Abstract
KLF8 regulates target genes by recruiting the p300 and PCAF co-activators via glutamines (Q) 118 and 248, the CtBP co-repressor to 86PVDLS90 or SUMO to lysine (K) 67. Here we examined how these interactions coordinate to regulate KLF8 transactivity. Mass spectrometry and immunoprecipitations determined that p300 and/or PCAF promoted KLF8 acetylation at K67, K93, and K95 and this acetylation was abolished in lysine-to-arginine (R) mutants. Treatment with HDAC inhibitors or expression of co-activators inhibited sumoylation at K67. K93R or K95R mutation exerted high levels of sumoylation while the acetylation mimetic mutations K93Q and K95Q blocked the sumoylation. Interestingly, CtBP promoted sumoylation at K67 of wild-type but not AVALF mutant KLF8, and KLF8 interaction with CtBP was inhibited by treatment with the HDAC inhibitors, ectopic expression of the co-activators, or K93Q or K95Q mutation. Promoter reporter assays showed that CtBP inhibited KLF8 transactivity which was rescued by PCAF or p300 expresson. Finally, KLF8-mediated cyclin D1 protein expression and cell cycle progression were significantly decreased in the K93R and K95R but increased in the K93Q, K95Q, K67R or K67Q mutant. Taken together, these results identified a novel mechanism by which co-activators promote KLF8 transactivity by competing with SUMO for K67 modification and by acetylating K93 and K95 to inhibit CtBP-induced K67 sumoylation.
Keywords: Krüppel-like factor 8 (KLF8), histone acetyltransferase (HAT), small ubiquitin modifier (SUMO), p300, p300/CBP associated factor (PCAF), histone deacetylase (HDAC), C-terminal binding protein (CtBP), acetylation and sumoylation
Introduction
KLF8 belongs to the Krüppel-like transcription factor (KLF) family, is frequently overexpressed in cancer and regulates many cancer-related processes including cell cycle progression [1-8], oncogenic transformation [9], epithelial-to-mesenchymal transition, migration and invasion [7, 10, 11]. KLF8 functions as both a repressor [3, 10, 12-14] or activator [1, 3-6] by recruiting the C-terminal binding protein (CtBP) co-repressor via the 86PVDLS90 repression motif or the p300 and PCAF histone acetyltransferase (HAT) co-activators via the Q118 and Q248 residues to target gene promoters. Known KLF8 target genes include γ-globin [12, 13], KLF4 [3], E-cadherin [10] and cyclin D1 [1, 3-5].
The expression of KLF8 is positively regulated by a signaling cascade that involves FAK, PI3K, Src and Sp1 [1, 2, 15, 16]. It is also regulated by other KLF family members KLF1 (positively) and KLF3 (negatively) [17]. In addition, KLF8 function is also regulated posttranslationally by sumoylation at K 67 [3].
The p300 and PCAF co-activators catalyze lysine acetylation on histone tails [18] as well as the transcriptional activator [19] when recruited to a target gene promoter. Acetylation of the transcription factor can regulate its protein stability, DNA binding, gene expression, localization and/ or protein interactions [20]. Sumoylation occurs through a multi-enzymatic pathway analogous to ubiquitination, but with small ubiquitin-related modifier (SUMO)-specific E1, E2 Ubc9, and E3 ligases that target the ΨKXE consensus motif within the substrate protein [21]. Interestingly, acetylation and sumoylation often mutually regulate each other [22].
The CtBP co-repressor functions as a homodimer that binds the PXLDS motif within the repressor [23, 24] and recruits other regulators such as histone deacetylases (HDACs) or methylases to target gene promoters. Interestingly, CtBP also recruits the SUMO E2 or E3 to the repressor to enhance sumoylation [23, 24].
Whether KLF8 is directly acetylated and whether collectively regulated by interaction between the p300/PCAF, CtBP and SUMO-mediated posttranslational modifications have not been studied to date.
In this study we demonstrate that p300 and PCAF acetylate KLF8 at the K67 to directly inhibit sumoylation at this site or at the K93 and K95 to indirectly inhibit the sumoylation by interfering with the CtBP binding to KLF8. This novel mechanism of posttranslational switch between acetylation and sumoylation allows for proper regulation by KLF8 of the cyclin D1 expression and the cell cycle progression.
Materials and methods
Cell culture and transfection
HEK293 and T80 human ovarian epithelial cells [25] were maintained in DMEM supplemented with 10% fetal bovine serum. Transfections were performed using Lipofectamine 2000 according to the manufacturer's instructions.
Plasmid construction
The pKH3 and pKH3-KLF8 expression plasmids were previously described [1], and KLF8 mutants were generated as previously described [4]. Point mutation specific primers include KLF8-K93R (forward- TTT CAC AGG CCC AAG GC, reverse- TTG GGC CTG TGA AAG G), KLF8-K93Q (forward- TTT CAC CAG CCC AAG GC, reverse-TTG GGC TGG TGA AAG G), KLF8-K95R (forward - AAG CCC AGG GCT CC, reverse- AGG AGC CCT GGG CTT G), KLF8-K95Q (forward- AAG CCC CAG GCT CC, reverse- AGG AGC CTG GGG CTT G), KLF8-K67Q (forward- ATG ACA TCC AGA TTG AGC C, reverse- TCA ATC TGG ATG TC), pKH3-KLF8-K67R was previously generated [3]. Correct mutations were confirmed by DNA sequencing and protein expression and nuclear localization was verified. The pHAN-SUMO-1 and pKH3-KLF8-Q118N-Q248N were previously described [3, 4]. The pKH3-His-CtBP was generated using PCR where pQE32-CtBP was used as a template and primers were designed to generate a 5’ SalI restriction site (ACG CGT CGA CAT GAG AGG ATC G) and a 3’ EcoRI site (CGG AAT TCC TAC AAC TGG TCA C). This construct was then inserted into pKH3 at these sites. The pCX/Flag-PCAF, PCAFΔHATA (Δ579-608), PCAFΔHATB(Δ609-624), pCMVβ-HA-p300, pCL/p300(ΔH), pRc/ RSV/CBP, and pRc/RSV/CBP(HAT-) (F1541A mutation) were previously described [26-32].
