Abstract
Extensive alternative splicing and RNA editing have been documented for the transcript of DmNaV (formerly para), the sole sodium channel gene in Drosophila melanogaster. However, the functional consequences of these post-transcriptional modifications are not well understood. In this study we isolated 64 full-length DmNaV cDNA clones from D. melanogaster adults. Based on the usage of 11 alternative exons, 64 clones could be grouped into 29 splice types. When expressed in Xenopus oocytes, 33 DmNaV variants generated sodium currents large enough for functional characterization. Among these variants, DmNaV5-1 and DmNaV7-1 channels activated at the most hyperpolarizing potentials, whereas DmNaV1-6 and DmNaV19 channels activated at the most depolarizing membrane potentials. We identified an A-to-I editing event in DmNaV5-1 that is responsible for its uniquely low-voltage-dependent activation. The wide range of voltage dependence of gating properties exhibited by DmNaV variants represents a rich resource for future studies to determine the role of DmNaV in regulating sodium channel gating, pharmacology, and neuronal excitability in insects.
Keywords: Sodium channel, Alternative splicing, RNA editing, Drosophila melanogaster, Para, Xenopus oocyte expression system
1. Introduction
Voltage-gated sodium channels are integral transmembrane proteins responsible for the generation of action potentials across the membranes of excitable cells. Mammalian sodium channels consist of a large pore-forming α-subunit and one or more smaller auxiliary β-subunits. The α-subunit is composed of four homologous domains (I–IV), each containing six hydrophobic transmembrane segments (S1–S6). Mammals have nine α-subunit genes which encode sodium channel isoforms with different gating properties and different expression patterns in various cell types, tissues, and developmental stages, presumably to fulfill unique functions in specific neurons (Catterall, 2000; Goldin et al., 2000; Yu and Catterall, 2003). In contrast, insects appear to have only a single sodium channel gene, such as DmNaV (formerly para) in Drosophila melanogaster, encoding the equivalent of the α-subunit of the mammalian sodium channels (Loughney et al., 1989). The sodium channel β-subunits in insects are less well understood, except that a unique transmembrane protein, TipE, is required for robust expression of insect sodium channels in heterologous Xenopus oocytes (Feng etal., 1995; Warmke et al., 1997).
Functionally distinct sodium currents have been documented in various insect neurons, indicating existence of heterogeneous sodium channels with distinct gating properties in vivo (Byerly and Leung, 1988; Lapied et al., 1990, 1999; Saito and Wu, 1991, 1993; Schafer et al., 1994; O'Dowd, 1995; Le Corronc et al., 1999; Grolleau and Lapied, 2000; Wicher et al., 2001; Defaix and Lapied, 2005). A fundamental question is how insects produce heterogeneous sodium channels from a single gene. One possibility is that post-transcriptional modifications of the primary sodium channel transcript result in variations of sodium channel gating properties. Indeed, alternative splicing and RNA editing of the DmNaV transcript have been reported (O'Dowd and Aldrich, 1988; Loughney et al., 1989; Thackeray and Ganetzky, 1994, 1995; O'Dowd, 1995; Warmke et al., 1997; Palladino et al., 2000). The functional consequences of alternative splicing and/or RNA editing of the DmNaV transcript, however, remain largely unknown. Prior to this study, only one full-length DmNaV cDNA clone has been functionally characterized in Xenopus oocytes (Warmke et al., 1997).
Recent characterization of the German cockroach BgNaV sodium channel gene suggests that alternative splicing and RNA editing are two major mechanisms by which cockroaches produce functionally diverse sodium channels (Tan et al., 2002; Liu et al., 2004; Song et al., 2004; Dong, 2007). To determine whether alternative splicing- and RNA editing-mediated generation of diverse sodium channel gating properties is a common phenomenon in insects, we conducted a large-scale functional characterization of alternative splicing and RNA editing variants of the D. melanogaster DmNaV gene. Here, we report the isolation of 64 full-length DmNaV cDNA clones from D. melanogaster and functional characterization of these variants in Xenopus oocytes.
2. Materials and methods
2.1. Synthesis of first-strand cDNAs
Total RNA was isolated from about 100 whole adults of D. melanogaster strain W1118 using the Invitrogen TRIzol Reagent kit (Invitrogen, Rockville, MD). Further isolation of mRNA was performed using the Promega polyA + RNA isolation kit (Promega, Madison, WI). First-strand cDNA was synthesized from mRNA using a DmNaV-specific primer (D-3′–1: 5′-tac tca tgc taa tac tcg cg-3′) based on the sequence immediately downstream of the stop codon (Genbank accession number: M32078), and Super-Script II RNase H-reverse transcriptase (Invitrogen, Rockville, MD). Conditions for the first-strand cDNA synthesis reaction were: 42°C for 2 min followed by a 60 min incubation at 48°C. RNA was removed by 20 min incubation with RNaseH at 37°C.
