Abstract
Nucleotide excision repair (NER) removes damage from DNA in a tightly regulated multiprotein process. Defects in NER result in three different human disorders, xeroderma pigmentosum (XP), trichothiodystrophy (TTD) and Cockayne syndrome (CS). Two cases with the combined features of XP and CS have been assigned to the XP–D complementation group. Despite their extreme UV sensitivity, these cells appeared to incise their DNA as efficiently as normal cells in response to UV damage. These incisions were, however, uncoupled from the rest of the repair process. Using cell-free extracts, we were unable to detect any incision activity in the neighbourhood of the damage. When irradiated plasmids were introduced into unirradiated XP–D/CS cells, the ectopically introduced damage triggered the induction of breaks in the undamaged genomic DNA. XP–D/CS cells thus have a unique response to sensing UV damage, which results in the introduction of breaks into the DNA at sites distant from the damage. We propose that it is these spurious breaks that are responsible for the extreme UV sensitivity of these cells.
Keywords: Cockayne syndrome/DNA breaks/nucleotide excision repair/UV light/xeroderma pigmentosum
Introduction
The process of nucleotide excision repair (NER) removes many types of DNA damage from the genome. It is a highly conserved mechanism (Taylor and Lehmann, 1998) that involves >30 proteins acting in a tightly regulated manner. Damage in DNA is recognized either by the XPC–HR23B protein complex in so-called global genome repair, or by the transcribing RNA polymerase II in transcription-coupled repair, together with the XPA protein. After these recognition steps the two helicases XPB and XPD open up the DNA around the lesion allowing incisions to be introduced on both sides of the damage by the endonucleases XPF-ERCC1 and XPG. This is followed by removal of the damage-containing oligonucleotide and filling of the resulting gap by DNA polymerase δ or ɛ and a DNA ligase (for recent reviews, see Wood 1996; de Laat et al., 1999). To ensure the removal of damaged sites without disruption to the rest of the genome it is of paramount importance that the correct order of events is maintained.
Defects in NER can lead to three different human disorders, xeroderma pigmentosum (XP), trichothiodystrophy (TTD) and Cockayne syndrome (CS). These diseases show distinct clinical phenotypes. The hallmarks of XP are sun-sensitivity, pigmentation changes and an ∼2000–fold increased incidence of skin cancer (Kraemer et al., 1987). TTD is characterized by sulfur-deficient brittle hair together with growth and mental retardation (Itin and Pittelkow, 1990). CS also exhibits growth and mental retardation and a characteristic bird-like face as well as neurodegeneration and retinal degeneration (Nance and Berry, 1992). Neither TTD nor CS is cancer-prone.
Seven genes, XPA–G are associated with NER defects in XP. Whereas the features of TTD have not been found in combination with XP, mutations in three of the XP genes, XPB, XPD and XPG can give rise to combined symptoms of XP and CS (XP/CS). The XPB and XPD proteins are subunits of TFIIH, a transcription factor with dual functions in both basal transcription and NER (Hoeijmakers et al., 1996). The two known XP–D/CS patients, XP8BR (Broughton et al., 1995) and XP-CS-2 (Lafforet and Dupuy, 1978), are the subject of the current study. These patients both had extremely severe clinical features. Both are compound heterozygotes. The mutations in the XPD gene in XP8BR are G675R in one allele and deletion of nucleotide 2083 in the second allele, the latter causing a frameshift at codon 669 and a truncated 708 amino acid protein, which is likely to be completely non-functional (Broughton et al., 1995). In XP-CS-2, only one allele is expressed and contains the mutation G602D (Takayama et al., 1995).
Cellular studies showed that despite a level of unscheduled DNA synthesis (UDS) of ∼30% of normal, similar to or greater than that in other XP–D cells, both XP–D/CS cell strains were considerably more sensitive to killing by UV irradiation than XP–D cells. In fact they are the most sensitive fibroblasts that we have analysed (Broughton et al., 1995). We have constructed a series of mutations analogous to those found in various cell strains from the XP–D complementation group in the homologous rad15 gene of Schizosaccharomyces pombe. The mutation corresponding to that found in the XP–D/CS cell strain XP8BR was much the most sensitive of all the mutations tested (Berneburg et al., 2000).
The repair of different photoproducts was found to be indistinguishable in XP–D and XP–D/CS cells (van Hoffen et al., 1999). Elimination of cyclobutane pyrimidine dimers (CPD) was undetectable and removal of 6–4 photoproducts was inefficient in both. Nevertheless, the XP–D/CS cells failed to recover normal rates of RNA synthesis even after very low doses of UVC irradiation (2 J/m2), whereas RNA synthesis did recover after this dose in XP–D cells (van Hoffen et al., 1999).
To gain further insight into the underlying defect in XP/CS cells we have investigated the proficiency of XP–D/CS cells to introduce incisions into their DNA during NER. Our results show that XP–D/CS cells are able to detect DNA damage and appear to initiate incisions; however, these incisions are aberrant and distant from the sites of the damage.
Results
Extreme sensitivity of XP–D/CS cells to inhibition of gene expression by UV
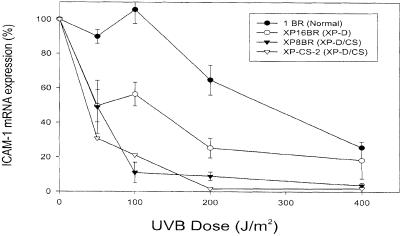
Details of the cell lines used in this paper are shown in Table I. Our previous work showed that XP–D/CS fibroblasts were extremely sensitive to killing by UV irradiation (Broughton et al., 1995). This was correlated with a reduced ability to restore overall RNA-synthesis levels to normal even after very low UV doses (van Hoffen et al., 1999). We have now measured the response to UV of a specific gene, which codes for intercellular adhesion molecule 1 (ICAM–1), whose expression in human fibroblasts can be induced by treatment with γ–interferon. We have shown previously that this upregulation is suppressed by irradiation with UVB (Krutmann et al., 1994). The reduction in expression of ICAM–1 by UVB treatment of XP–D cells is significantly greater than in normal cells (Ahrens et al., 1997). Based on these observations, we investigated post-UVB ICAM–1 expression in the two known XP–D/CS cell lines (Figure 1). Strikingly, both XP–D/CS cell lines revealed an extreme inhibition of the transcription of this immunologically relevant molecule. ICAM–1 expression was inhibited only above doses of 100 J/m2 in normal cells and was reduced to 50% in the XP–D cell line at 100 J/m2. In contrast, doses of UVB >50 J/m2 resulted in >80% inhibition of ICAM–1 transcription in both XP–D/CS lines. This inhibition is greater than that observed in any other XP cell line that we have studied. This end-point therefore correlates with the extreme sensitivity to killing by UV irradiation (Broughton et al., 1995), and the failure of RNA synthesis to recover even after very low doses of UV irradiation (van Hoffen et al., 1999).