Mass spectrometry analysis
HEK293 cells were transfected with HA-KLF8 (2 μg) and either p300 (10 μg), CBP (10 μg), or PCAF (8 μg). pKH3 empty vector was included in KLF8 alone or KLF8 + PCAF transfections so total DNA was equal to KLF8 + p300 or KLF8 + CBP. Cells were treated with HDAC inhibitors (5 μM trichostatin A (TSA), 20 μM sodium butyrate (NaBut), and 20 μM nicotinamide (NIA)) for 24 h. For each experiment confluent cells from 10 100-mm dishes were lysed with NP-40 buffer containing protease and HDAC inhibitors (1 mM Na3VO4, 1 mM PMSF, 20 μg/ml leupeptin, 0.06 TIU/ml aprotinin, 0.5 μM TSA, 2 μM NaBut, and 2 μM NIA). Pre-cleared cell lysates were incubated with anti-HA conjugated beads for 24 h. Immunoprecipitates were washed three times, eluted with SDS buffer, resolved on an SDS-PAGE gel and stained with Coomassie blue. Stained gel pieces were washed, reduced, alky-lated, and in-gel tryptic or chymotryptic digested. Proteolytic peptides were extracted from the gel, and peptides were concentrated and reconstituted in 10 μl of 5% formic acid followed by 5% liquid chromatography tandem mass spectrometry analysis using a Waters ESI Q-TOF2 system. In house MASCOT 2.2 from Matrix Science (London, UK) was used to assist in the interpretation of tandem mass spectra
Immunoprecipitation and western blotting
For KLF8 acetylation experiments HEK293 cells were co-transfected similarly as described in the Mass Spectrometry section. The cell lysates were incubated with HA rabbit antibody (HA-Y11, Santa Cruz Biotechnology) or control rabbit IgG overnight, followed by a 2 h rotation with Protein A/G beads. Western blotting using an antibody specific for acetylated lysines (Millipore, clone 4G12) or HA (HA-F7, Santa Cruz Biotechnology) were used. For KLF8 sumoylation experiments 0.2 μg HA-KLF8 was trans-fected with 0.2 Myc-SUMO-1 [3]. Cells were lysed with NP40-lysis buffer supplemented with 20 μM of the desumoylase inhibitor n-ethylmaleimide (Sigma) and immunoprecipitation and western blotting was performed as described above, except detection of Myc-SUMO-1 was done using a Myc specific antibody (9E10, Santa Cruz Biotechnology). Detection of endogenous cyclin D1 or actin was determined using a cyclin D1 (M-20, Santa Cruz Biotechnology) or actin (C4, Santa Cruz Biotechnology) specific antibody.
Promoter luciferase reporter assays
The experiments were performed similarly as previously described [4]. Briefly, T80 cells were grown to ∼75% confluence in a 12-well dish when the cyclin D1 promoter luciferase reporter vector (0.1 μg) was co-transfected into cells with 0.1 μg of the expression vectors encoding WT-or KLF8 mutants in combination with 0.5μg PCAF, 0.5 μg of p300, or 0.1 μg CtBP, along with the pRlSV40 Renilla luciferase reporter internal control plasmid (4 ng). Empty vector was trans-fected in order to ensure all samples were transfected with a total of 0.8 μg DNA. After 16 h, cell lysates were prepared and luciferase activity was analyzed using the Dual Luciferase Reporter Assay System (Promega) and the 20/20n Luminometer (Turner Biosystems).
BrdU incorporation assays
T80 cells were transfected with 0.5-μg pKH3 vector, WT-KLF8, or the KLF8 mutant constructs. BrdU incorporation rates were examined as previously described [2-4].
Results
KLF8 is acetylated at lysines 93, 95 and 67
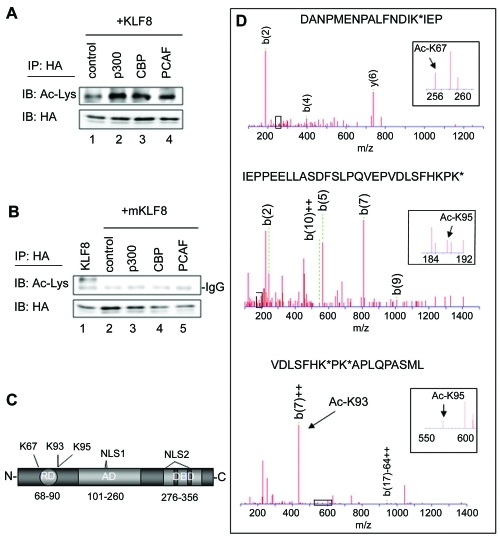
The KLF8 activation domain has been mapped to amino acids 101-260, where glutamines 118 and 248 are required for the recruitment of the p300, CBP, and PCAF co-activators [4]. Additionally, we have shown that p300 and PCAF are required for KLF8 mediated activation of the cyclin D1 promoter [4]. As is the case with many other transcription factors, we wondered whether these co-activators regulate KLF8 tran-scriptional activity by acetylating it. In order to test this, we transfected HA-KLF8 alone, or with p300, CBP, or PCAF and immunoprecipitated the cell lysates using an anti-HA antibody and blotted using an anti-acetyl lysine antibody. We found that KLF8 exerts a low basal level of ace-tylation, but co-expression of p300, CBP, or PCAF greatly increases KLF8 acetylation levels (Figure 1A, top panel, compare lanes 2, 3 and 4 with 1). In contrast, the co-activators do not exert an increase in acetylation when co-expressed with the KLF8 activation domain mutant (mKLF8) that cannot interact with the co-activators [4] (Figure 1B, top panel, compare lanes 2, 3, 4, and 5 with 1).
Figure 1.
