Abstract
Direct interaction between DNA polymerase δ and its processivity factor proliferating cell nuclear antigen (PCNA) is essential for effective replication of the eukaryotic genome, yet the precise manner by which this occurs is unclear. We show that the 54 kDa subunit of DNA polymerase δ from Schizosaccharomyces pombe interacts directly with Pcn1 (PCNA) both in vivo and in vitro. Binding is effected via a short sequence at the C–terminus of Cdc27 with significant similarity to the canonical PCNA binding motif first identified in the mammalian p21Cip1 protein. This motif is both necessary and sufficient for binding of Pcn1 by Cdc27 in vitro and is essential for Cdc27 function in vivo. We also show that the Pcn1 binding motif in Cdc27 is distinct from its binding site for Cdc1, the 55 kDa B-subunit of polymerase δ, and present evidence that Cdc27 can bind to Pcn1 and Cdc1 simultaneously. Finally, we show that Cdc27 performs at least two distinct essential functions, one of which is independent of Pcn1 binding.
Keywords: DNA polymerase/DNA replication/fission yeast/PCNA/protein–protein interactions
Introduction
Central to the chromosomal DNA replication apparatus in eukaryotic cells are three essential DNA polymerases, namely DNA polymerases α, δ and ɛ. Each of these enzymes is a multi-subunit entity, comprising a large catalytic subunit and a number of smaller subunits whose functions are obscure. Studies in yeast have demonstrated that each of the three catalytic subunits is essential for cell viability, as are many, although not all, of the smaller subunits (for a review, see MacNeill and Burgers, 2000). Given their central importance, it is not surprising that there is considerable interest in defining the precise function of each of the three DNA polymerases and in understanding how these enzymes interact with one another and with their various accessory factors.
On the basis of studies of SV40 viral DNA replication in vitro, DNA polymerase α (Pol α) is believed to play a key role in the initiation of leading strand synthesis as well as in the initiation of each Okazaki fragment on the lagging strand (Waga and Stillman, 1998). The four-subunit Pol α–primase complex synthesizes a short RNA–DNA segment that serves as a primer for elongation by either Pol δ or Pol ɛ. The precise roles of the latter two enzyme complexes in chromosomal replication are yet to be elucidated but models based on a variety of genetic and biochemical data suggest that Pol δ and Pol ɛ replicate different DNA strands and that Pol δ may be responsible for leading strand and Pol ɛ for lagging strand (Okazaki fragment) synthesis (reviewed by Burgers, 1996). However, in the budding yeast Saccharomyces cerevisiae it was recently shown that the catalytic activity of Pol ɛ is dispensable for replication (Kesti et al., 1999), indicating that at least under certain circumstances Pol δ is capable of replicating both leading and lagging strands.
To investigate these issues further, we have initiated a study of the role of Pol δ and its auxiliary factors in the genetically tractable fission yeast Schizosaccharomyces pombe. Highly purified Pol δ from S.pombe comprises four distinct subunits of 125, 55, 54 and 22 kDa (see Table I) (Zuo et al., 1997; Reynolds et al., 1998). The catalytic subunit is the Pol3 protein, while the 55, 54 and 22 kDa subunits have been identified as Cdc1, Cdc27 and Cdm1, respectively (Pignede et al., 1991; Park et al., 1993; MacNeill et al., 1996; Iino and Yamamoto, 1997; Reynolds et al., 1998). Interaction studies have indicated that Pol3 interacts directly with Cdc1, which in turn binds to Cdc27. No direct interaction between Pol3 and Cdc27 has been observed. Of the non-catalytic subunits, Cdc1 and Cdc27 are essential proteins while Cdm1 is not (MacNeill et al., 1996; Reynolds et al., 1998). The four-subunit structure of the S.pombe enzyme contrasts with that of Pol δ purified from mammalian cells, which is dimeric in nature, comprising homologues of Pol3 and Cdc1 only (the p125 and p48/p50 proteins), although a putative third subunit of the mammalian enzyme (p66) has recently been identified, a homologue of the Cdc27 protein (Hughes et al., 1999).
Table I. DNA Pol δ subunit structure across evolution.
| S.pombe | Human | S.cerevisiae | |
|---|---|---|---|
| Catalytic subunit | Pol3/Cdc6 | p125 | Pol3 |
| Small subunits | |||
| B-subunit | Cdc1 | p48/p50 | Pol31/Hys2 |
| Cdc27 | p66 | Pol32 | |
| Cdm1 | – | – | |
| PCNA | Pcn1 | PCNA | Pol30 |
Extensive biochemical studies have shown that Pol δ requires two additional factors for full functionality: replication factor C (RF-C) and proliferating cell nuclear antigen (PCNA), clamp loader and sliding clamp, respectively (Waga and Stillman, 1998). RF-C binds to the 3′ end of the short RNA–DNA primers synthesized by the Pol α–primase complex, then recruits PCNA and assembles it onto the DNA in an ATP-dependent manner. PCNA, a ring-shaped homotrimeric protein, encircles the DNA, forming a sliding clamp that tethers the polymerase complex to the DNA duplex to permit highly processive DNA synthesis. In addition to its role as a processivity factor, PCNA also appears to function as a protein targeting factor, localizing proteins such as DNA ligase I and DNA-cytosine-5-methyltransferase (MCMT) to the replication fork (Chuang et al., 1997; Montecucco et al., 1998).
In order to comprehend fully the function of the Pol δ complex, it is clearly necessary to understand the interactions between Pol δ and PCNA at the molecular level. The processivity of the heterodimeric mammalian Pol δ enzyme, comprising only the p125 catalytic subunit and the B-subunit p48/p50 (see Table I), can be stimulated by PCNA in vitro, indicating that no other protein factors are required for this to occur. The same is true of an artificial dimeric form of S.cerevisiae Pol δ, called Pol δ*, which comprises the Pol3 and Cdc1 homologues Pol3 and Pol31 only (Burgers and Gerik, 1998). Exactly how the interaction between PCNA and these dimeric enzymes is mediated remains to be seen; although there have been reports of direct contact between the catalytic subunit and PCNA (Brown and Campbell, 1993; S.J.Zhang et al., 1995; P.Zhang et al., 1999), it is now clear that the B-subunit is required for PCNA to stimulate processivity of the enzyme (Sun et al., 1997; Zhou et al., 1997), consistent with earlier data from a number of laboratories showing that the processivity of the catalytic subunit in isolation is not PCNA sensitive (see Hindges and Hübscher, 1995, and references therein).
Given that PCNA can stimulate the processivity of dimeric forms of Pol δ from organisms as diverse as humans and budding yeast, it is not unreasonable to expect that the processivity of a dimer of the fission yeast Pol3 and Cdc1 proteins will be stimulated in a similar manner, a hypothesis that is currently under investigation in a number of laboratories, including our own. In this paper, however, we describe an alternative route by which fission yeast Pol δ contacts PCNA. We identify the essential 54 kDa subunit of the Pol δ complex, the Cdc27 protein, as a new Pcn1 binding partner, and define a short Pcn1 binding region in Cdc27 that shares significant sequence similarity with the consensus PCNA binding motif previously identified in a variety of PCNA binding proteins such as p21Cip1, the Fen1 and XP-G nucleases, MCMT and DNA ligase I (reviewed by Warbrick, 1998). We show that deleting this motif from the Cdc27 protein abolishes Pcn1 binding in vitro and Cdc27 protein function in vivo, thereby providing the first demonstration that a p21Cip1-like PCNA binding motif can be essential for cell viability. We also show that Cdc27 is able to bind to Cdc1 and Pcn1 simultaneously, via distinct N- and C–terminal domains, and present genetic evidence indicating that Cdc27 has at least two distinct functions in the cell, one of which is independent of the requirement to bind PCNA. The implications of these results are discussed.