2.2. Polymerase chain reaction (PCR), cloning of full-length DmNaV cDNA and DNA sequencing
To amplify the complete coding region (6 kb) of DmNaV by PCR, a forward primer (D-DmNaV-KpnI: 5′-cgg ggt acc gcc acc atg gca gaa gat tcc gac tcg-3′) and a reverse primer (D-DmNaV-XbaI: 5′-gct cta gat act gct aat act cgc g-3′) were designed based on the 5′ and 3′ end sequences of the DmNaV open reading frame (ORF). The XbaI and KpnI recognition sequences (bold in the primer sequences) were added to the forward and reverse primers, respectively, to facilitate cloning. A Kozak sequence (bold-italic in the primer sequence) was added to primer D-DmNaV-KpnI for high-efficiency translation. The reaction mixture (50μl) contained 0.5μl cDNA, 50 pmol each primer, 200μM each dNTP, 1 U eLONGase (Invitrogen), 1.5mM MgCl2, and 1 × PCR reaction buffer. PCR was carried out as follows: one cycle of 94 °C for 1 min; 33 cycles of 94 °C for 30 s, 58 °C for 30 s, 68 °C for 8 min; and one cycle of 68 °C for 15 min. The PCR products were purified using the QIAEX II Gel Extraction Kit (QIAGEN Sciences, MD) and cloned into pGH19 (a Xenopus expression vector containing the 3′ and 5′ untranslated regions of the Xenopus β-globin gene at the 3′ end of T7 promoter and 5′ end of SP6 promoter, respectively; and was kindly provided by B. Ganetzky, University of Wisconsin). MAX Efficiency Stbl2-competent cells (Invitrogen) were used as host cells. The DNA sequence was determined at the Research Technology Support Facility on campus using primers that anneal to both strands. Overlapping sequences were assembled and all sequence data underwent standard quality control to remove ambiguity. Sequences of DmNaV inserts in cDNA clones were compared with the Genbank sequence (Accession number: M32078) using DNAStar (DNAStar Inc.).
2.3. Site-directed mutagenesis
Site-directed mutagenesis was performed by PCR using specific primers and Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). All mutagenesis results were verified by DNA sequencing.
2.4. Expression of DmNaV sodium channels in Xenopus oocytes
The procedures for oocyte preparation and cRNA injection were identical to those described previously (Tan et al., 2002). Plasmid DNA containing the DmNaV insert was linearized using the NotI restriction enzyme. The linearized DNA was used for in vitro synthesis of the DmNaV cRNA using in vitro transcription with T7 polymerase (mMESSAGE mMACHINE kit, Ambion). For robust expression of DmNaV sodium channels, DmNaV cRNA (0.25–10 ng) was co-injected into oocytes with the D. melanogaster tipE cRNA (1:1 molar ratio), which enhances the expression of insect sodium channels in oocytes (Feng et al., 1995; Warmke et al., 1997). More than one DmNaV and TipE cRNA preparations were tested for each clone with similar results. Oocytes carrying peak sodium currents of 0.5–2μA were used for functional analysis using two-electrode voltage clamp.
2.5. Electrophysiological recording and analysis
The voltage dependence of activation and inactivation were measured using the two-electrode voltage clamp technique. Methods for two-electrode recording and data analysis were similar to those described previously (Tan et al., 2002).
3. Results and discussion
3.1. Molecular analysis of 64 full-length cDNA clones
Prior to this study, molecular characterization of partial DmNaV cDNA sequences resulted in the identification of eleven alternative exons in the DmNaV transcript (Thackeray and Ganetzky, 1994; O'Dowd et al., 1995; Warmke et al., 1997, Lee et al., 2002). In order to determine the alternative splicing pattern of each full-length DmNaV transcript and to conduct functional expression of each splice type, it was necessary to isolate and characterize a large number of full-length cDNA clones. Using RT-PCR, we obtained 64 full-length DmNaV cDNA clones from adult D. melanogaster. PCR and/or sequencing allowed us to group the 64 clones into 29 splice types according to exon usage. Fig. 1 displays the 29 splice types (numbered 1–29) and number of clones (variants) in each splice type. Variants are named according to splice type. For example, variants in splice type 1 are designated as DmNaV1, and 13 variants belonging to this splice type are named DmNaV1-1 – DmNaV1-13.