Table I. Cell lines used in this study.
| Cell type | Phenotype | Cell lines | Mutation in XPDa |
|---|---|---|---|
| Primary fibroblasts | normal | 1BR, 48BR, C3PV | |
| XP–D | XP1BR | R683W/R683W | |
| XP16 BR | R683W/R616P | ||
| XP3NE | R683W/R683W | ||
| XP–D/CS | XP8BR | G675R/Fs669 | |
| XP-CS-2 | G602D | ||
| XP-G/CS | XP20BE | ||
| Transformed fibroblasts | normal | 1BR pre-crisisb | |
| XP–D/CS | XP8BR pre-crisisb | G675R/Fs669 | |
| Lymphoblastoid | XP-A | GM2345 (XP2OS) | |
| XP-B | GM2252 (XP11BE) | ||
| XP–D | XP7BE | R683W/Del exon 3 | |
| XP–D/CS | XP8BR | G675R/Fs669 | |
| XP-G | XPG83 (XP125LO) |
aThe amino acid changes resulting from mutations in the two alleles of the XPD gene are shown. Fs, frameshift; Del, deletion. Only one allele is expressed in XP-CS-2. Data from Taylor et al. (1997) (XP1BR, XP16BR, XP3NE), Broughton et al. (1995) (XP8BR), Takayama et al. (1995) (XP–CS-2) and our unpublished results (XP7BE).
bPre-crisis cells were derived from primary fibroblasts following transfection with pSV3neo (Mayne et al., 1986). The SV40 T-antigen expressed from this plasmid transforms the cells morphologically and extends their life-span, but the lines are not immortal.
Fig. 1. Inhibition of ICAM–1 expression by UVB. Normal, XP–D (XP16BR) or XP–D/CS fibroblasts were exposed to different doses of UVB irradiation and then treated with interferon–γ. Four hours later the levels of ICAM–1 mRNA were measured by semi-quantitative RT–PCR. Error bars represent the SEMs of three experiments.
XP–D/CS cells accumulate nicks after UV irradiation
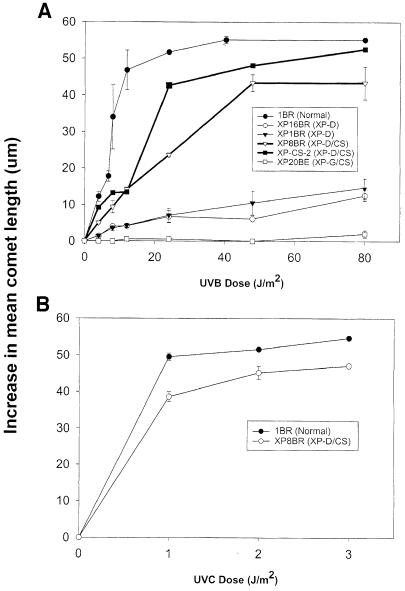
Previous measurements in XP–D/CS cells showed that, as in many other XP–D cell strains, the level of UV-induced UDS was ∼30% of that in normal cells, when measured either by autoradiography or liquid scintillation counting (Broughton et al., 1995), and that photoproduct removal was similarly deficient in both XP–D and XP–D/CS cells (van Hoffen et al., 1999). These studies did not therefore reveal any obvious differences in NER that could account for the much greater sensitivity of the XP–D/CS cells. In the following experiments we investigated another aspect of NER, the ability of XP–D/CS cells to incise DNA following UV exposure. To measure incision in XP–D/CS cells we employed the single cell gel electrophoresis (‘comet’) assay. The incision steps of NER are normally rate limiting and incised intermediates are not readily observable following UV irradiation. If, however, hydroxyurea (HU) and 1–β–d-arabinofuranosylcytosine (ara–C) are included in the post-irradiation medium, the subsequent repair steps of NER are inhibited, and incised intermediates accumulate (Squires et al., 1982) and can be measured using this assay (Green et al., 1992). We measured incision in normal, XP–D and XP–D/CS cells irradiated with either UVB (Figure 2A) or UVC (Figure 2B). As expected, the normal cell line 1BR generated increasing comet lengths, representing the accumulation of incised intermediates, with increasing UVB or UVC doses. As shown previously (Berneburg et al., 2000), in XP–D cells (XP1BR, XP16BR) comet formation was barely detectable with increasing UV dose, the incision efficiency being at least 10–fold lower than in normal cells (Figure 2A). These data are consistent with the very low rate of photoproduct removal in these cells. In striking contrast, however, both XP–D/CS cell lines (XP8BR and XP-CS-2) appeared to generate incision intermediates efficiently with a dose–response for both UVB and UVC only slightly lower than that of normal cells.
Fig. 2. Accumulation of incised intermediates after UV irradiation of normal and XP–D/CS cells. Fibroblasts were irradiated with different doses of UVB (A) or UVC (B) and incubated for 1 h in the presence of HU and ara–C. Incised intermediates were measured using the comet assay. The average increase in length of the comet tails is plotted as a function of UV dose.
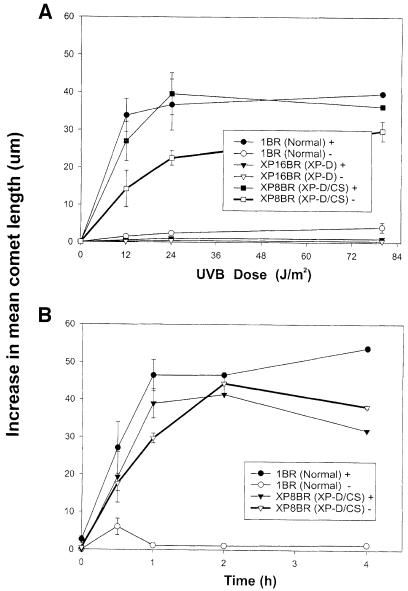
To see if this phenomenon was observed in XP/CS cells from other complementation groups, we also examined the response of the XP-G/CS cell line XP20BE (Moriwaki et al., 1996). Results in Figure 2A show that incision intermediates were undetectable in this cell line. The generation of incision intermediates appeared therefore to be specific for XP–D/CS cells, and was most unexpected in view of the striking sensitivity of these cells to UV irradiation. We therefore considered the possibility that the XP–D/CS cells might be able to incise the damaged DNA, but not complete the subsequent steps in the NER process. If this were the case, then incision intermediates might accumulate even in the absence of the repair synthesis inhibitors, ara–C and HU. As anticipated, in normal 1BR cells, incision intermediates accumulated only in the presence of the inhibitors (•) but not in their absence (○) (Figure 3A). In the absence of ara–C and HU, the removal and repair synthesis steps occur too quickly in normal cells for incision intermediates to accumulate. As anticipated, no intermediates were detected in the XP–D cell line XP16BR, either in the presence (▾) or absence (▿) of the inhibitors. Strikingly, however, the XP–D/CS cell line XP8BR accumulated incisions even in the absence of the inhibitors (□). A time-course showed that incisions could be observed as early as 30 min after irradiation and reached a maximum after 3–4 h (Figure 3B). No incisions were detected in normal cells even after 12 h incubation (data not shown). We noted that the accumulation of breaks in XP8BR fibroblasts in the presence of the inhibitors was independent of their proliferation status, whereas incisions in the absence of the inhibitors could only be detected in non-proliferating cells.
