Abstract
Few structures of viral serine proteases, those encoded by the Sindbis and Semliki Forest viruses, hepatitis C virus (HCV) and cytomegalovirus, have been reported. In the life cycle of HCV a crucial role is played by a chymotrypsin-like serine protease encoded at the N–terminus of the viral NS3 protein, the solution structure of which we present here complexed with a covalently bound reversible inhibitor. Unexpectedly, the residue in the P2 position of the inhibitor induces an effective stabilization of the catalytic His–Asp hydrogen bond, by shielding that region of the protease from the solvent. This interaction appears crucial in the activation of the enzyme catalytic machinery and represents an unprecedented observation for this family of enzymes. Our data suggest that natural substrates of this serine protease could contribute to the enzyme activation by a similar induced-fit mechanism. The high degree of similarity at the His–Asp catalytic site region between HCV NS3 and other viral serine proteases suggests that this behaviour could be a more general feature for this category of viral enzymes.
Keywords: hepatitis C virus/α-ketoacid/NS3/serine protease/viral hydrolases
Introduction
The hepatitis C virus (HCV) infects ∼3% of the world population, and since it causes chronic liver disease it is considered a major health problem worldwide (World Health Organization, 1999). Patients with chronic infection can develop liver cirrhosis and are at high risk of developing hepatocellular carcinoma (Avital, 1998). Neither a vaccine against viral infection nor effective therapy has been developed to date. HCV represents the most widely spread and challenging viral infection to block.
HCV is a positive-sense, single-stranded RNA virus and belongs to the Flaviviridae family. It consists of ∼9.6 kb, which in infected cells are translated into a polyprotein of ∼3011 amino acids. The genome organization comprises the structural proteins C, E1 and E2, and the non-structural proteins NS2, NS3, NS4A, NS4B, NS5A and NS5B, which are released by action of both host cell and virally encoded proteases (Neddermann et al., 1997; Bartenschlager, 1999).
The N–terminal domain of the HCV NS3 protein contains a serine protease, belonging to the chymotrypsin family (Lesk and Fordham, 1996), which is responsible for the proteolytic cleavage at the NS3/NS4A, NS4A/NS4B, NS4B/NS5A and NS5A/NS5B junctions of the viral polyprotein (Neddermann et al., 1997). The NS3 protease thus plays a pivotal role in the maturation of the viral polyprotein. Consequently, the activity of this enzyme has been studied under a broad range of conditions in view of its potential as a target for antiviral therapy (Bartenschlager, 1999; De Francesco and Steinkühler, 1999). Some enzymatic and structural features make this viral enzyme unique among the serine protease family: the serine protease domain is covalently attached to an RNA helicase possessing NTPase activity, it requires unusually long substrates (P6–P4′) for effective cleavage and possesses a solvent-accessible structural zinc-binding site (De Francesco and Steinkühler, 1999). In addition, the action of a virus-encoded protein cofactor, NS4A, is required for some but not all of the NS3-dependent proteolytic cleavage events. Thus, NS4A is necessary for the proteolytic processing of the NS4A/NS4B and NS4B/NS5A cleavage sites, whereas it only enhances the NS3 protease activity observed on the NS5A/NS5B cleavage site (Bartenschlager, 1999). NS4A functions as an activator of the NS3 serine protease by forming a non-covalent complex. The crystallographic (Love et al., 1996) and NMR solution (Barbato et al., 1999) structures of the uncomplexed enzyme, and the crystallographic structure of a complex with a peptide spanning the core domain of NS4A (Kim et al., 1996; Yan et al., 1998), have been solved. The interaction with NS4A requires the 22 N–terminal residues of NS3 and a 12–residue sequence in the centre of NS4A, which can be supplied as a synthetic peptide without loss of activation function (Bartenschlager, 1999). Comparative analysis of the crystallographic three-dimensional structures of the NS3 protease suggested a possible mechanism for the activation of the enzyme (see below). However, this model is not entirely satisfactory since it does not explain all the available biochemical data. In particular, the current model of action does not account for the proteolytic activity observed on the NS4A-independent substrates.
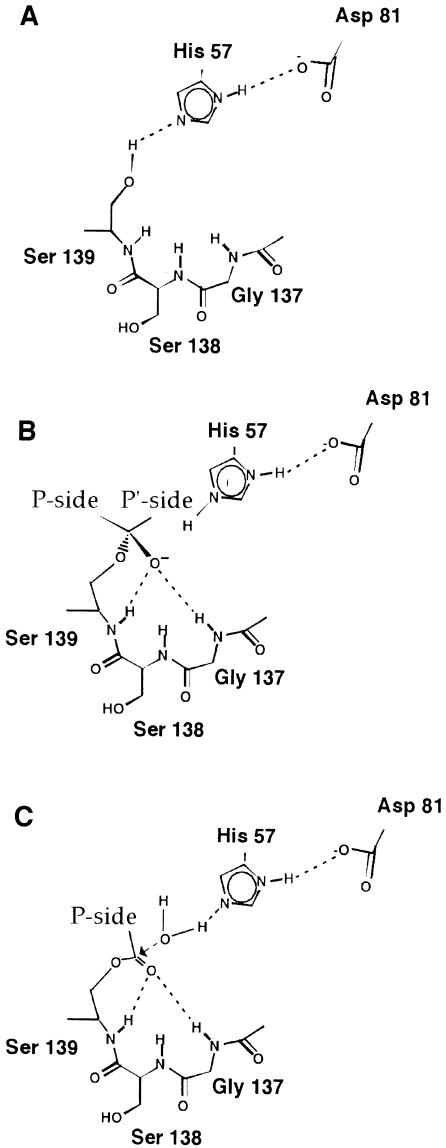
Figure 1 schematizes the basic steps of the current general model of action of the serine protease family (Fersht, 1985; Polgar, 1989; Phillips and Fletterick, 1992). The energy implications and the precise role of each catalytic residue in this model are currently under debate (Cleland et al., 1998; Warshel, 1998). However, all authors agree that a stable network of hydrogen bonds (Figure 1A) is required for a fully active enzyme and for the nucleophilic attack that leads to the tetrahedral intermediate of Figure 1B and subsequent hydrolysis of the acyl-enzyme (Figure 1C). The NS3 mechanistic model of action proposed to date is based on the observation that, in the crystal structure obtained in the absence of NS4A, the position of the catalytic aspartate (Asp81) significantly deviates from the configuration required for proteolysis, making the formation of a hydrogen bond with the catalytic histidine (His57) impossible. Conversely, the three catalytic residues, His57, Asp81 and Ser139, acquire the canonical serine protease conformation in the crystals obtained in the presence of the NS4A cofactor. On this basis it has been proposed that binding of NS4A to the N–terminal NS3 barrel results in spatial re-organization of the serine protease catalytic triad, ultimately leading to the formation of an active enzyme (Love et al., 1998). This model has been gaining favour, as documented by a recent review (Bartenschlager, 1999). While interesting, this model does not explain how the NS3 protease can be active on substrates such as the NS5A/NS5B junction in the absence of NS4A. A model where the uncomplexed NS3 serine protease may work via the formation of a catalytic ‘dyad’ is not supported by cell culture experiments; it has in fact been shown that mutation of each single residue of the catalytic triad results in a totally uncleaved polyprotein (Hijikata et al., 1993). Thus, if in the absence of NS4A the Asp81 is not participating in the catalytic triad, as suggested by the model, the NS3 protease should be an inactive enzyme.
