Abstract
IBS is a common disorder that has been shown to aggregate in families, to affect multiple generations, but not in a manner consistent with a major Mendelian effect. Relatives of an individual with IBS are two to three times as likely to have IBS, with both genders being affected. The estimated genetic liability ranges between 1–20%, with heritability estimates ranging between 0–57%. Although the role of childhood events such as nasogastric tube placement, poor nutrition, abuse, and other stressors have been clearly associated with IBS, these factors have not been studied in families and are unlikely to completely explain the clustering of bowel dysfunction observed in family studies. Furthermore, the familial clustering of IBS does not appear to be explained by psychological traits, based on family studies as well as candidate gene studies of functional variants associated with other psychiatric disorders. To date, over a hundred genetic variants in over 60 genes from various pathways have been studied in a number of candidate gene studies with several positive associations reported. These findings suggest that there may be distinct, as well as shared, molecular underpinnings for IBS and its subtypes. Much new and confirmatory work remains to be performed to elucidate the role of specific genetic variants in IBS development, as well as the specific ways the genes and environment interact to result in IBS susceptibility.
Keywords: Irritable bowel syndrome, IBS, genetics, gene, complex genetic disease
Introduction
As discussed in previous chapters, Irritable Bowel Syndrome (IBS) is a chronic disorder characterized by abdominal pain or discomfort and diarrhea and/or constipation that can be associated with altered gastrointestinal motility and visceral sensation. The chronicity of IBS symptoms and the lack of a cure or effective treatments result in loss of work and school productivity and impaired personal and health-related quality of life.1 Patients, their relatives, and friends frequently ask difficult questions, such as why they developed IBS. Although diet, psychological factors, infection, and gut flora may attenuate IBS symptoms, there is no simple answer to this question. As other family members may concurrently have bowel disturbances, the role of genes in disease development—and the sense of personal destiny it invokes--is somewhat intuitive, easy to accept, but also difficult to refute with facts or objective findings arguing the contrary.
Great advances have been made in the field of genetics in recent years. As the efficiency, ease, and cost of genotyping has decreased, our understanding of DNA sequence, structure, and function has improved dramatically. Previously, laboratory-based study of the human genome was often restricted to known genes and coding exon regions and study of a handful of genetic variants. Now, high-throughput technology allows genotyping of thousands to a million genetic markers across the genome and sequencing of nearly the entire [coding] human genome is now possible. Furthermore, advances in data storage and analysis of these large quantities of data has led to the discovery of susceptibility loci for a number of diseases including, notably, Crohn’s disease. These amazing scientific strides have led many to believe that “Virtually every human ailment, except perhaps trauma, has some basis in our genes.” (www.genome.gov). This premise has appealed to many IBS researchers and several have commenced in trying to identify an IBS gene, or set of genes.
Despite the acceptance by some that genes may cause—or at least contribute to the development of—IBS, careful examination of the body of literature is necessary to determine whether there is sound basis for this theory as gene discovery still requires considerable time, effort, and thus, financial resources. Certainly, alternative hypotheses—such as environmental exposures—exist and can not be ignored as important players in IBS. This article will provide a summary of the studies around the competing hypotheses of “gene v. environment” or “nature v. nurture” in IBS. An overview of family studies, candidate gene studies, and alternative hypotheses for IBS will be covered to provide the reader an overview of the works conducted in this area.
IBS as a complex genetic disorder
Classic Mendelian genetics diseases are typically caused by a few highly penetrant genetic defects on a single gene and are transmitted in a typical pattern through families. Mendelian diseases follow an autosomal dominant, recessive, co-dominant, or X-linked pattern of transmission through pedigrees. Recent genetic studies have focused less on Mendelian disorders but on complex genetic diseases. A “complex genetic disease” is defined as a multifactorial genetic disorder resulting from multiple genetic variants on several genes (i.e. polygenic) with contributions from environment and lifestyle. These genetic effects are modest in that the presence of a specific variant is rarely sufficient on its own to result in disease development. Complex genetic diseases are still heritable in that they tend to aggregate in families, but not in the same predictable fashion as classic Mendelian disorders.
Many common diseases of significant public health interest are thought to be complex disorders. Heart disease, hypertension, diabetes, obesity, autism, and mood disorders are a few of the diseases and disorders under study by geneticists and genetic epidemiologists. In gastroenterology, Crohn’s disease is the best example of a complex genetic disorder with successful susceptibility loci identification. Through a combination of family-based linkage studies, candidate gene and fine-mapping studies, as well as genome-wide association studies have led to the identification of several genes—such as NOD2, ATG16L1, IL23, IL12B, STAT3, NKX2-3—found to be consistently associated with Crohn’s disease.2 These discoveries offer the promise that if gene discovery was possible in this complex gastrointestinal disorder, that successful gene discovery is feasible in other multifactorial disorders and diseases including IBS.
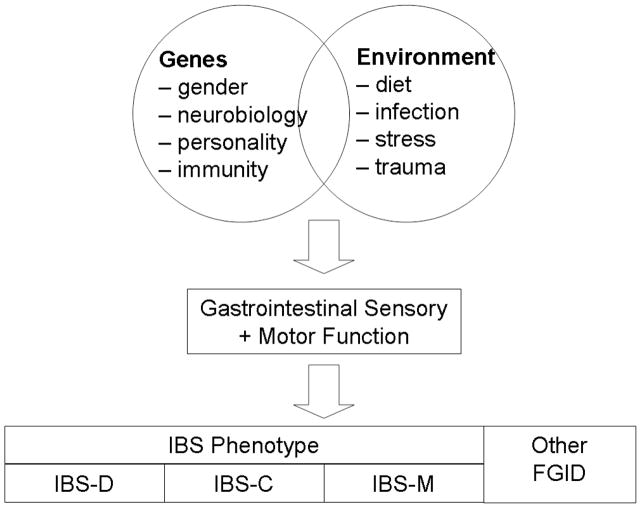
Despite the discovery of several genetic loci involved in Crohn’s disease development, it is clear that genes alone do not cause the disease and that environmental contributions—such as diet, smoking, early exposure to infectious organisms, and colonic microflora--are still important. A similar gene-environment paradigm could be proposed for IBS (Figure 1) where a combination of genetic factors and environmental factors result in the alterations in gastrointestinal sensation and motor function that ultimately result in symptom manifestation. Genetic variation in genes that encode proteins that regulate gender-based biological processes, control or modulate central or peripheral sensation and motility, or even regulate brain response to stress would be the obvious first candidates for IBS. These factors, interacting with environmental factors such as diet, infection, early life trauma and stress, are likely responsible for the overall IBS phenotype. However, the specific combinations of genetic variants and environmental factors likely can in part explain the clinical heterogeneity of IBS. Hence, it is conceivable and likely that there are different genes responsible for diarrhea than those responsible for constipation, and other genetic variants that predispose an individual to developing or sensing abdominal pain required for IBS that is not generally present in the non-painful functional disorders such as functional constipation and functional diarrhea. Exploring gene-environment interactions will be important if one postulates that IBS is a multifactorial, polygenic complex genetic disorder.
Figure 1.
Gene-environment paradigm in IBS development. A gene-environment paradigm supports the importance of both genes and environment in the development of IBS for both familial and sporadic IBS. A number of combinations of individual genetic and environmental risk factors are possible, with each specific combination resulting in specific alterations in gastrointestinal motor and sensory function, and ultimately symptom presentation and the final clinical phenotype.
IBS and heritability
Genetic diseases—complex or Mendelian—must run in families. Several studies of patients with IBS suggest that this disorder aggregates in families, and thus, appears potentially heritable. Children with persistent recurrent abdominal pain who were found to report IBS-like symptoms as adults were almost three-fold as likely to have at least one sibling with similar symptoms when compared to children-now-adults without IBS-like symptoms (40% v. 6%, p<0.05).3 Another study of IBS outpatients showed that one-third reported another family member with IBS, even in patients without a concurrent psychiatric diagnosis.4 In another study, having a first-degree relative with IBS was one of the characteristics found to be predictive of IBS over other gastrointestinal disorders among outpatients consulting their general practitioner for chronic abdominal pain.5
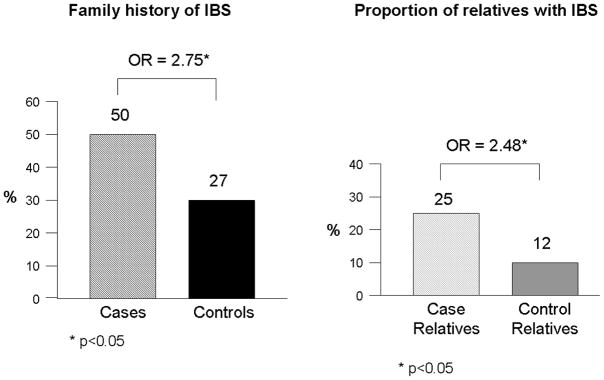
The majority of original studies evaluating familial clustering of IBS has been based predominantly on patient report of having another affected family member with IBS—proxy reporting. However, one study by our group has shown that accuracy of patient reporting on a specific relative’s IBS status is poor.6 As a consequence, a large family case-control study was performed which collected bowel symptom and medical history data directly from cases, controls, and their first-degree relatives. This study of 477 cases, 1492 case-relatives, 297 controls, and 936 control-relatives found that 50% of case- and 27% of control-families had at least one other relative with IBS (p<0.05) (Figure 2).7 The magnitude of this effect was an odds ratio of 2.75 (95%CI: 2.01–3.76). When looking at the absolute proportion of relatives affected, 25% of case-relatives reported having IBS compared to 12% of control-relatives (p<0.05). Furthermore, the magnitude of familial aggregation did not vary by gender of the proband with males and females being equally likely to have an affected relative or set of relatives. Although confidence intervals overlapped, there was a trend for the strength of aggregation to be greatest among probands with diarrhea, followed by constipation, and then mixed bowel habits. Prior intestinal infections, abuse, and depression or anxiety were more common among cases than controls, and among affected relatives than unaffected relatives. The overall prevalence of these risk factors were 9% of affected relatives reporting a prior infection (v. 5% among controls), 35% reporting abuse (v. 25%), and 44% reporting depression or anxiety (v. 22%). Thus, a family member of an individual with IBS is 2–3 times more likely to have IBS, familial clustering is present irrespective of predominant bowel pattern, and the known environmental risk factors for IBS are also common in IBS families.
