth of N.gonorrhoeae, we constructed an ssrANg/ssrAEc heterodiploid strain (C102) that has the ssrAEc gene located in the non-essential IgA1 protease gene (iga) (Figure 6A). Northern blot analysis, employing a probe specific for E.coli tmRNA, showed that the level of tmRNA of E.coli origin expressed in the heterodiploid gonococcal strain was approximately equal to that expressed in the E.coli strain K37 (Figure 6B). We then determined if the endogenous ssrANg gene in the heterodiploid strain could be replaced with an ssrANg allele insertionally inactivated by an m-Tncm transposon without affecting viability. For this purpose, two GCtm derivatives with m-Tncm inserted at different locations were employed, B12 within the ssrANg gene and C12 immediately 3′ of the ssrANg gene (see Figure 3).
Fig. 6. Substitution of the N.gonorrhoeae tmRNA with E.coli tmRNA in N.gonorrhoeae. (A) Insertion of ssrAEc in the N.gonorrhoeae genome at the iga locus. PCR fragments containing ssrAEc and the ermC gene (encoding resistance to erythromycin) were cloned into the gonococcal non-essential iga gene and the entire fragment was cloned in plasmid pUP6. The resulting plasmid, pIGA::ssrAEc, was trans- formed into N400 and erythromycin-resistant (Emr) transformants were isolated. These Emr colonies, shown to be heterodiploid for ssrA (ssrANg/ssrAEc), were formed by a double crossover within the iga sequences flanking the ssrAEc-ermC insert. Strain C102 is one of these heterodiploid derivatives of N400 that was chosen for further study. Two derivatives of GCtm carrying m-Tncm insertions, B12 (in ssrANg) and C12 (downstream of ssrANg), were used to transform C102 to determine if E.coli tmRNA is sufficient to support gonococcal viability. (B) Northern blot analysis to identify E.coli tmRNA in N.gonorrhoeae. RNA was isolated from bacterial strains whose genotypes are listed at the top of each lane. Two probes were used, one specific for E.coli tmRNA and the other specific for 16S rRNA; the latter served as a control to indicate the relative amounts of RNA loaded in each lane.
Although Cmr recombinants were obtained following transformation of the heterodiploid strain C102 with DNA carrying the B12 insertion, these transformants (ssrANg::cat/ssrAEc) were observed only after ∼3 days of incubation (Table III). This was in contrast to the results of transformations either with DNA carrying the B12 insertion into the homodiploid strain C101 (ssrANg/ssrANg) or with DNA carrying the C12 insertion into the heterodiploid strain C102 (ssrANg/ssrAEc), all of which produced Cmr colonies in ∼1 day (Table III). Surprisingly, once isolated, the heterodiploid transformants with the B12 insertion (e.g. C103) now grow normally, i.e. form colonies in ∼1 day. One explanation for the initial delay in colony formation is that transformants expressing only ssrAEc are viable but grow very slowly. More rapid growth occurs when they acquire some type of mutation(s) that allows the foreign tmRNA to function in N.gonorrhoeae either by making a change in E.coli tmRNA per se or by altering the expression or activity of an N.gonorrhoeae function(s). Alternatively, a mutation(s), by altering the expression or activity of a gene product(s) normally regulated by tmRNA, could eliminate the requirement for tmRNA.
Table III. Use of allelic replacement to study the requirement for an ssrA allele in N.gonorrhoeae.
| Selective marker used in allelic replacement | Antibiotic selection marker | Recipient strain and ssrA allele(s) |
|||
|---|---|---|---|---|---|
| N400 (ssrANg) | C101 (ssrANg/ssrANg | C102 (ssrANg/ssrAEc)a | C103 (ssrANg::cat/ssrAEc)a | ||
| m-Tncm in ssrANg (B12) | Cmr | − | + | +b | NA |
| m-Tncm downstream of ssrANg (C12) | Cmr | + | + | + | NA |
| knr in iga | Knr | NA | NA | + | − |
aThe ssrAEc gene is located in the iga gene.
bThese Cmr recombinant colonies appeared ∼3 days following plating, while other recombinant colonies appeared ∼1 day following plating.
(+) indicates that antibiotic-resistant recombinants were isolated.
(–) indicates that antibiotic-resistant recombinants were not isolated.
NA, not applicable.