Fig. 1. Schematic representation of the general mechanism of action of serine proteases. (A) Hydrogen bond network needed for a fully active enzyme. The current mechanistic model proposes that the histidine acts as a general base during catalysis, accepting a proton from the serine as it forms a bond with the substrate carbonyl carbon, giving rise to a tetrahedral intermediate (B). The negative charge on the oxygen atom is stabilized by the oxyanion hole formed by the amide protons of the catalytic serine and of a glycine residue two positions before the serine. The H-bond between the histidine δ–NH and the aspartate carboxyl groups ensures that the imidazole ring is in the correct tautomeric form to accept the serine proton. (C) The final step of the catalysis proceeds through hydrolysis of the acyl-enzyme by a water molecule. Note that for the substrate the nomenclature of Schecter and Berger (1967) is used in designating the cleavage sites as Pn-Pn–1-P1-P1′-Pn–1′-Pn′ with the scissile bond between P1 and P1′ and the C–terminus of the substrate on the prime site.
Recently, the solution structure of the NS3 protease has been published (Barbato et al., 1999), where the relative position of the catalytic triad, even in the absence of the NS4A cofactor, is compatible with that of a fully active serine protease. The disagreement between the crystal and solution structures is likely to be due to the fact that the former lacks the extremely important helix α3 (Figure 2B), probably due to distortions induced by crystal packing forces. This helix, present in solution, is crucial for the correct packing of the strand F1 that positions the catalytic Asp81. The NMR evidence therefore suggests that NS4A is likely to play a different and more subtle role with respect to NS3 serine protease activation. The solution structure of the free enzyme, however, also poses a new question, in fact the hydrogen bond between the carboxylate group of Asp81 and the δ–NH of the His57 is not stable since the whole region is completely solvent exposed (Barbato et al., 1999). This is also reflected in the unusually high temperature factors of the backbone atoms of the loop E1–F1 (including Asp81) in the crystallographic structures published to date, also in the presence of NS4A, indicating an intrinsic mobility at this site. A stable network of hydrogen bonds as schematized in Figure 1A is, however, essential in order to have an active enzyme. In all chymotrypsin-like enzymes this is ensured by burying the Asp–His side chain hydrogen bond and making it solvent inaccessible (McGrath et al., 1992). Thus, for NS3, either with or without the activating NS4A cofactor, one has to raise the issue of how the Asp–His hydrogen bond can be stabilized in a region that is solvent accessible and affected by mobility.
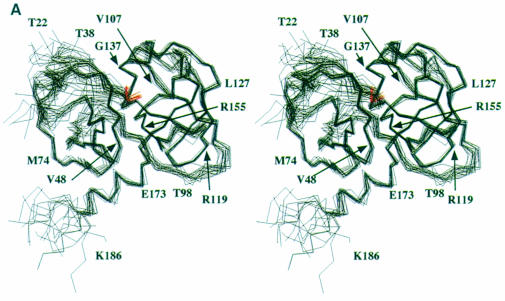
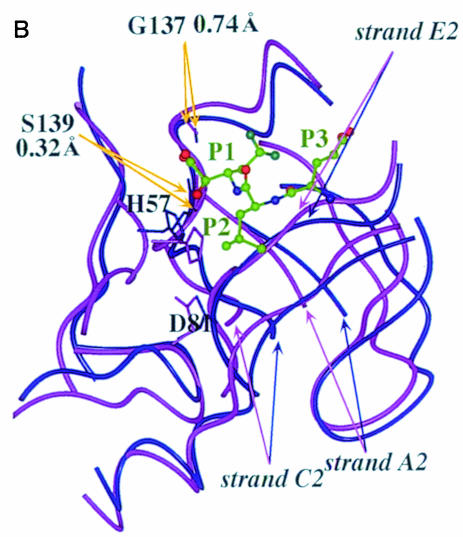
Fig. 2. (A) Stereoview of the 20 minimum energy structures. The superposition has been obtained using the structurally conserved regions (SCRs) as defined previously in Barbato et al. (1999). The inhibitor α–C atoms are shown in red. The N–terminal 21 residues were not included in the structure calculations, since their conformation is ill defined. (B) Molscript (Kraulis, 1991) representation of the NS3 protease domain, the chymotrypsin-like common SCRs are coloured in blue and the structural element labels follow the chymotrypsin-like nomenclature. The residues forming the secondary structural elements are: strands A1 (34–37), B1 (41–44), C1 (51–55), D1 (63–66), E1 (69–78), F1 (82–86), A2 (104–108), B2 (122–126), C2 (141–145), D2 (150–152), E2 (155–159), F2 (166–170); helices α1 (55–59), α2 (132–136), α3 (173–180).