Figure 2.
Familial aggregation of IBS. Fifty percent of cases with IBS will have at least one other family member with IBS, but the overall proportion of first-degree relatives with IBS is 25%. Overall, case-relatives are two to three times as likely to have IBS than control-relatives.
Study of the pattern of IBS transmission through families has yielded additional interesting observations. A recent study of children with functional gastrointestestinal disorders (FGIDs),—but not IBS exclusively-- their parents, and their child-aged siblings was performed and it showed that all mothers, fathers, and siblings of cases were more likely to be affected with another FGID than matching control-relatives.8 Mothers were the most likely to have a FGID (49.6% case-mothers v. 21.4% control-mothers, p<0.05), followed by their child-aged siblings (26.4% v. 17.6, N.S.), then fathers (13.6% v. 9.3%, N.S.). In contrast, our study of adult patients with IBS showed that adult-aged case-siblings had the highest risk of concurrent IBS, followed by their adult-aged children, and then parents.7 Although female relatives were more likely to be affected with IBS, both genders were still at risk for IBS compared to control- relatives. When formal segregation analysis was performed to determine whether the pattern of transmission of IBS through pedigrees is consistent with Mendelian models (e.g. autosomal dominant, recessive, X-linked, etc.), IBS transmission was not consistent with the sporadic model, meaning IBS appeared consistently in families and in parents, arguing against random mutations causing IBS.9 More importantly, there was general lack of convergence of the data with segregation analysis. This finding suggests that IBS does not result from a single, major locus, that there was incomplete penetrance, or that IBS could be the result of genetic and environmental factors. The findings are indicative of an underlying genetic basis for IBS.
Although IBS appears to run in families, the previously cited studies suggest that there may be individuals with “sporadic” or non-familial IBS as well as individuals with a familial form of IBS. This observation begs the question as to whether there are clinical characteristics suggesting a distinct mechanistic basis for the sporadic and familial forms of IBS. Genetic diseases may present at an earlier age or present with a more specific form of disease (e.g. colon cancer). Our study found that cases with a stronger family history of IBS had more severe pain, had concurrent fibromyalgia, heartburn, asthma, and reported symptoms of loose stools, urgency constipation, and pain compared to those with less of a family history of IBS (p<0.05).10 Age of onset was not predicted by family history of IBS thus demonstrating that patients with a family history did not experience IBS symptom onset at a younger age than patients without a family history of IBS. Although at a univariate level of analysis, somatization level, and personal or family history of psychiatric history or abuse were more common among individuals (probands and relatives) with IBS, these factors were not predictive of familial IBS with univariate or with multivariate analyses, suggesting that familial clustering was not related to psychological or psychiatric disease. Kanazawa, et al. has shown that among IBS patients reporting a parental history of bowel problems, these individuals exhibit higher levels of psychological distress.11 These studies suggest that familial forms of IBS are characterized by greater pain severity and greater comorbidity including psychological distress.
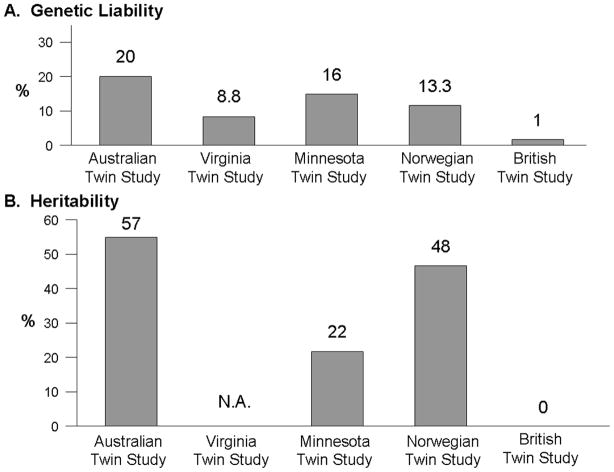
Twin studies also support the concept that IBS may be a complex disorder with genetic as well as environmental contributors. To date, there have been at least five twin studies of IBS or functional bowel disorders that estimate that the genetic liability ranges between 1–20% and heritability estimates ranging between 0–57% (Figure 2).12–16 In all but one study, the concordance rates for monozygotic twins (who are genetically identical) were higher than the concordance rates for dizygotic twins, suggesting an underlying genetic etiology. Nonetheless, Mohammed, et al. found that concordance rates for monozygotic and dizygotic twins were similar for IBS using a validated questionnaire and the Rome II diagnostic criteria to make the diagnosis of IBS, thus arguing that genes are not a major factor in IBS development, and certainly not a major gene with a strong effect. Studies with twins reared apart, which would permit greater discernment of the influence of different environments in genetically identical twins, are unfortunately lacking.
Heritability (h2) is another statistical estimate to quantitate the relative genetic contribution to a trait, relative to its environmental contributors. Heritability is the amount or proportion of phenotypic variance of the disease of interest in the population that is inherited through genetic factors. The twin studies show that the heritability estimates range from 0% in the British Twin Study, 22% in the Minnesota Twin Study, 48% in the Norwegian Twin Study, to 57% in the Australian Twin study (Figure 3). Using data collected from families and not twin pairs, when the IBS-Rome criteria were converted into a quantitative trait based on number of Rome symptoms endorsed and severity, the heritability estimates for IBS ranged from 0.19–0.35. depending on the weighting factors for each score. In summary, these studies suggest that the genetic contribution to IBS development is reasonably high and comparable to other diseases for which a genetic basis has been found.
Figure 3.
Estimated genetic liability and heritability in IBS twin studies. Five twin studies have evaluated the concordance of IBS between twin pairs. Genetic liability of IBS has been estimated to range between 1–20% and heritability estimates in the best fit model ranged between 0–57%.
Environmental factors and familial IBS
Despite the consistent observation that IBS clusters in families, this could still be explained by shared environmental contributors, or less likely, individual environmental exposures. Individual environmental contributors that are not likely to be shared among family members or across generations will not be discussed in detail in this review. These exposures include early and later life experiences such as nasogastric tube placement at birth,17 other painful stimuli experienced during infancy, maternal separation,18 socioeconomic status,19, 20 military deployment,21 and microflora.22 These individual environmental factors are still important to potentially include when gene-environment interactions may be required for disease development. This section will discuss shared household exposures that could represent an alternative mechanism besides an underlying genetic basis for the familial aggregation observed in the families of patients with IBS. Environmental exposures that could have been shared between multiple family members include verbal abuse, physical abuse, sexual abuse, shared household stressor/s (e.g. ill family member, unemployment, catastrophe, etc.), parenting style, and learned illness behavior.
Verbal, physical, and sexual abuse during childhood has been reported to be more common in patients with IBS than in matched controls in several studies. Up to 50% of patients with IBS may report a history of lifetime victimization.6, 23–26 The mechanism by which abuse is associated with, or possibly leads to development of IBS may be the result of psychological factors rather than a fundamental change in gastrointestinal motor and sensory function. Heitkempker et al. has shown that among those with IBS, few differences were observed in characteristics compared between women with and those without an abuse history.27 Furthermore, when four groups of patients with and without IBS and with and without sexual abuse histories had rectal distension studies performed, although IBS again reported greater sensitivity to rectal distension, sexual abuse did not attenuate results.28 Ringel, et al. has also recently shown that the presence or absence of abuse does not alter rectal sensation or lead to increased rectal pain sensation.29 In contrast, a population-based study found that although childhood abuse was associated with IBS, the abuse was no longer associated for IBS when neuroticism was accounted for and included in the analyses.30 In the only family study examining the role of abuse, our group has shown that in family relatives, the association between abuse and IBS disappeared after inclusion of somatization in the multivariate model. This suggests that either somatization leads to over-reporting of abuse, or perhaps more likely, abuse leads to somatization that ultimately results in IBS symptom development and reporting.7 Furthermore, as the majority of patients with IBS report no abuse history, abuse is clearly not required for the development of IBS. The familial clustering of IBS is unlikely to be explained by shared household exposure to an abuser, as the prevalence of abuse is the same among familial and non-familial, sporadic IBS.10
Besides household exposure to abuse, other shared stressors experienced in a family environment, particularly during childhood, could include adverse life events such as loss of a parent or close relative, significant illness in a household relative, singular or recurrent unemployment, and natural disasters. However, adverse childhood events appear very common—approximately 60% of 17,000 adults from an HMO reported one or more events during childhood, and no secular trends were observed across four birth cohorts.31 Another study specifically comparing the role of early adverse life events among IBS patients and controls with respect to salivary cortisol measurements before and after a visceral stressor found that early adverse life events were present in nearly 50% of both patients with IBS as well as their matched controls, and that IBS was equally common among individuals reporting adverse life events as compared to individuals reporting no early life events.32 Although cortisol levels were higher among individuals reporting adverse childhood events compared to those without early events, IBS patients did not have different salivary cortisol responses to stress than controls, and there was only a trend for an interaction between IBS patients who had reported life events and higher cortisol levels compared to controls (p=0.056). Furthermore, although adverse life events appear to be common across time and thus, possibly across generations, they are unlikely to be shared across multiple generations making the hypothesis that they could contribute to familial clustering of IBS less viable.