Fig. 1.
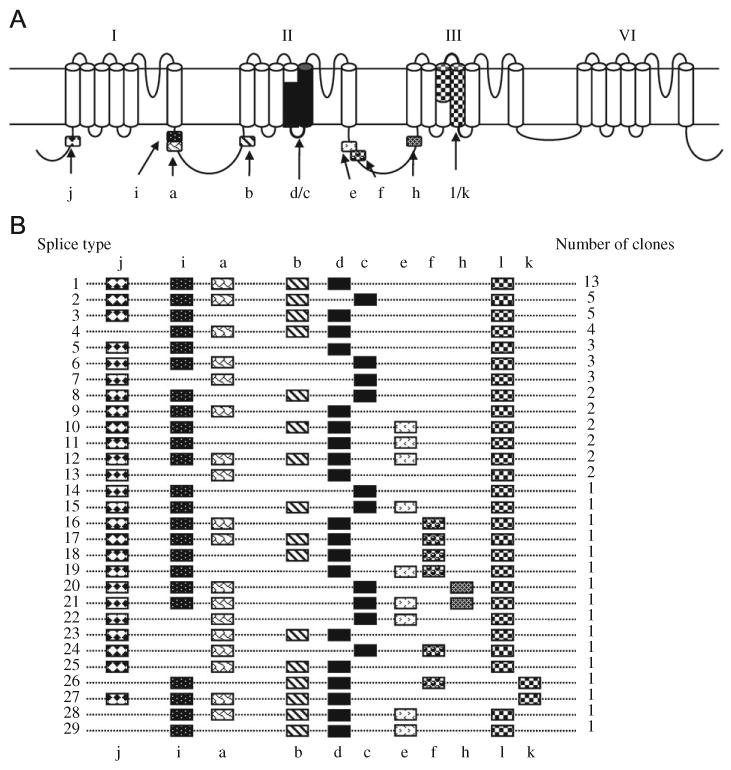
Identification of 29 alternative splice types of the DmNaV transcript. (A) A schematic diagram of the DmNaV sodium channel protein topology, with the positions of alternative exons indicated by patterned boxes. Exons, a, b, i, j, e, f and h are optional, whereas exons c/d and l/k are mutually exclusive. Exons l and k correspond to exons G1 and G2, respectively, in the cockroach BgNav sodium channel (Tan et al., 2002). (B) Schematic presentation of the usage of alternative exons in 64 full-length cDNA clones. The number of clones (i.e., variants) of each splice type is indicated.
We found that exons a, b, d, i, j and l were used in greater than 60% of the DmNaV variants (both D. melanogaster), whereas usage of exons c, e, f, h and k was less frequent (Fig. 1). This observation is consistent with earlier analysis of partial cDNAs from D. melanogaster and D. virilis (O'Dowd et al., 1995; Thackeray and Ganetzky, 1994, 1995). No positive or negative associations of these alternative exons with each other were observed.
3.2. Distinct gating properties of DmNaV variants expressed in Xenopus oocytes
For functional analysis of DmNaV variants, we coexpressed each variant with TipE in Xenopus oocytes and recorded sodium current from oocytes. Roughly half of the variants, 33, produced sodium currents large enough (0.7–2.5μA 1–8 days after cRNA injection) for further analysis. Four variants produced currents that were too small (<0.5μA) for functional analysis. Twenty-seven variants did not generate any detectable sodium current even 8 days after cRNA injection.