KLF8 is acetylated by p300, CBP and PCAF at lysines 67, 93 and 95. (A) p300, CBP, and PCAF promote KLF8 acetylation. HEK293 cells were transfected with HA-KLF8 and either empty vector control, p300, CBP, or PCAF. Lysates were immunoprecipitated (IP) using an anti-HA antibody and blotted (IB) using an anti-acetyl lysine or anti-HA antibody. (B) KLF8 cannot be acetylated when it does not interact with co-activators. Experiments were performed similarly as in A, but the Q118N-Q248N-KLF8 mutant that cannot bind to the co-activators (mKLF8) was transfected with empty vector control or with p300, CBP, or PCAF. Expression of the transfected constructs was verified by whole cell lysate blotting (data not shown). (C) Schematic diagram of KLF8, C-terminal DNA binding domain (DBD), non-classical nuclear localization signals (NLS1&2), N-terminal repression domain (RD), and a centrally located activation domain (AD). The amino acid location of each domain is indicated below. The location of lysines 67, 93, and 95 is also indicated. (D) Mass spectrometry identification of acetylated residues. Immunoprecipitated HA-KLF8 was resolved on an SDS-PAGE gel, stained with Coomassie (data not shown), digested with chymotrypsin or trypsin and prepared for mass spectrometry analysis as described in Materials and Methods. N-terminal b ions or C-terminal y ions were analyzed using the Mascot software. The identified peptide sequence and its MS/MS spectrum of fragmentation when cells were co-transfected with KLF8 and PCAF (top and middle) or KLF8 and p300 (bottom) are depicted. Inset: peaks that represent a lysine with the 42 dalton additional mass representative of an acetylated state. *=acetylated lysines, m/z=massto charge ratio.
Next we wanted to determine the specific lysines on KLF8 that are acetylated by each co-activator. We performed mass spectrometry where we co-expressed HA-KLF8 with p300, CBP, or PCAF; immunoprecipitated for HA-KLF8; resolved on an SDS-PAGE gel (data not shown); and subjected the sample to mass spectrometry analysis. Interestingly, we found that the lysines 67, 93 and 95 are major acetylation targets among numerous lysines acetylated, and the co-activators show both overlapping and distinct patterns of acetylation. The KLF8 sumoylation site lysine 67 was found to be acetylated by PCAF (Figure 1D, top), lysine 93 was identified as a site that is acetylated by p300 and lysine 95 as a site that is acetylated by both PCAF and p300 (Figure 1D, middle & bottom). These lysines are important because they are adjacent to the CtBP binding motif of KLF8 (Figure 1C), and acetylation in the proximity of such a motif has been shown to inhibit CtBP interactions with other transcription factors [33-37].
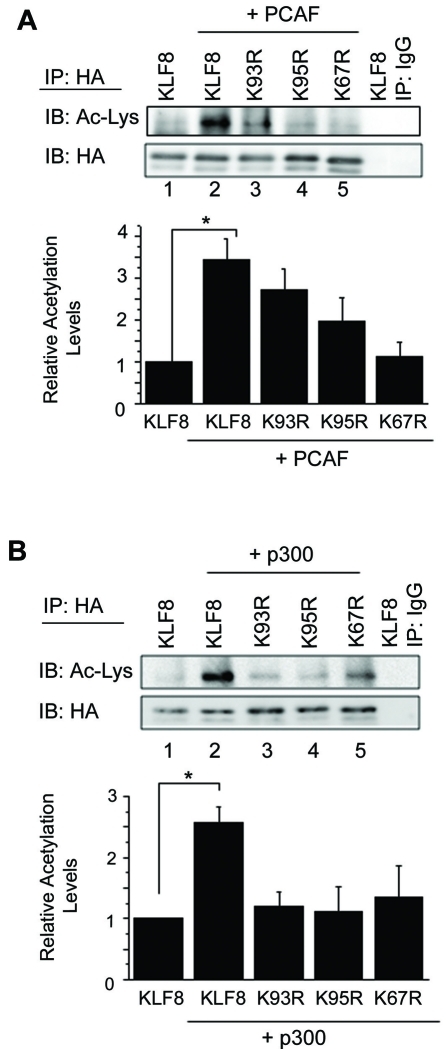
In order to verify the mass spectrometry results we generated HA-KLF8 point mutants where lysine 93, 95 or 67 was mutated to an arginine (K-to-R). These constructs were transfected with PCAF or p300, and immunoprecipitation for HA-KLF8 and western blotting for acetyl-lysine was performed. We found that co-expression of PCAF with WT or K93R KLF8 causes an increase in acetylation compared to KLF8 alone (Figure 2A, compare lanes 2 and 3 with 1), but PCAF cannot induce an increase in acetylation when co-expressed with the K95R or K67R mutants (Figure 2A, compare lanes 4 and 5 with 2). This suggests that lysines 95 and 67 are in fact targets for PCAF mediated KLF8 acetylation as indicated in the mass spectrometry results. Next, we found that the co-expression of p300 can induce acetylation of KLF8 (Figure 2B, compare lane 2 with 1), and surprisingly, this increase was dramatically reduced when each of the three lysines was mutated (Figure 2B, compare lanes 3, 4 and 5 with 2). While the mass spectrometry results only identified lysines 93 and 95 as p300 acetylation sites, the fact that we do not see an increase in acetylation in the K67R mutant when p300 is co-expressed suggests that lysine 67 may be a p300 acetylation site, or that p300 is required for PCAF mediated acetylation of lysine 67.
Figure 2.
Lysine 93 is acetylated by p300 and lysines 95 and 67 are acetylated by p300 and PCAF. (A) HEK293 cells were co-transfected with PCAF and KLF8 or its KàR mutants as indicated. Lysates were subjected to immunoprecipitation using an anti-HA antibody and were blotted using an anti-acetyl-lysine or anti-HA antibody. The histogram depicts acetyla-tion levels relative to KLF8, which was normalized to 1. Results represent mean ± S.E. where n > 3. *, p<0.05. (B) Experiments were performed similarly as in A except that p300 was co-transfected as indicated. Expression of the transfected constructs was verified by whole cell lysate blotting (data not shown).