To determine if a mutation(s) located in the ssrAEc gene is responsible for this normal growth phenotype, we constructed a new heterodiploid strain by introducing the ssrAEc gene from strain C103 into strain N400. Although this recombinant, C104, has the ssrAEc gene from strain C103, we still observed a delay in colony formation (∼3 days) following a transformation in which the wild-type ssrANg gene was replaced with the B12 insertion.
The next set of experiments tested whether a mutation(s) at a second site on the gonococcal chromosome eliminates the requirement for tmRNA. This was done by determining if we could replace the ssrAEc of strain C103 located in iga successfully by a kanamycin resistance gene (knr) cassette similarly cloned in iga, yielding a strain without a functional ssrA gene. Two ssrA heterodiploid strains were used in the experiment; strain C102 with both ssrA alleles (ssrANg and ssrAEc) intact and strain C103 with ssrAEc as the only functional ssrA allele. As shown in Table III, the ssrAEc gene could be replaced with iga::knr in C102, where there still was a functional ssrANg gene, but ssrAEc could not be replaced in C103 where the ssrANg gene had already been inactivated insertionally by a cat gene. Therefore, if the normal growth phenotype when ssrAEc is the only functional ssrA allele is due to a second site mutation(s), that mutation(s) does not eliminate the requirement for an intact ssrA gene.
Finally, we addressed the question of whether a mutation somewhere in the N.gonorrhoeae genome is required for effective action of the E.coli tmRNA in that bacterium. To do this, we reconstructed the heterodiploid strain by replacing the inactivated ssrANg::cat allele in C103 with the wild-type ssrANg allele, creating N.gonorrhoeae derivatives heterodiploid for ssrA (ssrANg/ssrAEc) (see Materials and methods). When these heterodiploids, such as C105, were transformed with DNA carrying the B12 insertion, Cmr recombinants were obtained in ∼1 day, not the ∼3 days observed following transformation of the original heterodiploid strain C102, a result expected if the C103 strain had acquired a mutation(s) that allows normal colony growth when E.coli tmRNA is the only functional tmRNA.
Discussion
In this study, we report the identification of the ssrA gene of N.gonorrhoeae and show that this gene, which shares 59% homology with the ssrA gene of E.coli, encodes a tmRNA essential for N.gonorrhoeae growth.
It has been proposed that the tagging of the C–terminus of unfinished polypeptides by tmRNA, through trans-translation, serves to direct those polypeptides to a degradative pathway (Keiler et al., 1996). Our studies show that although tagging for proteolysis is unlikely to be the essential function of tmRNA in N.gonorrhoeae, trans-translation is likely to be an essential function. To target polypeptides effectively for proteolysis, it was shown in E.coli that the tag requires specific amino acids (ALAA) at its C–terminus (Keiler et al., 1996). Since the tag encoded by ssrANg varies by only one amino acid from that of ssrAEc, it seems plausible to assume that the action of tmRNA as well as the associated processes are the same in the two organisms. By changing the tag sequence to one that either terminates translation upstream of the important terminal amino acids (ssrANg/ochre) or to one with changes in the terminal amino acids known to result in a faulty signal (ssrANg/DD) in E.coli, tmRNA should no longer provide a mechanism for targeting polypeptides for proteolysis (Parsell et al., 1990; Keiler et al., 1996). The fact that we could construct viable derivatives of N.gonorrhoeae that have ssrANg genes with either the ochre or DD tagging sequences as their sole intact ssrA gene suggests that the essential role of tmRNA in N.gonorrhoeae is in a step prior to degradation of the tagged polypeptide. On the other hand, attempts to construct derivatives of N.gonorrhoeae in which the wild-type ssrANg gene is replaced with a mutant allele that encodes a tmRNA defective in amino acid charging (ssrANg/UG) were unsuccessful. Thus, it appears that N.gonorrhoeae requires charged tmRNA, even though it does not require that tmRNAs encode the signal appropriate to direct polypeptide degradation.