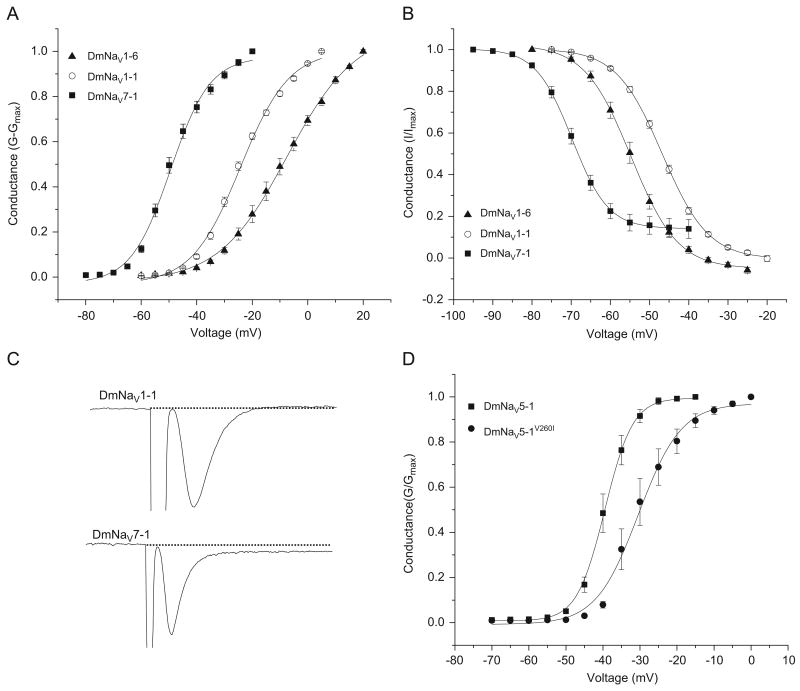
Analysis of the 33 functional variants revealed a broad range of voltage dependence for channel activation and inactivation (Table 1). Figs. 2A and B display the extent of variation in voltage dependence among functional DmNaV clones. The voltages for half-maximal activation (V1/2) of most variants (76%) were within 10 mV range, between −20 and −30 mV. Six variants, DmNaV19, DmNaV15, DmNaV1-6, DmNaV13-2, DmNaV1-3 and DmNaV2-3, required more positive membrane potentials for activation, whereas two variants, DmNaV5-1 and DmNaV7-1, activated at more negative membrane potentials (Table 1). In particular, DmNaV7-1 activated at voltage of <−60 mV (Fig. 2A). The so-called background sodium currents have been reported in cockroach dorsal unpaired median (DUM) neurons in terminal abdominal ganglia, which is believed to play a critical role in providing the depolarizing force needed to maintain pacemaker activity of these neurons (Lapied et al., 1990). The specific sodium channels behind such currents remain elusive. The extremely low-voltage-gating properties of DmNaV5-1 and DmNaV7-1 make them strong candidates for conducting background currents.
Table 1. Gating properties of 33 functional DmNav variants.
| Variant | Activation | Inactivation | ||
|---|---|---|---|---|
| V1/2 (mV) | k (mV) | V1/2 (mV) | k (mV) | |
| DmNaV19 | −6.7±0.5 | 5.2±0.2 | −50.1±0.3 | 6.3±0.1 |
| DmNaV15 | −11.4±0.6 | 7.3±0.2 | −34.9±0.9 | 5.5±0.3 |
| DmNaV1-6 | −12.0±2.1 | 9.8±0.7 | −49.9±0.5 | 5.6±0.9 |
| DmNaV13-2 | −15.5±0.4 | 8.2±0.3 | −45.5±0.4 | 7.5±0.2 |
| DmNaV1-3 | −16.2±0.5 | 8.0±0.2 | −42.3±0.6 | 7.3±0.6 |
| DmNaV2-3 | −20.5±1.6 | 8.0±0.5 | −46.7±0.2 | 4.8±0.2 |
| DmNaV2-2 | −20.6±0.4 | 7.4±0.2 | −39.8±0.4 | 6.1±0.2 |
| DmNaV4-1 | −21.6±0.5 | 6.3±0.2 | −41.1±0.2 | 5.2±0.1 |
| DmNaV3-1 | −22.6±0.9 | 6.6±0.4 | −46.4±0.4 | 5.0±0.1 |
| DmNaV17 | −23.1±1.0 | 7.0±0.3 | −47.4±0.6 | 5.1±0.2 |
| DmNaV28 | −23.3±0.8 | 5.5±0.3 | −39.9±0.5 | 5.4±0.2 |
| DmNaV3-2 | −23.4±0.5 | 6.4±0.2 | −43.8±0.4 | 5.4±0.1 |
| DmNaV1-2 | −23.4±0.8 | 7.5±0.3 | −45.1±0.4 | 5.5±0.1 |
| DmNaV26 | −23.6±0.5 | 3.7±0.2 | −39.7±0.2 | 4.4±0.1 |
| DmNaV10-2 | −23.7±0.6 | 4.5±0.2 | −40.1±0.3 | 4.5±0.2 |
| DmNaV2-1 | −23.8±0.9 | 6.7±0.2 | −48.3±0.5 | 5.3±0.1 |
| DmNaV1-1 | −24.6±0.8 | 7.6±0.3 | −47.3±0.3 | 5.6±0.1 |
| DmNaV23 | −24.9±0.7 | 6.6±0.4 | −46.1±0.4 | 5.2±0.1 |
| DmNaV13-1 | −25.3±0.9 | 4.7±0.3 | −42.9±0.3 | 4.8±0.1 |
| DmNaV2-4 | −25.7±0.6 | 5.6±0.4 | −46.1±0.8 | 4.9±0.1 |
| DmNaV29 | −25.9±1.2 | 4.0±0.3 | −38.9±0.3 | 4.8±0.1 |
| DmNaV14 | −26.0±0.7 | 3.0±0.1 | −32.1±0.3 | 4.0±0.1 |
| DmNaV5-3 | −26.1±0.9 | 5.9±0.3 | −35.1±0.6 | 4.9±0.1 |
| DmNaV20 | −26.2±0.9 | 5.0±0.3 | −45.1±0.3 | 4.9±0.1 |
| DmNaV1-5 | −26.7±0.8 | 4.9±0.2 | −43.5±0.2 | 4.8±0.1 |
| DmNaV10-1 | −26.9±1.1 | 8.1±0.7 | −44.8±1.1 | 4.8±0.8 |
| DmNaV1-4 | −27.2±0.7 | 5.8±0.2 | −45.2±0.4 | 5.4±0.0 |
| DmNaV24 | −27.2±0.8 | 4.9±0.2 | −41.9±0.6 | 5.0±0.1 |
| DmNaV22 | −28.4±1.1 | 4.6±0.2 | −42.0±0.3 | 5.0±0.1 |
| DmNaV27 | −29.1±0.6 | 3.6±0.2 | −42.9±0.4 | 4.5±0.1 |
| DmNaV5-2 | −29.5±0.7 | 10.4±0.4 | −49.2±0.5 | 6.5±0.3 |
| DmNaV5-1 | −37.1±0.6 | 3.4±0.1 | −44.4±0.7 | 4.5±0.1 |
| DmNaV7-1 | −47.7±0.6 | 6.3±0.2 | −66.4±0.2 | 4.5±0.0 |
Note: A minimum of eight oocytes was used to calculate V1/2 and SEM values.
Fig. 2.
Gating properties of representative DmNaV variants. (A) Voltage dependence of activation of DmNaV1-1, DmNaV1-6, and DmNaV7-1. (B) Voltage dependence of steady-state inactivation of DmNaV1-1, DmNaV1-6, and DmNaV7-1. (C) Peak sodium current traces of DmNaV1-1 and DmNaV7-1 indicating a persistent current in DmNaV7-1. (D) Voltage dependence of activation of DmNaV5-1 and DmNaV5-1V260I.
