Abstract
A phenylalanine residue (Phe1519) in the sixth transmembrane segment of domain III (IIIS6) of the cockroach BgNav sodium channel is required for the binding and action of pyrethroids. However, whether or not other residues in IIIS6 participate in the action of pyrethroids remains to be determined. In the present study, we conducted a systematic analysis of 20 residues in IIIS6 of the BgNav channel using alanine-scanning mutagenesis. Our results show that alanine substitutions of four residues, Ile1514, Gly1516, Phe1518 and Asn1522, altered sodium channel sensitivity to pyrethroid insecticides. Whereas the G1516A, F1518A and N1522A substitutions diminished sodium channel sensitivity to all seven pyrethroids examined, including four type I (lacking the α-cyano group at the phenoxybenzyl alcohol) and three type II (containing the α-cyano group) pyrethroids, the I1514A substitution enhanced sodium channel sensitivity to four type I and type II pyrethroids that contain the phenoxybenzyl alcohol only. We also show that alanine/lysine substitutions of Leu1521 and Ser1517 affected the action of BTX (batrachotoxin), but not pyrethroids. In the Kv1.2-based homology model of the open sodium channel, side chains of Ile1514, Phe1518 and Asn1522 are exposed towards helix IIS5 and linker IIS4–IIS5, which contain previously identified pyrethroid-interacting residues, whereas Ser1517 and Leu1521 face the inner pore where the BTX receptor is located. Thus the present study provides further evidence for structural models in which pyrethroids bind to the lipid-exposed interface formed by helices IIIS6, IIS5 and linker helix IIS4–IIS5, whereas BTX binds to the pore-exposed side of the IIIS6 helix.
Keywords: batrachotoxin, cockroach sodium channel, molecular modelling, pyrethroid insecticide, sixth transmembrane segment of domain III (IIIS6), Xenopus oocyte expression system
INTRODUCTION
Voltage-gated sodium channels are essential for the initiation and generation of action potentials in almost all excitable cells [1]. Sodium channel proteins contain four homologous domains (I–IV), each formed by six membrane-spanning segments (S1–S6) connected by intracellular and extracellular loops. The four positively charged S4 segments function as voltage sensors. In response to membrane depolarization, the S4 segments move outwards to initiate activation (i.e. channel opening) [1]. Transmembrane segments S5 and S6 of each domain and the membrane-re-entrant P-loops connecting the S5 and S6 segments that dip into the transmembrane region form the narrow outer pore and ion-selectivity filter [1]. The short linker connecting domains III and IV contains a sequence motif that is critical for fast inactivation of sodium channels [1].
Sodium channels are well-known targets for a variety of neurotoxins including BTX (batrachotoxin) and pyrethroid insecticides [2]. Pyrethroids are derived structurally from pyrethrins, the insecticidal components of pyrethrum extract from Chrysanthemum species [3]. They represent one of the most important and relatively safe classes of insecticides currently on the market for insect pest control. Previous pharmacological and electrophysiological studies show that pyrethroids bind to a unique receptor site on the sodium channel and cause prolonged sodium channel opening by inhibiting deactivation and inactivation [4,5]. However, the molecular basis of the pyrethroid interaction with sodium channels remained elusive until the identification of multiple point mutations in insect sodium channel genes from pyrethroid-resistant field insect populations [6–8]. Understanding how these point mutations reduce insect sodium channel sensitivity to pyrethroids has begun to shed light on the molecular determinants of the pyrethroid receptor site on the sodium channel. It appears that mutations in IIS5, IIIS6 and linker IIS4–IIS5 could drastically reduce or completely abolish insect sodium channel sensitivity to structurally diverse pyrethroids, suggesting that some (or all) of them may be involved in pyrethroid binding [9–12]. Homology modelling based on the crystal structure of the open potassium channel Kv1.2 [13] predicted that the pyrethroid-binding site in the sodium channel is located within a hydrophobic cavity formed by the transmembrane helices IIS5 and IIIS6, as well as the linker IIS4–IIS5 [14].
The involvement of IIIS6 in the binding and action of pyrethroids was first documented by the association of the F1519I mutation in IIIS6 of the southern cattle tick sodium channel with high-level pyrethroid resistance [15]. Interestingly, the F1519I substitution in the cockroach sodium channel completely abolishes sodium channel sensitivity to structurally diverse pyrethroids [11] and reduces the sensitivity of mammalian sodium channels to pyrethroids [16]. Furthermore, an aromatic residue at position 1519 is required for the binding and action of pyrethroids [11]. To determine whether residues adjacent to Phe1519 also have effects on pyrethroid action, either by providing additional contact sites for the pyrethroid molecule or by changing the side-chain conformation of Phe1519, we conducted a systematic alanine-scanning mutagenesis of the 20 residues flanking Phe1519 in IIIS6. This analysis uncovered a cluster of four additional residues, Ile1514, Gly1516, Phe1518 and Asn1522, in IIIS6, which are critical for the action of pyrethroids, and also revealed the involvement of two residues, Ser1517 and Leu1521, in the action of BTX. Homology modelling suggests that residues important for pyrethroid and BTX action are located on opposite sides of IIIS6, supporting the hypothesis that a single transmembrane helix can contribute to the distinct binding sites of two different classes of sodium channel toxins.
EXPERIMENTAL
Site-directed mutagenesis
The cockroach sodium channel BgNav1-1A cDNA (wild-type) [17] was used to generate mutant constructs. Site-directed mutagenesis was performed by PCR using mutant primers and Pfu Turbo DNA polymerase (Stratagene). All mutagenesis results were verified by DNA sequencing.
Expression of BgNav1-1A sodium channels in Xenopus oocyte
Procedures for oocyte preparation and cRNA injection were identical with those described previously [18]. For expression of the cockroach BgNav1-1A sodium channel, BgNav1-1A cRNA was co-injected into oocytes with cRNA of Drosophila melanogaster tipE (1:1 molar ratio) which is known to enhance the expression of insect sodium channels in oocytes [19,20].
Electrophysiological recording and analysis
Methods for electrophysiological recording and data analysis were similar to those described previously [21]. Sodium currents were recorded by using standard two-electrode voltage clamping. The voltage-dependence of activation and fast inactivation was determined using the protocols described previously [18,21]. The data were fitted with a Boltzmann equation to generate V0.5, the midpoint of the activation or inactivation curves, and k, the slope factor.