Based on the results obtained from studying the role of tmRNA in growth of certain λimmP22 phages, Withey and Friedman (1999) proposed that the major activity of tmRNA was removal of stalled ribosomes from mRNA. They postulated that some sequence or structure in the mRNA causes ribosome stalling, resulting in a block in translation. If this block is not removed, presumably by tmRNA, expression from the affected mRNA is reduced and if this, in turn, affects the synthesis of a protein(s) that is normally present in limited amounts, phage viability is impaired. It is plausible to assume that an analogous scenario occurs in N.gonorrhoeae. Accordingly, ribosome stalling results in lower than normal levels of an essential protein(s) normally synthesized in limited amounts, to levels below those needed for viability. Thus, release of the ribosome from the mRNA would be the important function of tmRNA, with degradation having, at most, an ancillary role. Consistent with this idea is the recent finding that the presence of rare codons in mRNA coupled with a scarcity of the cognate tRNA initiates the trans-translation mechanism (Roche and Sauer, 1999).
Relief of stalling could be important for another reason; by reducing sequestration of ribosomes, it could ensure that ribosomes are recycled adequately. The number of ribosomes in the bacterial cell appears ultimately to be regulated by the level of rRNA transcription (Sarmientos et al., 1983; Gourse et al., 1986). Hence, the number of rRNA operons (rrn) in a bacterium is likely to influence the nature of the response to a reduction in free ribosomes. Neisseria gonorrhoeae has four rrn operons (Dempsey and Cannon, 1994) compared with seven in E.coli (Kiss et al., 1977) and 10 in B.subtilis (Bott et al., 1984; LaFauci et al., 1986), ssrA being essential in N.gonorrhoeae but not in the latter strains. Thus, N.gonorrhoeae, with a relatively low number of rrn operons, may be more heavily dependent on ribosome recycling to maintain an adequate level of free ribosomes than those bacteria with a higher copy number of rRNA genes.
A major consideration in assessing the function of tmRNA is the fact that the tagging sequences of all known ssrA genes are very similar and have terminal hydrophobic amino acids (Williams and Bartel, 1996; Williams, 1999). The E.coli and N.gonorrhoeae tagging sequences differ at only one codon, while there is less homology in other regions of these two tmRNAs. These observations are consistent with the argument that the tagging sequences are likely to be central to the activity of tmRNAs. Thus, it was surprising to find that results from two of our experiments suggested that there could be some level of specificity of the bacterium for its own tmRNA.
The first indication of such specificity derived from the results of the complementation assays testing phage plating in E.coli when the only tmRNAs are the tmRNA variants encoded either by ssrANg/ochre or ssrANg/DD. Previous studies from our laboratory had shown that the ssrAEc variants that encode tmRNAs with the ochre and DD tag sequences complemented the E.coli strain with the insertionally inactivated ssrAEc gene for supporting the growth of λimmP22hy25. Although the tmRNA encoded by a wild-type ssrANg gene supports growth of λimmP22hy25, the products encoded by the ssrANg ochre and DD mutant alleles fail to support the growth of this phage. Moreover, as also reported here, the ochre and DD variants of ssrANg support growth of N.gonorrhoeae. The second indication of such specificity derived from our observation that colonies of N.gonorrhoeae derivatives that had ssrAEc as the sole functioning ssrA allele on the chromosome appeared to have acquired a mutation(s) during their initial growth that increased growth rate and led to colony formation. Northern blot analysis showed that the levels of E.coli tmRNA expressed in N.gonorrhoeae were comparable to levels expressed in K37 (an E.coli strain wild-type for ssrAEc) or to levels of gonococcal tmRNA expressed in gonococcal strain N400. Therefore, it seems unlikely that differences in levels of tmRNA can explain the apparent need for a mutation(s) for E.coli tmRNA to function fully in N.gonorrhoeae. It is more likely that there is some qualitative difference between these two tmRNAs that explains the inability of E.coli tmRNA to support gonococcal growth effectively without additional changes. Our studies suggest that if this increase in growth rate is due to a mutation(s), the mutation(s) is not located in the ssrAEc gene, but somewhere in the N.gonorrhoeae genome.
In considering possible targets for such a mutation, one obvious candidate is the smpB gene. SmpB protein is required for tmRNA activity both in E.coli (Karzai et al., 1999) and N.gonorrhoeae (J.Withey, C.Huang and D.I.Friedman, in preparation). These SmpB proteins, like their cognate tmRNAs, share significant sequence identities (45% amino acid identity for the SmpB proteins), but obviously have differences. Nevertheless, a mutant SmpB cannot explain our results since DNA sequence analysis of the smpB gene of gonococcal strain C103 revealed no differences from the sequence of the wild-type gonococcal smpB gene (data not shown). This result does not rule out the possibility that a mutation affecting another protein influences SmpB–tmRNA interaction.