Voltages for half-maximal inactivation (V1/2) were less variable compared with those for activation, with V1/2 values ranging from −33 to −66 mV (Table 1 and Fig. 2B). Interestingly, the V1/2 of inactivation of DmNaV7-1 was shifted significantly in the hyperpolarizing direction compared to other variants. Inactivation was complete by the end of a 20 ms depolarization for most variants including DmNaV1-1. However, five variants, DmNaV22, DmNaV14, DmNaV5-1 and DmNaV7-1 and DmNaV5-3, exhibited a persistent current about 10% of the peak current remained at the end of the 20 ms depolarizing pulse, as shown for DmNaV7-1 (Fig. 2C). This persistent current was also evident in the steady-state inactivation curve of DmNaV7-1, where the foot of the curve is lifted at depolarizing potentials (Fig. 2B).
Surprisingly, among functional DmNaV variants, there was no apparent correlation between splice types and gating properties (Table 1, Figs. 2A and B). For example, the gating properties of six splice type 1 variants (DmNaV1-1–DmNaV1-6) are significantly different (P> 0.05) based on a one-way ANOVA with Scheffe's analysis. This finding suggests that other post-transcriptional modifications (e.g., RNA editing) might play a primary role in determining the voltage dependence of activation and inactivation of DmNaV variants.
3.3. Nonfunctional DmNaV variants
Although not characterized further in this study, the 31 “nonfunctional” or poorly functional clones may be a potential source for future in-depth structure–function studies of the DmNaV channel. Several explanations can be proposed for the production of nonfunctional or poorly functional variants. First, some nonfunctional variants may contain premature stop codons. While the role of truncated, nonfunctional sodium channels are not understood, specific alternative splicing events have been identified in both mammals and insects that result in nonfunctional, truncated two-domain variants (Plummer et al., 1997; Tan et al., 2002; Wang et. al., 2003). Second, some variants may contain exon b, which was previously shown in several cockroach variants to suppress sodium channel expression in Xenopus oocytes (Song et al., 2004). However, our analysis showed that 18 of the 32 functional variants contain exon b (Table 1), suggesting that this exon is not the sole determinant of channel expression in Xenopus oocytes, but that other yet-to-be-identified sequence features are also involved. Identification of these other sequence features should improve our understanding of the mechanisms of intramolecular regulation of sodium channel gating. Third, it is also possible that some of these nonfunctional variants could be artifacts of the cloning, such as introduction of stop codon during RT-PCR cloning, or deletion of a stretch of sequence in the coding region. Finally, we used TipE as an auxiliary protein in the expression of DmNaV variants in Xenopus oocytes following the protocol of Warmke et al. (1997). It is possible that some variants require an auxiliary protein that is different from TipE for functional expression in this system. Recently, four TipE-homologous genes (TEH1-4) were identified in D. melanogaster (Derst et al., 2006). Interestingly, co-expression of three of these genes (TEH1-3) with a DmNaV variant in Xenopus oocytes elevated sodium current expression (Derst et al., 2006). In future it would be interesting to determine whether functional expression of some “nonfunctional” DmNaV variants requires a TEH protein, instead of TipE.