Pyrethroids were gifts from Klaus Naumann and Ralf Nauen (Bayer AG, Leverkusen, Germany). The method for application of pyrethroids in the recording system was identical with that described by Tan et al. [21]. The effects of pyrethroids were measured 10 min after toxin application. The pyrethroid-induced tail current was recorded during a 100-pulse train of 5-ms depolarization from − 120 to 0 mV with a 5-ms interpulse interval [10]. Percentages of channels modified by pyrethroids were calculated using the equation M = {[Itail/(Eh − ENa)]/[INa/(Et − ENa)]} × 100 [22], where Itail is the maximal tail current amplitude, Eh is the potential to which the membrane is repolarized, ENa is the reversal potential for sodium current determined from the current–voltage curve, INa is the amplitude of the peak current during depolarization before pyrethroid exposure, and Et is the potential of step depolarization. Dose–response curves were fitted to the Hill equation: M = Mmax/{1+(EC50/[pyrethroid])h}, in which [pyrethroid] represents the concentration of pyrethroid, EC50 represents the concentration of pyrethroid that produced the half-maximal effect, h represents the Hill coefficient, and Mmax is the maximal percentage of sodium channels modified.
Computer modelling
To visualize sodium channel residues involved in binding of pyrethroids, we have built a Monte Carlo-minimized Kv1.2-based model of the open sodium channel BgNav1-1 using the ZMM program [23,24] and the methodology described by Tikhonov and Zhorov [25,26]. The sequences of potassium and sodium channels were aligned as proposed previously [14,27].
RESULTS
Effects of alanine substitutions of 20 residues in IIIS6 on sodium channel gating
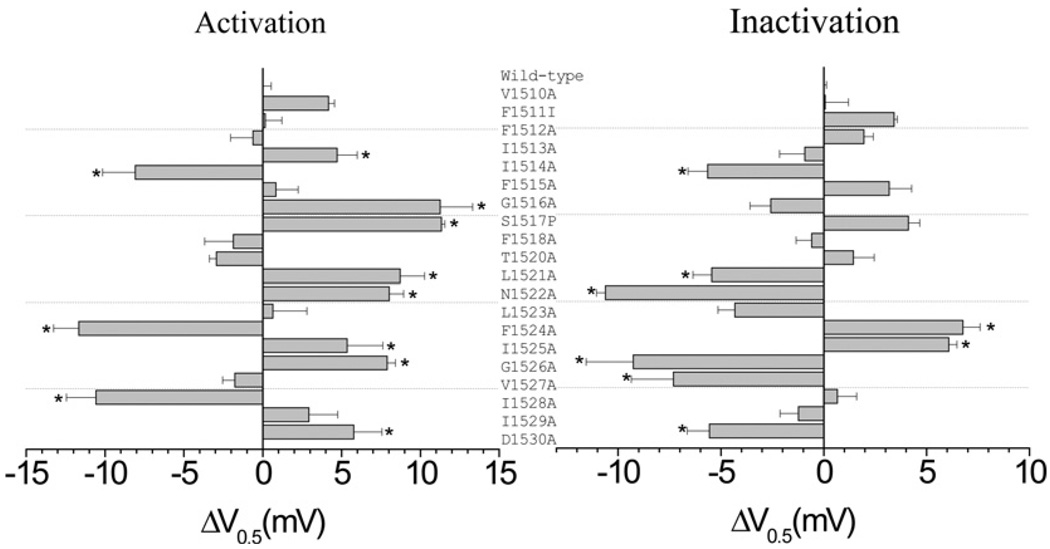
To determine whether residues in IIIS6, besides Phe1519, influence the action of pyrethroids on the sodium channel, we conducted alanine-substitution mutagenesis of residues in IIIS6 from Val1510 to Phe1518 (upstream of Phe1519) and from Thr1520 to Asp1530 (downstream of Phe1519) (Figure 1) in BgNav1-1A (a wild-type variant), which is highly sensitive to pyrethroids. We chose alanine substitution because alanine changes the size and chemical properties of the residues, but has minimal effects on protein secondary structures. The resulting channels were individually expressed in Xenopus oocytes. Sodium currents were recorded between days 3 and 7 after cRNA injection. Most of the mutant channels generated sufficient sodium currents for functional analysis, except for the F1511A and S1517A channels. We therefore made two additional mutant channels, containing F1511I and S1517P respectively. F1511I was chosen because an isoleucine residue is present in the majority of mammalian sodium channel proteins at the corresponding position. We chose S1517P because the same change was found in a cockroach functional full-length cDNA clone. Both F1511I and S1517P channels produced detectable currents. All mutant channels inactivated completely except for the F1524A and I1525A channels that exhibited a persistent current, 10–20%of the peak current.
Figure 1. Effects on channel gating of alanine substitutions of 20 residues flanking Phe1519 in IIIS6 of BgNav1-1A.
Voltages of half-maximal activation or steady-state inactivation (V0.5) of mutant channels were compared with that of the wild-type sodium channel. The histograms show the differences between voltage for the half-maximal activation or inactivation of wild-type and each mutant sodium channel. V0.5 values were obtained from Boltzmann fits, as described in the Experimental section. Asterisks indicate significant differences from the wild-type channel as determined by Student’s t test (P <0.05).
Eight substitutions (V1510A, F1511I, F1512A, F1515A, F1518A, T1520A, L1523A and I1529A) did not alter the voltage-dependence of either activation or inactivation. Twelve substitutions (I1513A, I1514A, G1516A, S1517P, L1521A, N1522A, F1524A, I1525A, G1526A, V1527A, I1528A and D1530A) altered the voltage-dependence of activation and/or inactivation (Figure 1). The shifts in voltage-dependence caused by these substitutions were within 10 mV in either the hyperpolarizing or depolarizing directions (Figure 1 and Supplemental Table S1 at http://www.BiochemJ.org/bj/419/bj4190377add.htm). For example, the I1514A substitution shifted both activation and inactivation in the hyperpolarizing direction by 6–10 mV. The I1525A substitution shifted both activation and inactivation in the depolarizing direction. The L1521A, N1522A, G1526A and D1530A substitutions shifted the activation in the depolarizing direction, but inactivation in the hyperpolarizing direction. The I1513A, G1516A and S1517P substitutions shifted the voltage-dependence of activation in the depolarizing direction, whereas I1528A substitution shifted in the hyperpolarizing direction, but none of them had any effect on channel inactivation. Finally, the V1527A substitution did not alter the voltage-dependence of activation, but caused a 7 mV hyperpolarizing shift in inactivation.