Besides childhood or early adulthood exposures and stressors, others have suggested that the familial clustering of IBS may be due to learned illness behavior modeled from parents. Whitehead et al. surveyed a sample of the general population in metropolitan Cincinnati and found that individuals with IBS symptoms exhibited somatic behavior (e.g. reporting more complaints, consulting physicians for minor complaints) but importantly, reported being given gifts or special foods as children during illness—findings not found in those in acute diseases such as peptic ulcer patients and the asymptomatic general population.33 Another study also confirmed that patients with IBS were more likely to miss school and have more doctor visits as children than individuals without IBS, suggesting that their parents paid greater attention to their illnesses reinforcing chronic illness behaviors.34 Levy et al. conducted an interview study of mothers with IBS and their children and compared them to mothers without IBS and their children.35 She found that children of mothers with IBS have more gastrointestinal as well as non-gastrointestinal symptoms and have more school absences and physician visits. The investigators found that mother-IBS status was independent from maternal solicitousness (reinforcement of illness behavior) where parent-IBS status influenced children’s perceptions of symptom severity but not their perception of the seriousness of gastrointestinal symptoms, whereas parental solicitousness appeared to influence the children’s reporting of seriousness of symptoms but not severity. Ultimately, the study supported the role of learned illness behavior, although the authors could not distinguish genetic and environmental contributors to gastrointestinal symptoms.
In summary, shared household environmental factors—whether abuse, other early adverse life events, or learned illness behavior from parental modeling—could explain a subset of cases with IBS, but the degree to which they contribute to familial clustering of IBS still remains to be fully elucidated.
Genetic diseases that mimic IBS
One of the great challenges of studying the genetics of IBS is that there are a number of disorders with symptoms similar to IBS, and a group of these may also have an underlying genetic basis. These diseases include inflammatory bowel disease (e.g. Crohn’s disease, ulcerative colitis) and celiac sprue. These genetic traits may also co-exist with IBS-- lactose or fructose intolerance, for example—whose presence does not necessarily preclude an IBS diagnosis. As these disorders are not always ruled out prior to inclusion in family studies, there will always be some degree of uncertainty regarding the potential role of these disorders in the familial aggregation of IBS. Despite this concern of another genetic disorder mimicking IBS, we found that in our group of over 500 IBS case that their IBS diagnoses endured over inflammatory conditions after extensive chart review as well as serological testing for celiac sprue.7 Celiac sprue was found in only 1% of IBS cases and in a comparable proportion of controls. When Villani et al. screened loci associated with Crohn’s disese and ulcerative colitis in this sample, IBD loci were not observed in the IBS group.36 Similarly, the prevalence of lactase nonpersistance was no different between IBS cases and their controls (15% v. 14%,37 suggesting that this autosomal recessive trait is unlikely to explain IBS let alone explain the familial aggregation of IBS. Nonetheless, as a mimicker, testing for and adjustment for lactose intolerance, inflammatory bowel disease, and celiac disease may be necessary in gene studies of IBS.
In addition to gastrointestinal diseases, the genetics of other non-gastrointestinal disorders such as psychiatric diseases and traits may be important to account for in family studies of IBS as psychiatric disorders are common in patients with IBS and select antidepressants can be used to treat IBS. A number of studies have shown that there is a link between IBS and psychiatric illness, including in families. Sullivan, et al. reported that among outpatients receiving their first diagnosis of IBS, the prevalence of depression among blood relatives was higher than in controls (5.7% v. 1.8%, p<0.05), at a level comparable with a group of individuals with major depression (5%).38 Although the proportion of relatives with depression was higher in IBS patients, the numbers were still quite low, but the authors argue that self-report of family history is less reliable and likely represents an underestimate of the true prevalence of depression. Woodman, et al. conducted interviews of relatives of patients with IBS and patients who had undergone a laparoscopic cholecystectomy and found that the relatives of IBS patients were more likely to report depressive disorders (33.3% v. 17.3%, p=0,05) and anxiety disorders (41.7 v. 18.7, p<0.05), but not somatoform (2.1 v. 1.8%, p>0.05) or substance use disorders (18.8 v. 32.0%, p>0.05).39 Hudson, et al. performed a similar study interviewing probands with and without major depressive disorder (MDD) and found that IBS was more common in relatives of probands with MDD than in relatives of probands without MDD (16 v. 7%). In short, there has been suggestion that IBS is part of a larger genetic psychiatric disorder. However, some could argue that because psychiatric co-morbidity is related to health-care seeking but not a common feature in community IBS,40 that it is unlikely that a broader psychiatric trait is the heritable component in IBS. Our large family case-control study did not find that a personal history of psychiatry disease, a family history of psychiatric disease, or a family history of alcohol abuse was predictive of familial IBS.10 And as discussed in the next section, we did not observe that common genetic variants associated with mood disorders were associated with IBS in the absence of psychiatric disease.41 Thus, although it is possible there is a genetic variant responsible for a psychiatric trait linked to IBS, there is likely a separate genetic and biological mechanism to explain the remaining symptoms of IBS.
Candidate Gene and Pathway Studies
To date, nearly 60 genes have been evaluated to determine whether specific genetic variants may be associated with IBS. The genes, the genetic markers, the findings, and references for these studies are summarized on Table 1. The genes studied lay in the following main pathways: serotonin, adrenergic, inflammation, intestinal barrier, and psychiatric. The genes studied were selected because of their proteins’ putative role in gastrointestinal motor or sensory function or because of their potential role in resistance or response to microbial organisms. As gastrointestinal motility and sensation are abnormal in a subset of IBS patients, infection is a risk factor for IBS development, and antibiotic therapy can attenuate IBS symptoms, the genes encoding proteins and peptides related to gastrointestinal function pose an attractive target of study. Because the number of genetic markers and genes is too great to review in entirety, this section will focus primarily on genetic pathways and positive findings reported by investigators.
Table 1.
List of candidate genes tested in IBS patients
| Gene | SNP RSID# | Variant | Associated with IBS? | Reference |
|---|---|---|---|---|
| Serotonin pathway genes | ||||
| SLC6A4 | Rs4795541 | 5-HTTLPR (44bp ins/del) | No LS, LL α IBS-C | 36, 44, 50, 64 |
| STin2 VNTR | No | 73–75 | ||
| rs25531 | - | G allele | 75 | |
| Rs3813034 | G>T | No | 50 | |
| HTR2A | rs6313 | 102T>C | No | 50, 76 |
| rs6311 | −1438G>A | No | 50, 76 | |
| rs6314 | H452Y | No | 50 | |
| HTR3A | - | −42C>T | CT α IBS-D | 46 |
| - | −25C>T | No | 46 | |
| - | *70C>T | No | 46 | |
| - | *503C>T | No | 46 | |
| HTR3B | - | 386A>C | AC/CC | 48 |
| HTR3C | - | 489C>A | CC α IBS-D | 77 |
| HTR3E | rs62625044 | *76G>A | GA α IBS-D | 46 |
| - | *115T>G | No | 46 | |
| - | *138C>T | No | 46 | |
| - | −189A>G | No | 46 | |
| TPH1 | rs1800532 | 218A>C | No | 50 |
| rs1799913 | 779A>C | No | 50 | |
| TPH2 | rs11178997 | −473T>A | No | 50 |
| rs4570625 | −703G>T | No | 50 | |
| rs4565946 | 90A>G | No | 50 | |
| Inflammatory/Immune pathway genes | ||||
| IL10 | rs1800896 | −1082G>A | A α IBS; No; G α IBS; GA α IBS | 36, 50–52, 78 |
| - | −819C>T | No | 52 | |
| rs1800872 | −592A>C | No | 50 | |
| TNFα | - | −308G>A | GA α IBS; No; No | 52, 79, 80 |
| - | −238G>A | G α IBS | 79 | |
| TGF β1 | rs1982073 | +869T>C | No | 51 |
| rs1800469 | −509C>T | No | 50 | |
| NOD2 | rs2066847 | Leu1007insC | No | 50 |
| rs2066844 | R702W | No | 50 | |
| rs2066845 | G908R | No | 50 | |
| rs2076756 | - | No | 50 | |
| NOD1 | rs6958571 | ND1+32656*1 | No | 50 |
| CARD8 | rs2043 | Cys10Stop | No | 50 |
| TLR4 | rs4986790 | D299G | No | 50 |
| TLR5 | rs5744168 | 1174C>T | No | 50 |
| TLR9+ | rs5743836 | −1237T>C | T α PI-IBS | 50 |
| rs352139 | 2848G>A | A α PI-IBS | 50 | |
| CD14 | rs2569190 | −159T>C | No | 50 |
| NFKB1 | rs28362491 | −94delATTG | No | 50 |
| ATG16L1 | rs2241880 | T300A | No | 50 |
| PTGER4 | rs4495224 | - | No | 50 |
| rs6313763 | - | No | 50 | |
| rs9292777 | - | No | 50 | |
| IRGM | rs4958847 | - | No | 50 |
| rs13361189 | - | No | 50 | |
| NKX2-3 | rs10883365 | - | No | 50 |
| rs7078219 | - | No | 50 | |
| PTPN2 | rs2542151 | - | No | 50 |
| rs2847297 | - | No | 50 | |
| IFNA | - | 874T>A | No | 80 |
| TNF | rs1800629 | −308G>A | No | 50 |
| rs1799724 | −857C>T | No | 50 | |
| rs1800630 | −863C>A | No | 50 | |
| rs1799964 | −1031T>C | No | 50 | |
| IL1A | - | −889T>C | No | 79 |