4. Sequence analysis of six DmNaV variants
A comprehensive sequence analysis of the 64 full-length DmNaV clones and site-directed mutagenesis of variant-specific amino acid changes are eventually required in order to identify those specific amino acid sequences that are responsible for the gating property differences reported in this paper. Furthermore, it would be important to determine whether these specific changes are caused by RNA editing, alternative splicing, or PCR errors introduced during RT-PCR cloning. Toward this goal, we have sequenced five functional variants (DmNaV5-1 and DmNaV7-1, DmNaV1-1, DmNaV1-6 and DmNaV19) that represent a wide range of gating properties (Table 2). As mentioned above, DmNaV5-1 and DmNaV7-1 are activated at low (i.e., more hyperpolarizing) voltages, DmNaV1-6 and DmNaV19 at high (i.e., more depolarizing) voltages, and DmNaV1-1 at an intermediate voltage (Fig. 2 and Table 1). Because we isolated the full-length clones by RT-PCR, we expected that some of nucleotide changes would result from random PCR errors. However, such random errors are highly unlikely if a nucleotide change occurs in independent clones that belong to different splice types. Based on this criterion, we could attribute a total of nine amino acid changes (in the upper part of Table 2) to A-to-I RNA editing. Whether these changes are the result of RNA editing remain to be determined because some of the sequence differences could be caused by single-nucleotide polymorphism between the strain (unspecified) in the Genbank and strain W1118 used in this study.
Table 2. Common and unique amino acid changes in five DmNaV variants.
| DmNaV1-1 | DmNaV7-1 | DmNaV5-1 | DmNaV19 | DmNaV1-6 | n.t. Δ | Location |
|---|---|---|---|---|---|---|
| R52Q | R52Q | R52Q | g–a* | N term. | ||
| E299Q | E299Q | E299Q | E299Q | E299Q | g–c | IS5–S6 |
| Q471R | Q471R | a–g* | L1 | |||
| R1296Q | R1296Q | g–a* | L2 | |||
| D1300N | D1300N | g–a* | IIIS1 | |||
| L1363F | L1363F | L1363F | L1363F | L1363F | g–c | IIIS2–S3 |
| I1661V | I1661V | I1661V | a–g | IVS2 | ||
| I1670M | I1670M | a–g | IVS2 | |||
| I1679V | I1679V | I1679V | a–g | IVS3 | ||
| I1691V | I1691V | I1691V | I1691V | a–g | IVS3 | |
| N1822S | N1822S | N1822S | a–g | IVS5–S6 | ||
| I260T | t–c | IS4–S5 | ||||
| I260V | a–g | IS4–S5 | ||||
| K303R | a–g | IS5–S6 | ||||
| W319R | t–c | IS5–S6 | ||||
| R437K | g–a | L1 | ||||
| L486P | t–c | L1 | ||||
| H487N | c–a | L1 | ||||
| D656G | a–g | L1 | ||||
| M846V | a–g | IIS1–S2 | ||||
| A859T | g–a | IIS2 | ||||
| F885L | t–c | IIS3 | ||||
| S922P | t–c | IIS4 | ||||
| V1420A | t–c | IIIS4–S5 | ||||
| V1436A | t–c | IIIS5 | ||||
| Q1450R | a–g | IIIS5 | ||||
| N1463Y | a–t | IIIS5–S6 | ||||
| I1519V | a–g | IIIS5–S6 | ||||
| S1587N | g–a* | L3 | ||||
| H1676R | a–g | IVS2–S3 | ||||
| A1731V | c–t | IVS3 | ||||
| H1772Y | c–t | IVS5 | ||||
Note: Amino acid sequences of these variants were compared with the GenBank sequence (Genbank accession number: M32078). Nucleotide bases that correspond to these in the DmNaV genomic sequence (FlyBase ID FBgn 003036) are in bold. Asterisks (*) represent the RNA editing events identified by Palladino et al. (2000). L1, L2, and L3 represent the three intracellular linkers connecting domains I and II, domains II and III, and domains III and IV, respectively.