Histone acetyltransferases negatively regulate KLF8 sumoylation
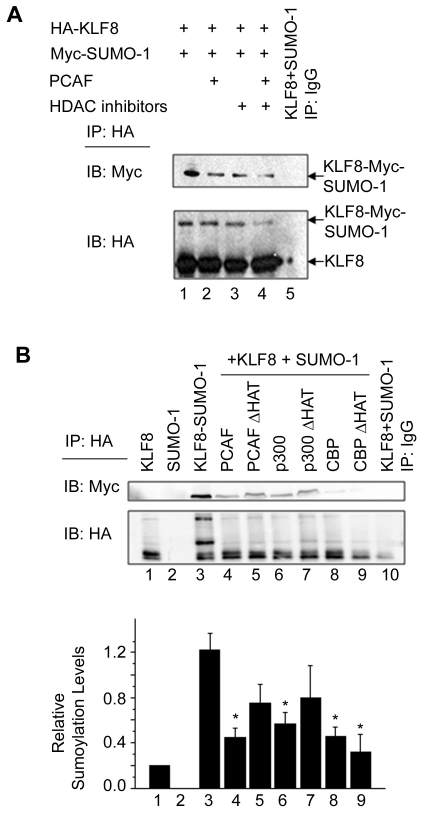
Since we identified lysine 67 as a site for both sumoylation and acetylation, we wondered whether these post-translational modifications compete with each other. In order to test this, we co-transfected HA-KLF8 and Myc-SUMO-1, and either transfected PCAF, treated with HDAC inhibitors, or both. Immunoprecipitation of HA-KLF8 followed by western blottingof Myc-SUMO-1 shows that co-expression of PCAF or treatment with HDAC inhibitors causes a decrease in KLF8 sumoylation levels (Figure 3A, top panel, compare lanes 2 and 3 with 1), and PCAF expression with treatment of HDAC inhibitors causes an even greater decrease in KLF8 sumoylation levels (Figure 3A, top panel, compare lane 4 with 2 and 3).
Figure 3.
KLF8 sumoylation is negatively regulated by histone acetyltransferases. (A) PCAF or HDAC inhibitors can inhibit KLF8 sumoylation. HEK293 cells were co-transfected with expression vectors coding for HA-KLF8, Myc-SUMO-1, or PCAF, and treated with HDAC inhibitors (trichostatin A, sodium butyrate, and nicotinamide) as indicated. Lysates were collected with NP-40 lysis buffer supplemented with 20 μM of the desumoylase inhibitor NEM followed by immunoprecipitation (IP) using an anti-HA antibody, and western blotting (IB) with either anti-Myc or anti-HA. Shown is a representative experiment where n=3. (B) Co-activators inhibit sumoylation. HEK293 cells were transfected with expression vectors coding for HA-KLF8, Myc-SUMO-1, PCAF, p300, CBP, or the HAT deficient mutants of PCAF, p300, or CBP as indicated. Lysates were collected with NP-40 lysis buffer supplemented with 20 μM of the desumoylase inhibitor NEM and immunoprecipitation experiments were performed using an anti-HA antibody where eluates were western blotted (IB) with either anti-Myc or anti-HA. Bottom, the relative expression of Myc-SUMO-1 as compared to HA-KLF8 is depicted. Results represent mean ±S.E. where n=4. *, p<0.05 compared to KLF8+SUMO-1 (column 3). Expression of the transfected constructs was verified by whole cell lysate blotting (data not shown).
The fact that we see a greater decrease in KLF8 sumoylation when cells transfected with PCAF and treated with HDAC inhibitors suggests that other co-activators may also be regulating KLF8 sumoylation in an acetylase dependent manner. To test this, WT or HAT deficient PCAF, p300, or CBP was transfected with HA-KLF8 and Myc-SUMO-1, and immunoprecipitation for HA-KLF8 was performed. We show that WT-PCAF and WT -p300 can significantly inhibit KLF8 sumoylation (Figure 3B, compare lanes or columns 4 and 6 with 3), whereas the HAT defective mutants are less effective in doing so (Figure 3B, compare lanes or columns 5 and 7 with 3). In contrast, CBP can cause a significant decrease in sumoylation but in a HAT independent manner (Figure 3B, compare lanes or columns 8 and 9 with 3), possibly through recruiting with PCAF to KLF8 as previously suggested [18].
Acetylation at lysines 93 and 95 negatively regulates KLF8 sumoylation
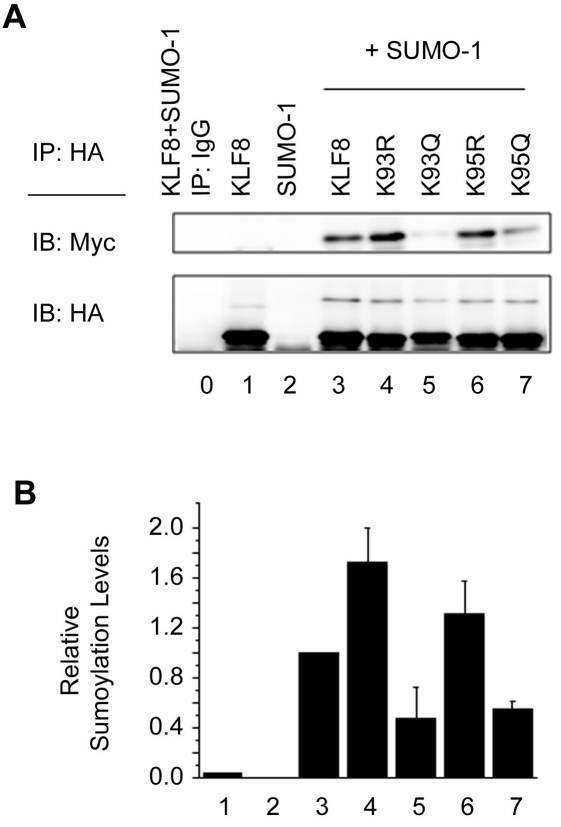
To examine whether the acetylation status at R93 and R95 can regulate subsequent KLF8 sumoylation at R67, we transfected WT-KLF8, K93R, K95R, K93Q or K95Q with Myc-SUMO-1 and performed immunoprecipitation experiments. We found that immunoprecipitation of K93R or K95R and western blotting of Myc-SUMO-1 demonstrates greater sumoylation levels compared to WT-KLF8 (Figure 4A and 4B, compare lanes or columns 4 and 6 with 3), whereas K93Q or K95Q causes a decrease in sumoylation lower than WT-KLF8 (Figure 4A and 4B, compare lanes or columns 5 and 7 with 3). These results suggest that acetylation at lysines 93 and 95 can inhibit sumoylation at lysine 67.
Figure 4.
KLF8 sumoylation is negatively regulated by acetylation at lysines 93 & 95. (A) HEK293 cells were co-transfected with expression vectors coding for Myc -SUMO-1, HA-KLF8, or HA-mutant KLF8 (KàR or KàQ). Lysates were collected with NP-40 lysis buffer supplemented with 20 μM of the desumoylase inhibitor NEM followed by immunoprecipitation (IP) using an anti-HA antibody, and western blotting (IB) with either anti-Myc or anti-HA antibodies. Expression of the transfected constructs was verified by whole cell lysate blotting (data not shown). (B) The expression of SUMOylated KLF8 relative to total expression of KLF8 that is normalized to 1. Results represent mean ±S.E. where n=3.