Since the E.coli and gonococcal tmRNAs have nearly identical tag sequences, sequences outside of the region encoding the tag may influence tmRNA function. Obviously, the tmRNA structure is important (Komine et al., 1994; Williams and Bartel, 1996; Felden et al., 1997; Zwieb et al., 1999), and our studies with the ssrANg/UG mutant show that for growth of both λimmP22hy25 and N.gonorrhoeae, amino acid charging is necessary for tmRNA function. Since both E.coli and N.gonorrhoeae ssrA encode similar signals for amino acid charging, we must look elsewhere to explain the functional differences between these two tmRNAs.
It is important to note for this consideration that the gonococcal ssrANg/ochre and ssrANg/DD mutant genes are functional in N.gonorrhoeae. This suggests that these variants of ssrANg may function more effectively in N.gonorrhoeae than in E.coli. We can offer three explanations for such an effect of the bacterial environment. Firstly, a difference in the sequence outside the tag region could influence some interaction of tmRNA. Secondly, a sequence difference could result in differences in structure that could influence such an interaction. Thirdly, the variant tmRNAs could function similarly in E.coli and N.gonorrhoeae, but the required levels of tmRNA activity necessary for N.gonorrhoeae viability may be less than those required for phage growth.
Why is tmRNA essential for N.gonorrhoeae but not for E.coli? There are two obvious differences between these bacteria. First, the N.gonorrhoeae genome is about half the size of the E.coli genome (Roe et al.). Secondly, N.gonorrhoeae only has one niche, the human host, while E.coli has two, existing within as well as without an animal host (Britigan et al., 1985). These two characteristics are likely to be related. Because it need only encode functions for survival in one niche, N.gonorrhoeae is likely to require a smaller array of gene products and, thus, have fewer redundant functions. Based on these considerations, we suggest three possible reasons why ssrA is essential in N.gonorrhoeae. (i) The activity supplied by tmRNA is essential and, unlike E.coli, a function capable of substituting for ssrA is not present to maintain proper levels of some essential protein(s) normally present in limited amounts. (ii) Neisseria gonorrhoeae is missing a function(s) that can substitute for the protein(s) that are present at sub-physiological levels in the absence of tmRNA. (iii) Ribosome stalling is more severe in N.gonorrhoeae.
Even though previous studies have shown that those bacteria in which the ssrA gene is inactivated are viable, the nearly universal presence of tmRNA (Williams, 1999) indicates that tmRNA provides an important survival advantage to bacteria. Thus, a bacterium such as N.gonorrhoeae, in which ssrA is an essential gene, should prove to be extremely useful in determining the critical role or roles of tmRNA.
Materials and methods
Bacterial strains and phage
Bacterial strains are listed in Table IV. λimmP22hy25 was constructed by an in vivo cross between λ and P22 (Hilliker and Botstein, 1976).
Table IV. Bacterial strains.
| Strain | Genus | Parental strain | Plasmid | Relevant genotype | Source/reference |
|---|---|---|---|---|---|
| K37 | E.coli | ssrAEc | this laboratory | ||
| K8619 | E.coli | K37 | ssrAEc::cat | Withey and Friedman (1999) | |
| K8745 | E.coli | K8619 | pGCtm | ssrANg | this work |
| K9101 | E.coli | K8619 | pGSmu1 | ssrANg/–StyI | this work |
| K9102 | E.coli | K8619 | pGSmu2 | ssrANg/UG | this work |
| K9335 | E.coli | K8619 | pGSmu3 | ssrANg/ochre | this work |
| K9336 | E.coli | K8619 | pGSmu4 | ssrANg/DD | this work |
| K9337 | E.coli | K8619 | PGSmu5 | ssrANg/HindIII | this work |
| VD300 | N.gonorrhoeae | ssrANg | Koomey et al. (1987) | ||
| N400 | N.gonorrhoeae | VD300 | ssrANg recA6(tetM)a | Freitag et al. (1995) | |
| C101 | N.gonorrhoeae | N400 | ssrANg/ssrANg | this work | |
| C102 | N.gonorrhoeae | N400 | ssrANg/ssrAEc | this work | |
| C103 | N.gonorrhoeae | N400 | ssrANg::cat/ssrAEc | this work | |
| C104 | N.gonorrhoeae | N400 | ssrANg/ssrAEcb | this work | |
| C105 | N.gonorrhoeae | C103 | ssrANg/ssrAEc | this work |
aThe expression of the recA gene is IPTG inducible.
bThis ssrAEc gene is derived from that of C103.