In this work we present the first structure of the complex of the NS3 protease domain with a covalently bound α–ketoacid peptidic inhibitor (see inset in Figure 3). Since the NS3 protease is involved in the maturation process of the viral polyprotein, this structure is relevant for the design of novel therapeutic agents active against HCV. The inhibitor used is a second generation inhibitor, which is based on the previous optimization of product inhibitors (Ingallinella et al., 1998). It has already been shown that the mode of binding of P region-derived product inhibitors is similar to the ground state binding of the corresponding substrates (Ingallinella et al., 1998). The C–terminal ketoacidic moiety only provides additional binding energy (Narjes et al., 2000). The details of the NS3–inhibitor complex structure are then used as a tool to gain information also on the substrate interaction itself. Our findings support a plausible role of the substrate itself in enzyme activation. The mechanism of substrate-induced protease activation provides a framework to explain how the NS3 protease could at least be partially active even in the absence of the NS4A cofactor. Thus, we propose a new model of mechanistic action for the NS3 protease that we believe reconciles the available experimental evidence presented to date. This mechanism of substrate-induced enzyme activation appears to be unique among the chymotrypsin-like proteases.
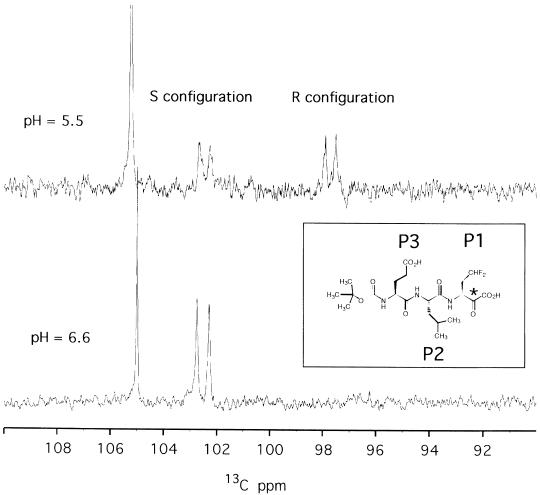
Fig. 3. The inset shows the reversible covalently bound inhibitor α–ketoacid Boc-Glu-Leu-(γdi-fluoro)Abu; the corresponding positions in the substrate-like nomenclature (P1, P2 and P3) are also indicated. The asterisk labels the activated carbonyl moiety, which acts as binding group and becomes the hemiketal chiral carbon upon complex formation. The spectra show regions of the 1D 13C experiment where the hemiketal carbon of the complex resonates. At pH 6.6 (bottom) only one doublet resonance is visible at 102.6 p.p.m., corresponding to the S configuration; at pH 5.5 (top), two resonances of similar intensity appear at 102.6 and 97.6 p.p.m. for the R and S configurations, respectively. The hemiketal signals are doublets since they are coupled with the α–C, which is also 13C labelled, while the carboxyl atom is unlabelled. The intense singlet at ∼105 p.p.m. represents a buffer resonance.
Results and discussion
NS3 protease overall topology
In Figure 2A a stereoview of the backbone bundle is presented. The structures were calculated excluding the first 21 residues, which, as in the case of the free enzyme (Barbato et al., 1999), are mobile and ill defined in solution (the constraints database and the statistical analysis of the structure quality are reported in Table I). Structurally conserved regions (SCR) that are common to all the known chymotrypsin-like proteases (Greer, 1990) are very well conserved in the complex (Figure 2B). NS3 protease is a relatively small protein (180 residues) and belongs to the sub-class of small chymotrypsin-like proteases (Bazan and Fletterick, 1988). As such it makes an economical use of loops, since it lacks a series of connecting elongations that are a common feature of the longer cellular proteases. The solution structure of the complex is similar to the solution structure of the free enzyme (Barbato et al., 1999), with a root mean square deviation (r.m.s.d.) of 1.05 Å for the SCR residues of the minimized average structures. The significant differences are located at the catalytic triad and in the C–terminal domain S–site where the inhibitor is bound (Figure 2A). The direct participation of the activating cofactor NS4A in the recognition or binding of the S–site inhibitors can be ruled out on structural grounds: NS4A binds at the N–terminal barrel, remote from the region of interest, and does not appear to affect the conformation at the active site directly. This conclusion is in agreement with the previously reported mapping of the interaction with substrate-derived inhibitors (Cicero et al., 1999), and is also confirmed by comparison with the crystal structure of the NS3/NS4A–inhibitor ternary complex (S.Di Marco and M.Sollazzo, personal communication).
Table I. Experimental restraints and structural statistics.
| NMR constraints | ||
| NOE | total | 3023 |
| intra | 905 | |
| inter short distance (<i + 3) | 986 | |
| inter long range (>i + 3) | 1132 | |
| Generic | total | 86 |
| H-bond | 81 | |
| Zn-binding site | 5 | |
| Dihedral | total | 177 |
| φ | 91 | |
| χ1 | 86 | |
| Stereospecific | methylene groups | 92/219 |
| methyl groups | 62/68 | |
| Structure statistics | ||
| R.m.s. deviations from experimental constraintsa | ||
| distance (Å) | 0.056 ± 0.002 | |
| dihedral(°) | 3.499 ± 0.642 | |
| Deviations from idealized geometry | ||
| bonds (Å) | 0.005 ± 0.0002 | |
| angles (°) | 0.812 ± 0.015 | |
| impropers (°) | 1.461 ± 0.008 | |
| Coordinates precision referred to mean structure (Å) residues SCR + helicesb | ||
| backbone | 0.487 ± 0.111 | |
| all heavy atoms | 0.860 ± 1.161 | |
| all residuesc | ||
| backbone | 0.738 ± 0.124 | |
| all heavy atoms | 1.120 ± 0.191 | |
| Ramachandran analysisd | ||
| % of residues in most favoured regions | 71.8 ± 2.6 | |
| % of residues in allowed regions | 21.5 ± 2.9 | |
| % of residues in generously allowed regions | 4.5 ± 1.3 | |
| % of residues in disallowed regions | 2.2 ± 1.4 |
aNone of the structures exhibited distance violations >0.5 Å or dihedral angle violations >5°.