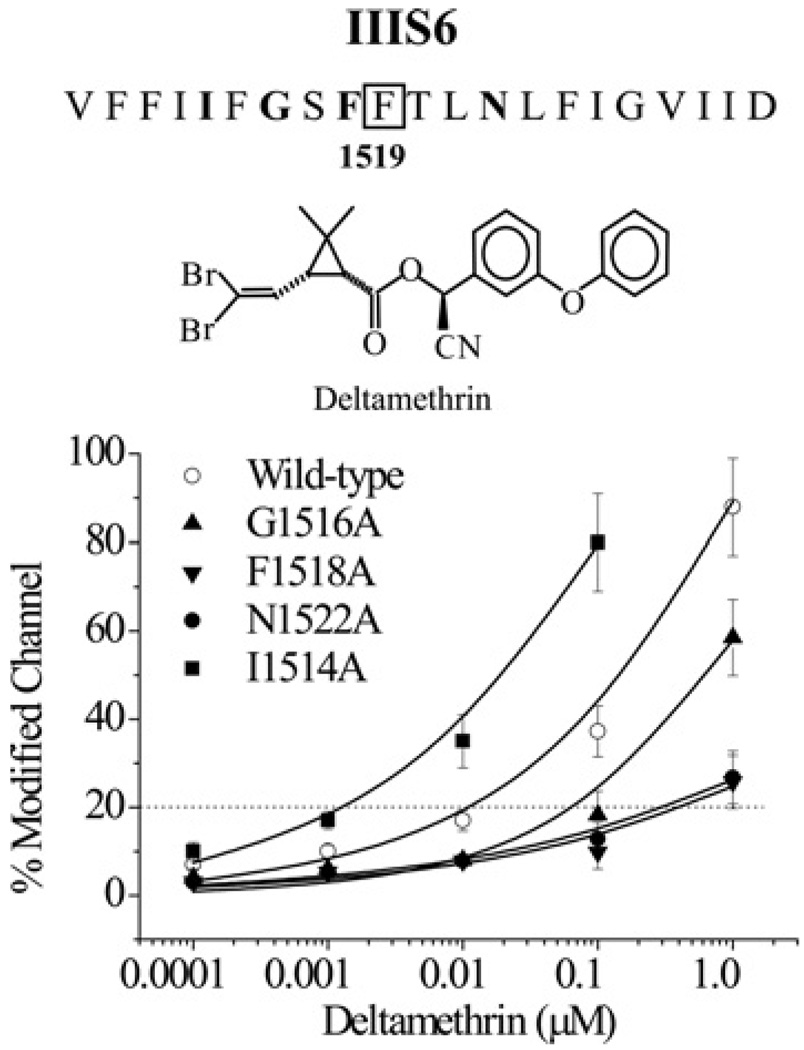
G1516A, F1518A, N1522A and I1514A substitutions alter sodium channel sensitivity to deltamethrin
Deltamethrin, a potent type II pyrethroid containing an α-cyano group at the phenoxybenzyl alcohol (Figure 2), was used to examine the pyrethroid sensitivity of all 20 mutant channels. Deltamethrin prolongs the opening of sodium channels, resulting in the induction of a tail current associated with repolarization [22]. This effect is state-dependent, with deltamethrin preferably binding to channels in the open state [9,10]. We applied a 100-pulse train of 5-ms depolarizations from −120 mV to −10 mV to detect deltamethrin-induced tail currents upon repolarization. At 1 µM, deltamethrin drastically modified the gating of wild-type BgNav1-1A channels as evident from the induction of a tail current with a bi-exponential decay (see Supplemental Table S2 at http://www.BiochemJ.org/bj/419/bj4190377add.htm). Alanine substitutions of Gly1516, Phe1518 and Asn1522 (located in the middle of IIIS6) significantly reduced the effect of deltamethrin, and higher concentrations of deltamethrin (10 µM) were required to induce tail currents. Furthermore, substitutions F1518A and N1522A converted the bi-exponential decay of the deltamethrin-induced tail current into a mono-exponential decay (Supplemental Table S2). In contrast, alanine substitution of Ile1514 enhanced the effect of deltamethrin; a detectable tail current was elicited at 1 nM. Based on the percentage of channel modification by deltamethrin, the I1514A channel was 10-fold more sensitive to deltamethrin than the wild-type channel, whereas the G1516A, F1518A and N1522A channels were more resistant to deltamethrin than the wild-type by 5-, 20- and 20-fold respectively (Figure 2). Alanine substitutions of the remaining 16 residues in IIIS6 did not alter the deltamethrin effect.
Figure 2. Identification of four new residues in IIIS6 that contribute to sodium channel interaction with deltamethrin.
The amino acid sequence of the 20 residues flanking Phe1519 in IIIS6 of BgNav1-1A is shown. The boxed Phe1519 was identified previously to be critical for the binding and action of pyrethroids [11]. The four residues identified in the present study to be required for normal pyrethroid action are marked in bold. Responses to deltamethrin of wild-type channels and the four mutant channels (I1514A, G1516A, F1518A and N1522A) with altered sensitivities to pyrethroids are shown. The percentage of channel modification by deltamethrin was determined using the method described in the Experimental section. G1516A, F1518A and N1522A reduced channel sensitivity to deltamethrin 5-, 20- and 20-fold respectively, whereas I1514A increased channel sensitivity to deltamethrin 10-fold.