Results
Identification of Pcn1 (PCNA) binding proteins using the two-hybrid method
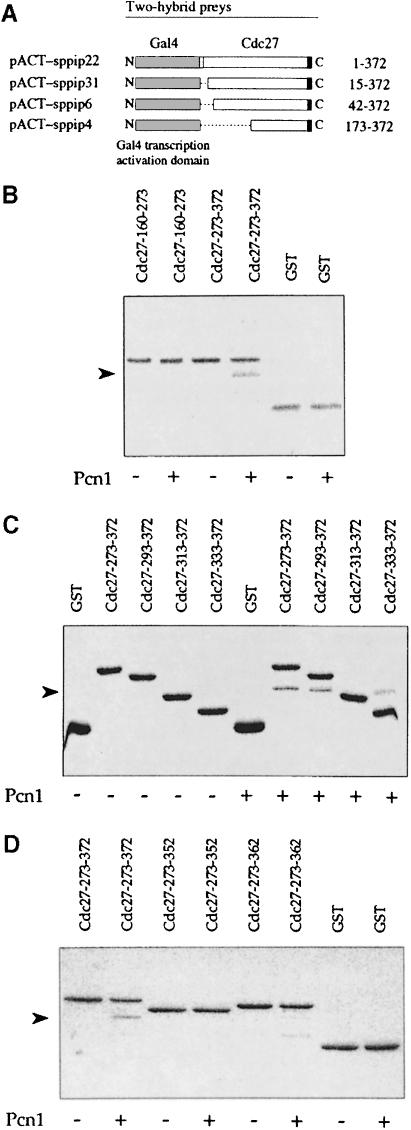
To identify proteins that interact with Pcn1 we performed a two-hybrid screen using the full-length S.pombe Pcn1 protein as bait. Cells containing bait plasmid pAS-PCNA-Sp were transformed with an S.pombe cDNA library in the prey vector pACTII (Warbrick et al., 1995). A total of ∼2 × 106 individual transformants were screened and eight positive clones identified. Two of these were derived from the pcn1+ gene itself, consistent with the fact that PCNA forms a homotrimeric complex in vivo. Two clones encoded a protein that has not previously been identified in fission yeast and that will be the subject of a future report (our unpublished results). The remaining four clones were derived from the cdc27+ gene (Hughes et al., 1992; MacNeill et al., 1996). Figure 1A shows that these clones encode a full-length Cdc27 protein and three proteins in which varying regions of the N–terminus are absent. The smallest clone has only residues 173–372 of Cdc27 fused to the Gal4 activation domain.
Fig. 1. Cdc27 binds Pcn1 in vivo and in vitro. (A) Gal4 AD–Cdc27 prey plasmids recovered from a two-hybrid screen using pAS-PCNA-Sp as bait: pACT-sppip22 (expresses full-length Cdc27 protein with an additional 13 N–terminal amino acids fused to the Gal4 AD), pACT-sppip31 (residues 15–372), pACT-sppip6 (residues 42–372) and pACT-sppip4 (residues 173–372). (B–D) In vitro binding of recombinant Pcn1 to GST–Cdc27 proteins. The GST fusion proteins shown were incubated with or without Pcn1 as indicated, before being pulled down using glutathione–Sepharose beads. The beads were washed extensively and bound Pcn1 was visualized by PAGE Blue 90 staining following SDS–PAGE. Specific binding of GST–Cdc27 to Pcn1 was observed at NaCl concentrations of 0–100 mM (results not shown). The arrowheads indicate the position of Pcn1. In (D), the GST–Cdc27-313–372 protein failed to bind Pcn1 despite the presence of an intact binding domain; the reasons for this are unclear.
In vitro binding of Cdc27 to Pcn1
To confirm the interaction between the Pcn1 and Cdc27 proteins we investigated whether the two proteins interacted in vitro. To this end we expressed recombinant N–terminally histidine-tagged Pcn1 in Escherichia coli, purified the protein to apparent homogeneity by affinity chromatography using Ni-NTA–agarose and tested for its ability to bind to a range of glutathione S-transferase (GST)–Cdc27 fusion proteins in vitro (see Table II and Materials and methods). These included GST–Cdc27-160–372, which comprises the C–terminal 213 amino acids of Cdc27 fused to GST and corresponds closely to the smallest fragment of Cdc27 identified in the two-hybrid screen. Pcn1 bound to GST–Cdc27-160–372 in this assay but not to GST alone (Table II). We also tested whether Pcn1 would bind to GST–Cdc27-1–169 but were unable to detect any binding (Table II), suggesting that stable binding to Pcn1 is effected solely via the ∼200 amino acid C–terminal region of Cdc27.
Table II. In vitro binding of Cdc27 fusion proteins to Pcn1.
| Fusion protein | Ability to bind Pcn1 |
|---|---|
| GST | − |
| GST–Cdc27 | + |
| GST–Cdc27-1–169 | − |
| GST–Cdc27-160–372 | + |
| GST–Cdc27-160–273 | − |
| GST–Cdc27-273–372 | + |
| GST–Cdc27-293–372 | + |
| GST–Cdc27-313–372 | − |
| GST–Cdc27-333–372 | + |
| GST–Cdc27-273–352 | − |
| GST–Cdc27-273–362 | − |
| GST–Cdc27-273–372-Q362A | + |
| GST–Cdc27-273–372-Q362V | + |
| GST–Cdc27-273–372-Q362D | + |
| GST–Cdc27-273–372-Q362E | + |
| GST–Cdc27-273–372-Q362N | + |
| GST–Cdc27-273–372-Q362K | + |
| PP | − |
| PP–Cdc27-353–372 | + |
GST, glutathione S-transferase; PP, PinPoint. See the legend to Figure 1 for additional details.
In order to map further the Pcn1 binding region in Cdc27 we constructed and tested a series of truncated Cdc27 proteins as GST fusions. Initial experiments focused on two proteins: GST–Cdc27-160–273 and GST–Cdc27-273–372. We found that GST–Cdc27-273–372 bound Pcn1 with similar affinity to GST–Cdc27-160–372, but that GST–Cdc27-160–273 did not bind Pcn1 at all in this assay (Figure 1B; Table II). We conclude from this that the Pcn1 binding site is located within the last 100 amino acids of the Cdc27 protein. Further mapping using GST–Cdc27 fusions localized the Pcn1 binding site to within the last 40 amino acids of Cdc27; we found that GST–Cdc27-333–372 bound Pcn1 as well as GST–Cdc27-273–372 (Figure 1C; Table II).
In vivo Cdc27–Pcn1 interaction
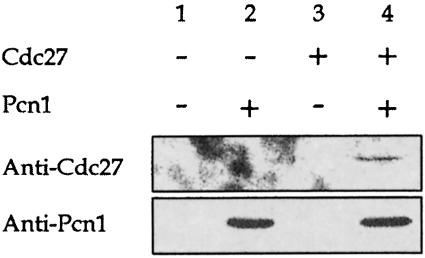
We also confirmed that the Cdc27 and Pcn1 proteins interacted in S.pombe. To do this we expressed tagged versions of the two proteins in the yeast: Cdc27 was C–terminally tagged with the 12CA5 haemagglutinin (HA) epitope sequence (MacNeill et al., 1996) and Pcn1 was N–terminally tagged with the sequence MRGS(His6), as described above. Both proteins are fully functional in S.pombe (MacNeill et al., 1996; our unpublished results). Protein extracts prepared from transformed cells under non-denaturing conditions were incubated with Ni–agarose resin, to pull down Pcn1, and the bound fractions subjected to SDS–PAGE and blotted with anti-HA antibodies to detect the presence of Cdc27. The results are shown in Figure 2. We found that Cdc27 was retained on the resin only in the presence of Pcn1 (Figure 2, compare lanes 3 and 4), indicating that the two proteins bind to one another in vivo in S.pombe as well as in vitro.
Fig. 2. Co-precipitation of Cdc27 and Pcn1 from S.pombe. Protein extracts prepared under non-denaturing conditions from cells transformed with the following expression vectors were incubated with Ni–agarose resin, to pull down Pcn1, and the bound fractions subjected to SDS–PAGE and blotted with anti-HA antibodies to detect the presence of Cdc27tag. Lane 1, vectors pREP3X, pREP4X; lane 2, pREP3X, pREP4XH6-Pcn1; lane 3, pREP3X-Cdc27tag, pREP4X; lane 4, pREP3X-Cdc27tag, pREP4XH6-Pcn1. Note that the cells were grown in minimal medium without thiamine, to derepress the nmt1 promoter in the pREP vectors.
Identification of a putative PCNA binding motif at the C–terminus of Cdc27
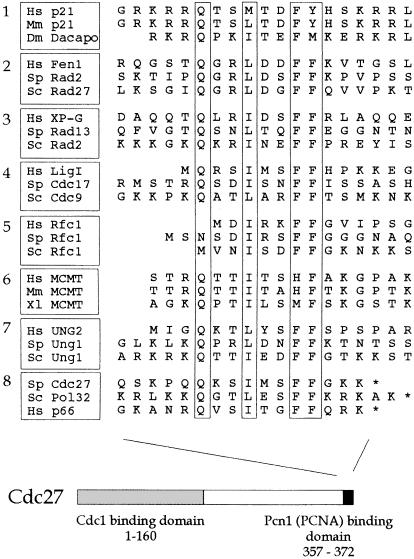
Previously, studies have identified a consensus PCNA binding motif in a variety of higher eukaryotic PCNA binding proteins, including p21Cip1 and Fen1 (reviewed by Warbrick, 1998). Inspection of the Cdc27 sequence revealed the presence of a related motif at the extreme C–terminus of the protein (Figure 3), in the sequence Q–I–FF (residues 362–369). To test the importance of these residues for Pcn1 binding we tested the effect of deleting these C–terminal residues on Pcn1 binding in vitro (Figure 1D). Two C–terminally truncated proteins were tested: GST–Cdc27-273–352 and GST–Cdc27-273–362. Neither was able to bind to Pcn1 (Figure 1D; Table II), indicating that an intact C–terminus, including the consensus PCNA binding motif, is essential for Pcn1 binding in vitro.