bResidues: 34–37, 41–44, 51–59, 63–66, 69–78, 82–86, 104–108, 122–126, 131–145, 150–152, 155–159, 166–170, 173–180.
cResidues: 33–95, 104–180, 188–190.
dThe program PROCHECK (Laskowski et al., 1993) was used to assess the overall quality of the structures. All the residues 22–186 and the inhibitor 188–190 (in total 167 residues) were used for the Ramachandran plot statistical evaluations.
Inhibitor binding stereochemistry
The carbon of the activated keto-group (see Figure 3, inset) of the inhibitor becomes a hemiketal quaternary carbon upon binding to the γ–O of the catalytic serine. Both R and S configurations are possible at this chiral centre. By using a sample with a selectively labelled 13C quaternary carbon, we could observe that in the pH interval 5.3–5.7 both chiral forms are present together with a small amount of the ketoacid form (non-covalently bound), whereas at pH values >6.0 only one configuration is dominant (Figure 3). This behaviour has already been observed by NMR on the complexes of chymotrypsin–N–acetyl-phenylalaninal (Shah et al., 1984) and trypsin–leupeptin (Ortiz et al., 1991). The X–ray structure of the Streptomyces griseus protease A bound to chymo– statin (pH 4.1) showed the simultaneous presence of both configurations (Delbaere and Brayer, 1985). Although our structural data (collected at pH 6.6) do not allow an unambiguous assignment of the stereochemistry, cogent arguments are presented below in favour of the S hemiketal carbon configuration being dominant at physiological pH. We did, however, perform structural calculations in parallel for both configurations (Figure 4A and B).
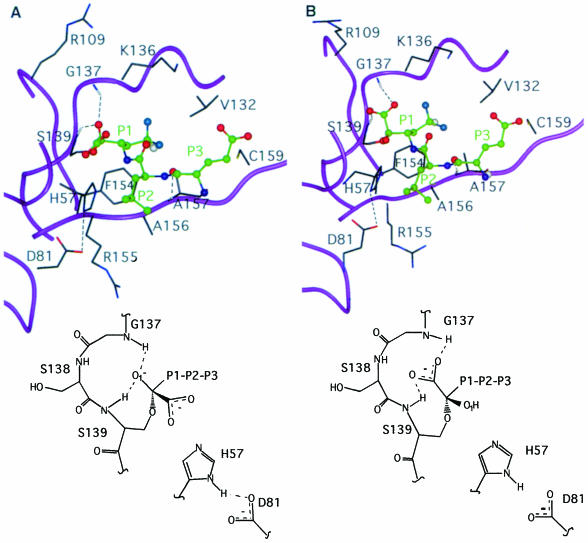
Fig. 4. (A and B) Selected region of the minimized average structure of the set of NMR structures assuming the R or S configuration at the hemiketal quaternary carbon, respectively. The protein backbone is shown in magenta ribbon representation, the inhibitor is represented in ball-and-stick representation with carbons in green. The relevant protein residues involved in the interaction are in stick representation with carbons in dark grey. The three positively charged residues (Arg109, Lys136 and Arg155) surrounding the catalytic site are shown and labelled. At the bottom of the figure a schematic representation of the covalent bond and the inhibitor interaction with the oxyanion hole is presented.
Inhibitor binding site: P1
An expanded view of the inhibitor-bound structure, for the hemiketal carbon R and S configurations, respectively (Figure 4A and B), reveals that the specificity pocket is occupied by the di–fluoro-Abu side chain, with the γ–H proximal to Phe154. In fact the γ–H experiences a downfield shift (Δδ = 0.3 p.p.m.), which may be the result of its proximity to the deshielding zone of the Phe154 aromatic ring. The positioning of the fluorine atoms was obtained from 1H-19F NOE data. The refined structures assuming the R or S configuration appear very similar (r.m.s.d. = 0.11 Å for the averaged minimized structures). As noted also by others (Delbaere and Brayer, 1985) the hemiketal complexation causes remarkably little movement in the positions of the catalytic residues. For the R configuration (Figure 4A) the carboxyl group is oriented towards the His57 and is solvent exposed, while the hemiketal oxygen O1 is involved in H–bonds with the oxyanion hole amide groups of Ser139 and Gly137 (dHN–O = 2.5 and 3.2 Å, respectively). The R configuration, with the carboxylate moiety directed towards the His57 ring, is likely to be favoured by the protonation of the imidazole ring, which takes place below pH 5.8, as clearly shown by pH titration data (Figure 7C). Also, if the R configuration were stable at high pH, the hemiketal oxygen O1 (Figure 4A) would exhibit a lowered pKa value, since its acidic form is stabilized by the interactions in the oxyanion hole, whereas no titration effect has been observed up to pH 9.5. In the case of the S configuration (Figure 4B), the carboxylate group points towards the oxyanion hole and forms direct H–bonds with HN Ser139 and Gly137 (dHN–O = 2.5 and 2.6 Å, respectively), while the hemiketal oxygen O1 is oriented towards the His57 and is solvent exposed. In the S configuration the carboxylate in the oxyanion hole is very close to the H donor groups, which accounts for the similar and large (>2 p.p.m.) downfield shifts observed for both NH protons. Thus, on the basis of all the previous direct and indirect evidence, one can argue that the hemiketal carbon at pH 6.6 adopts the S configuration, as illustrated in Figure 4B.