Plasmids
pUP6 (Wolfgang et al., 1999) contains two gonococcal uptake sequences (GCU) required for N.gonorrhoeae transformation and two NotI sites that flank both the polylinker and the uptake sequences. Cleavage at the NotI sites provided fragments for transformation. Plasmid pGCtm was constructed by cloning the ssrANg-containing GCtm PCR product into the pUP6 vector. Plasmids pGSmu1–GSmu5 are derivatives of pGCtm that, respectively, carry the mutant alleles ssrANg/StI–, ssrANg/UG, ssrANg/ochre, ssrANg/DD and ssrANg/HindIII (see ‘Construction of ssrANg mutant alleles’ for details). Plasmid pIGA, a derivative of pUP6 with a 4 kb fragment containing the gonococcal IgA1 protease gene (iga) cloned into the HindIII site of the vector, served as the precursor for construction of two plasmids. Each of these plasmids carries an insert cloned into the SacI site linked to an ermC gene in the iga gene; pIGA::ssrAEc with the ssrAEc gene and pIGA::knr with a gene encoding resistance to kanamycin (knr).
Media
Escherichia coli strains were grown on LB plates or in LB broth (Sambrook et al., 1989). The N.gonorrhoeae strains were grown on clear solid medium (Gc agar) at 37°C in 5% CO2 or liquid Gc medium pre-incubated in 5% CO2 (Gc broth) (Koomey et al., 1987). Antibiotics were used at the following final concentrations: 50 μg/ml of kanamycin for both E.coli and N.gonorrhoeae; 12.5 and 10 μg/ml of chloramphenicol for E.coli and N.gonorrhoeae, respectively; 300 and 8 μg/ml of erythromycin for E.coli and N.gonorrhoeae, respectively.
Cloning procedures
The GCtm fragment containing ssrANg was isolated for cloning by PCR, using Gc1 (5′–CGTTCGAGCATATCGGTTC–3′) and Gc4 (5′–CCGAGATACTGAAAGGTGCG–3′) as primers, and genomic DNA of strain N400 as the template.
The ssrAEc gene was isolated by PCR using Ec/Gc1 (5′–GATCGAGCTCGGCTATCACATCCGACAC–3′) and Ec/Gc2 (5′–GATCGAGCTCGCTTACTGCCACTGGACTT–3′) as primers, and E.coli genomic DNA as the template. The synthesized fragment was digested with SacI and, as described above, cloned into the SacI site of plasmid pIGA.
To introduce the ssrA gene of E.coli into the chromosome of N.gonorrhoeae, the non-essential gonococcal IgA1 protease (iga) gene cloned into pUP6 plasmid was used as the crossover sequence. The ermC gene, encoding erythromycin resistance and located adjacent to the cloned ssrAEc gene, served as the selective marker (Figure 6).
In vivo assay for tmRNA function
Functional activities of the cloned N.gonorrhoeae ssrA wild-type and mutant genes were assessed by testing for support of growth of λimmP22hy25. Plasmids with the cloned ssrA genes to be tested were transformed into K8619, a derivative of our standard E.coli strain K37 that carries the ssrA::cat allele (Withey and Friedman, 1999). Overnight cultures of the K8619 derivatives carrying the plasmid constructs to be tested were used to form lawns on LB plates containing, when indicated, the appropriate antibiotic. Phage growth was assessed by measuring efficiency of plating (EOP) as described by Bear et al. (1984).
Northern blot analysis and DNA sequencing
Northern blot analysis was performed essentially as described by Sambrook et al. (1989). Total bacterial RNA samples were prepared using the Qiagen RNA kit (Qiagen). One probe that hybridizes tmRNAs from E.coli and N.gonorrhoeae was synthesized employing the ssrAEc gene as a template and labeled with 32P using the DNA random labeling kit from Boehringer Mannheim. The other probe that specifically detects E.coli tmRNA was an oligonucleotide, end-labeled with 32P using T4 polynucleotide kinase (New England Biolabs), whose sequence (5′–GTCAGTCTTTACATTCGCTTG–3′) corresponds to a region of ssrAEc that differs from ssrANg (see Figure 1).