Fig. 3. PCNA binding motif at the C–terminus of Cdc27. Aligned sequences of the PCNA binding motifs identified in eight classes of PCNA binding protein as follows: 1, p21 and p21-like proteins; 2, Fen1 nucleases; 3, XP-G nucleases; 4, DNA ligase I; 5, large subunit of RF-C; 6, MCMT methyltransferases; 7, uracil-N-glycosylase; 8, Cdc27 plus its S.cerevisiae homologue Pol32 and the recently identified human homologue p66. Hs, human; Sp, S.pombe; Sc, S.cerevisiae; Mm, Mus musculus; Dm, Drosophila melanogaster; Xl, Xenopus laevis. See Warbrick et al. (1998) for further details.
In the case of p21Cip1, it has been shown that the conserved glutamine within the canonical PCNA binding motif plays an important, although not indispensable role in PCNA binding (Warbrick et al., 1995). In order to test whether this was also true of Glu362 in Cdc27 we tested a number of mutant Cdc27 proteins (expressed as GST–Cdc27-273–372 fusions) in which the glutamine was substituted with alanine, valine, aspartate, glutamate, asparagine or lysine. In each case, binding of the mutant protein to recombinant Pcn1 in vitro was indistinguishable from binding of the non-mutant protein (Table II). We conclude from this that Glu362 is not absolutely required for PCNA binding by Cdc27. Consistent with this, it has recently become clear that the glutamine is not found in every protein that binds PCNA via a p21Cip1-like motif. In the case of the large subunit of RF-C for example, where the PCNA binding motif is located at the extreme N–terminus of the protein (Figure 3), the glutamine is entirely absent (Montecucco et al., 1998).
The final 20 amino acids of Cdc27 are sufficient for Pcn1 binding in vitro
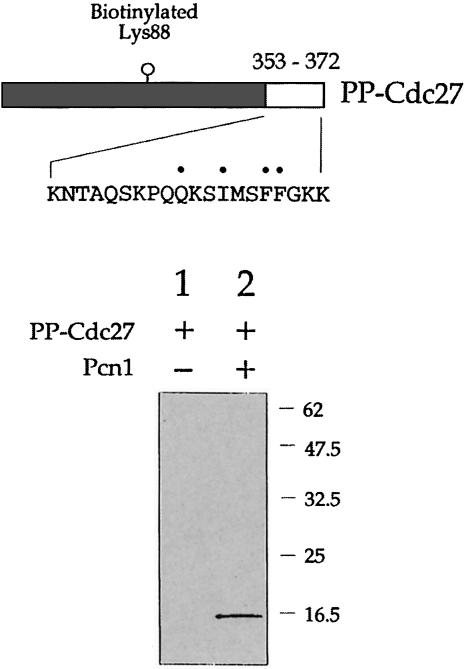
In order to test whether the last 20 amino acids of Cdc27 were sufficient as well as necessary for Pcn1 binding we expressed these residues in E.coli as a biotinylated fusion protein using the PinPoint™ system. The fusion protein produced in this way, designated PP–Cdc27, comprised 136 amino acids of the biotinylated protein (with biotinylation occurring in vivo at Lys88) fused to 20 amino acids from the C–terminus of Cdc27. PP–Cdc27 was expressed in a soluble form and purified using streptavidin–agarose. To test binding of PP–Cdc27 to Pcn1, the PP–Cdc27 protein was mixed with recombinant His-tagged Pcn1 and the Pcn1 pulled down using Ni–agarose. The bound proteins were then electrophoresed and the SDS–PAGE gel probed using streptavidin-coupled alkaline phosphatase to detect the presence of the (biotinylated) PP–Cdc27 protein (Figure 4). Using this method we found that the PP–Cdc27 protein was precipitated only in the presence of Pcn1; in the absence of Pcn1, PP–Cdc27 was not detectable, indicating that retention of PP–Cdc27 is Pcn1 dependent. Although specific, the strength of this interaction was much reduced compared with that seen with the various GST–Cdc27 fusion proteins described above (there was insufficient PP–Cdc27 bound to Pcn1 for this to be visualized by direct staining of the SDS–PAGE gel, thus the need to visualize the co-precipitated PP–Cdc27 using streptavidin-coupled alkaline phosphatase), suggesting that the final 20 amino acid residues of Cdc27 may not comprise the entire Pcn1 binding site.
Fig. 4. In vitro Pcn1 binding assay using biotinylated PP–Cdc27 fusion protein (PP–Cdc27). Upper part: schematic representation of PP–Cdc27 showing the location of the biotinylated lysine residue and, at the C–terminus, residues corresponding to 353–372 in Cdc27. Lower part: following mixing of the PP–Cdc27 protein with recombinant Pcn1, proteins binding to Pcn1 were isolated by Ni–NTA affinity chromatography and subjected to SDS–PAGE. The bound proteins were then transferred to a PVDF membrane and probed using streptavidin-labelled alkaline phosphatase to detect the presence of the biotinylated PP–Cdc27 protein. Retention of the PP–Cdc27 protein was dependent upon the presence of Pcn1 (lane 2). See the text for details.
To analyse further the binding of the C–terminal 20 amino acids of Cdc27 to Pcn1, peptides were synthesized corresponding to this region from both Cdc27 and its S.cerevisiae homologue Pol32. A p21Cip1-derived peptide whose interaction with PCNA has been characterized previously was analysed in parallel (Warbrick et al., 1995; Gulbis et al., 1996). The peptides were linked to biotin through an SGSG linker at their N–terminus, and in each case a peptide with the conserved glutamine residue substituted with alanine (p21-A, Cdc27-A and Pol32-A) was also tested.
In order to determine whether the peptides were capable of binding to Pcn1, each was conjugated to streptavidin–agarose beads and incubated with S.pombe cell extracts. After extensive washing, the amount of peptide-bound Pcn1 was tested using SDS–PAGE followed by Western blotting with the anti-PCNA monoclonal antibody PC10 (Figure 5A) (Waseem et al., 1992). All three non-mutant peptides bound S.pombe Pcn1 efficiently in this assay (Figure 5A). Binding was also seen with the alanine substitution peptides, although in the case of p21-A and Cdc27-A the extent of binding was somewhat reduced compared with the non-mutant peptide. The observation that the Cdc27-A peptide is still able to bind to Pcn1 is consistent with results obtained with the GST–Cdc27-Q362A fusion protein described above (Table II) and with the Cdc27-Q362A protein being able to function in S.pombe (see below).
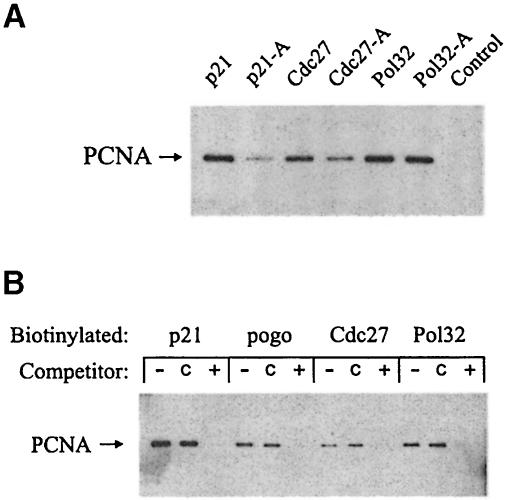
Fig. 5. Peptide–Pcn1 interactions. (A) Peptides were conjugated to streptavidin–agarose beads and incubated with S.pombe cell extracts. Following recovery and extensive washing of the beads, the bound PCNA was analysed by SDS–PAGE followed by Western blot analysis with the monoclonal anti-PCNA antibody PC10. The peptides used are described in Materials and methods, and represent previously described PCNA binding peptides from human p21Cip1 (p21), and C–terminal 20 amino acid sequences derived from Cdc27 (Cdc27) and Pol32 (Pol32). In each case, peptides were also tested in which the conserved glutamine was substituted with alanine (peptide-A). A peptide of unrelated sequence was used as a control for non-specific binding. (B) The ability of immobilized peptides to bind to PCNA was tested in the presence of either the p21Cip1-derived peptide (+), an unrelated control peptide (c) or the solvent DMSO (–). These were added to diluted S.pombe cell extracts before incubation with the immobilized, biotinylated peptides. The competitor p21Cip1 peptide is known to compete with the peptide pogo for binding to PCNA (Warbrick et al., 1998).
As noted above, the conservation of the proposed PCNA binding motifs in Cdc27 and Pol32 suggests that they may share a common binding site on PCNA with p21Cip1. Peptide competition experiments using a p21Cip1 peptide were carried out to investigate this (Figure 5B). The results show that the non-biotinylated p21Cip1 peptide was able to compete effectively for the binding of Pcn1 to the immobilized biotinylated Cdc27- and Pol32-derived peptides. These results suggest that these peptides share a common binding site on PCNA.