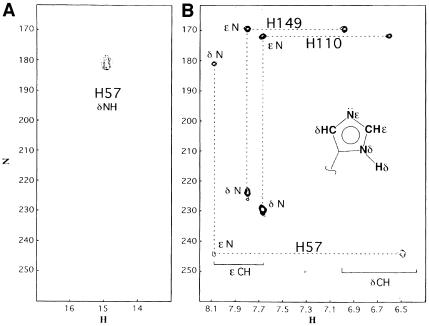
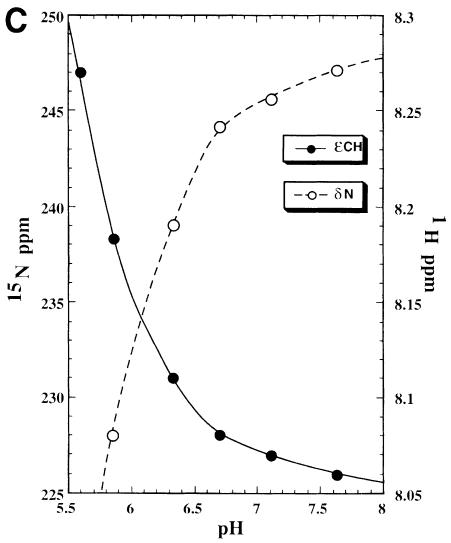
Fig. 7. (A) Selected region of the 1H–15N HMQC spectrum of the NS3–inhibitor complex, the experiment has been performed at pH 6.7 and 288 K with a jump and return type pulse and z–gradient pulses to reduce the intense water signal. The signal correlating the resonance at 1H 14.9 p.p.m. with 15N 180.9 p.p.m. arises from the direct correlation of the δN–δH atoms of the His57 residue. (B) 1H–15N long range correlation HSQC, acquired at pH 6.7 and 288 K, the delay during which 1H–15N antiphase is produced was set to 22 ms (1/2J) to refocus single-bond correlations. The cross peak pattern and the assignment are shown for all three histidines. His57 exhibits the typical L–shaped pattern, characteristic for the δ–NH tautomer, while the remaining two histidines, which exhibit the reverse pattern, are in the ɛ–NH tautomeric form. All histidines are in the uncharged form. In the inset there is a schematic representation of the nomenclature used for the histidine side chain atoms. (C) Titration of His57, followed by 1H–15N long range 2D HSQC and 1D-type experiments. The scale on the left refers to the 15N chemical shift (○), while that on the right refers to the 1H shift (•). It has not been possible to obtain a complete pH titration curve, since below pH 5.8 the relevant 15N signals disappear, indicating an intermediate exchange situation, and below pH 5.2 the sample instability does not allow determination of the His57 protonated state resonances. However, at pH 5.6 the 1H signal was still visible after 32 k scans in the 1D experiment.
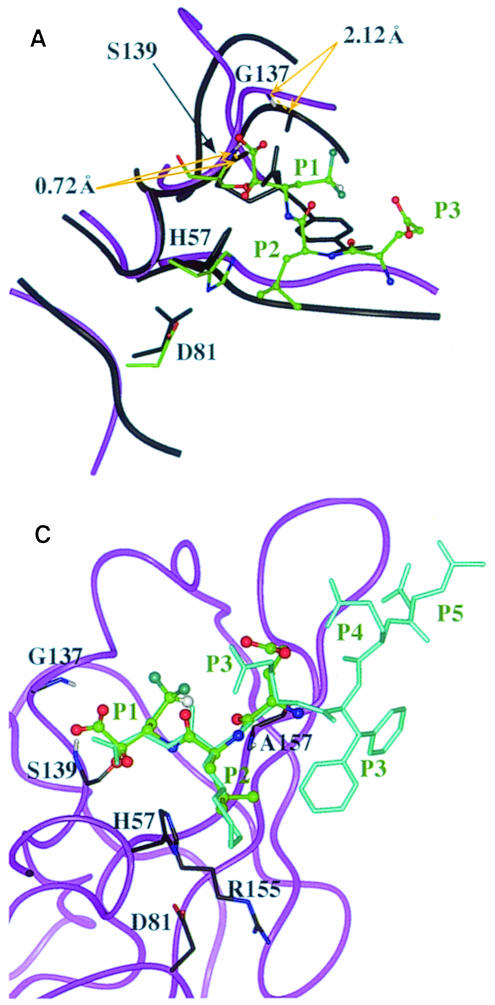
Two structures of chymotrypsin-like enzymes complexed with ketoacids have been published. Figure 5A shows the comparison between the NS3–inhibitor complex and the p-amidinophenyl pyruvic acid (APPA)–trypsin complex (Walter and Bode, 1983), by aligning the SCR residues. It is evident that the oxyanion loop formed in the NS3–inhibitor complex is wider than that in the APPA–trypsin complex. The distances between the equivalent backbone N atoms of the oxyanion hole residues are 0.7 and 2.1 Å (Figure 5A), indicating that although the catalytic serine NH position is rather similar, the oxyanion loop tends to widen as we follow the backbone of the NS3–inhibitor complex. Similar results are obtained by aligning the complex thrombin–APPA (Chen et al., 1995) and the NS3–inhibitor complex. The structure implies that in the NS3–inhibitor complex Gly137 NH is distant from the substrate P1 carbonyl oxygen (>3 Å). This requires that during catalysis either some local rearrangement reduces the distance to allow the anchorage needed to stabilize the tetrahedral intermediate and/or other factors, like electrostatics, contribute to the stabilization of the intermediate. In fact three positive charges, provided by Arg109, Lys136 and Arg155 (Figure 4B), can be localized in the vicinity. A similar stabilizing effect has been reported for subtilisin (Jackson and Fersht, 1993).