DNA sequencing was performed as previously described (Sanger et al., 1977). Gonococcal colonies were used as the source of template DNA to confirm appropriate allelic replacement. In other cases, purified plasmid or chromosome DNA was used as template DNA.
Shuttle mutagenesis
The procedures as described by Seifert et al. (1986) were employed to generate a library of transposon insertion mutations along the length of the GCtm fragment. The positions of the m-Tncm transposon insertions were identified by PCR and confirmed by DNA sequencing.
Transformation of N.gonorrhoeae
See Table IV for a description of the strains. Transformation of N.gonorrhoeae was performed essentially as described by Freitag et al. (1995). Since recA is under the control of the plac promoter in N400 and its derivatives, isopropyl-β-d-thiogalactopyranoside (IPTG) was used to induce the transient expression of RecA during transformation. Strain C101, which is diploid for ssrANg, was generated by transforming N400 with pGCtm and selecting for kanamycin-resistant transformants. Since the bacterium does not support replication of this plasmid (Drake et al., 1997), stable transformants will only be formed if the plasmid integrates into the gonococcal chromosome. Strain C102 was constructed by allelic replacement of the iga gene with the iga::ssrAEc allele by transforming the iga::ssrAEc fragment released from pIGA::ssrAEc into the N400 strain and selecting for erythromycin-resistant transformants (Figure 6A). C104, ssrANg/ssrAEc, was made by transforming the genomic DNA of C103 into N400 and isolating erythromycin-resistant colonies. C105 was constructed from C103 by replacing the ssrANg::cat allele with the wild-type ssrANg allele. C103 cells were placed on Gc plain plates (no antibiotics added) pre-impregnated with IPTG to induce RecA expression, then a drop of pGCtm DNA solution was placed on the bacteria. Bacteria from the pGCtm DNA-treated area were transferred to fresh Gc plain plates to isolate single colonies. Colonies were tested for growth on Gc plain plates and Gc plates containing chloramphenicol. Colonies that had become sensitive to chloramphenicol were tested by PCR to determine if the wild-type ssrANg allele had replaced the ssrANg::cat allele.
Construction of ssrANg mutant alleles
Mutant ssrANg alleles were constructed using splicing by overlap extension (SOE) (Horton et al., 1989). The following are brief descriptions of the GCtm variants (nucleotide numbering is based on the sequence of the deduced mature tmRNA). (i) ssrANg/UG has nucleotide substitutions that change the G:U base pair at the acceptor stem of tmRNA and a substitution that eliminates a StyI restriction site near the 5′ terminus of the ssrANg gene. The UG change was engineered by two substitutions, G3T and T357G. The StyI change, five nucleotides away from the G3T substitution, was engineered by a nucleotide substitution at position 10 (a change in the recognition site from CCTTGG to CGTTGG) (Figure 4). (ii) ssrANg/ochre has a nucleotide substitution that changes the fourth codon of the putative tagging sequence (GAA) to a nonsense codon (TAA) (Figure 4). (iii) ssrANg/DD has nucleotide substitutions that change codons 9 and 10 of the putative tagging sequences from alanine codons (GCA and GCT) to aspartic acid codons (GAC and GAT) (Figure 4). The GCtm variants with changes in the tagging sequence also have a substitution at nucleotide 13 of the tagging sequence that introduces a HindIII site (Figure 4). The changes creating the HindIII or eliminating the StyI sites do not affect tmRNA function; tmRNA expressed from genes having one or the other of those changes supported λimmP22hy25 growth in E.coli and N.gonorrhoeae growth (Tables I and II).
The C7 DNA used as template in the PCRs has the m–Tncm insertion located immediately downstream of ssrANg (Figure 3). Chloramphenicol resistance conferred by the transposon facilitated selection of allelic replacements. Cmr transformants were screened for the appropriate allelic replacement by restriction enzyme analysis of PCR products. DNA sequencing was employed to establish conclusively that the appropriate allelic replacement had occurred.
Acknowledgments
Acknowledgements
The authors thank Drs Victor DiRita, Frederick Neidhardt and Michele Swanson for helpful comments. This work was supported by Public Health Service Grant AI11459-10 (to D.I.F.), Public Health Service Grant A127837 (to J.M.K.) and GCRG Grant M01-RR0042 for sequence analysis (to University of Michigan). M.W. and J.W. acknowledge support from NIH training grant 2 T32 GM07315.