Fig. 3. Accumulation of incised intermediates in the absence of repair synthesis inhibitors after UV irradiation. Non-proliferating cells were (A) irradiated with different doses of UVB and incubated for 1 h, or (B) irradiated with 30 J/m2 UVB and incubated for different times prior to analysis of the DNA using the comet assay.
These data, showing the accumulation of DNA breaks in UV-irradiated XP8BR cells, led us to test the idea that XP–D/CS cells are able to initiate the NER process by incising the DNA, but might be unable to complete the process.
XP–D/CS cell extracts are completely defective in in vitro repair activities
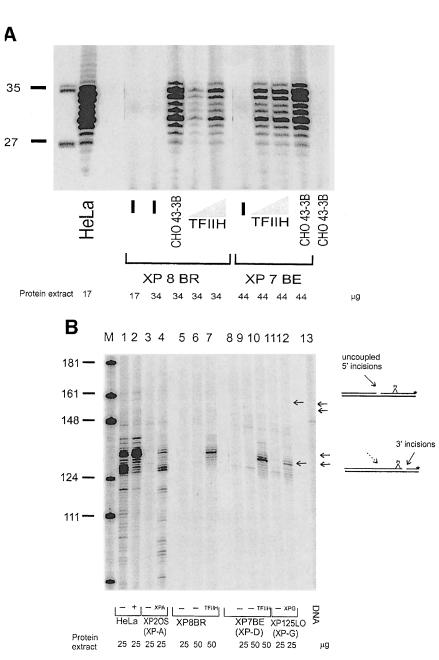
Various in vitro assays have been developed to measure the different steps in the NER process using cell-free extracts. In particular these assays are able to detect uncoupled incisions on a damaged template (Moggs et al., 1996; Evans et al., 1997). Incision on either the 3′ or 5′ side of the damage without incision on the other side has been reported for several cell lines (Evans et al., 1997). The high rates of incision without the actual removal of existing damage observed in XP–D/CS cells might be explained by uncoupled 3′ or 5′ incision. To test this idea, we carried out in vitro experiments in a reconstituted NER repair system. Whole-cell extracts derived from XP8BR lymphoblastoid cell lines were investigated for their ability to carry out dual or uncoupled incision as well as repair synthesis using a plasmid substrate containing a single cisplatin adduct (Moggs et al., 1996; Evans et al., 1997). Dual incisions result in the release of an ∼30 nucleotide fragment, which can be radioactively labelled and analysed on a denaturing polyacrylamide gel as described previously (Moggs et al., 1996). Figure 4A shows that HeLa cell extracts have efficient excision activity, whereas extracts from XP7BE (XP–D) or XP8BR (XP–D/CS) had no detectable activity. In both cases, activity could be restored by addition to the extracts of either purified TFIIH (which contains the XPD protein subunit) or extract of NER-defective 43-3B cells (defective in the NER protein ERCC1).
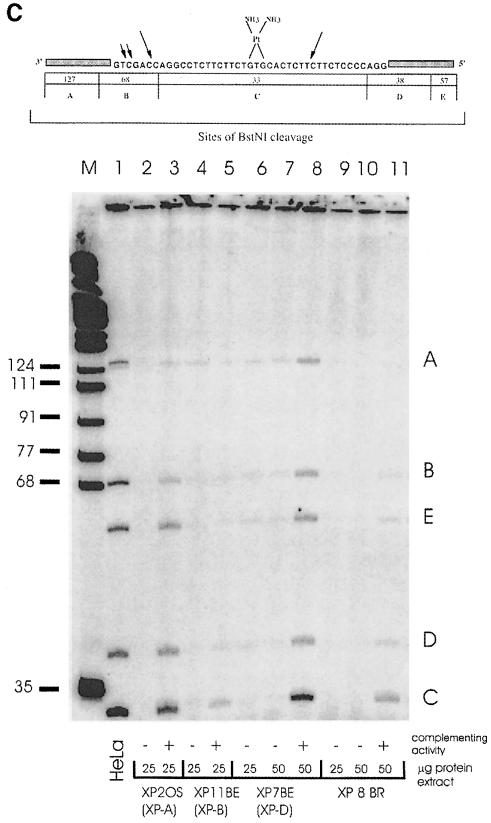
Fig. 4. Absence of NER in cell-free extracts of XP–D/CS cells. (A) Cell-free extracts of HeLa, XP7BE or XP8BR cells were incubated with plasmid DNA containing a single cisplatin lesion. Excised fragments were directly end-labelled with [32P]dCTP and separated on polyacrylamide gels. Different amounts of cell extract protein were used as indicated, and in some samples they were supplemented either with TFIIH or with 30 μg of protein extract from the NER-deficient Chinese hamster ovary cell line 43-3B. (B) To analyse incision activity of cell extracts, the amounts of extract protein indicated were incubated with substrate and a DNA fragment containing the lesion was isolated by restriction digestion and 3′-end-labelled. M, size marker. (C) Repair synthesis. Substrate was incubated with extracts and triphosphates including radioactive dCTP to label repair patches. The DNA was then digested with BstNI and the products separated on polyacrylamide gels.
The experiment shown in Figure 4B was designed to detect single or dual incisions in the DNA. The plasmid containing the single cisplatin lesion was cleaved with AvaII and end-labelled 140 nucleotides from the 3′ side of the site of the damage (Constantinou et al., 1999). It was then incubated with cell extracts and purified proteins as indicated. Incisions 3′ to the lesion will generate labelled fragments of ∼130 bases, whereas uncoupled incisions 5′ to the lesion will generate a 155 base fragment. HeLa cells efficiently incised the DNA 3′ to the adduct (Figure 4B, lanes 1 and 2), whereas extracts from XP-A (lane 3), XP-G (lane 11), XP–D (lanes 8 and 9) and XP–D/CS cells (lanes 5 and 6) were almost completely defective in incision ability. Addition of XPA, XPG or TFIIH proteins to the appropriate extracts restored 3′ incision activity (Figure 4B, lanes 4, 12, 10 and 7). Finally, in the experiments of Figure 4C, the ability of the extracts to carry out repair synthesis following excision of the cisplatin adduct was measured. Following incubation with the extracts in the presence of [32P]dNTP to label the repair patches, the DNA was extracted and digested with BstNI. Most of the repaired DNA is located in a 33 bp fragment, which contains the site of the lesion, and to a lesser extent in surrounding fragments of 38, 57, 68 and 127 bp. Repair activity was seen in HeLa cells in Figure 4C, lane 1. In contrast, activity was very low or absent in XP-A, XP-B, XP–D and XP8BR extracts (Figure 4C, lanes 2, 4, 6, 7, 9 and 10), but could be restored by addition of complementing activity (lanes 3, 5, 8 and 11). These results obtained with cell-free extracts therefore contrasted with our in vivo analysis and provided no evidence for uncoupled NER incision activity.