CtBP positively regulates KLF8 sumoylation
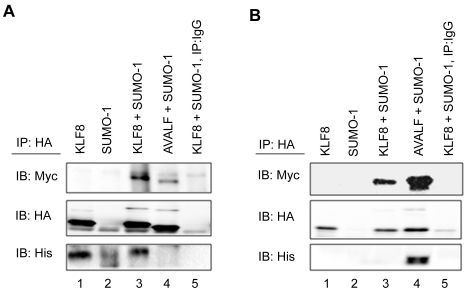
CtBP can regulate the transcriptional activity of its interacting proteins in both enzymatic dependent and independent manners [24]. Interestingly, CtBP has been shown to interact with E2 and E3 sumoylation ligases, and it has been suggested that CtBP may mediate sumoylation of CtBP-interacting proteins [23, 24]. Since it has previously been shown that KLF8 can interact with CtBP and that KLF8 is sumoylated, we wanted to determine whether CtBP could mediate this sumoylation. The HA-AVALF-KLF8 repression domain mutant (PVDLS to AVALF) that cannot interact with CtBP was transfected with Myc-SUMO-1 for immunoprecipitation analysis. We found that AVALF-KLF8 sumoylation levels significantly decrease compared to KLF8 (Figure 5A, top panel, compare lane 4 with 3). The loss of the mutant KLF8 interaction with CtBP was verified using an anti-CtBP antibody (Figure 5A, bottom panel, compare lane 4 with 3). Conversely, when His-tagged CtBP was overex-pressed with KLF8 we see an increase in KLF8 sumoylation levels (Figure 5B, top panel, compare lane 4 with 3). Taken together this suggests that CtBP interaction with KLF8 promotes KLF8 sumoylation.
Figure 5.
CtBP positively regulates KLF8 sumoyla-tion. (A) The KLF8 repression domain mutant cannot be sumoylated as efficiently as WT KLF8. HEK293 cells were transfected with HA-KLF8, HA-AVALF-KLF8 and Myc-SUMO-1 as indicated. Lysates were collected with NP-40 lysis buffer supplemented with 20 μM of the desumoylase inhibitor NEM followed by immunoprecipitation using an anti-HA antibody and western blotting (IB) using anti-Myc, anti-HA, and anti -CtBP antibodies. (B) Overexpression of CtBP promotes KLF8 sumoylation. HEK293 cells were transfected with HA-KLF8, Myc-SUMO, or His-CtBP as indicated. Experiments were performed as in (A). Shown are representative experiments where n=3. Expression of the transfected constructs was verified by whole cell lysate blotting (data not shown).
CtBP binding to KLF8 is negatively regulated by p300 and PCAF mediated acetylation
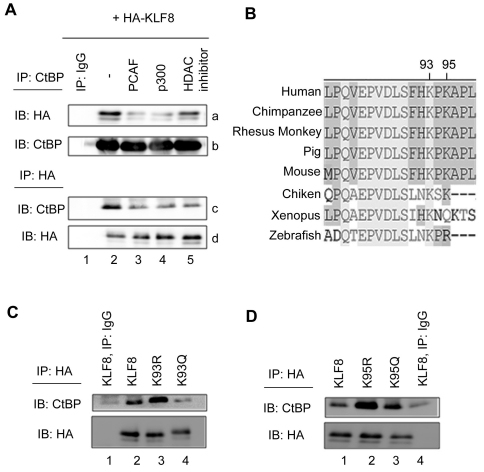
Others have suggested that acetylation may be a general mechanism to regulate CtBP interactions with other proteins [33-36]. In order to test whether p300 or PCAF mediated acetylation can regulate CtBP binding, we transfected KLF8 alone or with PCAF or p300 and performed co-immunoprecipitation experiments for KLF8 with endogenous CtBP. Immunoprecipitation of CtBP followed by blotting shows that PCAF or p300 expression causes a decrease in the interaction between KLF8 and CtBP (Figure 6A, panels a and b, compare lanes 3 and 4 with 2). Additionally, treatment with HDAC inhibitors can also cause a decrease in the interaction (Figure 6A, panels a and b, compare lane 5 with 2). Similar result were obtained when the reverse experiment was performed where we im-munoprecipitated HA-KLF8 and blotted for CtBP (Figure 6A, panels c and d, compare lanes 3, 4, and 5 with 2).
Figure 6.
The interaction between CtBP and KLF8 is negatively regulated by PCAF and p300 mediated acetylation. (A) PCAF, p300, or HDAC inhibitors inhibit KLF8 interaction with CtBP. HEK293 cells were co-transfected with expression vectors coding for HA-KLF8, PCAF, or p300 or treated with HDAC inhibitors (trichostatin A, sodium butyrate, and nicotinamide) as indicated. Endogenous CtBP was immunoprecipi-tated (IP) using an anti-CtBP antibody, and blotted (IB) with either anti-HA (panel a), or anti-CtBP antibodies (panel b). Conversely, KLF8 was immunopre-cipitated using anti-HA, and blotted using anti-CtBP (panel c) or anti-HA antibodies (panel d). (B) Alignment of KLF8 orthologs shows that the PVDLS motif and lysines 93 and 95 are highly conserved. (C & D), Acetylation at lysines 93 and 95 inhibits KLF8 interaction with CtBP. Experiments were performed similarly as in A except that K93R or K93Q (C) or K95R or K95Q (D) was transfected. Shown are representative experiments where n=3. Expression of the transfected constructs was verified by whole cell lysate blotting (data not shown).