Fig. 5. (A) Expanded view of the pairwise superposition of the NS3–inhibitor complex S configuration (average minimized structure backbone in magenta ribbon, the catalytic triad in stick represen– tation and the ketoacid inhibitor in ball-and-stick representation with atom type colour code) and APPA–trypsin complex (PDB accession code 1tpp, backbone in black ribbon and inhibitor in black stick representation), the SCR residues' backbone atoms have been used for the superposition. The resulting distances between equivalent nitrogen atoms forming the oxyanion hole are shown. (B) Pairwise super– position of the average minimized structures of the free enzyme, blue (PDB accession code 1bt7), and the complex, assuming the S configuration, magenta, with the inhibitor shown in ball-and-stick representation (green). The catalytic triad is reported in stick representation. The HN groups forming the oxyanion hole and their distances in the two structures are shown. The SCR strands that account mostly for the total r.m.s.d. observed (1.05 Å) are A2 and C2. (C) Pairwise superposition of the minimized average structure of the set assuming the S configuration, with inhibitor I in cyan stick representation (product of reaction inhibitor complex) (Cicero et al., 1999). The backbone heavy atoms of the P1–P3 residues have been used in the superposition.
We further observe that the positions of the residues forming the oxyanion hole (Gly137 and Ser139) are essentially invariant in the free enzyme (Barbato et al., 1999) and in the complexes; dNH Ser139 = 0.32 Å and Gly137 = 0.74 Å (r.m.s.d. of the α–C of residues 136–140 = 0.25 Å) (Figure 5B).
The structure of an enzyme-bound, non-covalent inhibitor (inhibitor I) was obtained with transfer NOEs (Cicero et al., 1999), and in Figure 5C its superposition with the covalently bound ketoacid inhibitor structure (S configuration) is shown. The r.m.s.d. for the P1–P3 backbone heavy atoms' superposition is 0.22 Å. Furthermore, as shown in Figure 5C, the carboxylic end of inhibitor I is only partially stabilized by the oxyanion hole interactions since the hydrogen bond distances are longer. This characteristic is probably also reflected by the low affinity observed in the inhibitor I type family of compounds (Ingallinella et al., 1998). Comparing the S configuration of the NS3–ketoacid complex (Figure 4B) with the tetrahedral intermediate (Figure 1B), it can be seen that the ketoacid, due to its one-carbon homologated chain, is able to place its carboxyl group more effectively in the oxyanion hole. As a result the required interactions can take place without any apparent need for rearrangement. These results substantially confirm that the ketoacidic moiety's role is to add binding energy without altering the general mechanism of the interaction.
Inhibitor binding site: P2
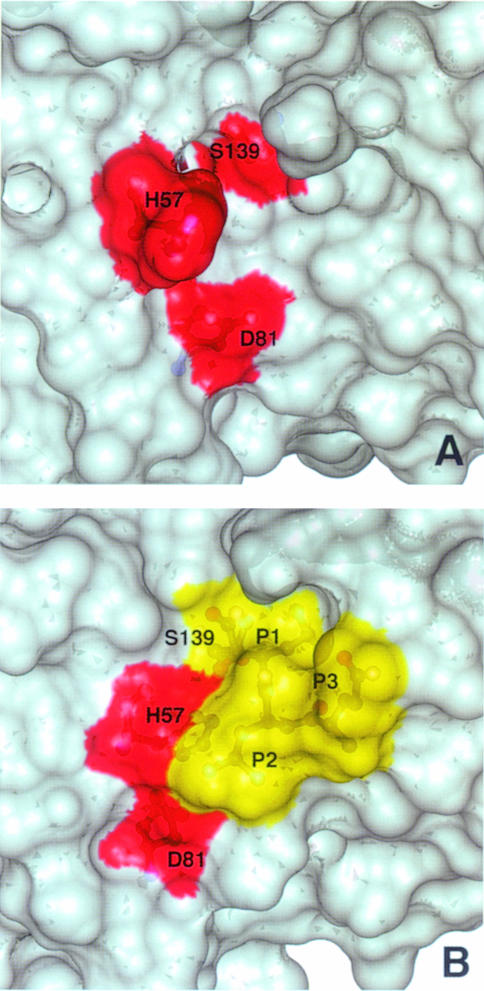
A very interesting feature is observed at the P2 Leu side chain. This side chain is positioned above the His57 imidazole moiety and also partially shields the Asp81 carboxylate moiety from the solvent (Figure 4B). The local hydrophobicity is further enhanced by the packing of the side chain methyl of Ala156 and the side chain methylenes of Arg155 in extended conformation and parallel to an ideal axis formed by the δ–NH of His57 and the COO– of Asp81. While the total exposed surface in this region is 86 Å2 in the free enzyme (Figure 6A), in the complex it is reduced to 34 Å2 (Figure 6B). For chymotrypsin-type enzymes the corresponding average value is ∼40 Å2, thus very close to the value found in the NS3–inhibitor complex. The solvent-exposed nature of this region in the free NS3 enzyme prevented the NMR detection of the signal of the histidine δ–NH hydrogen bonded to the aspartate carboxyl even at extreme temperature conditions (–8°C) (Barbato et al., 1999). This signal is commonly observed in other serine proteases (Markley, 1978; Bachovchin, 1985). The observation of this signal is an indication of the intact hydrogen bond between the catalytic Asp–His residues. It has been shown that this hydrogen bond becomes unstable when solvent exposed (Frey et al., 1994) and, on the contrary, is extremely stable when sheltered from the solvent (ΔG″ >10 kcal/mol) (Markley and Westler, 1996). The exclusion of solvent at this site upon complex formation allows the experimental observation of the signal arising from the δ–NH proton of His57. Figure 7A shows the typical downfield HN signal (1H, 14.9 p.p.m.; 15N, 180.9 p.p.m.) arising from the δ–NH of His57 (pH = 6.7, T = 288 K). Figure 7B shows the long range HSQC 1H–15N connectivities between the ɛ–CH (1H, 8.08 p.p.m.) and the δ–NH (15N, 180.9 p.p.m.) and ɛ–N (15N, 245.0 p.p.m.) of the His57. From the difference in the 15N chemical shifts of the δ- and ɛ–N one can derive that His57 is not protonated at pH 6.7. Further– more, the 15N chemical shift values show that the δ–N is acting as an H–bond donor while the ɛ–N is not involved in any hydrogen bonding (Bachovchin, 1986) (Figures 4B and 7B). The typical inverse L–shaped pattern of the connected peaks (Figure 7B) shows that His110 and 149 are in the ɛ–NH tautomeric state, while the L–shaped peak