XP8BR cells accumulate breaks in genomic DNA in the presence of ectopic damage
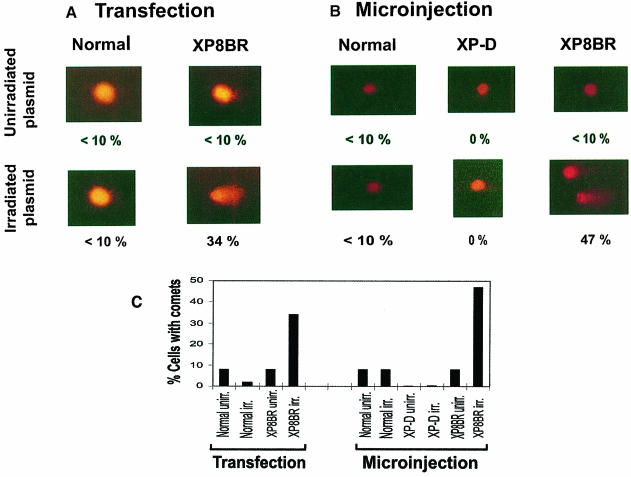
The in vitro studies did not provide any evidence for the induction of breaks in the proximity of the damage. We therefore considered the possibility that the breaks generated in the cellular DNA of XP8BR following UV irradiation (Figure 3) were not targeted to the sites of the damage. To test this hypothesis, we introduced irradiated plasmid DNA into the cells and used the comet assay to look for the ability of this damaged DNA to induce breaks in undamaged genomic DNA. We used two independent approaches, transfection and microinjection, to introduce the damaged DNA into the cells. In each case we introduced unirradiated or irradiated plasmid into both normal and XP8BR cells.
Transfection. Cells were cotransfected with the plasmid pcDNA3.1, either unirradiated or irradiated with a UVB dose of 15 000 J/m2 (photoproduct yield approximately equivalent to that produced by 500 J/m2 UVC) together with a second (unirradiated) plasmid, pHook–1. The latter expresses the gene phOx sFv, which encodes a single-chain antibody directed against phOx that is displayed on the cell surface. Cells expressing this antibody can be rapidly separated by binding to phOx hapten linked to magnetic beads. The molar ratio of pcDNA3.1 to pHook–1 was 4:1, and it was anticipated that many of the cells that had taken up the pHook–1 plasmid would also have incorporated the pcDNA3.1. With this procedure we were able to separate the transfected cells from the 99% of the population that remained untransfected. The transfected cells were then analysed using the comet assay. After cotransfection, the cells were incubated for 24 h to allow for the expression of the phOx antibody. The expressing cells were separated, embedded in agarose, and comets analysed as described above. Cells were considered to have comets when the comet length exceeded 20 μm. Less than 10% of normal cells had comets when transfected with either unirradiated or irradiated plasmid (Figure 5A, first column). Similarly, when XP8BR cells were transfected with unirradiated plasmid, <10% of the cells displayed comets. In striking contrast, when irradiated plasmid was transfected into XP8BR cells, comets were seen in ∼35% of the transfected cells (Figure 5A, second column). The length of the comet tails observed in XP8BR transfected with irradiated plasmid varied over a wide range from 20 to 50 μm. We conclude from these results, summarized in Figure 5C (left), that in the XP–D/CS cells, the presence of UV damage in the plasmid induced breaks in the genomic DNA.
Fig. 5. Introduction of irradiated plasmids causes breakage of genomic DNA in XP–D/CS cells. (A) Transfection. SV40-transformed 1BR.neo or XP8BR.neo cells were cotransfected with unirradiated or UV-irradiated pcDNA3.1 and pHook–1. Twenty-four hours later, the cultures were harvested and cells expressing the pHook–1 plasmid were separated with hapten-linked magnetic beads, embedded in agarose and the genomic DNA analysed using the comet assay. Typical cells are shown after electrophoresis in the comet assay, together with the percentage of cells with comet tails >20 μm. (B) Microinjection. Normal, XP–D (XP3NE) or XP8BR cells in a marked area on a dish were microinjected with UV-irradiated pcDNA3.1 and incubated for 6 h, before harvesting and analysis using the comet assay. Details as in (A). (C) The data are summarized as a percentage of cells with comet tails >20 μm under different conditions.
Microinjection. In order to provide further independent evidence for this conclusion, we microinjected irradiated and unirradiated plasmid molecules into the nuclei of normal, XP3NE (XP–D) and XP8BR fibroblasts. This procedure also had the advantage that we were able to use primary fibroblasts, whereas for the transfection experiments we were obliged to use transformed cells to obtain a workable transfection frequency. We injected ∼200–400 cells within a small area of the Petri dish containing a coverslip. After injection, the cells were incubated for 6 h, and the population of injected cells was harvested and subsequently analysed by the comet assay. The results were very similar to those generated with the transfection procedure. After a 6 h incubation following injection, very few or no comets were seen in normal (Figure 5B, left) or XP–D cells (Figure 5B, middle) or in XP8BR cells injected with unirradiated plasmid. In striking contrast, microinjection of irradiated plasmid into XP8BR cells led to comet formation in ∼45% of the injected cells (Figure 5B, right). The length of the comet tails ranged from 20 to 60 μm. Results of the microinjection experiments are summarized in Figure 5C (right).
These results indicate that in XP–D/CS cells, sensing of the damage is uncoupled from the incisions measured by the comet assay, which take place away from the site of the DNA damage. For this to take place the damage must transmit a signal to the machinery that generates the nick in the genomic DNA. We considered the possibility that this signal transduction might be mediated by p53. The transfection experiments described above, however, were carried out with fibroblasts transformed with SV40 T-antigen. Such cell lines are in general functionally p53–, because the T-antigen sequesters the p53 protein. In order to confirm that this was indeed the case, we measured levels of both p53 and its downstream effector p21 in irradiated and unirradiated cells by immunoblotting. When primary fibroblasts were used, both p53 and p21 levels were increased following irradiation. In the T-antigen-transformed cells, the constitutive levels of p53 were at least 10–fold higher than those in untransformed cells, but there was little increase on irradiation. In contrast, p21 levels were low in the transformed cells and showed little change on irradiation (results not shown). From these data we infer that the signal transduction is unlikely to be mediated by p53, since it occurs in cells in which the p53 appears to be functionally inactive.