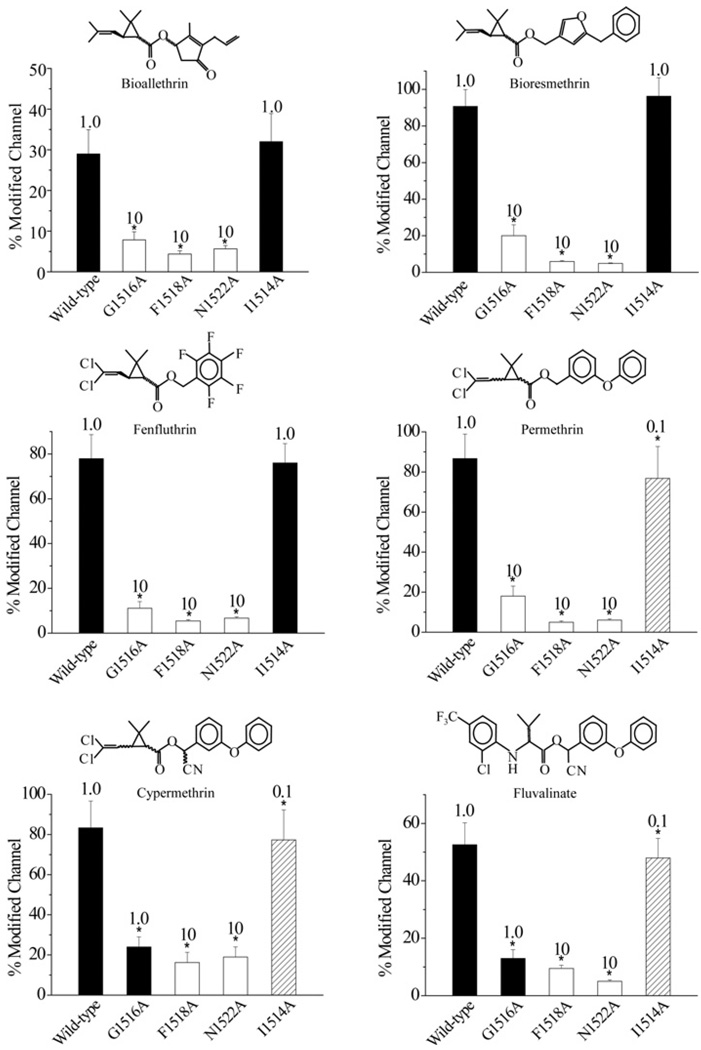
The I1514A substitution enhances sodium channel sensitivity to pyrethroids that contain the phenoxybenzyl alcohol moiety
To determine whether these substitutions also alter channel sensitivity to other pyrethroids, we examined the channel sensitivity to six additional structurally diverse pyrethroids (four type I pyrethroids: bioallethrin, bioresmethrin, fenfluthrin and permethrin; and two more type II pyrethroids: cypermethrin and fluvalinate). We found that the I1514A channel was also more sensitive to permethrin, cypermethrin and fluvalinate, but not to bioallethrin, bioresmethrin and fenfluthrin. Interestingly, deltamethrin, permethrin, cypermethrin and fluvalinate all contain a common phenoxybenzyl alcohol moiety, whereas this is lacking in bioallethrin, bioresmethrin and fenfluthrin (Figure 3).
Figure 3. I1514A, G1516A, F1518A and N1522A substitutions alter sodium channel sensitivity to a range of structurally different pyrethroid insecticides.
The percentage of channel modification by pyrethroids was determined using the method described in the Experimental section. Chemical structures of pyrethroids are shown above each histogram. The concentration of pyrethroid used in determining the percentage of channel modification was 0.1 µM (hatched bars), 1 µM (closed bars) or 10 µM (open bars), depending on the sensitivity of the mutant channels.
The G1516A, F1518A and N1522A substitutions made the channel more resistant to all six pyrethroids. Specifically, F1518A and N1522A substitutions almost completely abolished the channel sensitivity to all type I pyrethroids and drastically reduced the channel sensitivity to type II pyrethroids. The G1516A substitution also reduced the action of type I pyrethroids more drastically than that of type II pyrethroids (Figure 3).
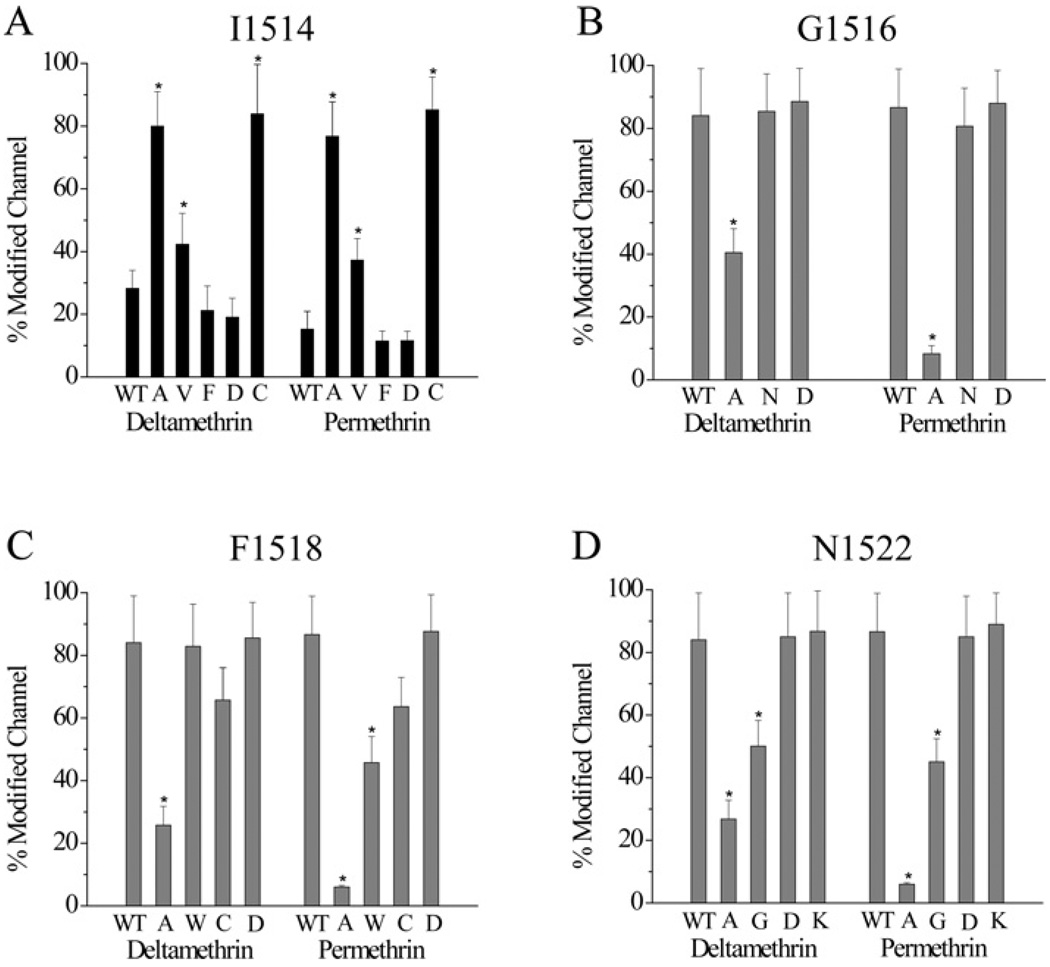
Effect of side chains at amino acid positions 1514, 1516, 1518 and 1522 on channel sensitivity to permethrin and deltamethrin
To determine how different side chains at the amino acid positions 1514, 1516, 1518 and 1522 might influence sodium channel gating properties and action of pyrethroids, we made substitutions using amino acids with side chains differing in size, charge or hydrophobicity from the original residue, and examined the resultant channels for gating and sensitivity to permethrin (a type I pyrethroid) and deltamethrin (a type II pyrethroid).