| IL1B | rs1143627 | −31C>T | No | 50 |
| rs1143634 | F105F | No | 50 | |
| rs16944 | −511C>T | No | 50, 79 | |
| - | +3962C>T | No | 79 | |
| IL1R | - | Pst-I 1970C>T | C α IBS | 79 |
| IL1RA | - | Mspa-I 11100T>C | No | 79 |
| IL4 | rs2243250 | −590C>T | No; C α IBS | 50, 78 |
| - | −33T | T α IBS | 78 | |
| IL4R | rs1801275 | Q576R | No | 50 |
| rs1805015 | S503P | No | 50 | |
| IL6 | rs1800795+ | −174G>C | C α PI-IBS; G α IBS | 50, 79, 80 |
| - | nt565 | No | 79 | |
| - | −1082A>G | No | 80 | |
| - | −819T>C | No | 80 | |
| - | −592A>C | No | 80 | |
| IL6R | rs4537545 | - | No | 50 |
| rs8192284/ | D358A | No | 50 | |
| rs2228145 | ||||
| IL7R | rs6897932 | T244I | No | 50 |
| IL8 | rs4073 | −251T>A | No | 50 |
| IL12B | rs10045431 | - | No | 50 |
| rs6887695 | - | No | 50 | |
| Il13 | rs20541 | R130Q | No | 50 |
| rs1800925 | −1112C>T | No | 50 | |
| IL23R | rs11209026 | R381Q | No | 50 |
| (3p21) | rs9858542 | - | No | 50 |
| (10q21.1) | rs224136 | - | No | 50 |
| rs224135 | - | No | 50 | |
| (5q31) | rs2188962 | - | No | 50 |
| Intestinal barrier genes | ||||
| SLC22A4 | rs1050152 | 1672C>T | No | 50 |
| SLC22A5 | rs2631367 | −207G>C | No | 50 |
| CDH1+ | rs16260 | −160C>A | A α PI-IBS | 50 |
| rs5030625 | −347G>A | No | 50 | |
| ICAM1 | rs5498 | K469E | No | 50 |
| rs1799969 | G241R | No | 50 | |
| DLG5 | rs1248696 | R30Q | No | 50 |
| MYO9B | rs1545620 | S1011A | No | 50 |
| ABCB1 | rs1045642 | 3435C>T | No | 50 |
| rs3789243 | - | No | 50 | |
| NR1I2 | rs3814055 | −25385C>T | No | 50 |
| rs6785049 | 7635A>G | No | 50 | |
| LTF | rs7645243 | Leu632Leu | No | 50 |
| BPI | rs4358188 | E216K | No | 50 |
| Adrenergic pathway genes | ||||
| NET | - | 237G>C | No | 81 |
| ADRA2A | rs1800035 | 753C>G | No | 50, 64, 81 |
| rs1800544 | −1291C>G | G α IBS-C; CG/GG α IBS-D; No | 50, 81, 82 | |
| ADRA2C | - | Del 332–335 | IBS-C | 81 |
| Psychiatric genes | ||||
| FKBP5 | rs3800373 | - | No | 41 |
| rs1360780 | - | No | 41 | |
| rs9470080 | - | No | 41 | |
| COMT | rs4680 | Val158Met | Met α IBS-C | 41 |
| NPY | rs16147 | - | No | 41 |
| BDNF | rs6265 | Val166Met | Met α psychiatric IBS | 41 |
| ANKK1 | rs1800497 | - | No | 41 |
| DRD2 | rs12364283 | Ser311Cys | No | 41 |
| OPRM1 | rs1799971 | 118A>G | G α IBS-M, IBS-D females | 41 |
| Other tested genes | ||||
| CCK | rs1800857 | 779T>C | No | 50 |
| CCK1R | - | Intron1 779T>C | C α IBS-C | 83 |
| Exon1 G>A | No | 83 | ||
| - | 266G>C | No | 84 | |
| - | 8641G>A | No | 84 | |
| - | 984T>C | No | 84 | |
| CNR1 | - | AAT repeats | Yes | 85 |
| FAAH | rs324420 | 385C>A | A α IBS-D, IBS-M. No | 41, 65 |
| GN β3 | rs5443 | 825C>T | No, but possible interaction with GI infection | 45, 50, 64, 86–88 |
| NPSR1 | rs2609234 | - | No | 66 |
| rs714588 | - | No | 66 | |
| rs1963499 | - | No | 66 | |
| rs2168890 | - | No | 66 | |
| rs2530547 | - | No | 66 | |
| rs887020 | - | No | 66 | |
| rs1379928 | - | No | 66 | |
| rs323917 | - | No | 66 | |
| rs323922 | - | No | 66 | |
| rs324377 | - | No | 66 | |
| Hopo546333 | - | No | 66 | |
| rs324396 | - | No | 66 | |
| rs10278663 | - | No | 66 | |
| rs740347 | - | No | 66 | |
| rs324981 | Ile107Asn | No | 66 | |
| rs727162 | Arg241Ser | No | 66 | |
| rs6972158 | Arg344Gln | No | 66 | |
| SCN5A | G298S | IBS-D | 89 | |
Serotonin has been well studied because of its presence in the intestinal tract and brain. Over 95% of the body’s serotonin is located in intestinal enterochromaffin cells, but this neurotransmitter is best known for its effect on mood. Selective serotonin reuptake inhibitors (SSRIs) are one the most effective anti-depressant therapies available. Furthermore, serotonin receptor agonists and antagonists are known to accelerate and slow down gastrointestinal transit and positive impact IBS symptoms.42, 43 5-HTT LPR--for serotonin transporter linked polymorphic region)—is the best studied functional variant in the psychiatric and IBS field. Eleven candidate gene association studies and a meta-analysis have been published, largely demonstrating no link between this variable number tandem repeat (VNTR) polymorphism and IBS and its subtypes.44 In contrast to the psychiatric literature in which a number of studies suggest gene-environment interactions between this variant and abuse or other stressors, no similar interaction was seen between this variant and environment factors such as abuse, parental psychiatric disease, and gastrointestinal infection.45
In addition to studies of other genetic variants on the serotonin transporter gene (SLC6A4), several studies have examined functional SNPs on the genes encoding serotonin receptors. These genes that encode various receptor subtypes include: HTR2A, HTR3A, HTR3B, HTR3C, and HTR3E. Three studies had positive findings. One study found that the HTR3A −42C>T C/T genotype was more common in IBS-D patients.46 Unfortunately, this finding was not replicated in a second cohort. In contrast, the *76G>A SNP on the HTR3E gene was found to be associated with IBS-D in two separate cohorts.46 This finding is intriguing because the 5-HT3E receptor is expressed in colon, small intestine, and stomach and in vitro studies show that there is increased expression of 5-HT3E subunits and thus may have direct effects on the function of 5-HT3 receptors.47 Fukudo et al. studied the 386A>C polymorphism on the HTR3B gene and found that regions of brain activation differed between AA subjects and AC and CC subjects.48 Our group has recently performed a comprehensive study of over 20 serotonin-related genes in 968 cases and controls and have observed the following associations: 1) 27 SNP associations on 9 genes with IBS with several associations on TDO2, HTR2A, and HTR7; 2) 32 SNPs on 15 genes with IBS-C with multiple associations on HTR4 and HTR7; 3) 26 SNPs on 7 genes with IBS-D with multiple associations on HTR2A; and 4) 32 associations on 9 genes with multiple associations on TPH2, DDC, TDO2, HTR1E, HTR2A, and HTR7.49 Thus, there are several lines of evidence to suggest that genes involved with serotonin processing are important in the pathophysiology of IBS.
Genes encoding intestinal barrier proteins have also been studied because of a subgroup of IBS patients whose symptoms began after an acute gastroenteritis, known as post-infectious IBS (PI-IBS). Villani et al. conducted an extensive study utilizing the Walkerton Health Study (WHS), a population cohort created after contamination of a municipal water supply in Walkerton, Canada that resulted in a large outbreak of acute E. Coli 0157:H7 and C. jejeuni gastroenteritis with over 2300 developing acute gastroenteritis.50 This cohort has undergone clinical assessments as well as intensive surveys to define past and current health and has been followed longitudinally to provide a unique resource to better understand the epidemiological features of PI-IBS. In this study, the investigators selected 79 functional variants in 51 candidate genes that either encoded proteins involved in intestinal epithelial barrier function, innate immune response, the serotonin pathways, as well as other genes postulated by others to be potentially associated with IBS, and tested these variants in 228 cases with PI-IBS and 581 controls. Four of the SNPs were associated with PI-IBS. Two variants were on the TLR9 gene encoding the toll-like receptor 9; one was on CDH1 gene encoding the tight junction protein, cadherin; and one variant was on IL6 which encodes the cytokine, interleukin-6. Fine-mapping was then performed with selection of additional SNPs in these 3 genomic regions including functional SNPs associated with inflammatory conditions, non-synonymous SNPs reported in dbSNP with minor allele frequencies >1%, SNPs disrupting transcription-factor, microRNA or enhancer binding sites, SNPs located in regulator regions, and tagging SNPs to cover the remaining regions of the three genes (defined as 10kb upstream and 10kb downstream of coding sequence). When multiple logistic regression to identify risk factors for PI-IBS was performed again, now including the 3 candidate polymorphisms from TLR9, IL6, and CDHI, the original risk factors (e.g. younger age, female gender, etc.) as well as these three genetic variants were found to be independent risk factors for development of PI-IBS. These findings appear to be specific to PI-IBS, because when the genetic variants were re-tested in a separate cohort of non-PI-IBS cases and controls, these findings were not reproduced.36
In addition to the genes cited above, other genes in the inflammation pathway have been studied because of reports of cytokine abnormalities in patients with IBS. IL10 is the gene that has been studied by three groups. Gonssalkorale et al. tested one SNP −1082G>A and found that IBS patients were less likely to have the high producer genotype.51 Van der Veek and colleagues subsequently observed no association between the same SNP and IBS, as well as no association between the −819C>T SNP and IBS.52 Villani et al. noted an association between two SNPs on IL10 with non-infectious IBS (−592A>C, −1082G>A).36 With further fine-mapping, 16 SNPs on the IL10 gene were genotyped and five additional SNPs were found to be associated with non-infectious IBS. Three of the five new associations could be explained by linkage disequilibrium with the two original variants, but two variants were still associated with non-infectious IBS suggesting that genetic variation in the IL10 promoter region may lead to IBS susceptibility. Nonetheless, the inconsistency in findings between study populations, particularly over the −1082G>A polymorphism, suggests that additional confirmatory work on the IL10 gene and cytokine is needed.