Incision observed by comet assay is independent from apoptosis
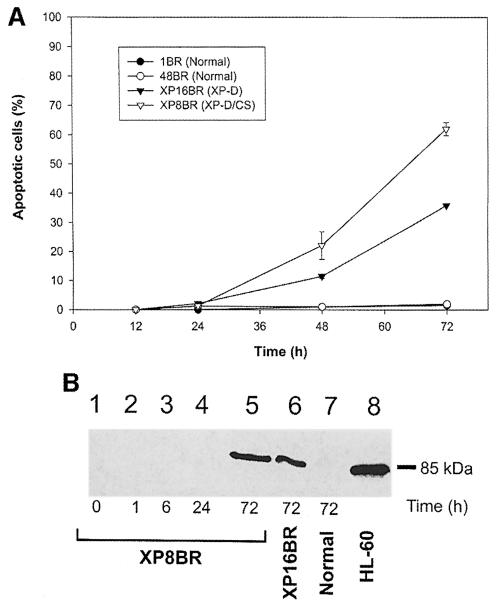
We considered the possibility that the DNA damage-induced incisions that we observed in XP8BR cells could be carried out by apoptotic pathways. To investigate this hypothesis, we measured apoptosis in normal, XPD and XP–D/CS primary fibroblasts as a function of time after irradiation. Apoptosis was measured by an end-labelling method and by cleavage of the PARP protein by caspase 3. Using the former procedure, apoptotic signals were detected 48 and 72 h after irradiation with 100 J/m2 in 15 and 35% of the XP–D cells (XP16BR), respectively, and in 45 and 66% of the XP–D/CS cells (Figure 6A). No apoptosis was observed in either of the normal cell strains investigated. Similar results were obtained with the PARP cleavage assay (Figure 6B). No apoptotic signals were observed at any of the earlier times, at which comet formation could be readily detected. These data show that although XP–D and XP–D/CS cells do undergo apoptosis after relatively high doses of UV irradiation, the incisions observed using the comet assay are unlikely to be generated by apoptotic pathways.
Fig. 6. Apoptosis in normal and XP8BR cells after UVB irradiation. Cells were irradiated with 100 J/m2 UVB and at various times after irradiation apoptosis was measured either using the Apoptag kit (A) or by the cleavage of PARP (B). In the latter experiments, sonicated cell lysates were electrophoresed in SDS–PAGE gels and the gels analysed by Western blotting using an antibody specific for the 85 kDa cleavage product of PARP.
Discussion
Our results have shown that in XP–D/CS cells, repair incisions appear to be generated following UV irradiation with an efficiency approaching that in normal cells (Figure 2), even though photoproduct removal is very inefficient in these cells (van Hoffen et al., 1999). In normal cells, however, incision intermediates can only be detected in the presence of repair synthesis inhibitors. In contrast, in the XP–D/CS cells, incisions accumulated even in the absence of the inhibitors (Figure 3), suggesting that they were uncoupled from the rest of the NER process and perhaps unrelated to NER.
Incisions in the absence of damage removal might have been explained by uncoupled incision on the 3′ or 5′ side of the damaged site. Uncoupled 5′ incisions have been reported previously in extracts of the XP-B cell line XP11BE or of catalytic site mutants of XP-G cells (Constantinou et al., 1999), and uncoupled 3′ incisions in extracts of XP-F and ERCC1 cells (Evans et al., 1997), none of which exhibited the features of XP/CS cells. However, dual incisions, uncoupled incisions and repair synthesis activity were all undetectable in the in vitro assays using extracts of XP8BR cells (Figure 4). Two possible explanations for these findings are: (i) the in vitro plasmid system is not an adequate model for cellular DNA in vivo in this particular circumstance; or (ii) the incisions detected by the comet assay in XP–D/CS cells occur somewhere distant from the actual sites of the damage. Evidence has been obtained in support of the latter by introducing irradiated plasmid DNA into undamaged cells (Figure 5). We have shown that incisions can be generated in undamaged genomic DNA in XP8BR as a result of this ectopic damage. It is most likely that this uncontrolled DNA breakage accounts for the extreme sensitivity of XP–D/CS cells to UV, as measured by cell killing (Broughton et al., 1995), recovery of RNA synthesis (van Hoffen et al., 1999) and inhibition of ICAM–1 upregulation (Figure 1).
The combined features of XP and CS are found in individuals in the XP-B, D and G complementation groups. Although the XP-G/CS cell line XP20BE is also very sensitive to cell killing by UV, incision intermediates did not accumulate in this cell line. This indicates a different underlying defect for the UV sensitivities of XP-G/CS and XP–D/CS cells.
XP–D/CS and TFIIH
The inactivating mutations in the two XP–D/CS lines are both in regions rich in mutations found in other patients. XP8BR contains the mutation Gly675Arg, only two amino acids away from a mutation Asp673Gly found in TTD6PV (Botta et al., 1998) and only eight amino acids from the Arg683Trp mutation found in 85% of XP–D patients (Taylor et al., 1997). Likewise, XP-CS-2 contains the mutation Gly602Asp, immediately adjacent to Arg601, which is mutated in two XP patients (Taylor et al., 1997; our unpublished observations). One of these patients has only very mild symptoms. There is therefore nothing obvious that distinguishes the region of the protein in which the XP/CS patients are mutated from other parts of the protein. It is perhaps significant that mutations in both XP–D/CS patients result in changes of a glycine residue to a charged amino acid, whereas the mutations found most commonly in XP and TTD patients result in changes of arginine residues (Taylor et al., 1997). Alterations of glycine residues often have profound effects, since the conversion of glycine to substantially larger residues can have significant steric effects on the tertiary structure of the protein.
The XPD protein is a subunit of transcription factor TFIIH, which apart from its role in basal transcription is a vital component of NER (Hoeijmakers et al., 1996). The XPD protein has a 5′–3′ helicase activity, which is stimulated ∼10–fold by its interaction with the p44 subunit of TFIIH (Coin et al., 1998). Coin et al. (1998) showed that purified XPD containing the Gly675Arg mutation found in XP8BR was unable to interact with p44, and by implication the stimulation of the helicase activity in its TFIIH context was abolished. This could account for the low levels of NER in XP8BR cells. However, the interaction of XPD with p44 was also abolished by other mutant proteins, in which the mutations corresponded to those found in XP and TTD patients. Furthermore, all these mutant proteins retained residual helicase activity in the absence of p44 that was only slightly lower than that of wild-type XPD (Coin et al., 1998). These biochemical findings do not therefore shed any light on the very distinctive consequences of the XPD/CS mutations.