The Pcn1 binding motif is required for Cdc27 function in vivo
To test whether the Pcn1 binding motif was required for the in vivo function of Cdc27 we constructed several mutant alleles of cdc27+ with mutations in or around the Pcn1 binding motif. Each mutant allele was cloned into plasmid pREP3X, 3′ to the regulatable nmt1 promoter, and the resulting plasmids transformed into a cdc27+/cdc27::his7+ diploid strain (see Materials and methods). Transformant colonies were then sporulated and the meiotic products examined following growth on minimal medium in the presence or absence of thiamine, i.e. with the nmt1 promoter either repressed or derepressed. The results of the experiments are summarized in Figure 6A and below. Note that to ensure that any effects seen were not the result of the mutant proteins acting as dominant negatives, equal numbers of spores were plated onto plates with and without histidine, to allow comparison of plasmid-carrying cdc27+ and cdc27Δ isolates. Although overproduction of wild-type Cdc27 leads to some cell elongation (MacNeill et al., 1996), and this is exacerbated when C–terminal sequences are deleted (see below), in no case did overproduction of the mutant proteins prevent colony formation in the cdc27+ background; under the conditions used >103 haploid colonies per plate were typically obtained.
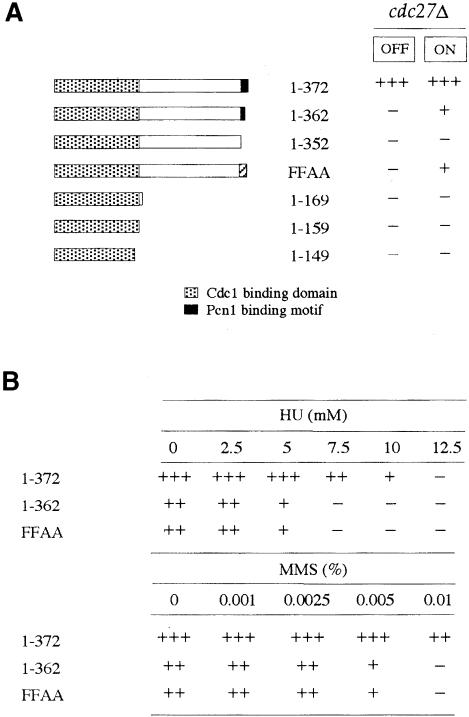
Fig. 6. The C–terminal 20 amino acid residues of Cdc27 are required for in vivo function. (A) pREP3X plasmids expressing the indicated mutant proteins (note that FFAA = F368AF369A) were transformed into cdc27+/cdc27Δ diploid cells. Transformant colonies were then sporulated and spores prepared by helicase treatment plated onto EMM medium as described in the text. After 3–4 days incubation at 30°C, the degree of rescue of the cdc27Δ phenotype was scored subjectively as follows: +++, excellent rescue, cell size at cell division equivalent to wild type; ++, good rescue, cell size at cell division typically in the range 16–20 μm compared with 14 μm for cells expressing the wild-type Cdc27; +, weak rescue, cell size at division typically >20 μm; –, no rescue, indistinguishable from cells transformed with pREP3X vector. (B) cdc27Δ cells expressing Cdc27-1–362 or Cdc27-F368A/F368A (FFAA) grown by necessity in EMM medium lacking thiamine were plated onto EMM medium containing either HU or MMS at the concentrations indicated and scored for growth after 4–6 days.
The mutant proteins could be divided into three distinct classes on the basis of their ability to rescue the cdc27Δ phenotype. The Cdc27-1–352 protein alone fell into the first class, as no cdc27Δ [pREP3X–Cdc27-1–352] haploids could be recovered following spore germination either in the presence or absence of thiamine (i.e. with nmt1 promoter in the plasmid repressed or derepressed). This indicates that the C–terminal 20 amino acids of Cdc27 comprising the Pcn1 binding site are essential for the in vivo function of the protein.
The second class comprised the Cdc27-1–363 and Cdc27-F368AF369A proteins, which could only rescue the cdc27Δ phenotype when expressed to a high level (nmt1 promoter fully derepressed) and even then the degree of rescue was much reduced compared with that conferred by the wild-type Cdc27 protein. In both cases the rescued cdc27Δ cells were highly elongated compared with cells expressing the wild-type protein (cell length at cell division ∼25 μm compared with ∼14 μm for cdc27Δ cells expressing wild-type Cdc27 from pREP3X–Cdc27). In addition, no rescue was observed in medium containing thiamine, when expression from the nmt1 promoter is repressed, indicating that high levels of expression are crucial for the observed phenotype.
To investigate the nature of the functional defect in the Cdc27-1–352 and Cdc27-F368AF369A proteins, we analysed the properties of haploid cdc27Δ cells expressing Cdc27-1–362 and Cdc27-F368AF369A (necessarily grown in the absence of thiamine). In parallel, we also analysed cdc27Δ cells expressing Cdc27-D362, Cdc27-E362, Cdc27-A362 and Cdc27-V362 (grown in the presence of thiamine, see below). Cells were tested for sensitivity to the DNA synthesis inhibitor hydroxyurea (HU), and to the DNA damaging agents methylmethanesulfonate (MMS) and UV. The results are summarized in Figure 6B. We found that cells expressing Cdc27-1–362 and Cdc27-F368AF369A were significantly more sensitive to low levels of HU and MMS than cells expressing wild-type Cdc27 (Figure 6B), but there was no difference in their sensitivity to UV (results not shown). These results suggest that the Cdc27–Pcn1 interaction does not have a significant part to play in the repair of UV damage in S.pombe, a process in which Pol δ is believed to play an important role (Giot et al., 1997). Conclusions regarding the role of Pcn1 binding in resistance to HU and MMS must be tempered by the fact that growth of Cdc27-1–352- and Cdc27-F368AF369A-expressing cells is poorer than that of wild type even in the absence of drug treatment.
In the third class were the proteins Cdc27-D362, Cdc27-E362, Cdc27-A362 and Cdc27-V362, all of which had properties that were indistinguishable from the wild-type Cdc27 protein under all conditions tested. We conclude therefore that Glu362 is not absolutely required for Cdc27 function in vivo, just as it is not required for Pcn1 binding in vitro (Table II). However, it should be noted that the level of these mutant Cdc27 proteins present in cells under conditions when the nmt promoter is turned off (i.e. during growth in thiamine) may still exceed the normal level of endogenous Cdc27 protein in wild-type cells, so that it is not possible to conclude that Cdc27-D362, Cdc27-E362, Cdc27-A362 and Cdc27-V362 have truly wild-type Cdc27 activity.
Overproduction of the C–terminus of Cdc27 causes cell cycle delay
Previously, it was shown that overproduction of the human p21Cip1 protein in S.pombe resulted in cell elongation brought about by delayed cell cycle progression (Tournier et al., 1996). This phenotype was ascribed to the PCNA binding function of p21Cip1, as cell elongation was not seen when a mutant p21Cip1 protein defective in PCNA binding was similarly expressed. To ask whether a similar phenotype would be seen when the C–terminus of Cdc27 was overproduced, a plasmid expressing amino acids 273–372 of Cdc27 as a histidine-tagged fusion protein was transformed into S.pombe and the properties of the transformant cells analysed microscopically (see Materials and methods for details of expression constructs). We found that overproduction of the last 100 amino acids of Cdc27 from the nmt1 promoter led to a marked increase in cell length at cell division compared with wild-type cells (22.4 versus 14.0 μm). This effect was dependent upon the presence of the Pcn1 binding motif, however, as cells expressing amino acids 273–352 were not significantly elongated (15.0 μm). Interestingly, we also found that while overproduction of this protein (Cdc27-273–352) did not bring about cell elongation, overproduction of Cdc27-1–352 did, presumably due to the amino acids in region 1–273 acting as a dominant negative (data not shown).
We also tested cells expressing point mutated variants of the last 100 amino acids corresponding to those tested for in vitro Pcn1 binding and found that these also failed to inhibit cell cycle progress (e.g. cell size at division for cells expressing Q362A was 14.2 μm). This indicates that Glu362 does contribute to Pcn1 binding in vivo, even though the Cdc27-Q362A protein still binds Pcn1 in vitro and is still functional in vivo.