pattern of the His57 is typical for the δ–NH tautomer (Pelton et al., 1993). In Figure 7C the pH titration profile of the δ–NH spin signals is reported. An upper bound limit for the pKa value is estimated at ∼5.7. This is at least 1 pH unit lower than the pKa of His57 in the free enzyme, which is 6.8 (Urbani et al., 1998), and ∼5 pH units lower than what has been found for other covalently complexed inhibitors such as trifluoromethyl ketones (Cassidy et al., 1997) (pKa in the range 10.5–12.0). These inhibitors also form acyl-enzyme complexes with the catalytic serine of the type described above. This observation appears unprecedented in the serine proteases of the chymotrypsin family. We propose that this effect is due to the lowering of the dielectric constant resulting from the exclusion of water molecules from the solvation sphere of the His ring. It is well known that when the imidazole moiety is located in a hydrophobic environment its pKa is lowered (Fersht, 1985; Yang et al., 1993). Moreover, it has been shown on subtilisin that the presence of positive charges in the vicinity (<10–15 Å away) can reduce the pKa of a His residue by ∼0.4–0.6 pH units (Russell and Fersht, 1987). Thus, in our case the large variation of the pKa could be due to the fact that in addition to the desolvation effect there are also three adjacent positive charges only 5–10 Å away from the catalytic His ring (Figure 4B). A similar argument has been invoked recently to explain the lowered pH value of maximum enzymatic activity of some mutants of Bacillus lentus subtilisin (DeSantis and Jones, 1998).
Fig. 6. (A and B) View of the surface of the catalytic triad (red) for the free (A) and complex (B) enzyme structures exposed to the solvent. The protein surface is shown in grey (A and B) and the inhibitor in yellow (B).
Inhibitor binding site: P3
The backbone HN and CO groups of the inhibitor P3 Glu and of protein Ala157 residues are involved in an intermolecular, anti-parallel β–sheet-like hydrogen bond pattern. This accounts for the lower deuterium exchange rate of the Ala157 HN residue as compared with the free enzyme. There are no NOEs, apart from the intra-residue ones, involving the inhibitor P3 Glu side chain. As a result, in the calculated structures the side chain of P3 does not seem to be in a defined conformation, since it is free to sample the space delimited by V132, C159 and Lys136 (Figure 4B). These results are not unexpected since the inhibitor P3 position is engaged in a backbone to backbone hydrogen bond, of the type found for the NS3–inhibitor complex, also in a number of known serine protease–inhibitor complexes (Bode et al., 1984, 1990; Navia et al., 1989; Hecht et al., 1991). Also in these complexes the P3 residue side chain is usually solvent exposed or non-interacting with the enzyme.
Mechanistic model of action for the NS3 serine protease
The first step in the formulation of a mechanistic model of action of NS3 is to explain on a structural basis the activity observed in the absence of the activating cofactor. The free enzyme structure is similar to that of all the other chymotrypsin-like proteins, except that it leaves the essential His–Asp interaction solvent exposed. Solvent interference is therefore a key feature for the activity of this enzyme, and solvent expulsion from the surface of the His–Asp interaction can be achieved upon substrate complex formation. Hence a stabilized hydrogen bond network such as that described in Figure 1A is eventually formed, without the presence of NS4A being required. This stabilization mechanism also holds true in the case of the complex with the cofactor NS4A, since the His–Asp pair also appears to be solvent exposed in this case (Kim et al., 1996; Yan et al., 1998).
The second step requires a model for the cofactor activation that accounts for all the available experimental evidence. Kinetic data have shown that the action of NS4A is exerted primarily at the Kcat level, and that it involves the S′ enzyme sub-site, thereby suggesting that its action facilitates the exit of the substrate leaving group after cleavage (Landro et al., 1997). From the crystal structure of the complex NS3/NS4A (Kim et al., 1996; Yan et al., 1998) it has been noted that interactions of the terminal residues of the prime side with NS4A are possible. Furthermore, both the crystal structure and the solution structure of the free enzyme have provided evidence that the whole region comprising residues 59–66 (which comprise the strand D1, Figure 2B) is mobile in the absence of NS4A. This region is actually part of the S′ sub-site. Thus, the primary NS4A action would be to stabilize the fold of these residues. The direct interaction of NS4A with the A1 strand (Figure 2B), moreover, would contribute to the re-shaping of the S′ sub-site in such a way that the interaction with the prime portion of the substrate facilitates the catalytic turnover. In addition it is known that NS4A targets the NS3 protein to the membrane, although the actual implications of this process for the enzymatic mechanism have still to be elucidated, since no detailed structural information for a membrane complex is available to date.