References
- Bear S.E., Court, D.L. and Friedman, D.I. (1984) An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J. Virol., 52, 966–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D. and Herskowits, I. (1974) Properties of hybrids between Salmonella phage P22 and coliphage lambda. Nature, 251, 584–589. [DOI] [PubMed] [Google Scholar]
- Bott K., Stewart,G.C. and Anderson,A.G. (1984) Genetic mapping of cloned ribosomal RNA genes. In Hoch,J.A. and Ganesan,A.T. (eds), Syntro Conference on Genetics and Biotechnology of Bacilli. Academic Press, New York, NY, pp. 19–34. [Google Scholar]
- Britigan B.E., Cohen, M.S. and Sparling, P.F. (1985) Gonococcal infection: a model of molecular pathogenesis. N. Engl. J. Med., 312, 1683–1694. [DOI] [PubMed] [Google Scholar]
- Chauhan A.K. and Apirion, D. (1989) The gene for a small stable RNA (10Sa RNA) of Escherichia coli. Mol. Microbiol., 3, 1481–1485. [DOI] [PubMed] [Google Scholar]
- Dempsey J.A. and Cannon, J.G. (1994) Locations of genetic markers on the physical map of the chromosome of Neisseria gonorrhoeae FA1090. J. Bacteriol., 176, 2055–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake S.L., Sandstedt, S.A. and Koomey, M. (1997) PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol. Microbiol., 24, 657–668. [DOI] [PubMed] [Google Scholar]
- Felden B., Himeno, H., Muto, A., McCutcheon, J.P., Atkins, J.F. and Gesteland, R.F. (1997) Probing the structure of the Escherichia coli 10Sa RNA (tmRNA). RNA, 3, 89–103. [PMC free article] [PubMed] [Google Scholar]
- Freitag N., Seifert, H.S. and Koomey, M. (1995) Characterization of the pilF–pilD pilus assembly locus of Neisseria gonorrhoeae. Mol. Microbiol., 16, 575–586. [DOI] [PubMed] [Google Scholar]
- Gemski P. Jr, Baron, L.S. and Yamamoto, N. (1972) Formation of hybrids between coliphage lambda and Salmonella phage P22 with a Salmonella typhimurium hybrid sensitive to these phages. Proc. Natl Acad. Sci. USA, 69, 3110–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Roche, E., Zhou, Y.N. and Sauer, R.T. (1998) The ClpXP and ClpAP proteases degrade proteins with C–terminal peptide tails added by the SsrA tagging system. Genes Dev., 12, 1338–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R.L., de Boer, H.A. and Nomura, M. (1986) DNA determinants of rRNA synthesis in E.coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell, 44, 197–205. [DOI] [PubMed] [Google Scholar]
- Herman C., Thévenet, D., Boulc, P., Walker, G.C. and D'Ari, R. (1998) Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev., 12, 1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker S. and Botstein, D. (1976) Specificity of the genetic elements controlling regulation of early functions in temperate bacterial phages. J. Mol. Biol., 106, 537–566. [DOI] [PubMed] [Google Scholar]
- Horton R.M., Hunt, H.D, Ho, S.N., Pullen, J.K. and Pease, L.R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene, 77, 61–68. [DOI] [PubMed] [Google Scholar]
- Hou Y.M. and Schimmel, P. (1988) A simple structure is a major determinant of the identity of a transfer RNA. Nature, 333, 140–145. [DOI] [PubMed] [Google Scholar]
- Karzai A.W., Susskind, M.M. and Sauer, R.T. (1999) SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J., 18, 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K.C., Waller, P.R.H. and Sauer, R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- Kirby J.E., Trempy, J.E. and Gottesman, S. (1994) Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J. Bacteriol., 176, 2068–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A., Sain, B. and Venetianer, P. (1977) The number of rRNA operons in Escherichia coli. FEBS Lett., 79, 77–79. [DOI] [PubMed] [Google Scholar]
- Komine Y., Kitabatake, M., Yokogawa, T. and Nishikawa, K. (1994) A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA, 91, 9223–9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y., Kitabatake, M., Yokogawa, T. and Inokuchi, H. (1996) 10Sa RNA is associated with 70S ribosome particles in Escherichia coli. J. Bacteriol., 119, 463–467. [DOI] [PubMed] [Google Scholar]