Cdc27 can bind to Cdc1 and Pcn1 simultaneously
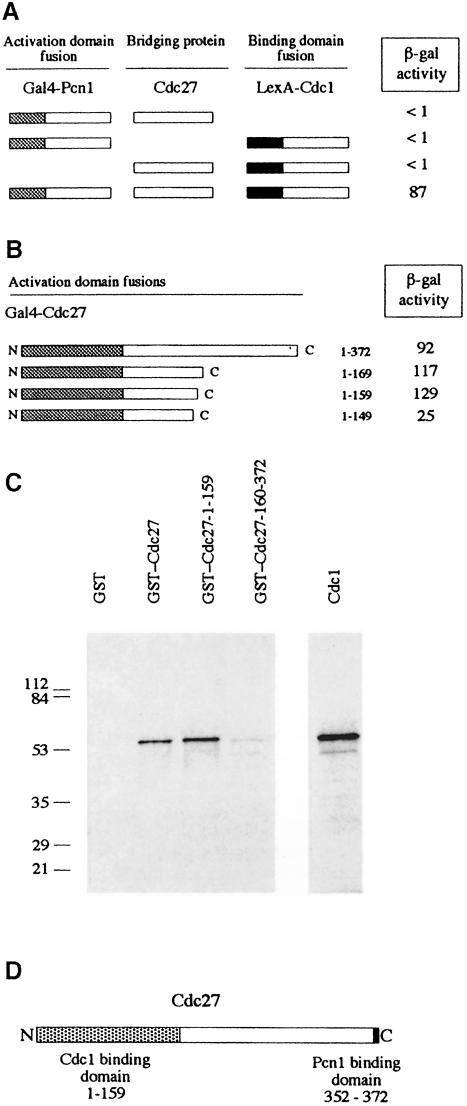
We have previously shown that Cdc27 binds to Cdc1, the 55 kDa B-subunit of Pol δ in fission yeast (MacNeill and Fantes, 1997; Zuo et al., 1997). To test whether Cdc27 could bind to Cdc1 and Pcn1 simultaneously, we asked whether Cdc1 and Pcn1 fusion proteins could be brought together in the two-hybrid system by independent high-level expression of Cdc27. We found that this was indeed the case, that Cdc27 was able to bridge Gal4–Pcn1 and LexA–Cdc1 fusion proteins and activate LacZ expression (Figure 7A). This indicates that the Cdc1 and Pcn1 binding domains in Cdc27 are non-overlapping.
Fig. 7. Distinct N- and C–terminal binding regions for Cdc1 and Pcn1 on Cdc27. (A) Three-component two-hybrid assay. Gal4 AD–Pcn1 and LexA BD–Cdc1 fusions are brought together by simultaneous co-expression of Cdc27. Schematic representation of Gal4 AD–Pcn1 fusions and LexA BD–Cdc1 fusions, with the results of the two-hybrid analysis shown to the right (β-galactosidase activity in liquid cultures expressed in Miller units). (B) Schematic representation of Gal4 AD–Cdc27 fusions used to map the Cdc1 binding domain, with the results of the two-hybrid analysis shown to the right (β-galactosidase activity in liquid cultures expressed in Miller units). (C) In vitro Cdc1–Cdc27 interactions. [35S]methionine-labelled in vitro synthesized Cdc1 was mixed with 5 μg of various GST fusion proteins or with GST alone. Following incubation at room temperature for 3 h, the GST and GST–Cdc27 proteins were re-isolated using glutathione–Sepharose beads. These were washed extensively before being boiled in sample buffer and the sample run on a 12.5% SDS–PAGE gel. Retention of Cdc1 was determined by fluorography. Molecular weight standards are shown to the left of the gel with molecular weights given in kilodaltons. (D) Schematic representation of the Cdc27 protein showing the Cdc1 and Pcn1 binding regions defined in this study.
To confirm this we mapped the Cdc1 binding domain using both the two-hybrid system and an in vitro Cdc1 binding assay. Results from the two-hybrid system are shown in Figure 7B; a series of truncated Cdc27 proteins were constructed and expressed in yeast as Gal4 activation domain fusions from plasmid pGAD2F (MacNeill et al., 1996). Cdc1 binding activity was measured using standard methods and the results expressed in Miller units (Miller, 1972). We found that removal of >200 amino acids from the C–terminus of Cdc27 did not reduce binding to Cdc1. Indeed, binding of the Cdc27-1–159 and Cdc27-1–169 proteins was consistently stronger than binding of the full-length Cdc27 protein, whereas removal of a further 10 residues from the C–terminus (Cdc27-1–149) reduced Cdc1 binding 5-fold.
Confirmation of these results was provided by testing the ability of various GST–Cdc27 fusion proteins to bind Cdc1 in vitro (Figure 7C). For this purpose Cdc1 was expressed in rabbit reticulocyte lysates in the presence of [35S]methionine. The lysate was then mixed with various GST–Cdc27 fusion proteins, or with GST alone, before the GST proteins were pulled down on glutathione beads and binding of Cdc1 assayed by SDS–PAGE and autoradiography (see Materials and methods). We found that GST–Cdc27-1–159 bound as well as, if not better than, full-length Cdc27 in this assay, in agreement with the results obtained using the two-hybrid system (Figure 7C). For Cdc27-1–149 the results of the two-hybrid and in vitro binding assays were also in agreement: in both cases removal of amino acids 150–159 led to a reduction in Cdc1 to a level below that seen with the full-length protein (data not shown). Some binding of Cdc1 to a GST–Cdc27-170–372 fusion was detected, but at low levels only (<10% of wild type). We conclude from these studies that Cdc27 binds to Cdc1 and Pcn1 via distinct domains at the N- and C–termini of the protein, respectively (Figure 7D).
Rescue of temperature-sensitive Cdc27 proteins by overproduction of the N–terminal domain of Cdc27
Three temperature-sensitive mutant alleles of cdc27 were identified in mutant screens (Nasmyth and Nurse, 1981), two of which (cdc27-P11 and cdc27-K3) have been characterized at the molecular level (Hughes et al., 1992). Both of these alleles can be rescued by Cdc1 overproduction (MacNeill et al., 1996) and both contain mutations at the same amino acid, Gly57, within the Cdc1 binding region described above. These observations suggest that the defect in these proteins may be a reduced ability to bind Cdc1. In support of this, when we tested the Cdc27-P11 protein for Cdc1 binding using the two-hybrid assay we were unable to detect any interaction (<1 Miller unit of β-galactosidase activity).
Prior to determining that the C–terminal Pcn1 binding region of Cdc27 was essential for rescue of cdc27Δ cells, we tested the ability of a series of truncated Cdc27 proteins to rescue cdc27-P11 and found that the N–terminal 159 amino acids of Cdc27 were sufficient for rescue (data not shown). This implies that Pcn1 binding via the C–terminal motif is not required for rescue of this allele. Expression of the first 149 amino acids was insufficient to rescue cdc27-P11, however, showing that residues between 150 and 160 are essential for the activity of the truncated protein. (Recall that deletion of these amino acids led to a 5-fold reduction in binding to Cdc1 in the two-hybrid system; Figure 7B.)
The ability of Cdc27-1–159 to rescue cdc27-P11 but not a cdc27Δ strain indicates (i) that the Cdc27-P11 protein retains some activity at the restrictive temperature, i.e. that it is not a complete null, and (ii) that the function that is impaired in cdc27-P11 cells is independent of the need for Cdc27 to bind via its p21Cip1-like motif to Pcn1. This in turn suggests that Cdc27 may have two distinct functions, only one of which is defective in cdc27-P11 cells, and that the Cdc27-P11 protein is still able to interact productively with Pcn1 at the restrictive temperature. It is unclear from this analysis, however, whether it is necessary for Cdc27-P11 protein to bind to Cdc1 to function at the restrictive temperature. If Cdc1 binding were not required for Cdc27-P11 function then it might be possible to express the two halves of the wild-type Cdc27 protein separately in vivo and observe rescue of a cdc27Δ mutant. To examine this possibility we expressed Cdc27-1–159 together with Cdc27-170–372 in a cdc27+/cdc27::his7+ (cdc27Δ) diploid (see Materials and methods for details). The diploid was then sporulated and the properties of the viable spore products analysed. No viable leu+ ura+ his+ haploids were obtained, indicating that the Cdc27 protein must be intact to allow rescue of cdc27::his7+. Logically, therefore, the N–terminal Cdc1 binding region of Cdc27-P11 must be required for the residual function of the mutant protein at high temperature.