Four of the nine A-to-I editing (Table 2) were previously reported in the DmNaV transcript (Palladino et al., 2000). However, these RNA editing events are detected in both low-voltage-activated and high-voltage-activated variants and are therefore not likely responsible for the unique gating properties of these variants. In addition to the A-to-I editing, E299Q and L1363F were found in all five sequenced variants; both were caused by C to G changes. Finally, at the amino acid position 260, two separate changes, one from I to V and the other from I to T, were found in DmNaV5-1 and DmNaV7-1, respectively. Further sequencing of the corresponding regions in other 59 full-length clones reveal one more clone, DmNaV4-2, which belongs to a different splice type, and also contains an I260 V change. DmNaV4-2 was not functional in Xenopus oocytes, presumably caused by another unidentified sequence(s) in this variant. The I260 V amino acid change results from an a(tc) to g(tc) nucleotide substitution. Because it is highly unlikely that two DmNaV cDNA clones from different splice types would have the PCR errors at the same nucleotide, we concluded that the I260 V change in DmNaV5-1 is caused by RNA editing. Examination of the genomic sequence of the DmNaV gene revealed the a(tc) codon at the corresponding position. Therefore, the I260 V change is the result of A-to-I editing. Next, we conducted experiments to determine whether the I260 V change in DmNaV5-1 is responsible for the low-voltage dependence of activation. We substituted V260 with I260 in DmNaV5-1. Interestingly, the V260I substitution shifted the voltage dependence of activation by 6 mV in the depolarizing direction (Fig. 2D), demonstrating that this A-to-I editing event is responsible for the low-voltage dependence of activation of DmNaV5-1.
In summary, we report here the functional characterization of the largest collection of insect sodium channel variants to date. Further site-directed mutagenesis experiments are currently underway to identify potentially all amino acid changes/RNA editing events that are responsible for variant-specific gating properties. This work lays a foundation for further studies to understand the role of diverse DmNaV variants in regulating neuronal excitability in insects.
Acknowledgments
We thank Dr. Kris Silver and anonymous reviewers for critical review of this manuscript. The work was supported by a National Science Foundation Grant (IBN 9808156).
References
- Byerly L, Leung HT. Ionic current of Drosophila neurons in embryonic cultures. J Neurosci. 1988;8:4379–4393. doi: 10.1523/JNEUROSCI.08-11-04379.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- Defaix A, Lapied B. Role of a novel maintained low-voltage-activated inward current permeable to sodium and calcium in pacemaking of insect neurosecretory neurons. Invert Neurosci. 2005;5:135–146. doi: 10.1007/s10158-005-0003-x. [DOI] [PubMed] [Google Scholar]
- Derst C, Walther C, Veh RW, Wicher D, Heinemann SH. Four novel sequences in Drosophila melanogaster homologous to the auxiliary DmNaV sodium channel subunit TipE. Biochem Biophys Res Commun. 2006;339:939–948. doi: 10.1016/j.bbrc.2005.11.096. [DOI] [PubMed] [Google Scholar]
- Dong K. Insect sodium channels and insecticide resistance. Invert Neurosci. 2007;7:17–30. doi: 10.1007/s10158-006-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Deák P, Chopra M, Hall LM. Cloning and functional analysis of TipE, a novel membrane protein that enhances Drosophila DmNaV sodium channel function. Cell. 1995;82:1001–1011. doi: 10.1016/0092-8674(95)90279-1. [DOI] [PubMed] [Google Scholar]
- Goldin A, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Netter YB, Noda M, Tamkun MM, Waxman SG, Wood JN, Catterall WA. Nomenclature of voltage-gated sodium channels. Neuron. 2000;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Grolleau F, Lapied B. Dorsal unpaired median neurones in the insect central nervous system: towards a better understanding of the ionic mechanisms underlying spontaneous electrical activity. J Exp Biol. 2000;203:1633–1648. doi: 10.1242/jeb.203.11.1633. [DOI] [PubMed] [Google Scholar]
- Lapied B, Malecot CO, Pelhate M. Patch-clamp study of the properties of the sodium current in cockroach single isolated adult aminergic neurons. J Exp Biol. 1990:387–403. [Google Scholar]