At position 1514, we made four more substitutions, to valine, cysteine, aspartic acid or phenylalanine. Like I1514A, the I1514V and I1514C channels became more sensitive to both deltamethrin and permethrin (Figure 4A). Neither the I1514D nor the I1514F channel has altered sensitivity to pyrethroids. All four substitutions shifted channel activation in the hyperpolarizing direction. Like I1514A, the I1514D and I1514F mutations also caused hyperpolarizing shifts in the voltage-dependence of inactivation (see Supplemental Table S3 at http://www.BiochemJ.org/bj/419/bj4190377add.htm).
Figure 4. Effects of amino acid substitutions at Ile1514, Gly1516, Phe1518 and Asn1522 on sodium channel sensitivity to pyrethroid insecticides.
Percentages of channel modification by deltamethrin and permethrin for mutant channels with different side chain substitutions for Ile1514 (A), Gly1516 (B), Phe1518 (C) or Asn1522 (D). A concentration of 0.1 µM (both deltamethrin and permethrin) was used for substitutions for Ile1514, whereas 1 µM was used for the rest of channels. WT, wild-type.
At position 1516, we introduced three additional substitutions: to proline, asparagine or aspartic acid. The G1516P channel failed to generate any detectable sodium current. The G1516N and G1516D channels produced sufficient sodium currents, but neither had altered channel sensitivity to either deltamethrin or permethrin (Figure 4B). The G1516D substitution shifted the voltage-dependence of activation in the hyperpolarizing direction and inactivation in the depolarizing direction (Supplemental Table S3). In comparison, the original G1516A substitution caused a depolarizing shift in activation and had no effect on inactivation (Supplemental Table S3). The G1516N substitution only shifted the voltage-dependence of inactivation in the depolarizing direction.
At position 1518, we generated three additional substitutions, to tryptophan, cysteine or aspartic acid. These substitutions did not alter channel sensitivity to deltamethrin and permethrin, except for the F1518W substitution which slightly reduced permethrin sensitivity (Figure 4C). The F1518W substitution caused a 14 mV hyperpolarizing shift in activation (Supplemental Table S3).
Finally, at position 1522, we introduced four additional substitutions, to glycine, phenylalanine, aspartic acid or lysine. The N1522F channel did not generate any detectable sodium current. Like N1522A, the N1522G channel was more resistant to both pyrethroids, whereas N1522D and N1522K channels were as sensitive to both pyrethroids as the wild-type channel (Figure 4D). The N1522K substitution shifted the activation in the hyperpolarizing direction, in contrast with the depolarizing shift caused by the original N1522A substitution (Supplemental Table S3). The N1522G and N1522D substitutions did not alter gating.
Ser1517 and Leu1521 are critical for the action of BTX
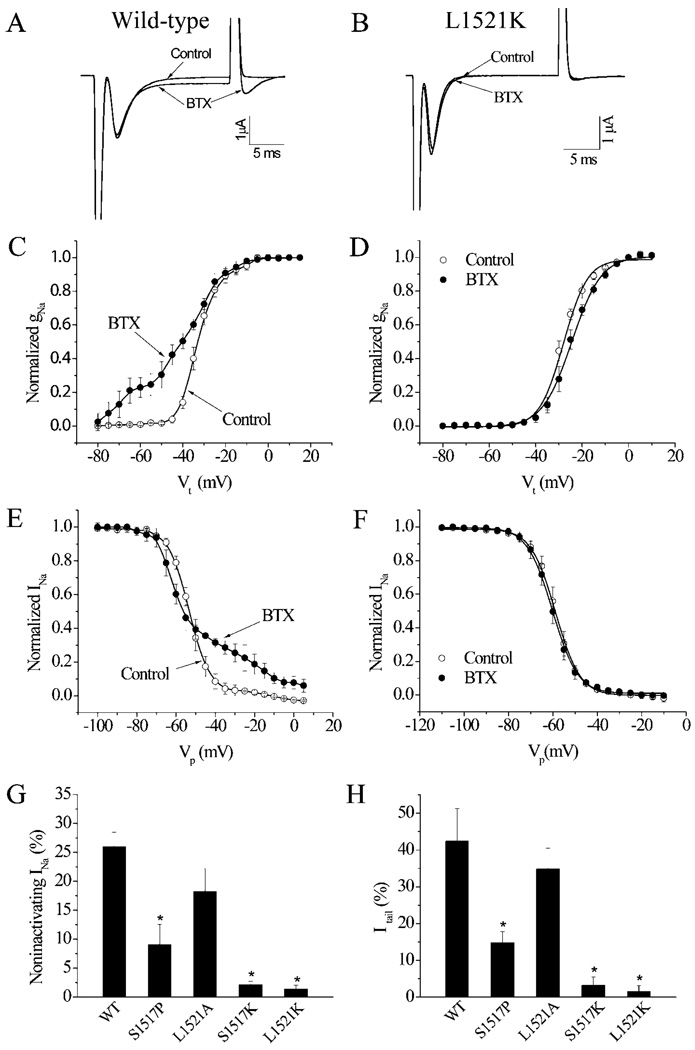
Similar to its effects on mammalian sodium channels, BTX shifted the voltage-dependence of activation in the hyperpolarizing direction, inhibited fast inactivation, as observed by the non-inactivating component of inactivation, and induced a tail current associated with repolarization of the BgNav1-1A channel (Figure 5A) [11]. Lysine substitutions of two residues in IIIS6 in the Nav1.4 sodium channel, corresponding to Ser1517 and Leu1521 in BgNav, completely abolished BTX effects [28]. Thus we made two additional substitutions, S1517K and L1521K, in BgNav. Consistent with the results from the Nav1.4 sodium channel, S1517K and L1521K channels were extremely resistant to BTX modification (Figure 5). The S1517P substitution reduced the action of BTX, but the L1521A substitution did not alter the activity of BTX (Figures 5G and 5H).
Figure 5. S1517K and L1521K substitutions abolished BTX action on the cockroach BgNav sodium channels.
(A and B) Sodium current traces from wild-type (A) and L1521K (B) sodium channels elicited, after 3000 repetitive pulses at a frequency of 10 Hz, by a 20-ms test pulse to −10 mV from a holding potential of −120 mV before and after the application of 500 nM BTX. (C and D) Curves of voltage-dependence of activation of wild-type (C) and L1521K (D) sodium channels. (E and F) Curves of steady-state inactivation of wild-type (E) and L1521K (F) sodium channels. (G) Percentages of non-inactivating current induced by BTX in wild-type (WT) and mutant sodium channels. (H) Percentages of tail current induced by BTX in wild-type (WT) and mutant sodium channels.