Discussion
DNA Pol δ in the fission yeast S.pombe is a multi-subunit complex comprising a large catalytic subunit and three smaller subunits, two of which are essential for cell viability (MacNeill et al., 1996; Zuo et al., 1997; Reynolds et al., 1998). We are interested in understanding how the four subunits of the enzyme complex contact one another and other components of the replication machinery, and in understanding the effects on Pol δ function of disrupting these protein–protein contacts. Previously, we have shown that Pol3 can bind directly to Cdc1 and that Cdc1 binds to Cdc27 (MacNeill et al., 1996). In this paper we show that Cdc27 interacts directly with the essential processivity factor Pcn1 (PCNA) through a binding domain located at the C–terminus of the Cdc27 protein, and that the Cdc1 and Pcn1 binding regions in Cdc27 are distinct and separable. The Pcn1 binding region comprises a short motif similar to the PCNA binding motifs found in a variety of eukaryotic proteins including p21Cip1, Fen1, XP-G, MCMT and DNA ligase I (Warbrick, 1998). We have shown that this motif is both necessary and sufficient for Pcn1 binding in vitro (Figures 1, 4 and 5) and is essential for Cdc27 function in vivo (Figure 6). Intriguingly, the location of the PCNA binding motif at the extreme C–terminus of Cdc27 parallels that of the sliding clamp binding peptide in the DNA polymerase of the T4-related bacteriophage RB69 (Shamoo and Steitz, 1999), suggesting the possibility of further structural and functional similarities between the single-subunit polymerase of RB69 and the multi-subunit eukaryotic DNA Pol δ. X–ray crystallographic studies have shown that this peptide binds the sliding clamp at a position identical to that of the p21Cip1 peptide binding to PCNA (Shamoo and Steitz, 1999). In addition to identifying the Pcn1 binding motif, the Cdc1 binding region on Cdc27 has also been mapped to an N–terminal region comprising amino acids 1–160 (Figure 7). Attempts to define the N–terminal Cdc1 binding site further by testing fusion proteins comprising 50 or 100 amino acid portions of the region 1–160 were unsuccessful (data not shown).
Evidence from mammalian and budding yeast systems has shown that dimeric forms of Pol δ comprising the catalytic and B-subunit only (homologues of S.pombe Pol3 and Cdc1, respectively) can have their processivity stimulated by PCNA in vitro (Sun et al., 1997; Zhou et al., 1997; Burgers and Gerik, 1998; Gerik et al., 1998). The processivity of the catalytic subunit is not stimulated in this way, however, indicating that the B-subunit is necessary for the observed effect. Whether the catalytic subunit can interact directly with PCNA in the absence of the B-subunit has been the subject of some debate. Recent studies in both S.pombe and S.cerevisiae failed to detect direct interactions between the catalytic subunit and PCNA in a variety of assays (Eissenberg et al., 1997; Tratner et al., 1997), yet in mammalian systems there is increasing evidence that PCNA is able to interact with a short region (the N2 region) close to the N–terminus of the catalytic subunit (S.J.Zhang et al., 1995; P.Zhang et al., 1999). We have tested whether the N2 region of S.pombe Pol3 can interact with recombinant Pcn1 in vitro, but could not detect any interaction (our unpublished results). While this may suggest that the interaction described above is confined to mammalian proteins, we cannot rule out the possibility that the interaction between the S.pombe proteins is weaker than that between the mammalian proteins, precluding its detection in these relatively insensitive assays. Further work will be required to distinguish between these possibilities.
Based on work in S.cerevisiae, where Pol δ purifies as a dimer of heterotrimers (Pol3⋅Pol31⋅Pol32), we might expect that the S.pombe enzyme will exist as a dimer of heterotetramers (Pol3⋅Cdc1⋅Cdc27⋅Cdm1). In the budding yeast system, the Cdc27 homologue Pol32 is required for dimerization of the heterotrimer, as in the absence of Pol32 the enzyme purifies as a simple Pol3⋅Pol31 heterodimer (Burgers and Gerik, 1998; Gerik et al., 1998). As noted above, the processivity of this form of the enzyme, measured on synthetic poly(dA)⋅oligo(dT) templates, can be stimulated by addition of PCNA. However, much more PCNA is required to make processive complexes than with Pol δ itself, suggesting that Pol32 contributes the ability of the enzyme to be stimulated by PCNA. Also, on singly primed single-stranded DNA templates, the Pol3⋅Pol31 dimer is more prone to pausing than is Pol δ, possibly due to the enzyme complex being destabilized by secondary structure elements. As noted by Burgers and co-workers, there are striking parallels between the behaviour of the Pol3⋅Pol31 dimer (termed Pol δ*) and the dimeric form of the mammalian enzyme, which is also prone to replication pausing and displays similar rates of synthesis to Pol δ*. This prompted speculation that the mammalian enzyme might have additional subunits yet to be identified; recent work suggests that this is indeed the case, as a putative third subunit of mammalian Pol δ (a homologue of Cdc27) has now been identified (Hughes et al., 1999).
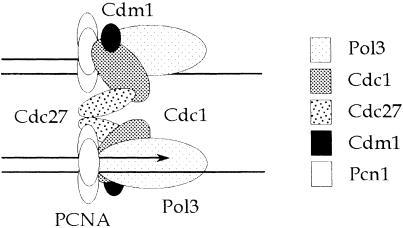
One possible interpretation of the genetic data presented here, indicating that Cdc27 has both Pcn1-dependent and Pcn1-independent functions, is that only one of the two Cdc27 molecules in a dimeric complex is required to interact with Pcn1 for the complex to be functional in vivo. If we assume that the dimeric Pol δ is able to function at the eukaryotic replication fork in a manner akin to that of Pol III in E.coli (reviewed by Stillman, 1996), we might predict that Cdc27–Pcn1 interaction is required for either leading- or lagging-strand replication, but not both. This is not inconsistent with the earlier observation that PCNA is required for both leading- and lagging-strand synthesis by Pol δ (Prelich and Stillman, 1988) as this may simply reflect the necessity of having PCNA interact with the catalytic and/or B-subunit of the complex for synthesis of both strands. Our speculative model for S.pombe Pol δ structure is shown in Figure 8. In this model, Pol δ is shown as a dimer of heterotetramers (Pol3⋅Cdc1⋅Cdc27⋅Cdm1) coordinately replicating the leading and lagging strands. Each tetramer is shown binding to Pcn1 via Pol3 and Cdc1, but only one of the two tetramers is shown binding to Pcn1 via Cdc27. Future work will focus on testing the validity of this model.
Fig. 8. A speculative model for the structure of fission yeast Pol δ at the replication fork, and its interactions with PCNA, based on the genetic results in this paper and the enzymology of others (Burgers and Gerik, 1998; Gerik et al., 1998). See the text for discussion.
Materials and methods
Yeast and bacterial strains
All the S.pombe strains used have been described previously (Nasmyth and Nurse, 1981; Apolinario et al., 1993; MacNeill et al., 1996). To construct the cdc27::his7+ deletion diploid (see below), haploid leu1-32 ura4-D18 his7-366 ade6-M210 h+ and leu1-32 ura4-D18 his7-366 ade6-M216 h– strains (Apolinario et al., 1993) were crossed and ade+ diploids selected by plating the mating mix (24 h after mating on malt extract medium at 25°C) on EMM supplemented with leucine, uracil and histidine only. Saccharomyces cerevisiae strains Y190 and CTY10-5d were used for two-hybrid analysis. Escherichia coli strain JM109 was used for general molecular cloning purposes; strain CJ236 (dut– ung–) was used for preparation of single-stranded DNA for in vitro mutagenesis (Bio-Rad); strain BL21 (DE3) [pLysS] was used for induction of GST–Cdc27 and PP–Cdc27 fusion proteins (Pharmacia); strain M15 [pREP4] was used for expression of MRGS-His6-Pcn1 (Qiagen); and strain JA226 was used for plasmid recovery from yeast.
Two-hybrid screening
Two-hybrid screening for proteins that interact with S.pombe PCNA was carried out using plasmid pAS-PCNA-Sp described previously (Warbrick et al., 1995) and an S.pombe cDNA library in pACTII (a gift of Dr Steve Elledge). To eliminate false positives, plasmids recovered from the screen were tested against pAS plasmids encoding unrelated Gal4 fusion proteins, including pAS-Snf1, pAS-p53, pAS-Cdk2 and pAS-lamin.
Bacterial expression and purification of Pcn1 fusion proteins
The pcn1+ open reading frame (ORF) was cloned into pQE32 (Qiagen) to permit expression of recombinant Pcn1 in E.coli strain M15 [pREP4]. The Pcn1 protein produced in this manner has the N–terminal sequence MetArgGlySer(His6)GlyIleProMet, i.e. the tag comprises 13 amino acids. Gel filtration of the recombinant Pcn1 protein (by FPLC over a Superdex 200 HR 10/30 column) indicated that trimerization was unaffected by the presence of the N–terminal tag, consistent with the results of others (Jonsson et al., 1995). Similarly, the tagged Pcn1 protein was fully functional in S.pombe (data not shown). Expression and purification conditions were essentially as described elsewhere (Jonsson et al., 1995; Arroyo et al., 1996). Further details are available from the authors on request.
Expression and purification of Cdc27 fusion proteins
Details of the construction of the plasmids used to express the Cdc27 fusion proteins described in this study, together with expression/purification strategies, are available from the authors on request.
In vitro protein–protein interaction assay
Routinely, 0.2 μg of His6-Pcn1 was mixed with 0.2 μg of GST–Cdc27 in a total volume of 500 μl of buffer A (50 mM Tris–HCl pH 7.5, 0.1% Triton X-100) and incubated for 60 min at room temperature on a rotating wheel. A 40 μl volume of a 50% (v/v) slurry of glutathione–Sepharose was then added and the incubation continued for a further 30 min, at which point the glutathione–Sepharose beads were washed extensively with buffer A. After the final wash, the supernatant was discarded, and the beads boiled in an equal volume of 2× SDS–PAGE gel sample buffer and electrophoresed on a 12.5% gel. Proteins were visualized by PAGE Blue G90 staining (Fluka).