- Lapied B, Stankiewicz M, Grolleau F, Rochat H, Zlotkin E, Pelhate M. Biophysical properties of scorpion alpha-toxin-sensitive background sodium channel contributing to the pacemaker activity in insect neurosecretory cells (DUM neurons) Eur J Neurosci. 1999;11:1449–1460. doi: 10.1046/j.1460-9568.1999.00554.x. [DOI] [PubMed] [Google Scholar]
- Le Corronc H, Hue B, Pitman RM. Ionic mechanisms underlying depolarizing responses of an identified insect motor neuron to short periods of hypoxia. J Neurophysiol. 1999;81:307–318. doi: 10.1152/jn.1999.81.1.307. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ingles PJ, Knipple DC, Soderlund DM. Developmental regulation of alternative exon usage in the house fly Vssc1 sodium channel gene. Invert Neurosci. 2002;4:125–133. doi: 10.1007/s10158-001-0014-1. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Song WZ, Dong K. Persistent tetrodotoxin-sensitive sodium current resulting from U-to-C RNA editing of an insect sodium channel. Proc Natl Acad Sci USA. 2004;101:11862–11867. doi: 10.1073/pnas.0307695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K, Kreber R, Ganetzky B. Molecular analysis of the DmNaV locus, a sodium channel gene in Drosophila. Cell. 1989;58:1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- O'Dowd D. Voltage-gated currents and firing properties of embryonic Drosophila neurons grown in a chemically defined medium. J Neurobiol. 1995;27:113–126. doi: 10.1002/neu.480270111. [DOI] [PubMed] [Google Scholar]
- O'Dowd DK, Aldrich RW. Voltage-clamp analysis of sodium channels in wild-type and mutant Drosophila neurons. J Neurosci. 1988;8:3633–3643. doi: 10.1523/JNEUROSCI.08-10-03633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- Plummer NW, McBurney MW, Meisler MH. Alternative splicing of the sodium channel SCN8A predicts a truncated two-domain protein in fetal brain and non-neuronal cells. J Biol Chem. 1997;272:24008–24015. doi: 10.1074/jbc.272.38.24008. [DOI] [PubMed] [Google Scholar]
- Saito M, Wu CF. Expression of ion channels and mutational effects in giant Drosophila neurons differentiated from cell-division arrested embryonic neuroblasts. J Neurosci. 1991;11:2135–2150. doi: 10.1523/JNEUROSCI.11-07-02135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Wu CF. Ionic channels in cultures Drosophila neurons. EXS. 1993;63:366–389. doi: 10.1007/978-3-0348-7265-2_20. [DOI] [PubMed] [Google Scholar]
- Schafer S, Rosenboom H, Menzel R. Ionic currents of Kenyon cells from the mushroom body of the honeybee. J Neurosci. 1994;14:4600–4612. doi: 10.1523/JNEUROSCI.14-08-04600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Liu Z, Tan J, Nomura Y, Dong K. RNA editing generates tissue-specific sodium channels with distinct gating properties. J Biol Chem. 2004;279:32554–32561. doi: 10.1074/jbc.M402392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JG, Liu ZQ, Nomura Y, Goldin AL, Dong K. Alternative splicing of an insect sodium channel gene generates pharmacologically distinct sodium channels. J Neurosci. 2002;22:5300–5309. doi: 10.1523/JNEUROSCI.22-13-05300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackeray J, Ganetzky B. Developmentally regulated alternative splicing generates a complex array of Drosophila DmNaV sodium channel isoforms. J Neurosci. 1994;14:2569–2578. doi: 10.1523/JNEUROSCI.14-05-02569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackeray J, Ganetzky B. Conserved alternative splicing patterns and splicing signals in the Drosophila sodium channel gene DmNaV. Genetics. 1995;141:203–214. doi: 10.1093/genetics/141.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Huang ZY, Dong K. Molecular characterization of an arachnid sodium channel gene from the varroa mite (Varroa destructor) Insect Biochem Mol Biol. 2003;33:733–739. doi: 10.1016/s0965-1748(03)00068-7. [DOI] [PubMed] [Google Scholar]
- Warmke J, Reenan RA, Wang P, Qian S, Arena JP, Wang J, Wunderler D, Liu K, Kaczorowski GJ, Van der Ploeg LH, Ganetzky B, Cohen CJ. Functional expression of Drosophila DmNaV sodium channels. Modulation by the membrane protein TipE and toxin pharmacology. J Gen Physiol. 1997;110:119–133. doi: 10.1085/jgp.110.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Walther C, Wicher C. Non-synaptic ion channels in insects—basic properties of currents and their modulation in neurons and skeletal muscles. Prog Neurobiol. 2001;64:431–525. doi: 10.1016/s0301-0082(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Yu F, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]