Computer modelling
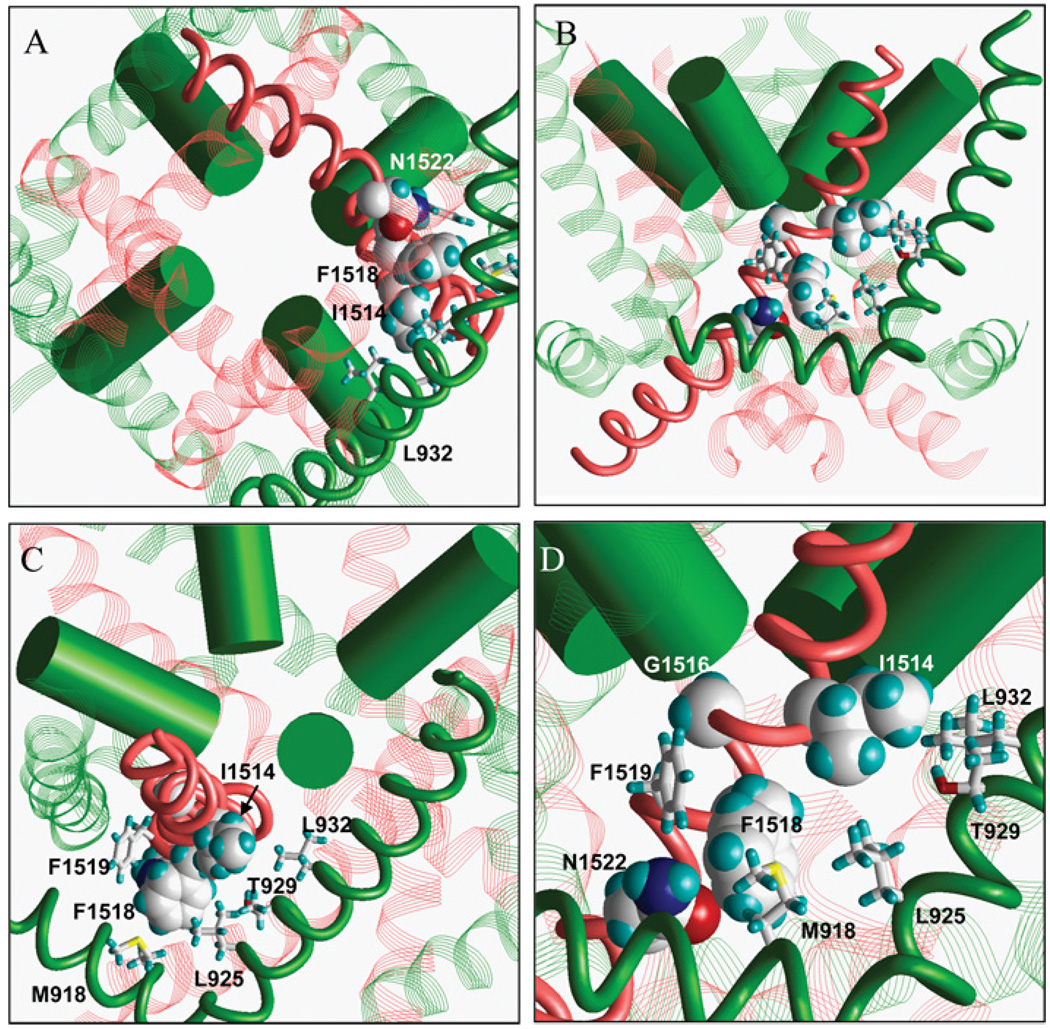
Mapping of Ile1514, Phe1518 and Asn1522 in the model of BgNav1-1A shows these residues exposed towards helix IIS5 and linker IIS4–IIS5 (Figure 6). These helices incorporate the pyrethroid-interacting residues Met918, Leu925, Thr929 and Leu932 (in the housefly sodium channel) and Phe1519 (in the cockroach sodium channel) identified previously as necessary for pyrethroid action and/or binding [6–8]. The two groups of residues form a cluster (Figure 6) in agreement with recent models [12,14] in which the pyrethroid receptor is located in the interface between helices IIIS6, IIS5 and the linker helix IIS4–IIS5.
Figure 6. A Kv 1.2-based model of the cockroach BgNav sodium channel in the open state.
The helices are shown as follows: IIIS6, red rod; IIS4–IIS5 and IIS5, green rods; pore helices, green cylinders; IS6, IIS6 and IVS6, red strands; linkers IIS4–IIS5 and IIS5 in domains I, III and IV, green strands. The side chains Ile1514, Gly1516, Phe1518 and Asn1522 in IIIS6, which were shown to be required for normal pyrethroid action in the present study, are space-filled. Sticks show side chains Met918 in the IIS4–IIS5 linker, Leu925, Thr929 and Leu932 in IIS5 (housefly sodium channel) and Phe1519 in IIIS6 (cockroach sodium channel) identified previously to be required for pyrethroid action and/or binding [6–8]. (A) Cytoplasmic view. (B) Side view. (C) View along the P-helix in domain II. (D) Enlarged view of (B) with the cluster of putative pyrethroid-interacting residues.
DISCUSSION
Pyrethroids and BTX act on the opposing sides of the IIIS6 helix
Residues in IIIS6 are predicted to form an α-helix and to be part of the inner pore of the sodium channel, with one side of the IIIS6 helix facing the lumen of the pore. Previous studies have shown that residues in S6 segments from all four domains form the receptor site of BTX [2] and computer modelling predicts that BTX binds within the inner pore of the sodium channel [29]. According to this model, the BTX-binding residues, including Ser1276 and Leu1280 (in Nav1.4) would face the pore. IIIS6 also plays an essential role in the interaction of the sodium channel with pyrethroids, as is evident from the requirement of Phe1519 for the binding and action of structurally diverse pyrethroids on the cockroach BgNav channel [11]. We have shown in the present study that Ser1517 and Leu1521 in BgNav (corresponding to Ser1276 and Leu1280 in Nav1.4 respectively) are important for BTX action on the cockroach sodium channel, but they are not involved in pyrethroid action. In contrast, Phe1519 [11] and Asn1522 (the present study) are important for pyrethroid action, but they are not involved in the action of BTX. Phe1519 and Asn1522 are predicted to be situated on the side of the IIIS6 α-helix which is opposite to the BTX-binding residues (Figure 6). Therefore these results provide experimental evidence supporting the notion that pyrethroids and BTX act on opposing sides of the IIIS6 helix.