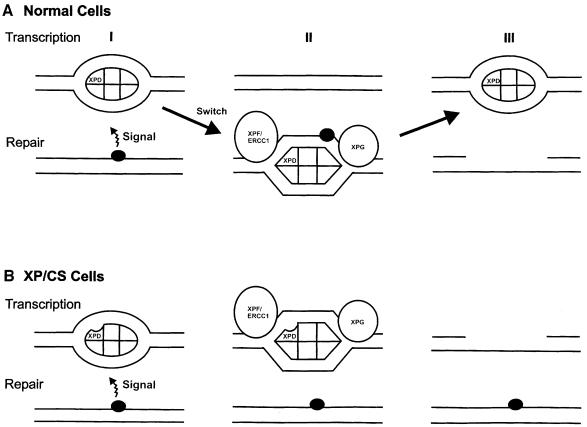
It is not immediately apparent why specific mutations in TFIIH should result in DNA breakage, since no nuclease activity is associated with TFIIH. However, TFIIH is undoubtedly involved in numerous, as yet poorly understood, protein–protein interactions to recruit it to sites of NER and transcription initiation. Following the introduction of DNA damage, it has not yet been established how TFIIH is recruited to the site of the damage, but it has been proposed that it exists in two different modes for transcription and repair (Svejstrup et al., 1995). It has also been suggested that the restoration of TFIIH from repair to transcription mode after completion of repair requires the CS proteins, and that the inhibition of RNA synthesis by DNA damage might not result from direct blockage of transcription elongation, but rather by inhibition of initiation of RNA synthesis (van Oosterwijk et al., 1996). Such mechanisms implicate signalling processes transmitted from the site of the damage to the site of transcription initiation, although the nature of such signals is unknown. We propose the following speculative model to account for our findings with XP/CS cells (Figure 7). We suggest that in normal cells, on receiving a signal from the damaged DNA, TFIIH becomes activated into its repair mode and recruited to the site of the damage (II in Figure 7A), repair is effected and TFIIH returns to its transcription mode (III). In the XP–D/CS cells, we suggest that the TFIIH is activated into repair mode, but remains attached to the DNA at the sites of transcription initiation (II in Figure 7B). This results in the recruitment of the nucleases XPG and XPF/ERCC1 or other nucleases to the sites of transcription initiation and the aberrant insertion of breaks at these sites instead of at the sites of damage (III in Figure 7B). These aberrant breaks at transcriptionally active sites result in the exquisite sensitivity of transcriptional responses to UV in these cells (Figure 1; van Hoffen et al., 1999), which in turn leads to extreme sensitivity of the cells to killing by UV. This model is very speculative and should be regarded as a working hypothesis on which to base future experiments.
Fig. 7. Model for the effects of the defect in XP–D/CS cells. (A) In normal cells (I), transcription initiation utilizes TFIIH in its transcription mode (depicted as an ellipse). A signal from a DNA damaged site recruits TFIIH and converts it into repair mode (depicted as a hexagon) (II) and NER takes place. After incision, TFIIH reverts to its transcriptional configuration (III). (B) In XP–D/CS cells, the signal from the damaged site converts TFIIH into its repair conformation (II), but is unable to recruit it to the site of the damage. Nicks are therefore generated at sites of transcription initiation, instead of at repair sites (III).
The breaks that we observe in cells are generated very soon after exposure of cells to UV damage and are distinct from the general breakdown of DNA that occurs during apoptosis of the cells many hours later.
Relationship to clinical features
Many of the clinical features of XP are readily attributable to UV-hypermutability of skin cells resulting in increased numbers of mutations in critical oncogenes or tumour suppressor genes. The features of CS are, however, much more difficult to explain. Current hypotheses include subtle transcriptional deficiencies (e.g. van Gool et al., 1997) and deficiencies in the repair of oxidative damage (Cooper et al., 1997); CS and XP/CS cells are specifically deficient in transcription-coupled repair of oxidative damage. Based on our observations, an alternative explanation is that, at least in XP–D/CS cells, different types of damage result, via a trans-acting signal, in the introduction of breaks into the genomic DNA at the sites of transcription initiation. This in turn results in reduced levels of gene expression.
Conclusions
We have shown that the introduction of DNA damage into XP–D/CS cells results in the introduction of breaks into the DNA at sites distant from the damage. We propose that these breaks are responsible for the failure of RNA synthesis to recover following even very low doses of UV irradiation (van Hoffen et al., 1999), and this in turn renders the cells very sensitive to killing by UV irradiation. It is possible that these cells exhibit similar responses to other types of DNA damage and this may account for some of the clinical features of these patients.
Materials and methods
Cell culture
Primary human fibroblasts from normal and DNA-repair-deficient individuals (Table I) were cultured in Eagle's minimum essential medium (Gibco-BRL) containing 15% fetal calf serum (FCS) and kept at 37°C in a humidified atmosphere with 5% CO2. For transfection experiments, SV40-transformed pre-crisis fibroblasts were cultured under the same conditions as primary cells but medium was supplemented with 200 μg/ml G418.
UV irradiation
For irradiation, medium was removed and cells were irradiated with the indicated doses of UVB from four Westinghouse FS20 sunlamps and UVC from a Philips 6 W germicidal lamp, which are known to emit primarily in the UVB and UVC ranges, respectively.
ICAM–1 expression
Detection of ICAM–1 mRNA following UVB irradiation has been described previously (Ahrens et al., 1997). In brief, immediately after UVB irradiation cells were washed, cultured in medium and stimulated with 500 U/ml recombinant human interferon–γ (Genzyme). After a 4 h incubation period cells were harvested, total RNA was isolated, and interferon–γ-induced ICAM–1 mRNA expression was determined by semi-quantitative differential RT–PCR, as described previously (Berneburg et al., 2000).
Single cell gel electrophoresis ‘comet’ assay
A layer of 0.7% agarose in phosphate-buffered saline (PBS) was prepared on frosted microscope slides. Fibroblasts were trypsinized for 5 min with trypsin/0.4% EDTA at 37°C, resuspended in PBS and mixed with an equal volume of previously melted low melting point agarose to give a final concentration of 4 × 105 cells/ml in 0.7% agarose. A 50 μl aliquot of the cell/agarose suspension (containing 2 × 104 cells) was added on top of the previously prepared agarose layer on each slide, and a coverslip placed over the cells. Slides were maintained on ice and coverslips were removed for irradiation. Immediately following irradiation, cells were incubated at 37°C in the dark with 100 μM araC and 10 mM HU to allow NER-mediated incisions to accumulate. The slides were subsequently immersed in lysis solution (2.5 M NaCl, 0.2 M NaOH, 100 mM EDTA–Na2, 10 mM Tris base, 10% dimethylsulfoxide and 1% Triton X-100 pH 10) for at least 1 h at 4°C. Slides were then electrophoresed in alkaline conditions (0.3 M NaOH, 1 mM EDTA–Na2) at 20 V for 24 min. Following electrophoresis, slides were washed with neutralizing buffer (0.4 M Tris–HCl pH 7.5) and stained by addition of 35 μl of ethidium bromide solution (20 μg/ml) onto the gel. Comet lengths were determined by fluorescence microscopy and measured employing the Casys system (Synoptics, Cambridge, UK).
For measuring incisions in the absence of inhibitors, cells were incubated in medium containing 0.5% FCS for 1 week to bring them into a non-proliferating state. Cells were then irradiated with the doses indicated, kept for the times indicated in the absence of araC and HU, and the comet assay was conducted as described above.