Because depression, anxiety, and somatoform disorders have been reported to occur in up to 94% of IBS patients,53 our group was interested in evaluating the relationship between functional SNPs that have been consistently associated with psychiatric disorders and clinical traits such as mood disorders and pain sensitivity. To this end, ten SNPs on eight genes (FKBP5, COMT, NPY, BDNF, ANKK1, DRD2, OPRM1, FAAH) were evaluated.41 None of the ten tested SNPs were observed to be associated with IBS overall. However, the COMT Val158Met variant was associated with IBS-C (OR=1.81, 95% CI: 1.01–3.25), the OPRM1 118A>G variant was associated with IBS-M (OR=1.57, 95% CI: 1.06–2.30) and IBS-D in females (OR=1.69, 95% CI: 1.00–2.85), and the BDNF Val166Met SNP was associated in IBS individuals with a psychiatric disorder. These findings are of interest as the COMT variant has been linked to anxiety, obsessive-compulsive disorder, panic disorder, and cognitive performance54–56 while the OPRM1 variant has been linked to pain sensitivity, opioid dependence, and social sensitivity.57, 58 The BDNF variant has also been linked to emotional reactivity and post-traumatic stress disorder, mood disorders, and schizophrenia.59–61
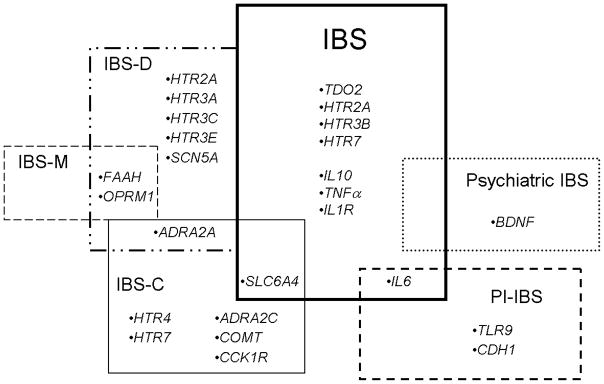
In conclusion, genes HTR2A, HTR3E, IL10, IL6 and other genes are intriguing potential candidate genes for IBS (Figure 4). Furthermore, distinct genes—IL6, CDH1, and TLR9—may represent susceptibility loci for post-infectious IBS, with genetic studies suggesting that there is a different genetic basis for sporadic, non-infectious IBS and post-infectious IBS. But clearly, the role of these genetic variants in IBS development still bears further replication in other patient cohorts, as well as confirmation through additional clinical and laboratory testing.
Figure 4.
Summary of positive gene associations, by IBS type and subtype. Positive gene associations that were shown in Table 1 are summarized by IBS phenotype. Some genetic associations are unique to specific IBS types and subtypes, where others were shown to overlap between IBS subtypes.
Endophenotypes in IBS
Clinical disease can be the result of an aberrant single physiological pathway, but can also be the result of multiple abnormalities in several biological pathways. In the latter case, understanding and discovering the individual pathophysiological mechanisms that result in disease presentation can be difficult, and can be even more difficult if the disease has heterogeneous clinical features that vary from individual to individual. The term, “endophenotype” originated in the psychiatric literature with the idea that an objective, measurable, and ideally reproducible phenotype of a more abstract or complex clinical phenotype would be more closely related to an underlying biological process. Use of endophenotypes in research implies that the complex biological processes causing disease are more easily discoverable if study of the disease can be parsed out into individual, smaller components. Endophenotypes may be a biomarker, or may be referred to as an intermediate phenotype or subclinical trait. In genetics research, endophenotypes should generally have the following characteristics: be associated with illness in the general population, be heritable, co-segregate with illness in families, manifest in an individual irrespective of whether the disease is active or inactive, and be found in affected and unaffected family members of an individual with disease at a higher rate than in the general population.62 Because of the lack of objective findings and the clinical heterogeneity of functional gastrointestinal disorders such as IBS, use of an endophenotype strategy in gene discovery for IBS has appeal. Although none have yet been validated as endophenotypes using the Gottesman criteria, at least two testing modalities have been proposed to be potential endophenotypes for IBS: gastrointestinal transit as well as brain imaging with visceral stimulation (Table 2).
Table 2.
List of genes evaluated in gene-endophenotype studies
| Endophenotype: Gastrointestinal transit | Endophenotype: Brain imaging |
|---|---|
Serotonin genes
|
Serotonin
|
Gastrointestinal transit as an intermediate phenotype has been studied extensively by Camilleri and colleagues. This group first postulated that because adrenergic and serotonergic mechanisms modulate gastrointestinal motility, an exploratory study was performed examining these four functional variants-- ADRA2A −1291C>G, ADRC2A Del 332–325, GNB3 825C>T, and 5-HTT LPR-- in 251 patients with FGIDs and healthy volunteers.63 No direct correlation was seen between these 4 genotypes and upper and lower gastrointestinal emptying and transit time, but when interactions between phenotype (e.g. IBS subtype) and genotype were evaluated, several specific interactions were observed. The GNβ3 CC genotype appeared to be predictive of faster gastric emptying among IBS-D patients, the ADRA2A CC genotype was predictive of faster gastric emptying among IBS-M patients, and the ADRA2C heterozygous genotype was associated with slower gastric emptying among IBS-M patient but faster gastric emptying among IBS-C patients. In a subsequent study, in addition to reporting associations with IBS phenotypes (also summarized in Table 1), this group studied the role of the same four genetic markers and rectal compliance and pain sensation and found an association between 5-HTTLPR LS/SS and increased pain sensation and rectal compliance.64 Besides these genetic markers, this group evaluated the role of FAAH 385C>A in a group of 482 individuals with FGIDs (predominantly IBS patients) with the underlying hypothesis that because cannabinoid receptors are located on brain stem, gastric, and colonic neurons, functional variants that affect endocannabinoid metabolism could affect gastrointestinal motility and/or sensation, and therefore potentially explain IBS-type symptoms.65 They did find that the A allele was associated with IBS-D and IBS-M (p<0.05) as well as accelerated transit in patients with IBS-D. In another study, Camilleri et al. proposed that 20 SNPs on the NPSR1 gene, which encodes a receptor for neuropeptide S which is localized in gut enteroendocrine cells, could potentially play a role in modulated gastrointestinal motor function and found that four SNPs were associated with colon transit time, with significance that persisted after false detection rate correction for multiple tests.66
The above studies suggest that the genetic mechanisms for gastrointestinal transit may be specific to IBS subtype. In addition to the previous studies evaluating a mixed group of IBS patients with subanalysis of IBS subtypes, other studies have looked at isolated subgroups. One study of IBS-C patients and seven variants in genes encoding cholecystokinin (CCK), CCKA receptors (CCKAR), and CCKB receptors (CCKBR) found that the intronic 779C>T polymorphism appeared to be associated with delayed gastric emptying in IBS-C patients, but not associated with small intestinal or colonic transit.67 A more recent study evaluated variations in genes controlling bile acid absorption, processing, and metabolism with colon transit in a clinical trial evaluating the effects of sodium chenodeoxycholate (CDC) on colon transit and symptoms in patients with IBS-C.68 Sixteen non-synonymous SNPs on 7 genes (NROB2, SC10A2, KLB, FGFR4, OSTα, OSTβ, CYP7A1) were genotyped and analyzed to determine whether they predicted colon transit time in 36 patient with IBS-C. One SNP on the FGFR4 gene was found to be associated with filling of the ascending colon (p=0.015) and borderline associations between a SNP on the CYP7A1 gene and colon transit (p=0.08). The same investigator has also reported an association between rs17618244 on the KLB gene and colon transit time in healthy volunteers as well as those with all IBS subtypes.69
Fewer studies have been published regarding the role that genetic variants play in modulating brain activation and processing of visceral stimuli. The only paper published as a full manuscript is a study from Fukudo et al. evaluating the role of the 5-HTT LPR polymorphism in brain processing of rectal stimuli by rectal barostat.70 The investigators postulated that since serotonin neurons in the brain are important in mood as well as anxiety-related behaviors, that 5-HTT LPR could potentially predict activation of prefrontal-limbic circuits. Specifically, they hypothesized that the s allele (resulting in lower transcription of the serotonin transporter, leading to greater synaptic quantities of serotonin) could lead to greater activation of the anterior cingulate cortex (ACC) and amygdala. The investigators found that individuals (not IBS patients) with the s/s genotype were more likely to activate left ACC, right parahippocampal gyrus, and left orbitofrontal cortex (OFC) on positron emission tomography (PET) imaging than those bearing the l allele. Since these regions are important in central emotional processing, these findings suggest that this variant may also be relevant in brain activation after colorectal distension in patients with IBS. Labus, et al. reported similar findings—that the s/s genotype appears to result in altered activation of brain regions regulating emotional arousal.71 In contrast to l carriers who exhibited the expected negative connectivity between the sACC and the amygdala, s/s carriers lacked the feedback inhibition of the amygdala, which they postulated could result in magnified perception of visceral sensation and thus, represent a “vulnerability factor” for IBS. The findings from these two studies suggest the 5-HTT LPR, although not a risk factor for the clinical IBS phenotype, could represent a susceptibility factor for components of IBS such as central processing of visceral events.