Peptide binding assays
Schizosaccharomyces pombe protein extracts were made from log phase, wild-type S.pombe cells grown in YE medium. Cells were recovered by centrifugation, washed twice in water and resuspended in a small volume of lysis buffer containing 25 mM NaCl, 0.1 mM EDTA, 1% Triton X–100 and 20 mM Tris–HCl pH 7.6. The cells were lysed by vortexing with glass beads. Insoluble material was removed by centrifugation at 14 000 r.p.m. in a benchtop centrifuge. The following 20 amino acid peptides were used, linked via residues SGSG at the N–terminus to biotin (Chiron Mimotopes, Australia). Each peptide was dissolved in dimethylsulfoxide (DMSO) to a final concentration of 5 mg/ml. Peptide sequences: p21 (KRRQTSMTDFYHSKRRLIFS), p21–A (KRRATSM– TDFYHSKRRLIFS), Cdc27 (KNTAQSKPQQKSIMSFFGKK), Cdc– 27–A (KNTAQSKPQAKSIMSFFGKK), Pol32 (SNKRLKKQGTLESF– FKRKAK), Pol32–A (SNKRLKKAGTLESFFKRKAK), Pogo (KLFNL– HINSAVLQKKITDYF) and control (PESVELKWSEPNEEELIKFM).
Peptide pull-down experiments. A total of ∼2.5 μg of each peptide was incubated with 10 μl of streptavidin–agarose beads (Sigma) in phosphate-buffered saline (PBS) for 1 h at room temperature; the beads were then washed extensively in PBS and recovered each time by centrifugation. A 20 ml volume of S.pombe protein extract diluted in PBS to a final concentration of 1 mg/ml was added to the washed beads and incubated with the beads on ice for 1 h. The beads were washed extensively in PBS containing 0.05% Tween-20, and bound proteins removed by boiling in SDS–PAGE loading buffer for 5 min. Proteins were separated on 15% SDS–PAGE gels and electrophoretically transferred to PVDF membrane (Amersham). The membranes were blocked in PBS containing 2% skimmed milk for 30 min, then incubated for 1 h with the monoclonal anti-PCNA antibody PC10 diluted 1 in 1000 in 2% skimmed milk–PBS. This antibody recognizes PCNA from a range of species including human and S.pombe (Waseem et al., 1992). After washing, blots were incubated with secondary horseradish peroxidase-conjugated rabbit anti–mouse antibodies (Dako) diluted 1 in 1000 in 2% skimmed milk–PBS for 1 h, followed by washing in PBS–0.05% Tween-20. Bound antibody was visualized using the ECL system according to the manufacturer's instructions (Amersham).
Peptide competition experiments. In competition experiments, non-biotinylated peptides were added to the diluted cell extracts before incubation with the immobilized, biotinylated peptides. The peptides used were a p21-derived peptide (KRRQTSMTDFYHSKRRLIFS) or an unrelated control peptide (KPVRLPSIQAIPCAP) added from a stock dissolved in DMSO at 20 mg/ml to a final concentration of 1 mg/ml. In each case a control reaction was carried out in which an equivalent amount of DMSO was added to control for the effects of the solvent.
Construction of the cdc27+/cdc27::his7+ strain
We replaced the entire cdc27+ ORF on one chromosome of a diploid strain with his7+ using standard methods. The cdc27::his7+ spores germinated normally and gave rise to a variable number, generally between 2 and 16, of elongated cells. This phenotype is indistinguishable from that seen with the cdc27+::ura4+ partial deletion strain described previously (MacNeill et al., 1996). Full details of the method used are available from the authors on request.
Analysis of Cdc27 mutants in the cdc27Δ strain
Plasmids (either pREP3X- or pREP3XH6-based) expressing various Cdc27 mutants were transformed into the cdc27+/cdc27::his7+ strain described above and transformants obtained on minimal medium (EMM) plates supplemented with uracil and thiamine. Following sporulation on malt extract medium, spores were plated onto EMM supplemented with uracil and adenine, and where possible haploid his+ (cdc27Δ) leu+ (pREP3X or pREP3XH6 plasmid) ade– ura– isolates were identified.
Overproduction of the C–terminus of Cdc27
BamHI fragments encoding various portions of the Cdc27 protein (as above) were cloned into plasmid pREP3XH6 to facilitate expression of the Cdc27 sequences fused to an MRGS(His6) tag (Gray and MacNeill, 2000). The resulting plasmids were transformed into an S.pombe leu1-32 h– strain for analysis. Cell lengths (>30 septated cells per culture) were determined using a micrometer.
Three-component two-hybrid assay
For three-component two-hybrid assays, the ADE2 marked plasmid pAA, a generous gift of Dr E.Chang, Cold Spring Harbor, NY, was used (Chang et al., 1994). To create pAA-Cdc27, pTZ19R-Cdc27 cDNA was mutagenized by oligonucleotide-directed in vitro mutagenesis to introduce a BamHI site at the 5′ end of the cdc27+ ORF (oligo sequence: 5′-GAAAAAGAAATTTCGGATCCAATGGAGGAATGGAGA-3′,BamHI site underlined). Then the cdc27+ ORF (BamHI fragment) was transferred to pAA to make pAA-Cdc27. (Note that the Cdc27 protein expressed from pAA is N–terminally tagged with the c-myc 9E10 monoclonal antibody epitope, amino acid sequence MEQKLISEEDDL.) Next, pAA-Cdc27 was transformed into S.cerevisiae CTY10-5d containing pBTM116-Cdc1 (MacNeill et al., 1996) and pACT-sppip35 (see above) and transformants obtained on SD agar plates supplemented with methionine and histidine only.
Two-hybrid analysis of Cdc1 versus Cdc27
Two-hybrid analysis of truncated Cdc27 proteins was performed in S.cerevisiae CTY10-5d using pBTM116-Cdc1 as bait as described previously (MacNeill et al., 1996). Plasmids for two-hybrid analysis of mutant Cdc27 proteins were constructed using pGAD2F and the same (BamHI) constructs as were used for the pGEX3X-Cdc27–GST fusion vectors described above.
In vitro Cdc1–Cdc27 binding
To monitor in vitro binding between Cdc1 and various GST–Cdc27 fusion proteins (see below), Cdc1 was expressed in rabbit reticulocyte lysates in the presence of 35S-labelled methionine from plasmid pGEM4Z-Cdc1tag as described previously (MacNeill et al., 1996). For the GST–Cdc27 binding assay, 7.5 μl of reticulocyte lysate (either unprogrammed or programmed with pGEM4Z-Cdc1tag) were added to 440 μl of TXB (50 mM Tris–HCl pH 7.5, 0.1% Triton X-100) containing 10 μg/ml RNase A, and incubated on ice for 10 min. To this was added 20 μl of glutathione–Sepharose 4B beads carrying either ∼400 μg of GST alone or of each GST fusion protein. Following mixing at room temperature for 1 h, the beads were washed four times with 500 μl of TXB, boiled in 20 μl of loading buffer (twice) for 3 min, electrophoresed on 10% SDS–PAGE gels and exposed overnight to BioMax film (Kodak).
Acknowledgments
Acknowledgements
We would like to thank our colleagues in Edinburgh and Dundee for their help during the course of this work. We are also grateful to Dr E.Chang (Cold Spring Harbor) for plasmid pAA, Nicola Preston and Vicky Clarke for automated DNA sequencing, and Dr Charles Hoffman (Boston College, Boston) for drawing our attention to the possibilities of gene deletion using his7+ and supplying the relevant materials. We are also grateful to Joan Davidson for media preparation. S.A.M. and N.R. are funded by the Wellcome Trust. E.W. is funded by the Cancer Research Campaign and the Association for International Cancer Research (AICR).