Human cell extracts and purified repair proteins
Whole-cell extracts were prepared as described previously (Wood et al., 1995) from cell lines listed in Table I. Preparation of purified XPA, XPC–HR23B, XPG, RPA was as described (Aboussekhra et al., 1995; Evans et al., 1997). TFIIH (Heparin-5PW fraction) was purified according to Hwang et al. (1996) and was kindly provided by Dr J.-M.Egly (Strasbourg).
Dual incision
Dual incision assays were carried out as described previously (Evans et al., 1997). Whole-cell extracts were incubated with purified, reconstituted protein. After a pre-incubation period of 10 min at 30°C, DNA substrate containing a single cisplatin residue (50 μg) was added and incubated for 90 min at 30°C. Reaction intermediates were labelled with [α-32P]dCTP and separated on a 14% denaturing SDS gel.
Uncoupled incision
Incisions 3′ and 5′ to the lesion were detected as described previously. The plasmid containing a single cisplatin lesion was cleaved with AvaII and the blunt-end filled with Escherichia coli DNA polymerase I (Klenow fragment) and [32P]dCTP to label the damaged strand 140 nucleotides from the 3′ side of the site of the damage (Constantinou et al., 1999). It was then incubated with different cell extracts.
Repair synthesis
In vitro repair was carried out as described previously (Evans et al., 1997). In brief, reaction mixtures (50 μl) contained 200 ng of platinated or control DNA substrate and whole-cell extract protein (100 μg unless otherwise indicated) in a buffer containing 45 mM HEPES–KOH pH 7.8, 70 mM KCl, 7.5 mM MgCl2, 0.9 mM DTT, 0.4 mM EDTA, 2 mM ATP, 20 μM each of dATP, dCTP, dGTP and dTTP, 22 mM phosphocreatine (di-Tris salt), 2.5 μg of creatine phosphokinase, 3.4% glycerol and 18 μg of bovine serum albumin (Wood et al., 1995). Cell extracts were pre-incubated with buffer at 30°C for 5 min, DNA substrate was added and reactions incubated at 30°C for 30 min or as indicated.
Plasmid preparation
For transfection and microinjection, pcDNA3.1 plasmid was irradiated in a 3 cm dish in 500 μl of H2O with 15 000 J/m2 of UVB. For microinjection the irradiated or an equal amount of unirradiated plasmid was precipitated and resuspended in H2O.
For transfection, irradiated or unirradiated plasmid was precipitated together with pHook–1 plasmid in order to increase cotransfection efficiency.
Cotransfection
The Capture-Tec system (Invitrogen) was used. SV40-transformed pre-crisis fibroblasts (2.5 × 105) were plated in three wells of a 6-well dish and transfected the next day with FuGENE 6 (Boehringer Mannheim) according to the manufacturer's protocol with the prepared mixture of pHook–1 (0.25 μg DNA/transfection) and either unirradiated or irradiated pcDNA3.1 (1 μg/transfection). After a 12 h incubation, cells were washed twice with PBS and then carefully detached with 3 mM EDTA in PBS. The cells were washed together with phOx hapten-linked magnetic beads, bound to the beads, and collected according to the manufacturer's protocol. Transfection efficiency ranged from 0.5 to 2%.
Microinjection
Normal, XP8BR or XP3NE (XP–D) primary fibroblasts were seeded into 3-cm Petri dishes containing a coverslip, previously marked with a 3 mm diameter circle, at 2 × 104 cells per dish and incubated in medium overnight. All of the ∼200 cells within the circle were microinjected using an Olympus inverted microscope equipped with a Narishige 1–MB microinjection apparatus (Narishige, Japan) with either unirradiated or irradiated plasmid at a concentration of 0.1 μg/μl. The cells were then incubated for a further 6 h. The medium was removed and the area containing microinjected cells was sealed off with silicone-covered colony-picking rings. The microinjected cells were covered with trypsin and surplus trypsin was removed immediately. Detached cells were taken off and used in the comet assay as described above.
Apoptosis
In situ detection. Cells (2.5 × 105) were plated in 10–cm Petri dishes, irradiated the next day with 100 J/m2 of UVB and left for the periods indicated. Cells were then trypsinized followed by fixation and staining according to the ApopTag™ Plus Kit (Oncor, Gaithersburg, MD). Apoptotic cells were scored under fluorescence light microscopy.
Western blot analysis for PARP. Cells (1 × 106) were treated as described for in situ apoptosis detection. Cells were washed in PBS three times and the cell pellets were resuspended in an equal volume of 2× SDS buffer and boiled. Standard Western blotting procedures were performed, using as probe anti-PARP p85 Fragment pAb (Promega, UK) specific for the p85 fragment of PARP (poly ADP-ribose polymerase) resulting from caspase cleavage of the intact 116 kDa molecule. An apoptosis positive control was performed by incubating 1 × 106 HL-60 suspension cells with 0.15 mM camptothecin (Sigma) for 4 h and then preparing extracts as described above.
p53 regulation
Primary and pre-crisis SV40-transformed 1BR and XP8BR fibroblasts were exposed to 20 J/m2 (1BR) or 5 J/m2 (XP8BR) UVC and incubated for 24 h. The cells were harvested and the levels of p53 and p21 were analysed by immunoblotting using anti-p53 and anti-p21 antibodies.
Acknowledgments
Acknowledgements
We are indebted to Sue Harcourt and Emily Capulas for assistance with cell culture and apoptosis experiments, and to Patricia Kannouche, Elaine Taylor and Penny Jeggo for helpful comments on the manuscript. This work was supported by DFG Fellowship Be 2005/1-1 to M.B., EC contract ENV4-CT95-0174 and Department of Health Grant 121/6439 to M.H.L.G., EC Concerted Action contract BMH4-CT98-3045 to A.R.L. and M.S., and a grant from the Associazione Italiana per la Ricerca sul Cancro grant to M.S.