In addition to studies of the much-studied 5-HTT LPR variant, two additional variants have been studied in the context of brain processing. Fukudo, et al. subsequently genotyped the rs1176744 polymorphism on the gene encoding the 5-HT3B receptor (HTR3B) using similar methodology as the previous study and found that those with the AC or CC genotype demonstrated more activation of the right amygdala, left insula, and left OFC—regions controlling negative emotion-- than AA subjects.48 Truong, et al. used a different strategy to evaluate amygdala-based responses by measuring acoustic startle response by threats (fear potentiation [FP]) and prepulse inhibition [PPI] in relation to the COMT gene, which encodes the catechol O-methyltransferase enzyme, and important regulator of synaptic dopamine levels.72 This group found that there was an interaction between genotype, diagnosis (IBS, fibromyalgia, controls) and threat condition where Met carriers in patients showed enhanced fear potentiation when faced with imminent threat. The authors speculate that reduced synaptic dopamine in these individuals impair downregulation of amygdala-based startle responses that may lead to hypervigilance to symptoms.
The Future of Genetic Studies in IBS
The field of genetics is rapidly evolving, with technological advances making it increasingly possible to identify common and rare variants on genomic DNA. Current approaches to gene discovery range considerably from the established candidate gene association studies, to the newer genome-wide association studies (GWAS), to newest techniques that involve deep sequencing to identify less common variants. Although the candidate genes have been studied in IBS, the newer methods have not yet been applied to the study of IBS for a number of reasons and limitations. Ultimately, the greatest constraint on IBS gene discovery is the IBS phenotype. IBS is a heterogeneous, unstable disorder, without a well-defined molecular pathway or established biomarkers whose development is clearly multifactorial. Twin studies suggest that there are genes of modest effect that in part explain IBS, but environmental exposures are clearly important in determining the clinical manifestations of IBS. The importance of these environmental factors—such as early or childhood trauma or stress—cannot be ignored as they may be crucial components to IBS vulnerability.
Though twin studies suggest a modest contribution of genetics to IBS and other functional disorders, such studies are limited by the same concerns regarding heterogeneous or even vague disease definitions. Although there does not appear to be a single or dominant Mendelian gene causing IBS, the fact that IBS clusters in families and through multiple generations suggests that there is likely a heritable component to IBS. Nonetheless, future genetic studies will need to study well-characterized, homogeneous groups, or subgroups, of IBS patients. Based on the candidate gene studies, it appears that there may be a different molecular basis for constipation versus diarrhea, sporadic IBS versus post-infectious IBS, and perhaps even for anxiety-associated IBS versus non-anxiety-associated IBS. In addition to studying IBS clinical phenotypes, validated endophenotypes—whether gastrointestinal transit, brain imaging, or another modality such as visceral sensation measures—will provide a complementary approach for successful gene identification. Because environment clearly impacts IBS development and symptom manifestation, these gene-environment interactions should be included in analyses of future genetic studies of IBS. Similarly, studies of epigenetics—environment-induced DNA methylation or histone modification that are heritable but do not result in DNA sequence changes—have not formally been performed but perhaps should be evaluated in future IBS genetic studies. Although genetic studies of IBS need not be family-based and can certainly be performed utilizing information collected from unrelated cases and controls, families represent a unique opportunity to discern the genetic and environmental contributors to the development of IBS. Much work still remains to be done in the field of IBS genetics, presenting an exciting opportunity to make great strides in our understanding in the underlying mechanisms of IBS.
Acknowledgments
This work was supported by National Institutes of Health grant, DK76797
Footnotes
The author has nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.El-Serag HB, Olden K, Bjorkman D. Health-related quality of life among persons with irritable bowel syndrome: a systematic review. Aliment Pharmacol Ther. 2002;16(6):1171–1185. doi: 10.1046/j.1365-2036.2002.01290.x. [DOI] [PubMed] [Google Scholar]
- 2.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8(6):458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 3.Pace F, Zuin G, Di Giacomo S, et al. Family history of irritable bowel syndrome is the major determinant of persistent abdominal complaints in young adults with a history of pediatric recurrent abdominal pain. World J Gastroenterol. 2006;12(24):3874–3877. doi: 10.3748/wjg.v12.i24.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whorwell PJ, McCallum M, Creed FH, et al. Non-colonic features of irritable bowel syndrome. Gut. 1986;27(1):37–40. doi: 10.1136/gut.27.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellentani S, Baldoni P, Petrella S, et al. A simple score for the identification of patients at high risk of organic diseases of the colon in the family doctor consulting room. The Local IBS Study Group. Fam Pract. 1990;7(4):307–312. doi: 10.1093/fampra/7.4.307. [DOI] [PubMed] [Google Scholar]
- 6.Saito YA, Zimmerman JM, Harmsen WS, et al. Irritable bowel syndrome aggregates strongly in families: a family-based case-control study. Neurogastroenterol Motil. 2008;20:790–797. doi: 10.1111/j.1365-2982.2007.1077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito YA, Petersen GM, Larson JJ, et al. Familial aggregation of irritable bowel syndrome: a family case-control study. Am J Gastroenterol. 2010;105(4):833–841. doi: 10.1038/ajg.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buonavolonta R, Coccorullo P, Turco R, et al. Familial aggregation in children affected by functional gastrointestinal disorders. J Pediatr Gastroenterol Nutr. 2010;50(5):500–505. doi: 10.1097/MPG.0b013e3181b182ef. [DOI] [PubMed] [Google Scholar]
- 9.Saito-Loftus Y, Larson J, Atkinson E, et al. Irritable bowel syndrome (IBS) is not a major gene, Mendelian disorder. Am J Gastroenterol. 2008;103(Suppl S):S472. [Google Scholar]
- 10.Saito Y, Zimmerman J, Almazar-Elder A, et al. Clinical characteristics of familial irritable bowel syndrome (IBS) differ from sporadic IBS. Gastroenterology. 2008;134(4 Suppl 1):A-30. [Google Scholar]
- 11.Kanazawa M, Endo Y, Whitehead WE, et al. Patients and nonconsulters with irritable bowel syndrome reporting a parental history of bowel problems have more impaired psychological distress. Dig Dis Sci. 2004;49(6):1046–1053. doi: 10.1023/b:ddas.0000034570.52305.10. [DOI] [PubMed] [Google Scholar]
- 12.Morris-Yates A, Talley NJ, Boyce PM, et al. Evidence of a genetic contribution to functional bowel disorder. Am J Gastroenterol. 1998;93(8):1311–1317. doi: 10.1111/j.1572-0241.1998.440_j.x. [DOI] [PubMed] [Google Scholar]
- 13.Levy RL, Jones KR, Whitehead WE, et al. Irritable bowel syndrome in twins: heredity and social learning both contribute to etiology. Gastroenterology. 2001;121(4):799–804. doi: 10.1053/gast.2001.27995. [DOI] [PubMed] [Google Scholar]
- 14.Lembo A, Zaman M, Jones M, et al. Influence of genetics on irritable bowel syndrome, gastro-oesophageal reflux and dyspepsia: a twin study. Aliment Pharmacol Ther. 2007;25(11):1343–1350. doi: 10.1111/j.1365-2036.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- 15.Mohammed I, Cherkas LF, Riley SA, et al. Genetic influences in irritable bowel syndrome: a twin study. Am J Gastroenterol. 2005;100(6):1340–1344. doi: 10.1111/j.1572-0241.2005.41700.x. [DOI] [PubMed] [Google Scholar]
- 16.Svedberg P, Johansson S, Wallander MA, et al. Extra-intestinal manifestations associated with irritable bowel syndrome: a twin study. Aliment Pharmacol Ther. 2002;16(5):975–983. doi: 10.1046/j.1365-2036.2002.01254.x. [DOI] [PubMed] [Google Scholar]
- 17.Anand K, Runeson B, Jacobson B. Gastric suction at birth associated with long-term risk for functional intestinal disorders in later life. J Pediatr. 2004;144(4):449–454. doi: 10.1016/j.jpeds.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Barreau F, Ferrier L, Fioramonti J, et al. New insights in the etiology and pathophysiology of irritable bowel syndrome: contribution of neonatal stress models. Pediatric Research. 2007;62(3):240–245. doi: 10.1203/PDR.0b013e3180db2949. [DOI] [PubMed] [Google Scholar]