Recent studies have shown that acetylation at a lysine immediately downstream of the PXDLS repression motif has specifically been shown to inhibit the protein interaction with CtBP. A PXDLSXXK motif has been shown to be highly conserved among multiple proteins [33], suggesting that acetylation at the lysine downstream of the PXDLS motif may be a general mechanism to regulate CtBP binding. Importantly, KLF8 possesses this motif where lysine 93 is the lysine within this motif and lysine 95 is immediately downstream. These lysines are highly conserved among orthologs of KLF8 (Figure 5B), and we have shown that they are acetylated by p300 or PCAF (Figure 1 and 2). To test whether acetylation at these specific sites regulates CtBP binding to KLF8, we transfected WT-KLF8, K93R or K95R (acetylation deficient), or K93Q or K95Q (constitutively acetylated mimetic) KLF8 mutants and performed similar experiments as in Figure 5A. We found that K93R causes a strong increase in KLF8-CtBP interaction, where K93Q-KLF8 does not interact with CtBP as efficiently as WT-KLF8 (Figure 6C, compare lanes 3 or 4 with 2). Similar results were seen in the case of lysine 95 (Figure 6D, compare lanes 2 or 3 with 1). Overall these results suggest that when KLF8 cannot be acetylated at lysines 93 or 95 (K93R & K95R) the interaction with CtBP is strong, whereas mutations that mimic constitutive acetylation of lysines 93 or 95 (K93Q & K95Q) inhibit KLF8 interaction with CtBP, suggesting that CtBP interaction with KLF8 can be dynamically regulated by acetylation.
Acetylation at lysines 93 and 95 blocks the inhibitory effect of CtBP on KLF8 transactivity
We have previously shown that sumoylation at lysine 67 can inhibit the ability of KLF8 to activate the cyclin D1 promoter [3]. Here we have shown that CtBP positively regulates KLF8 sumoylation. Therefore, we wondered whether CtBP could also negatively regulate KLF8 tran-scriptional activity at the cyclin D1 promoter.
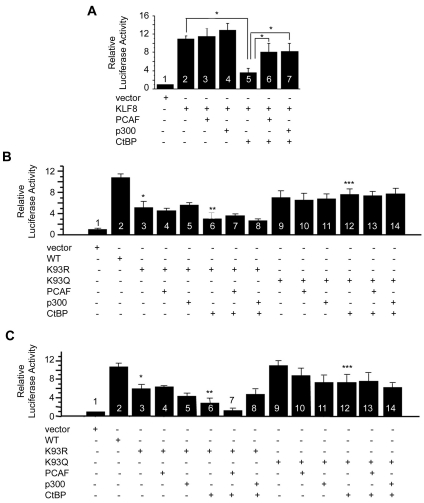
Using luciferase assays we found that co-expression of CtBP with KLF8 does in fact inhibit the ability of KLF8 to activate the cyclin D1 promoter (Figure 7A, compare column 5 with 2). However, the addition of PCAF or p300 can rescue promoter activity (Figure 7A, compare columns 6 and 7 with 2 and 5), suggesting that the histone acetyltransferases compete with CtBP to regulate KLF8 transactivity.
Figure 7.
KLF8 acetylation at lysines 93 and 95 positively regulates cyclin D1 promoter activity. (A) PCAF or p300 can rescue CtBP mediated inhibition of KLF8 transcriptional activity. Luciferase activity was measured in T80 human ovarian epithelial cells that were co-transfected with the cyclin D1 promoter reporter and either the empty vector, KLF8, PCAF, p300, or CtBP in the indicated combinations. After 16 h, cells were harvested for luciferase assays as described in Materials and Methods. Results were normalized to vector control. *, p<0.05 compare column 5 with 2, or 6 and 7 with 5. (B) Acetylation at lysine 93 inhibits CtBP mediated inhibition of KLF8 transcriptional activity. Similar experiments were performed using WT-KLF8 or the KLF8-K93R and K93Q mutants. *, p<0.01 compare column 3 with 2. **, p<0.01 compare column 6 with 2. ***, p<0.05 compare column 12 with 6. (C) Acetylation at lysine 93 inhibits CtBP mediated inhibition of KLF8 transcriptional activity. Similar experiments were performed using WT-KLF8 or the KLF8-K95R and K95Q mutants. *, p<0.01 compare column 3 with 2. **, p<0.01 compare column 6 with 2. ***, p<0.01 compare column 12 with 6. All experiments were done in triplicate and the results represent mean ± S.E. where n>3.
Next we wanted to test the role of lysines 93 and 95 in regulating KLF8 transactivity. We found that the K93R mutant causes a significant decrease in cyclin D1 promoter activity compared to WT-KLF8 (Figure 7B, compare columns 3 and 2), and the addition of PCAF or p300 cannot cause an increase in promoter activity (Figure 7B, compare columns 4 and 5 with 2). CtBP again caused a decrease in promoter activity compared to WT-KLF8 and K93R-KLF8 (Fig 7B, compare column 6 with 2 and 3), but unlike the result we see with WT-KLF8, the addition of PCAF or p300 cannot rescue the inhibitory effects of CtBP (Figure 7B, compare columns 7 and 8 with 6). In contrast, there is not a significant decrease in cyclin D1 promoter activity in the K93Q-KLF8 mutant compared to WT-KLF8, and the addition of CtBP is not able to inhibit its transactivity (Figure 7B, compare columns 9, 12, 13, and 14 with 2 or 6). We performed the same experiments with the K95R and K95Q mutants and found similar results as seen using the K93R and K93Q KLF8 mutants (Figure 7C). Taken together, these results suggest that the acetylation status at lysines 93 and 95 regulates the ability of CtBP to inhibit KLF8 mediated activation of the cyclin D1 promoter.
The acetylation status at lysines 93 and 95 regulates KLF8 mediated cyclin D1 protein expression and cell cycle progression
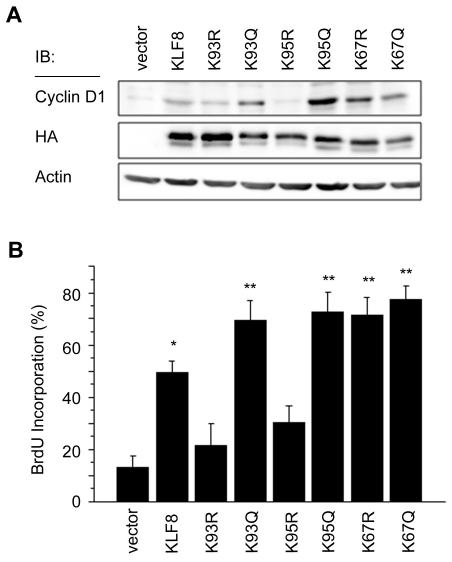
We have previously shown that KLF8 can promote cyclin D1 expression and cell cycle progression [1, 3-5]. In order to determine whether acetylation at lysines 93, 95, and 67 affects the ability of KLF8 to promote these events we transfected WT-KLF8, the acetylation deficient, or the acetylation mimetic mutants of K93, K95, and K67 into T80 human ovarian epithelial cells and measured cyclin D1 protein expression by western blotting or cell cycle progression using BrdU incorporation assays. We found that while K93R and K95R cannot induce cyclin D1 expression or cell cycle progression as effectively as WT, the K93Q, K95Q, K67R, and K67Q mutants increase cyclin D1 expression and cell cycle progression compared to WT (Figure 8A and B). The fact that the K67R and K67Q mutation both induce cyclin D1 expression and promote cell cycle progression further asserts that acetylation at this site functions specifically to inhibit sumoylation at this site. Overall, these results suggest that acetylation at lysines 93, 95, and 67 promotes the ability of KLF8 to activate cyclin D1 expression and promote cell cycle progression.