References
- Apolinario E., Nocero, M., Jin, M. and Hoffman, C.S. (1993) Cloning and manipulation of the Schizosaccharomyces pombe his7+ gene as a new selectable marker for molecular genetic studies. Curr. Genet., 24, 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo M.P., Downey, K.M., So, A.G. and Wang, T.S.F. (1996) Schizosaccharomyces pombe proliferating cell nuclear antigen mutations affect DNA polymerase δ processivity. J. Biol. Chem., 271, 15971–15980. [DOI] [PubMed] [Google Scholar]
- Brown W.C. and Campbell, J.L. (1993) Interaction of proliferating cell nuclear antigen with yeast DNA polymerase δ. J. Biol. Chem., 268, 21706–21710. [PubMed] [Google Scholar]
- Burgers P.M.J. (1996) Enzymology of the replication fork. In Blow,J.J. (ed.), Eukaryotic DNA Replication. IRL Press, Oxford, UK, pp. 1–28. [Google Scholar]
- Burgers P.M.J. and Gerik, K.J. (1998) Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem., 273, 19756–19762. [DOI] [PubMed] [Google Scholar]
- Chang E.C., Barr, M., Wang, Y., Jung, V., Xu, H.P. and Wigler, M.H. (1994) Cooperative interaction of S.pombe proteins required for mating and morphogenesis. Cell, 79, 131–141. [DOI] [PubMed] [Google Scholar]
- Chuang L.S.H., Ian, H.I., Koh, T.W., Ng, H.H., Xu, G.L. and Li, B.F.L. (1997) Human DNA (cytosine-5) methyltransferase PCNA complex as a target for p21Waf1. Science, 277, 1996–2000. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., Ayyagari, R., Gomes, X.V. and Burgers, P.M.J. (1997) Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol. Cell. Biol., 17, 6367–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik K.J., Li, X.Y., Pautz, A. and Burgers, P.M.J. (1998) Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem., 273, 19747–19755. [DOI] [PubMed] [Google Scholar]
- Giot L., Chanet, R., Simon, M., Facca, C. and Faye, G. (1997) Involvement of the yeast DNA polymerase δ in DNA repair in vivo. Genetics, 146, 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray F.C. and MacNeill,S.A. (2000) The Schizosaccharomyces pombe rfc3+ gene encodes a homologue of the human hRFC36 and S.cerevisiae Rfc3 subunits of replication factor C. Curr. Genet., in press. [DOI] [PubMed] [Google Scholar]
- Gulbis J.M., Kelman, Z., Hurwitz, J., Odonnell, M. and Kuriyan, J. (1996) Structure of the C–terminal region of p21Waf1/Cip1 complexed with human PCNA. Cell, 87, 297–306. [DOI] [PubMed] [Google Scholar]
- Hindges R. and Hübscher, U. (1995) Production of active-mouse DNA polymerase δ in bacteria. Gene, 158, 241–246. [DOI] [PubMed] [Google Scholar]
- Hughes D.A., MacNeill, S.A. and Fantes, P.A. (1992) Molecular cloning and sequence-analysis of cdc27+ required for the G2–M transition in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 231, 401–410. [DOI] [PubMed] [Google Scholar]
- Hughes P., Tratner, I., Ducoux, M., Piard, K. and Baldacci, G. (1999) Isolation and identification of the third subunit of mammalian DNA polymerase δ by PCNA-affinity chromatography of mouse FM3A cell extracts. Nucleic Acids Res., 27, 2108–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y. and Yamamoto, M. (1997) The Schizosaccharomyces pombe cdc6 gene encodes the catalytic subunit of DNA polymerase δ. Mol. Gen. Genet., 254, 93–97. [DOI] [PubMed] [Google Scholar]
- Jonsson Z.O., Podust, V.N., Podust, L.M. and Hubscher, U. (1995) Tyrosine-114 is essential for the trimeric structure and the functional activities of human proliferating cell nuclear antigen. EMBO J., 14, 5745–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti T., Flick, K., Keranen, S., Syvaoja, J.E. and Wittenberg, C. (1999) DNA polymerase ɛ catalytic domains are dispensable for DNA replication, DNA repair and cell viability. Mol. Cell, 3, 679–685. [DOI] [PubMed] [Google Scholar]
- MacNeill S.A. and Burgers,P.M.J. (2000) Chromosomal DNA replication in yeast: enzymes and mechanisms. In Fantes,P.A. and Beggs,J.D. (eds), Frontiers in Molecular Biology: The Yeast Nucleus. Oxford University Press, Oxford, UK, in press. [Google Scholar]
- MacNeill S.A. and Fantes, P.A. (1997) Genetic and physiological analysis of DNA replication in fission yeast. Methods Enzymol., 283, 440–459. [DOI] [PubMed] [Google Scholar]
- MacNeill S.A., Moreno, S., Reynolds, N., Nurse, P. and Fantes, P.A. (1996) The fission yeast Cdc1 protein, a homolog of the small subunit of DNA polymerase δ, binds to Pol3 and Cdc27. EMBO J., 15, 4613–4628. [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Montecucco A., et al. (1998) DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J., 17, 3786–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. and Nurse, P. (1981) Cell-division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 182, 119–124. [DOI] [PubMed] [Google Scholar]
- Park H., Francesconi, S. and Wang, T.S.F. (1993) Cell-cycle expression of 2 replicative DNA polymerases α and δ from Schizosaccharomyces pombe. Mol. Biol. Cell, 4, 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignede G., Bouvier, D., Derecondo, A.M. and Baldacci, G. (1991) Characterization of the pol3 gene-product from Schizosaccharomyces pombe indicates interspecies conservation of the catalytic subunit of DNA polymerase δ. J. Mol. Biol., 222, 209–218. [DOI] [PubMed] [Google Scholar]
- Prelich G. and Stillman, B. (1988) Coordinated leading and lagging strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell, 53, 117–126. [DOI] [PubMed] [Google Scholar]
- Reynolds N., Watt, A., Fantes, P.A. and MacNeill, S.A. (1998) Cdm1, the smallest subunit of DNA polymerase δ in the fission yeast Schizosaccharomyces pombe, is non-essential for growth and division. Curr. Genet., 34, 250–258. [DOI] [PubMed] [Google Scholar]
- Shamoo Y. and Steitz, T.A. (1999) Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell, 99, 155–166. [DOI] [PubMed] [Google Scholar]
- Stillman B. (1996) Comparison of DNA replication in cells from prokarya and eukarya. In DePamphilis,M.L. (ed.), Eukaryotic DNA Replication. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 435–460. [Google Scholar]
- Sun Y.B., Jiang, Y.Q., Zhang, P., Zhang, S.J., Zhou, Y., Li, B.Q., Toomey, N.L. and Lee, M. (1997) Expression and characterization of the small subunit of human DNA polymerase δ. J. Biol. Chem., 272, 13013–13018. [DOI] [PubMed] [Google Scholar]
- Tournier S., Leroy, D., Goubin, F., Ducommun, B. and Hyams, J.S. (1996) Heterologous expression of the human cyclin-dependent kinase inhibitor p21Cip1 in the fission yeast, Schizosaccharomyces pombe reveals a role for PCNA in the chk1+ cell-cycle checkpoint pathway. Mol. Biol. Cell, 7, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratner I., Piard, K., Grenon, M., Perderiset, M. and Baldacci, G. (1997) PCNA and DNA polymerase δ catalytic subunit from Schizosaccharomyces pombe do not interact directly. Biochem. Biophys. Res. Commun., 231, 321–328. [DOI] [PubMed] [Google Scholar]
- Waga S. and Stillman, B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- Warbrick E. (1998) PCNA binding through a conserved motif. BioEssays, 20, 195–199. [DOI] [PubMed] [Google Scholar]
- Warbrick E., Lane, D.P., Glover, D.M. and Cox, L.S. (1995) A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21Waf1 and proliferating cell nuclear antigen. Curr. Biol., 5, 275–282. [DOI] [PubMed] [Google Scholar]
- Warbrick E., Heatherington, W., Lane, D.P. and Glover, D.N. (1998) PCNA binding proteins in Drosophila melanogaster: the analysis of a conserved PCNA binding domain. Nucleic Acids Res., 26, 3925–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseem N.H., Labib, K., Nurse, P. and Lane, D.P. (1992) Isolation and analysis of the fission yeast gene encoding polymerase δ accessory protein PCNA. EMBO J., 11, 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Mo, J.Y., Perez, A., Leon, A., Liu, L., Mazloum, N., Xu, H. and Lee, M. (1999) Direct interaction of proliferating cell nuclear antigen with the p125 catalytic subunit of mammalian DNA polymerase δ. J. Biol. Chem., 274, 26647–26653. [DOI] [PubMed] [Google Scholar]
- Zhang S.J., Zeng, X.R., Zhang, P., Toomey, N.L., Chuang, R.Y., Chang, L.S. and Lee, M. (1995) A conserved region in the amino–terminus of DNA polymerase δ is involved in proliferating cell nuclear antigen binding. J. Biol. Chem., 270, 7988–7992. [DOI] [PubMed] [Google Scholar]
- Zhou J.Q., He, H., Tan, C.K., Downey, K.M. and So, A.G. (1997) The small subunit is required for functional interaction of DNA polymerase δ with the proliferating cell nuclear antigen. Nucleic Acids Res., 25, 1094–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo S.J., Gibbs, E., Kelman, Z., Wang, T.S.F., O'Donnell, M., MacNeill, S.A. and Hurwitz, J. (1997) DNA polymerase δ isolated from Schizosaccharomyces pombe contains five subunits. Proc. Natl Acad. Sci. USA, 94, 11244–11249. [DOI] [PMC free article] [PubMed] [Google Scholar]