- 19.Howell S, Talley N, Quine S, et al. The irritable bowel syndrome has origins in the childhood socioeconomic environment. Am J Gastroenterol. 2004;99(8):1572–1578. doi: 10.1111/j.1572-0241.2004.40188.x. [DOI] [PubMed] [Google Scholar]
- 20.Mendall M, Kumar D. Antibiotic use, childhood affluence and irritable bowel syndrome (IBS) Eur J Gastroenterol Hepatol. 1998;10(1):59–62. doi: 10.1097/00042737-199801000-00011. [DOI] [PubMed] [Google Scholar]
- 21.Gray GC, Reed RJ, Kaiser KS, et al. Self-reported symptoms and medical conditions among 11,868 Gulf War-era veterans: the Seabee Health Study. Am J Epidemiol. 2002;155(11):1033–1044. doi: 10.1093/aje/155.11.1033. [DOI] [PubMed] [Google Scholar]
- 22.Gasbarrini A, Lauritano EC, Garcovich M, et al. New insights into the pathophysiology of IBS: intestinal microflora, gas production and gut motility. Eur Rev Med Pharmacol Sci. 2008;12 (Suppl 1):111–117. [PubMed] [Google Scholar]
- 23.Drossman DA. Irritable bowel syndrome and sexual/physical abuse history. Eur J Gastroenterol Hepatol. 1997;9(4):327–330. doi: 10.1097/00042737-199704000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Walker EA, Katon WJ, Roy-Byrne PP, et al. Histories of sexual victimization in patients with irritable bowel syndrome or inflammatory bowel disease. Am J Psychiatry. 1993;150(10):1502–1506. doi: 10.1176/ajp.150.10.1502. [DOI] [PubMed] [Google Scholar]
- 25.Talley NJ, Fett SL, Zinsmeister AR, et al. Gastrointestinal tract symptoms and self-reported abuse: a population-based study. Gastroenterology. 1994;107(4):1040–1049. doi: 10.1016/0016-5085(94)90228-3. [DOI] [PubMed] [Google Scholar]
- 26.Talley NJ, Fett SL, Zinsmeister AR. Self-reported abuse and gastrointestinal disease in outpatients: association with irritable bowel-type symptoms. Am J Gastroenterol. 1995;90(3):366–371. [PubMed] [Google Scholar]
- 27.Heitkemper M, Jarrett M, Taylor P, et al. Effect of sexual and physical abuse on symptom experiences in women with irritable bowel syndrome. Nursing Res. 2001;50(1):15–23. doi: 10.1097/00006199-200101000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Whitehead WE, Crowell MD, Davidoff AL, et al. Pain from rectal distension in women with irritable bowel syndrome: relationship to sexual abuse. Dig Dis Sci. 1997;42(4):796–804. doi: 10.1023/a:1018820315549. [DOI] [PubMed] [Google Scholar]
- 29.Ringel Y, Whitehead WE, Toner BB, et al. Sexual and physical abuse are not associated with rectal hypersensitivity in patients with irritable bowel syndrome. Gut. 2004;53(6):838–842. doi: 10.1136/gut.2003.021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talley NJ, Boyce PM, Jones M. Is the association between irritable bowel syndrome and abuse explained by neuroticism? A population based study. Gut. 1998;42(1):47–53. doi: 10.1136/gut.42.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dube SR, Felitti VJ, Dong M, et al. The impact of adverse childhood experiences on health problems: evidence from four birth cohorts dating back to 1900. Preventive Medicine. 2003;37(3):268–277. doi: 10.1016/s0091-7435(03)00123-3. [DOI] [PubMed] [Google Scholar]
- 32.Videlock EJ, Adeyemo M, Licudine A, et al. Childhood trauma is associated with hypothalamic-pituitary-adrenal axis responsiveness in irritable bowel syndrome. Gastroenterology. 2009;137(6):1954–1962. doi: 10.1053/j.gastro.2009.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitehead W, Winget C, Fedoravicius A, et al. Learned illness behavior in patients with irritable bowel syndrome and peptic ulcer. Dig Dis Sci. 1982;27(3):202–208. doi: 10.1007/BF01296915. [DOI] [PubMed] [Google Scholar]
- 34.Lowman B, Drossman D, Cramer E, et al. Recollection of childhood events in adults with irritable bowel syndrome. J Clin Gastroenterol. 1987;9(3):324–330. doi: 10.1097/00004836-198706000-00017. [DOI] [PubMed] [Google Scholar]
- 35.Levy R, Whitehead W, Walker L, et al. Increased somatic complaints and health-care utilization in children: effects of parent IBS status and parent response to gastrointestinal symptoms. Am J Gastroenterol. 2004;99(12):2442–2451. doi: 10.1111/j.1572-0241.2004.40478.x. [DOI] [PubMed] [Google Scholar]
- 36.Villani A, Saito Y, Lemire M, et al. Validation of genetic risk factors for post-infectious irritable bowel syndrome (IBS) in patients with sporadic IBS. Gastroenterology. 2009;136(5 Suppl 1):289. [Google Scholar]
- 37.Chang J, Locke G, Talley N, et al. Comparison of lactase variant MCM6–13910C>T testing and self-report of dairy sensitivity in patients with irritable bowel syndrome. Am J Gastroenterol. 2010;105(Suppl 1):S499. doi: 10.1097/MCG.0000000000001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan G, Jenkins PL, Blewett AE. Irritable bowel syndrome and family history of psychiatric disorder: a preliminary study. Gen Hosp Psychiatry. 1995;17(1):43–46. doi: 10.1016/0163-8343(94)00064-k. [DOI] [PubMed] [Google Scholar]
- 39.Woodman CL, Breen K, Noyes R, Jr, et al. The relationship between irritable bowel syndrome and psychiatric illness. A family study. Psychosomatics. 1998;39(1):45–54. doi: 10.1016/S0033-3182(98)71380-5. [DOI] [PubMed] [Google Scholar]
- 40.Whitehead WE, Bosmajian L, Zonderman AB, et al. Symptoms of psychologic distress associated with irritable bowel syndrome. Comparison of community and medical clinic samples. Gastroenterology. 1988;95(3):709–714. doi: 10.1016/s0016-5085(88)80018-0. [DOI] [PubMed] [Google Scholar]
- 41.Saito Y, Larson J, Atkinson E, et al. A candidate gene association study of functional “psychiatric” polymorphisms in irritable bowel syndrome. Gastroenterology. 2010;138(5, Suppl 1):348. [Google Scholar]
- 42.Agonists Ford AC, Brandt LJ, Young C, et al. Efficacy of 5-HT3 Antagonists and 5-HT4 in Irritable Bowel Syndrome: Systematic Review and Meta-analysis. Am J Gastroenterol. 2009;104:1831–43. doi: 10.1038/ajg.2009.223. [DOI] [PubMed] [Google Scholar]
- 43.Tack J, Muller-Lissner S, Bytzer P, et al. A randomised controlled trial assessing the efficacy and safety of repeated tegaserod therapy in women with irritable bowel syndrome with constipation. Gut. 2005;54(12):1707–1713. doi: 10.1136/gut.2005.070789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Kerkhoven LA, Laheij RJ, Jansen JB. Meta-analysis: a functional polymorphism in the gene encoding for activity of the serotonin transporter protein is not associated with the irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26(7):979–986. doi: 10.1111/j.1365-2036.2007.03453.x. [DOI] [PubMed] [Google Scholar]
- 45.Saito Y, Larson J, Atkinson E, et al. The role of 5-HTT LPR and GNbeta3 825C>T polymorphisms and gene environment interactions in irritable bowel syndrome. Gastroenterology. 2009;136(5 Suppl 1):289. doi: 10.1007/s10620-012-2319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kapeller J, Houghton L, Monnikes H, et al. First evidence for an association of functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17(19):2967–2977. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- 47.Niesler B, Frank B, Kapeller J, et al. Cloning, physical mapping and expression analysis of the human 5-HT3 serotonin receptor-like genes HTR3C, HTR3D and HTR3E. Gene. 2003;310:101–111. doi: 10.1016/s0378-1119(03)00503-1. [DOI] [PubMed] [Google Scholar]
- 48.Fukudo S, Ozaki N, Watanabe S, et al. Impact of serotonin-3 receptor gene polymorphism on brain activation by rectal distention in human. Gastroenterology. 2009;136(5 Suppl 1):A-170. [Google Scholar]
- 49.Saito Y, Larson J, Atkinson E, et al. A serotonin-pathway candidate gene association study of irritable bowel syndrome (IBS) Gastrenterology. 2010;138(5, Suppl 1):348. [Google Scholar]
- 50.Villani A-C, Lemire M, Thabane M, et al. Genetic risk factors for post-infectious irritable bowel syndrome following a waterborne outbreak of gastroenteritis. Gastroenterology. 2010;138(4):1502–1513. doi: 10.1053/j.gastro.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 51.Gonsalkorale WM, Perrey C, Pravica V, et al. Interleukin 10 genotypes in irritable bowel syndrome: evidence for an inflammatory component? Gut. 2003;52(1):91–93. doi: 10.1136/gut.52.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Veek PP, van den Berg M, de Kroon YE, et al. Role of tumor necrosis factor-alpha and interleukin-10 gene polymorphisms in irritable bowel syndrome. Am J Gastroenterol. 2005;100(11):2510–2516. doi: 10.1111/j.1572-0241.2005.00257.x. [DOI] [PubMed] [Google Scholar]
- 53.Whitehead WE, Palsson O, Jones KR. Systematic review of the comorbidity of irritable bowel syndrome with other disorders: what are the causes and implications? Gastroenterology. 2002;122(4):1140–1156. doi: 10.1053/gast.2002.32392. [DOI] [PubMed] [Google Scholar]
- 54.Dickinson D, Elvevag B. Genes, cognition and brain through a COMT lens. Neuroscience. 2009;164(1):72–87. doi: 10.1016/j.neuroscience.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Domschke K, Deckert J, O’Donovan MC, et al. Meta-analysis of COMT val158met in panic disorder: ethnic heterogeneity and gender specificity. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(5):667–673. doi: 10.1002/ajmg.b.30494. [DOI] [PubMed] [Google Scholar]