Figure 8.
Acetylation at lysines 93, 95 and 67 regulates KLF8 mediated cyclin D1 protein expression and cell cycle progression. (A) T80 cells were trans-fected with vector, KLF8 or its mutants as indicated. 16 h post transfection lysates were collected and subjected to western blot analysis using anti-cyclin D1, anti-HA, or anti-actin antibodies. Shown is a representative experiment where n=3. (B) T80 cells were transfected with empty vector, KLF8 or the indicated mutants. Cell cycle progression through G1 was determined by measuring BrdU incorporation as described in Materials and Methods. *p < 0.05 as compared to the vector control. **p<0.05 as compared to WT-KLF8.
Discussion
Post-translational modifications such as lysine acetylation and sumoylation play a crucial role in the regulation of many critical transcription factors [21]. Importantly, co-regulator proteins can either promote or inhibit these different modifications. Here we show the first evidence that KLF8 is tightly regulated by a posttranslational switch between acetylation and sumoylation mediated by p300 and PCAF co-activators and the CtBP co-repressor. Firstly, we show that KLF8 is acetylated by the p300 and PCAF and that acetylation at the K67 directly competes for sumoylation at the same site, while acetylation at K93 and K95 also inhibits sumoylation at K67. Additionally, CtBP interaction with KLF8 enhances sumoylation of KLF8. Fourthly, acetylation at K93 and K95 inhibits the CtBP interaction with KLF8 and that the acetylation-mimetic K93Q and K95Q mutation causes a decrease in the interaction with CtBP. Finally, acetyaltion at K67, K93, and K95 is required for KLF8 to promote the cyclin D1 expression and the cell cycle progression.
We have previously demonstrated that sumoylation at the K67 site inhibits the ability of KLF8 to transactivate the cyclin D1 promoter and promote the cell cycle progression [3]. The fact that both the K67R and K67Q mutations increase cyclin D1 expression and cell cycle progression to similar levels suggests that it is that prevention of sumoylation rather than acetylation per seat this site that is critical for KLF8 function.
Consistent with our findings, others have reported that competition between sumoylation and acetylation to modify the same lysine residue regulates the function of MEF2 and HIC1 where HDAC-promoted sumoylation inhibits transcription factor activity and acetylation blocks these inhibitory effects [38-40]. Interestingly, the acetylation/sumoylation switch on MEF2 is further regulated by its phosphorylation status [39] and it will be worthwhile to determine whether the KLF8 acetylation/sumoylation switch is also regulated by a phosphorylation event.
The CtBP co-repressor has been demonstrated to physically interact with the SUMO E2 (Ubc9) and E3 ligases (HPC2 and PIAS1) [23, 41, 42] allowing CtBP to act as a platform for sumoylation of the transcription factor ZEB1 [23]. Since we have shown that both Ubc9 and PIAS1 can enhance KLF8 sumoylation [3], it is plausible that a similar mechanism may also apply to the sumoylation of KLF8 by CtBP.
Our data clearly shows that the interaction between KLF8 and CtBP can be inhibited at least partially by acetylation at K93 and K95 (see Figure 6). This result is in agreement with previous observations that acetylation at a lysine immediately downstream of the CtBP interacting motif PLDLS of adenovirus E1A inhibits its interaction with CtBP and consequently the CtBP-mediated repression possibly by altering the secondary structure of E1A [35, 36]. Whether or not acetylation also alters the secondary structure of KLF8 to inhibit the interaction between KLF8 and CtBP is another interesting future study. This inhibitory mechanism has also been reported with other transcription factors including RIP140, SATB1 and EVI1 [33, 34, 43] and many other proteins seem to possess this similar motif where the PXDLS is flanked by lysine residues [33, 43]. This suggests that acetylation -regulated CtBP binding may be a general regulatory mechanism that controls transcription factor function. Interestingly, a few of the proteins whose interaction with CtBP is regulated by acetylation are also sumoylated. Like KLF8, these proteins could be regulated by a similar switch between co-activator-mediated acetylation and CtBP-mediated sumolylation.
In summary, this study provides new insights into the posttranslational regulation of KLF8 by the interplay between HAT co-activator mediated acetylation and CtBP promoted sumoylation. As KLF8 regulates many critical processes particularly in cancer progression, our results highlight the importance of the tight regulation of KLF8 function beyond the control of its expression and open new opportunities for effective KLF8-targeted cancer therapies in the future.
Acknowledgments
We thank our colleagues Drs. Dorina Avram, Kathleen Bove, C. Michael DiPersion, Susan LaFlamme and Chunhong Yan for critical and helpful comments. We also thank Dr. Steven M. Frisch of University of West Virginia for kindly providing the pQE32-CtBP construct. This research was supported by grants from NCI (CA132977), the American Cancer Society (#RSG CCG-111381) and Susan G. Komen for Cure (KG090444 and KG080616) to JZ. X.W. is a Komen postdoctoral fellow.