References
- Aboussekhra A., et al. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified components. Cell, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- Ahrens C., et al. (1997) Photocarcinogenesis correlates with immunosuppression in DNA-repair-defective individuals. Proc. Natl Acad. Sci. USA, 94, 6837–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berneburg M., Clingen, P.H., Harcourt, S.A., Lowe, J.E., Taylor, E.M., Green, M.H.L., Krutmann, J., Arlett, C.F. and Lehmann, A.R. (2000) The cancer-free phenotype in trichothiodystrophy is unrelated to its repair defect. Cancer Res., 60, 431–438. [PubMed] [Google Scholar]
- Botta E., Nardo, T., Broughton, B.C., Marinoni, S., Lehmann, A.R. and Stefanini, M. (1998) Analysis of mutations in the XPD gene in Italian patients with trichothiodystrophy: site of mutation correlates with repair deficiency but gene dosage appears to determine clinical severity. Am. J. Hum. Genet., 63, 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton B.C., et al. (1995) Molecular and cellular analysis of the DNA repair defect in a patient in xeroderma pigmentosum complementation group D with the clinical features of xeroderma pigmentosum and Cockayne syndrome. Am. J. Hum. Genet., 56, 167–174. [PMC free article] [PubMed] [Google Scholar]
- Coin F., Marinoni, J.C., Rodolfo, C., Fribourg, S., Pedrini, A.M. and Egly, J.M. (1998) Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nature Genet., 20, 184–188. [DOI] [PubMed] [Google Scholar]
- Constantinou A., Gunz, D., Evans, E., Lalle, P., Bates, P.A., Wood, R.D. and Clarkson, S.G. (1999) Conserved residues of human XPG protein important for nuclease activity and function in nucleotide excision repair. J. Biol. Chem., 274, 5637–5648. [DOI] [PubMed] [Google Scholar]
- Cooper P.K., Nouspikel, T., Clarkson, S.G. and Leadon, S.A. (1997) Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science, 275, 990–993. [DOI] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers, N.G. and Hoeijmakers, J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- Evans E., Moggs, J.G., Hwang, J.R., Egly, J.M. and Wood, R.D. (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J., 16, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.H.L., Lowe, J.E., Harcourt, S.A., Akinluyi, P., Rowe, T., Cole, J., Anstey, A.V. and Arlett, C.F. (1992) UV-C sensitivity of unstimulated and stimulated human lymphocytes from normal and xeroderma pigmentosum donors in the comet assay: a potential diagnostic technique. Mutat. Res., 273, 137–144. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H.J., Egly, J.-M. and Vermeulen, W. (1996) TFIIH: a key component in multiple DNA transactions. Curr. Opin. Genet. Dev., 6, 26–33. [DOI] [PubMed] [Google Scholar]
- Hwang J.R., Moncollin, V., Wermeulen, W., Seroz, T., van Vuuren, H., Hoeijmakers, J.H.J. and Egly, J.M. (1996) A 3′–5′ XPB helicase defect in repair/transcription factor TFIIH of xeroderma pigmentosum group B affects both DNA repair and transcription. J. Biol. Chem., 271, 15898–15904. [DOI] [PubMed] [Google Scholar]
- Itin P.H. and Pittelkow, M.R. (1990) Trichothiodystrophy: review of sulfur-deficient brittle hair syndromes and association with the ectodermal dysplasias. J. Am. Acad. Dermatol., 20, 705–717. [DOI] [PubMed] [Google Scholar]
- Kraemer K.H., Lee, M.M. and Scotto, J. (1987) Xeroderma pigmentosum. Cutaneous, ocular and neurologic abnormalities in 830 published cases. Arch. Dermatol., 123, 241–250. [DOI] [PubMed] [Google Scholar]
- Krutmann J., Bohnert, J.E. and Jung, E.G. (1994) Evidence that DNA damage is a mediate in ultraviolet B radiation-induced inhibition of human gene expression: ultraviolet B radiation effects on intercellular adhesion molecule-1 (ICAM–1) expression. J. Invest. Dermatol., 102, 428–432. [DOI] [PubMed] [Google Scholar]
- Lafforet D. and Dupuy, J.-M. (1978) Photosensibilite et reparation de l'ADN: possibilite d'une parente nosologique entre xeroderma pigmentosum et syndrome de Cockayne. Arch. Fr. Pediatr., 35, 65–74. [PubMed] [Google Scholar]
- Mayne L.V., Priestley, A., James, M.R. and Burke, J.F. (1986) Efficient immortalization and morphological transformation of human fibroblasts by transfection with SV40 DNA linked to a dominant marker. Exp. Cell Res., 162, 530–538. [DOI] [PubMed] [Google Scholar]
- Moggs J.G., Yarema, K.J., Essigmann, J.M. and Wood, R.D. (1996) Analysis of incision sites produced by human cell extracts and purified proteins during nucleotide excision repair of a 1,3-intrastrand d(GpTpG)-cisplatin adduct. J. Biol. Chem., 271, 7177–7186. [DOI] [PubMed] [Google Scholar]
- Moriwaki S.-I., et al. (1996) DNA repair and ultraviolet mutagenesis in cells from a new patient with xeroderma pigmentosum group G and Cockayne syndrome. J. Invest. Dermatol., 107, 647–653. [DOI] [PubMed] [Google Scholar]
- Nance M.A. and Berry, S.A. (1992) Cockayne syndrome: review of 140 cases. Am. J. Med. Genet., 42, 68–84. [DOI] [PubMed] [Google Scholar]
- Squires S., Johnson, R.T. and Collins, A.R.S. (1982) Initial rates of DNA incision in UV-irradiated human cells. Differences between normal, xeroderma pigmentosum and tumour cells. Mutat. Res., 95, 389–404. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q., Wang, Z., Feaver, W.J., Wu, X., Bushnell, D.A., Donahue, T.F., Friedberg, E.C. and Kornberg, R.D. (1995) Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell, 80, 21–28. [DOI] [PubMed] [Google Scholar]
- Takayama K., Salazar, E.P., Lehmann, A.R., Stefanini, M., Thompson, L.H. and Weber, C.A. (1995) Defects in the repair and transcription gene ERCC2 in the cancer-prone disorder xeroderma pigmentosum. Cancer Res., 55, 5656–5663. [PubMed] [Google Scholar]
- Taylor E.M. and Lehmann, A.R. (1998) Conservation of eukaryotic DNA repair mechanisms. Int. J. Radiat. Biol., 74, 277–286. [DOI] [PubMed] [Google Scholar]
- Taylor E.M., et al. (1997) Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc. Natl Acad. Sci. USA, 94, 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool A.J., van der Horst, G.T.J., Citterio, E. and Hoeijmakers, J.H.J. (1997) Cockayne syndrome: defective repair of transcription. EMBO J., 16, 4155–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoffen A., Kalle, W.H., Jong-Versteeg, A., Lehmann, A.R., Zeeland, A.A. and Mullenders, L.H. (1999) Cells from XP–D and XP–D-CS patients exhibit equally inefficient repair of UV-induced damage in transcribed genes but different capacity to recover UV-inhibited transcription. Nucleic Acids Res., 27, 2898–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterwijk M.F., Versteeg, A., Filon, R., van Zeeland, A.A. and Mullenders, L.H.F. (1996) The sensitivity of Cockayne's syndrome cells to DNA-damaging agents is not due to defective transcription-coupled repair of active genes. Mol. Cell. Biol., 16, 4436–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood R.D. (1996) DNA repair in eukaryotes. Annu. Rev. Biochem., 65, 135–167. [DOI] [PubMed] [Google Scholar]
- Wood R.D., Biggerstaff, M. and Shivji, M.K.K. (1995) Detection and measurement of nucleotide excision repair synthesis by mammalian cell extracts in vitro. Methods, 7, 163–175. [Google Scholar]