- 56.Hosak L. Role of the COMT gene Val158Met polymorphism in mental disorders: a review. Eur Psychiatry. 2007;22(5):276–281. doi: 10.1016/j.eurpsy.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Way BM, Lieberman MD. Is there a genetic contribution to cultural differences? Collectivism, individualism and genetic markers of social sensitivity. Soc Cogn Affect Neurosci. 2010;5(2–3):203–211. doi: 10.1093/scan/nsq059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan EC, Lim EC, Teo YY, et al. Ethnicity and OPRM variant independently predict pain perception and patient-controlled analgesia usage for post-operative pain. Mol Pain. 2009;5:32. doi: 10.1186/1744-8069-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frielingsdorf H, Bath KG, Soliman F, et al. Variant brain-derived neurotrophic factor Val66Met endophenotypes: implications for posttraumatic stress disorder. Ann N Y Acad Sci. 1208:150–157. doi: 10.1111/j.1749-6632.2010.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cirulli F, Berry A, Bonsignore LT, et al. Early life influences on emotional reactivity: evidence that social enrichment has greater effects than handling on anxiety-like behaviors, neuroendocrine responses to stress and central BDNF levels. Neurosci Biobehav Rev. 2010;34(6):808–820. doi: 10.1016/j.neubiorev.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 61.Fan J, Sklar P. Genetics of bipolar disorder: focus on BDNF Val66Met polymorphism. Novartis Found Symp. 2008;289:60–72. doi: 10.1002/9780470751251.ch5. discussion 72–63, 87–93. [DOI] [PubMed] [Google Scholar]
- 62.Gottesman, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 63.Grudell AB, Camilleri M, Carlson P, et al. An exploratory study of the association of adrenergic and serotonergic genotype and gastrointestinal motor functions. Neurogastroenterol Motil. 2007 doi: 10.1111/j.1365-2982.2007.01026.x. [DOI] [PubMed] [Google Scholar]
- 64.Camilleri M, Busciglio I, Carlson P, et al. Candidate genes and sensory functions in health and irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2008;295(2):G219–225. doi: 10.1152/ajpgi.90202.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camilleri M, Carlson P, McKinzie S, et al. Genetic variation in endocannabinoid metabolism, gastrointestinal motility, and sensation. Am J Physiol Gastrointest Liver Physiol. 2008;294(1):G13–19. doi: 10.1152/ajpgi.00371.2007. [DOI] [PubMed] [Google Scholar]
- 66.Camilleri M, Carlson P, Zinsmeister AR, et al. Neuropeptide S receptor induces neuropeptide expression and associates with intermediate phenotypes of functional gastrointestinal disorders. Gastroenterology. 2010;138(1):98–107. e104. doi: 10.1053/j.gastro.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cremonini F, Camilleri M, McKinzie S, et al. Effect of CCK-1 antagonist, dexloxiglumide, in female patients with irritable bowel syndrome: a pharmacodynamic and pharmacogenomic study. Am J Gastroenterol. 2005;100(3):652–663. doi: 10.1111/j.1572-0241.2005.41081.x. [DOI] [PubMed] [Google Scholar]
- 68.Rao AS, Wong BS, Camilleri M, et al. Chenodeoxycholate in females with irritable bowel syndrome-constipation: a pharmacodynamic and pharmacogenetic analysis. Gastroenterology. 2010;139(5):1549–1558. 1558, e1541. doi: 10.1053/j.gastro.2010.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong B, Camilleri M, Carlson P, et al. Klotho-beta gene polymorphism is associated with colonic transit in health and lower functional gastrointestinal disorders. Gastroenterology. 2010;138(5 Suppl 1):S-624. [Google Scholar]
- 70.Fukudo S, Kanazawa M, Mizuno T, et al. Impact of serotonin transporter gene polymorphism on brain activation by colorectal distention. NeuroImage. 2009;47(3):946–951. doi: 10.1016/j.neuroimage.2009.04.083. [DOI] [PubMed] [Google Scholar]
- 71.Labus JS, Mayer EA, Hamaguchi T, et al. 5-HTTLPR gene polymorphism modulates activity and connectivity within an emotional arousal network of healthy control subjects during visceral pain. Gastroenterology. 2008;134(4, Suppl 1):A121. doi: 10.1371/journal.pone.0123183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Truong T, Kilpatrick L, Naliboff B, et al. COMT genetic polymorphism is associated with alterations in attentional processing in patients with IBS and other functional pain syndromes. Gastrenterology. 2009;136(Suppl 1):A-74. [Google Scholar]
- 73.Pata C, Erdal ME, Derici E, et al. Serotonin transporter gene polymorphism in irritable bowel syndrome. Am J Gastroenterol. 2002;97(7):1780–1784. doi: 10.1111/j.1572-0241.2002.05841.x. [DOI] [PubMed] [Google Scholar]
- 74.Niesler B, Kapeller J, Fell C, et al. 5-HTTLPR and STin2 polymorphisms in the serotonin transporter gene and irritable bowel syndrome: effect of bowel habit and sex. Eur J Gastroenterol Hepatol. 2009 June 25; doi: 10.1097/MEG.0b013e32832e9d6b. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 75.Kohen R, Jarrett M, Cain K, et al. The serotonin transporter polymorphism rs25531 is associated with irritable bowel syndrome. Dig Dis Sci. 2009 Jan 1; doi: 10.1007/s10620-008-0666-3. (Epub online early) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pata C, Erdal E, Yazc K, et al. Association of the -1438 G/A and 102 T/C Polymorphism of the 5-HT2A Receptor Gene with Irritable Bowel Syndrome 5-HT2A Gene Polymorphism in Irritable Bowel Syndrome. J Clin Gastroenterol. 2004;38(7):561–566. doi: 10.1097/00004836-200408000-00005. [DOI] [PubMed] [Google Scholar]
- 77.Kapeller J, Houghton L, Walstab J, et al. A coding variant in the serotonin receptor 3C subunit is associated with diarrhea-predominant irritable bowel syndrome. Gastroenterology. 2009;136(5 Suppl 1):A-155–156. [Google Scholar]
- 78.Barkhordari E, Rezaei N, Mahmoudi M, et al. T-helper 1, T-helper 2, and T-regulatory cytokines gene polymorphisms in irritable bowel syndrome. Inflammation. 2010;33(5):281–286. doi: 10.1007/s10753-010-9183-6. [DOI] [PubMed] [Google Scholar]
- 79.Barkhordari E, Rezaei N, Ansaripour B, et al. Proinflammatory cytokine gene polymorphisms in irritable bowel syndrome. J Clin Immunol. 2010;30(1):74–79. doi: 10.1007/s10875-009-9342-4. [DOI] [PubMed] [Google Scholar]
- 80.Santhosh S, Dutta A, Samuel P, et al. Cytokine gene polymorphisms in irritable bowel syndrome in Indian population--a pilot case control study. Trop Gastroenterol. 2010;31(1):30–33. [PubMed] [Google Scholar]
- 81.Kim HJ, Camilleri M, Carlson PJ, et al. Association of distinct alpha(2) adrenoceptor and serotonin transporter polymorphisms with constipation and somatic symptoms in functional gastrointestinal disorders. Gut. 2004;53(6):829–837. doi: 10.1136/gut.2003.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sikander A, Rana SV, Sharma SK, et al. Association of alpha 2A adrenergic receptor gene (ADRa2A) polymorphism with irritable bowel syndrome, microscopic and ulcerative colitis. Clinica Chimica Acta. 2010;411(1–2):59–63. doi: 10.1016/j.cca.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 83.Park S, Rew J, Lee S, et al. Association of CCK(1) receptor gene polymorphisms and irritable bowel syndrome in Korean. J Neurogastroenterol Motil. 2010;16(1):71–76. doi: 10.5056/jnm.2010.16.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Colucci R, Ghisu N, Bellini M, et al. Analysis of cholecystokinin type 1 receptor gene polymorphisms in patients with irritable bowel syndrome. Gastroenterology. 2009;136(5 Suppl 1):A-225. [Google Scholar]
- 85.Park J, Choi M, Cho Y, et al. Cannabinoid Receptor 1 gene polymorphism and irritable bowel syndrome in the Korean population: a hypothesis-generating study. J Clin Gastroenterol. 2010 doi: 10.1097/MCG.0b013e3181dd1573. [DOI] [PubMed] [Google Scholar]
- 86.Holtmann G, Siffert W, Haag S, et al. G-protein beta 3 subunit 825 CC genotype is associated with unexplained (functional) dyspepsia. Gastroenterology. 2004;126(4):971–979. doi: 10.1053/j.gastro.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 87.Andresen V, Camilleri M, Kim HJ, et al. Is there an association between GNbeta3-C825T genotype and lower functional gastrointestinal disorders? Gastroenterology. 2006;130(7):1985–1994. doi: 10.1053/j.gastro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 88.Saito YA, Locke GR, 3rd, Zimmerman JM, et al. A genetic association study of 5-HTT LPR and GNbeta3 C825T polymorphisms with irritable bowel syndrome. Neurogastroenterol Motil. 2007;19(6):465–470. doi: 10.1111/j.1365-2982.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- 89.Saito YA, Strege PR, Tester DJ, et al. Sodium channel mutation in irritable bowel syndrome: evidence for an ion channelopathy. Am J Physiol Gastrointest Liver Physiol. 2009;296(2):G211–218. doi: 10.1152/ajpgi.90571.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]