Potential roles of the four IIIS6 residues in their interaction with pyrethroids
Mapping of other IIIS6 residues (Ile1514, Gly1516 and Phe1518) required for normal pyrethroid action in the BgNav1-1A channel shows that Ile1514 and Phe1518, but not Gly1516, are exposed towards IIS5 and the linker IIS4–IIS5. This is intriguing because IIS5 and linker IIS4–IIS5 contain several residues that are required for pyrethroid action, including the well-known Met918 in the linker IIS4–IIS5 and Thr929 and Leu932 in IIS5 [30,31]. Altogether, residues involved in pyrethroid action in IIIS6, IIS5 and linker IIS4–IIS5 form a cluster (Figure 6) in agreement with a recent model [12,14] in which the putative pyrethroid receptor is located at the lipid-exposed interface between IIIS6, IIS5 and linker IIS4–IIS5.
The contribution of Ile1514 to sodium channel interaction with pyrethroids seems to be unique. First, Ile1514 appears to define one edge of the IIIS6 amino acid cluster required for normal pyrethroid action. Secondly, in contrast with Gly1516, Phe1518, Phe1519 and Asn1522, alanine substitutions of which drastically reduce the action of all pyrethroids tested, the I1514A substitution actually enhances sodium channel sensitivity to pyrethroids. Furthermore, this enhanced sensitivity seems to depend on the chemical structure of pyrethroids. Specifically, the I1514A substitution enhanced the action of three type II pyrethroids (containing the α-cyano group) and one type I pyrethroid permethrin (lacking the α-cyano group), but did not have an effect on the action of the remaining three type I pyrethroids bioallethrin, bioresmethrin and fenfluthrin. Interestingly, the four pyrethroids whose action is affected by the I1514A substitution share a common structural feature: a phenoxybenzyl alcohol moiety (Figure 3). In fact, fenfluthrin differs from permethrin only at the alcohol moiety; fenfluthrin has a fluorinated single benzene ring instead of a phenoxybenzyl alcohol as in permethrin. These results suggest that the involvement of Ile1514 in pyrethroid action depends on the presence of a phenoxybenzyl alcohol group in a pyrethroid. It would be interesting in the future to expand the analysis of the structural compatibility between amino acid side changes at position 1514 and diverse chemical structures of pyrethroids.
The hydrogen side chain of Gly1516 is predicted to be away from either the pore-oriented BTX-binding site (Ser1517 and Leu1521) or the putative lipid-bound pyrethroid-binding pocket (Phe1519) illustrated in Figure 6. Thus Gly1516 is not likely to be involved directly in pyrethroid binding. Interestingly, Gly1516 has been predicted to be a gating hinge residue critical for the rotational movement of the inner helix in response to membrane depolarization, based on analysis of the open configuration of MthK channels [32] and site-directed mutagenesis of a bacterial sodium channel [33]. Therefore substitution of this residue could affect orientation of the cytoplasmic half of the IIIS6 helix and hence proper orientation of the pyrethroid-interacting residues.
Phe1518, like Gly1516, is also predicted to not be in BTX- and pyrethroid-binding pockets. Furthermore, unlike Phe1519, for which only aromatic residue substitutions restore sodium channel sensitivity to deltamethrin, an aromatic residue is not essential at position 1518. For example, neither tryptophan nor cysteine substitution for Phe1518 affected BgNav1-1A channel sensitivity to deltamethrin (Figure 4C). Phe1518 probably plays an indirect role in pyrethroid action, similarly to Gly1516, by properly orienting residues important for sensitivity to pyrethroids (e.g. Phe1519 and possibly Asn1522).
Asn1522 is predicted to be situated near Phe1519 on the same side of the IIIS6 α-helix. Interestingly, like F1519A, the N1522A substitution also almost completely abolished the action of all seven pyrethroids. However, substitutions of a similar size of side chain regardless of negative or positive charge (i.e. asparagine, aspartic acid or lysine) did not affect the action of pyrethroids. These results suggest that substitution of similarly sized amino acids (aspartic acid or lysine) for asparagine at 1522 re-establishes a critical contact point, lacking in the N1522A mutant, for pyrethroid activity in voltage-gated sodium channels.
In conclusion, our systematic site-directed mutagenesis of IIIS6 of the cockroach sodium channel residues revealed a cluster of amino acid residues that are essential for the action of pyrethroids. Together with the previous finding of Phe1519 as a key residue for the binding of pyrethroid insecticides [11], this study highlights the importance of IIIS6 in pyrethroid action. It is worth mentioning that these residues may not be the only ones in IIIS6 that are involved the interaction with pyrethroids. Our alanine-scanning analysis could miss some pyrethroid-interacting residues simply because alanine substitutions of these residues do not alter the action of pyrethroids. Nevertheless, it is clear that pyrethroid binding and action likely require amino acid residues from different regions (IIIS6, IIS4–IIS5 and IIS5) of the sodium channel. Further site-directed mutagenesis and modelling studies are necessary to visualize involvement of pyrethroid-interacting residues in the action of different pyrethroids, and to understand how structurally diverse pyrethroids interact with these pyrethroid-interacting residues at the atomic level.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Kris Silver for a critical review of this manuscript before submission. Computations were performed using the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET http://www.sharcnet.ca).
FUNDING
This study was supported by grants to K.D. from National Institutes of Health [grant number GM057440] and National Science Foundation [grant number IBN0224877] and by the grant to B. S. Z. from the Canadian Institutes of Health Research [grant number MOP-53229].