- Koomey M., Gotschlich, E.C., Robbins, K., Bergstrom, S. and Swanson, J. (1987) Effects of recA mutations on pilus antigenic variation and phase transitions in N.gonorrhoeae. Genetics, 117, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFauci G., Widom, R.L., Eisner, R.L., Jarvis, E.D. and Rudner, R. (1986) Mapping of rRNA genes with integrable plasmids in Bacillus subtilis. J. Bacteriol., 165, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W.H. and Foss, K. (1988) Changing the identity of a tRNA by introducing a G–U wobble pair near the 3′ acceptor end. Science, 240, 793–796. [DOI] [PubMed] [Google Scholar]
- Muto A., Ushida, C. and Himeno, H. (1998) A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci., 23, 25–29. [DOI] [PubMed] [Google Scholar]
- Oh B.-K. and Apirion, D. (1991) 10Sa RNA, a small stable RNA of Escherichia coli, is functional. Mol. Gen. Genet., 221, 52–56. [DOI] [PubMed] [Google Scholar]
- Oh B.-K., Chauhan, A.K., Isono, K. and Apirion, D. (1990) Location of a gene (ssrA) for a small, stable RNA (10Sa RNA) in the Escherichia coli chromosome. J. Bacteriol., 172, 4708–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D.A., Silber, K.R. and Sauer, R.T. (1990) Carboxy-terminal determinants of intracellular protein degradation. Genes Dev., 4, 277–286. [DOI] [PubMed] [Google Scholar]
- Ray B.K. and Apirion, D. (1979) Characterization of 10S RNA: a new stable RNA molecule from Escherichia coli. Mol. Gen. Genet., 174, 25–32. [DOI] [PubMed] [Google Scholar]
- Retallack D.M., Johnson, L.L. and Friedman, D.I. (1994) Role for 10Sa RNA in the growth of λ-P22 hybrid phage. J. Bacteriol., 176, 2082–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche E.D. and Sauer, R.T. (1999) SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J., 18, 4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe B.A., Lin,S.P., Song,L., Yuan,X., Clifton,S., Ducey,T., Lews,L. and Dyer,D.W. Neisseria gonorrhoeae genome sequencing–strain FA1090. URL: http://dna1.chem.ou.edu/gono.html
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sanger F., Nicklen, S. and Coulson, A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmientos P., Sylvester, J.E., Contente, S. and Cashel, M. (1983) Differential stringent control of the tandem E.coli ribosomal RNA promoters from the rrna operon expressed in vivo in multicopy plasmids. Cell, 32, 1337–1346. [DOI] [PubMed] [Google Scholar]
- Seifert H.S., Chen, E.Y., So, M. and Heffron, F. (1986) Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 83, 735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch M.A., Baumann, M., Friedman, D.I. and Baron, L.S. (1986) Identification and characterization of mutations in Escherichia coli that selectively influence the growth of hybrid λ bacteriophages carrying the immunity region of bacteriophage P22. J. Bacteriol., 167, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadaki T., Fukushima, M., Ushida, C., Himeno, H. and Muto, A. (1996) Interaction of 10Sa RNA with ribosomes in Escherichia coli. FEBS Lett., 399, 223–226. [DOI] [PubMed] [Google Scholar]
- Tu G.-F., Reid, G.E., Zhang, J.-G., Moritz, R.L. and Simpson, R.J. (1995) C–terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem., 270, 9322–9326. [DOI] [PubMed] [Google Scholar]
- Withey J. and Friedman, D.I. (1999) Analysis of the role of trans-translation in the requirement of tmRNA for λimmP22 growth in Escherichia coli. J. Bacteriol., 181, 2148–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.P. (1999) The tmRNA website. Nucleic Acids Res., 27, 165–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.P. and Bartel, D.P. (1996) Phylogenetic analysis of tmRNA secondary structure. RNA, 2, 1306–1310. [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M., van Putten, J.P.M., Hayes, S.F. and Koomey, M. (1999) The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol., 31, 1345–1357. [DOI] [PubMed] [Google Scholar]
- Zwieb C., Wower, I. and Wower, J. (1999) Comparative sequence analysis of tmRNA. Nucleic Acids Res., 27, 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]