References
- 1.Zhao J, Bian ZC, Yee K, Chen BP, Chien S, Guan JL. Identification of transcription factor KLF8 as a downstream target of focal adhesion kinase in its regulation of cyclin D1 and cell cycle progression. Mol Cell. 2003;11:1503–1515. doi: 10.1016/s1097-2765(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Urvalek AM, Liu J, Zhao J. Activation of KLF8 transcription by FAK in human ovarian epithelial and cancer cells. J Biol Chem. 2008;283:13934–13942. doi: 10.1074/jbc.M709300200. [DOI] [PubMed] [Google Scholar]
- 3.Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, Zhao J. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J Biol Chem. 2006;281:16664–16671. doi: 10.1074/jbc.M513135200. [DOI] [PubMed] [Google Scholar]
- 4.Urvalek AM, Wang X, Lu H, Zhao J. KLF8 recruits the p300 and PCAF co-activators to its amino terminal activation domain to activate transcription. Cell Cycle. 2010;9:601–611. doi: 10.4161/cc.9.3.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta TS, Lu H, Wang X, Urvalek AM, Nguyen KH, Monzur F, Hammond JD, Ma JQ, Zhao J. A unique sequence in the N-terminal regulatory region controls the nuclear localization of KLF8 by cooperating with the C-terminal zinc-fingers. Cell Res. 2009;19:1098–1109. doi: 10.1038/cr.2009.64. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd JA. KLF8 sets the pace for the cell cycle through interactions with p300 and PCAF. Cell Cycle. 2010;9:650–651. [PubMed] [Google Scholar]
- 7.Fu WJ, Li JC, Wu XY, Yang ZB, Mo ZN, Huang JW, Xia GW, Ding Q, Liu KD, Zhu HG. Small interference RNA targeting Kruppel-like factor 8 inhibits the renal carcinoma 786-0 cells growth in vitro and in vivo. J Cancer Res Clin Oncol. 2010 doi: 10.1007/s00432-010-0776-0. [DOI] [PubMed] [Google Scholar]
- 8.Evans PM, Liu C. New insights into KLF8-mediated transactivation. Cell Cycle. 2010;9:649–650. [PubMed] [Google Scholar]
- 9.Wang X, Zhao J. KLF8 transcription factor participates in oncogenic transformation. Onco-gene. 2007;26:456–461. doi: 10.1038/sj.onc.1209796. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, Zhao J. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res. 2007;67:7184–7193. doi: 10.1158/0008-5472.CAN-06-4729. [DOI] [PubMed] [Google Scholar]
- 11.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 12.van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu JH, Navas P, Cao H, Stamatoyannopoulos G, Song CZ. Systematic RNAi studies on the role of Sp/KLF factors in globin gene expression and erythroid differentiation. J Mol Biol. 2007;366:1064–1073. doi: 10.1016/j.jmb.2006.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, Lloyd JA. A functional screen for Kruppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells Mol Dis. 2005;35:227–235. doi: 10.1016/j.bcmd.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Cox BD, Natarajan M, Stettner MR, Gladson CL. New concepts regarding focal adhesion kinase promotion of cell migration and proliferation. J Cell Biochem. 2006;99:35–52. doi: 10.1002/jcb.20956. [DOI] [PubMed] [Google Scholar]
- 16.DingQ, Grammer JR, Nelson MA, Guan JL, Stewart JE Jr. and Gladson CL. p27Kip1 and cyclin D1are necessary for focal adhesion kinase regulation of cell cycle progression in glioblastoma cells propagated in vitro and in vivo in the scid mouse brain. J Biol Chem. 2005;280:6802–6815. doi: 10.1074/jbc.M409180200. [DOI] [PubMed] [Google Scholar]
- 17.Eaton SA, Funnell AP, Sue N, Nicholas H, Pearson RC, Crossley M. A network of Kruppel-like Factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J Biol Chem. 2008;283:26937–26947. doi: 10.1074/jbc.M804831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 19.Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 2008;90:306–312. doi: 10.1016/j.biochi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Bio-chem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol Life Sci. 2007;64:3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuppuswamy M, Vijayalingam S, Zhao LJ, Zhou Y, Subramanian T, Ryerse J, Chinnadurai G. Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol Cell Biol. 2008;28:269–281. doi: 10.1128/MCB.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. IntJ Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, Patterson A, Marquez RT, Auersperg N, Yu Y, Hahn WC, Mills GB, Bast RC., Jr A genetically defined model for human ovarian cancer. Cancer Res. 2004;64:1655–1663. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, Bieker JJ. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci U S A. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Kadam S, Emerson BM, Bieker JJ. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol Cell Biol. 2001;21:2413–2422. doi: 10.1128/MCB.21.7.2413-2422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 29.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 30.Kwok RP, Laurance ME, Lundblad JR, Goldman PS, Shih H, Connor LM, Marriott SJ, Goodman RH. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature. 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 31.Harton JA, Zika E, Ting JP. The histone acetyl-transferase domains of CREB-binding protein (CBP) and p300/CBP-associated factor are not necessary for cooperativity with the class II transactivator. J Biol Chem. 2001;276:38715–38720. doi: 10.1074/jbc.M106652200. [DOI] [PubMed] [Google Scholar]
- 32.Jiang H, Lu H, Schiltz RL, Pise-Masison CA, Ogryzko VV, Nakatani Y, Brady JN. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol Cell Biol. 1999;19:8136–8145. doi: 10.1128/mcb.19.12.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vo N, Fjeld C, Goodman RH. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol Cell Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purbey PK, Singh S, Notani D, Kumar PP, Limaye AS, Galande S. Acetylation-dependent interaction of SATB1 and CtBP1 mediates transcriptional repression by SATB1. Mol Cell Biol. 2009;29:1321–1337. doi: 10.1128/MCB.00822-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Yao H, Vo N, Goodman RH. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc Natl Acad Sci U S A. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molloy D, Mapp KL, Webster R, Gallimore PH, Grand RJ. Acetylation at a lysine residue adjacent to the CtBP binding motif within adenovirus 12 E1A causes structural disruption and limited reduction of CtBP binding. Virology. 2006;355:115–126. doi: 10.1016/j.virol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 37.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and on-cogenesis. Mol Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacety-lase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 40.Stankovic-Valentin N, Deltour S, Seeler J, Pinte S, Vergoten G, Guerardel C, Dejean A, Leprince D. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27:2661–2675. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 42.Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 43.Shimanara A, Yamakawa N, Nishikata I, Morishita K. Acetylation of lysine564 adjacent to the CTBP-binding motif in EVI1 is crucial for transcriptional activation of GATA2. J Biol Chem. 2010 doi: 10.1074/jbc.M110.102046. [DOI] [PMC free article] [PubMed] [Google Scholar]