Abbreviation used
- BTX
batrachotoxin
REFERENCES
- 1.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 2.Wang SY, Wang GK. Voltage-gated sodium channels as primary targets of diverse lipid-soluble neurotoxins. Cell. Signalling. 2003;15:151–159. doi: 10.1016/s0898-6568(02)00085-2. [DOI] [PubMed] [Google Scholar]
- 3.Elliott M. Synthetic pyrethroids. In: Elliott M, editor. Synthetic Pyrethroids (ACS Symposium Series No. 42); American Chemical Society; Washington, DC. 1977. pp. 1–28. [Google Scholar]
- 4.Narahashi T. Neuroreceptors and ion channels as the basis for drug action: past, present, and future. J. Pharmacol. Exp. Ther. 2000;294:1–26. [PubMed] [Google Scholar]
- 5.Zlotkin E. The insect voltage-gated sodium channel as target of insecticides. Annu. Rev. Entomol. 1999;44:429–455. doi: 10.1146/annurev.ento.44.1.429. [DOI] [PubMed] [Google Scholar]
- 6.Soderlund DM. Sodium channels. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Oxford: Elsevier; 2005. pp. 1–18. [Google Scholar]
- 7.Davies TGE, Field LM, Usherwood PNR, Williamson MS. A comparative study of voltage-gated sodium channels in the Insecta: implications for pyrethroid resistance in anopheline and other neopteran species. Insect Biochem. Mol. Biol. 2007;16:361–375. doi: 10.1111/j.1365-2583.2007.00733.x. [DOI] [PubMed] [Google Scholar]
- 8.Dong K. Insect sodium channels and insecticide resistance. Invert. Neurosci. 2007;7:17–30. doi: 10.1007/s10158-006-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vais H, Atkinson S, Pluteanu F, Goodson SJ, Devonshire AL, Williamson MS, Usherwood PNR. Mutations of the para sodium channel of Drosophila melanogaster identify putative binding sites for pyrethroids. Mol. Pharmacol. 2003;64:914–922. doi: 10.1124/mol.64.4.914. [DOI] [PubMed] [Google Scholar]
- 10.Vais H, Williamson MS, Goodson SJ, Devonshire AL, Warmke JW, Usherwood PNR, Cohen CJ. Activation of Drosophila sodium channels promotes modification by deltamethrin: reductions in affinity caused by knock-down resistance mutations. J. Gen. Physiol. 2000;115:305–318. doi: 10.1085/jgp.115.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan J, Liu Z, Wang R, Huang ZY, Chen AC, Gurevitz M, Dong K. Identification of amino acid residues in the insect sodium channel critical for pyrethroid binding. Mol. Pharmacol. 2005;67:513–522. doi: 10.1124/mol.104.006205. [DOI] [PubMed] [Google Scholar]
- 12.Usherwood PNR, Davies TGE, Mellor IR, O’Reilly AO, Peng F, Vais H, Khambay BP, Field LM, Williamson MS. Mutations in DIIS5 and the DIIS4–S5 linker of Drosophila melanogaster sodium channel define binding domains for pyrethroids and DDT. FEBS Lett. 2007;581:5485–5492. doi: 10.1016/j.febslet.2007.10.057. [DOI] [PubMed] [Google Scholar]
- 13.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 14.O’Reilly AO, Khambay BP, Williamson MS, Field LM, Wallace BA, Davies TGE. Modelling insecticide-binding sites in the voltage-gated sodium channel. Biochem. J. 2006;396:255–263. doi: 10.1042/BJ20051925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He H, Chen AC, Davey RB, Ivie GW, George JE. Identification of a point mutation in the para-type sodium channel gene from a pyrethroid-resistant cattle tick. Biochem. Biophys. Res. Commun. 1999;261:558–561. doi: 10.1006/bbrc.1999.1076. [DOI] [PubMed] [Google Scholar]
- 16.Wang SY, Barile M, Wang GK. A phenylalanine residue at segment D3–S6 in Nav1.4 voltage-gated Na+ channels is critical for pyrethroid action. Mol. Pharmacol. 2001;60:620–628. [PubMed] [Google Scholar]
- 17.Song W, Liu Z, Tan J, Nomura Y, Dong K. RNA editing generates tissue-specific sodium channels with distinct gating properties. J. Biol. Chem. 2004;279:32554–32561. doi: 10.1074/jbc.M402392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan J, Liu Z, Nomura Y, Goldin AL, Dong K. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J. Neurosci. 2002;22:5300–5309. doi: 10.1523/JNEUROSCI.22-13-05300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng G, Deák P, Chopra M, Hall LM. Cloning and functional analysis of TipE, a novel membrane protein that enhances Drosophila para sodium channel function. Cell. 1995;82:1001–1011. doi: 10.1016/0092-8674(95)90279-1. [DOI] [PubMed] [Google Scholar]
- 20.Warmke JW, Reenan RA, Wang P, Qian S, Arena JP, Wang J, Wunderler D, Liu K, Kaczorowski GJ, Van der Ploeg LH, et al. Functional expression of Drosophila para sodium channels: modulation by the membrane protein TipE and toxin pharmacology. J. Gen. Physiol. 1997;110:119–133. doi: 10.1085/jgp.110.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan J, Liu Z, Tsai T-D, Valles S, Goldin AL, Dong K. Novel sodium channel gene mutations in Blattella germanica reduce the sensitivity of expressed channels to deltamethrin. Insect Biochem. Mol. Biol. 2002;32:445–454. doi: 10.1016/s0965-1748(01)00122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatebayashi H, Narahashi T. Differential mechanism of action of the pyrethroid tetramethrin on tetrodotoxin-sensitive and tetrodotoxin-resistant sodium channels. J. Pharmacol. Exp. Ther. 1994;270:595–603. [PubMed] [Google Scholar]
- 23.Zhorov BS. Vector method for calculating derivatives of energy of atom-atom interactions of complex molecules according to generalized coordinates. J. Struct. Chem. 1981;22:4–8. [Google Scholar]
- 24.Zhorov BS. Vector method for calculating derivatives of the energy deformation of valence angles and torsion energy of complex molecules according to generalized coordinates. J. Struct. Chem. 1983;23:649–655. [Google Scholar]
- 25.Tikhonov DB, Zhorov BS. Sodium channels: ionic model of slow inactivation and state-dependent drug binding. Biophys. J. 2007;93:1557–1570. doi: 10.1529/biophysj.106.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tikhonov DB, Zhorov BS. Benzothiazepines in L-type calcium channel: insights from molecular modeling. J. Biol. Chem. 2008;283:17594–17604. doi: 10.1074/jbc.M800141200. [DOI] [PubMed] [Google Scholar]
- 27.Zhorov BS, Tikhonov DB. Potassium, sodium, calcium and glutamate-gated channels: pore architecture and ligand action. J. Neurochem. 2004;88:782–799. doi: 10.1111/j.1471-4159.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- 28.Wulff H, Zhorov BS. Potassium, sodium, calcium and glutamate-gated channels: pore architecture and ligand action. Chem. Rev. 2008;108:1744–1773. doi: 10.1111/j.1471-4159.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- 29.Tikhonov DB, Zhorov BS. Sodium channel activators: model of binding inside the pore and a possible mechanism of action. FEBS Lett. 2005;579:4207–4212. doi: 10.1016/j.febslet.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Soderlund DM, Knipple DC. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem. Mol. Biol. 2003;33:563–577. doi: 10.1016/s0965-1748(03)00023-7. [DOI] [PubMed] [Google Scholar]
- 31.Vais H, Williamson MS, Devonshire AL, Usherwood PNR. The molecular interactions of pyrethroid insecticides with insect and mammalian sodium channels. Pest Manag. Sci. 2001;57:877–888. doi: 10.1002/ps.392. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Y, Yarov-Yarovoy V, Scheuer T, Catterall WA. A gating hinge in Na+ channels: a molecular switch for electrical signaling. Neuron. 2004;41:859–865. doi: 10.1016/s0896-6273(04)00116-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.