Conclusions
In the present paper we analyse the structure of the NS3 serine protease in complex with a ketoacid peptidic inhibitor. The structure is relevant from a pharmaceutical point of view since it can be useful in the process of rational design of inhibitors as an approach towards drug discovery. Moreover, on the assumption that from this structure we can infer information on the substrate interaction we propose a novel mechanism of NS3 serine protease activation, whereby the conformation of the enzyme active site is stabilized by the solvent shielding effect exerted by the substrate. Our findings provide a framework to explain how the NS3 protease of HCV might be partially activated in an NS4A-independent manner with certain substrates. Our results substantially confirm our earlier hypothesis about the role of the P2 substrate position in the stabilization of the hydrogen bond between His57 and Asp81 (Barbato et al., 1998). Although observed in the absence of the NS4A cofactor, substrate-induced stabilization of the enzyme's catalytic triad is also bound to occur in the presence of NS4A since the catalytic site is similar in both situations. Recently it has been reported that NS4A complexation induces an NS3 structure that is already, but not entirely, pre-organized for substrate binding (Bianchi et al., 1999). In agreement with this observation we propose that NS4A could activate the NS3 protease indirectly, by modifying the S′ sub-site stabilizing the conformation of its boundaries in a way that pre-organizes the site to the binding process and allows the immediate recognition of the different substrates favouring their turnover. This hypothesis is in line with the observation that the formation of a binding site for the P′ portion of the substrate is induced by the presence of NS4A (Landro et al., 1997).
To our knowledge this is the first reported case in which a chymotrypsin-like protease is shown to require binding of the substrate to stabilize its catalytic machinery in the correct geometry. However, the extensive similarity at the catalytic site of the structures of the NS3–inhibitor complex and the bifunctional capsid protease proteins of Semliki Forest virus (Choi et al., 1997) and Sindbis virus (Tong et al., 1993) suggest that this finding might not be unique to HCV. These other viral proteases exert only a single autolytic cleavage at their C–terminus, resulting in an inhibited enzyme with the C–terminal residues of the protein acting as auto-inhibitors. The reported structures show that in the absence of the equivalent inhibitory residues, the His–Asp pair would be solvent exposed in an identical fashion to HCV NS3 protease. Solvent expulsion in these viral proteins is again an effect of the position P2 Glu residue side chain (packed on the His ring) and the residue Leu235 (Semliki virus) or 231 (Sindbis virus), the latter of which is equivalent to Arg155 in NS3. Although the cytomegalovirus serine protease is not a chymotrypsin-like serine protease (in the catalytic triad the Asp is mutated to His), the equivalent hydrogen bond His–His is also solvent exposed and is again protected by the P2 residue upon substrate binding (Tong et al., 1998). It is therefore tempting to speculate that this mode of stabilization of the catalytic machinery could be a more general characteristic of the viral serine protease class. It could represent a case of an evolutionary convergent mechanism developed by the viruses to enhance selectivity for their viral substrate targets against the cellular serine protease substrates.
Materials and methods
Protein expression and inhibitor synthesis
15N or 15N/13C samples of the protease domain of the NS3 protein were expressed in Escherichia coli cells BL21(DE3) using a plasmid encoding residues 1–180 of NS3BK strain and a solubilizing tail (ASKKKK residues) inserted at the C–terminus; the plasmid was under the control of pT7.7 polymerase. The cells were grown in M9 minimum medium supplemented with 1 g/l (15NH4)2SO4 and 2 g/l [13C]glucose (Sigma); a further addition of 6.8 mg/l ZnCl2 was necessary since NS3 is a zinc-binding protein. The [15N]Asp, Asn and Thr selectively labelled sample was obtained by incorporating 0.33 g/l each of the labelled residues in the M9 modified medium. Expression was achieved by incubating the cells with 0.4 mM isopropyl-β–d–thiogalactopyranoside (IPTG) at an A600 of 0.6 for 3 h at 30°C. A sample expressed in a medium containing 10% [13C]glucose was produced for the stereospecific assignments of the Val and Leu methyl groups (Neri et al., 1989). The remaining protein expression and purification protocol was carried out as previously described (De Francesco et al., 1996). The design, synthesis and characterization of the α–ketoacid [tBut-Glu-Leu-(di–fluoro)Abu] has also been described (Narjes et al., 2000). A sample with the inhibitor doubly labelled at the Leu, and at the backbone atoms of the (di–fluoro)Abu (including the carbonyl group, but excluding the carboxyl) was also synthesized. The final buffer contained 20 mM sodium phosphate, 3 mM dithiothreitol, 10 mM β–octyl-glucoside, 5 mM NaN3 pH 6.6. The concentration of the complex was 0.6–0.8 mM. The monomeric state of the protein in the samples was verified with T2 15N relaxation experiments (not shown).
NMR spectroscopy
The NMR experiments were acquired at 298 and 310 K on Bruker Avance 500 and 600 MHz machines equipped with z–shielded gradient triple resonance probes. Backbone assignments of the enzyme were obtained using HNCA, HNCO, HN(CO)CA, CBCA(CO)NH, CBCANH (Bax and Grzesiek, 1993) and HACACO (Bazzo et al., 1995) experiments at 28 and 37°C. Side chain assignments were obtained using H(C)CH and (H)CCH TOCSY (16 and 24 ms mixing time) and COSY experiments together with a CC(CO)NH TOCSY (Grzesiek et al., 1993a) (24 ms mixing time) at 37°C.
Distance restraints were obtained from a number of different 3D-NOESY type experiments: 15N NOESY-HSQC (120 and 80 ms mixing time); 15N ROESY-HSQC (40 ms); 13C NOESY-HSQC (120 and 80 ms mixing time); and several different filtered type 2D- and 3D–NOESY experiments, including 1H–19F NOEs, were performed to detect selectively intra- or inter-molecular NOEs, detecting the inhibitor signals. Two different 13C 3D–NOESY-HSQC (80 ms mixing) experiments were used optimized on the aromatic 1H–13C 1J coupling, using 1.2 and 1.0 ms for the transfer (1/4J).
Angular restraints were obtained using HNHA (Kuboniwa et al., 1994), HA(CA)HB-COSY (Grzesiek et al., 1995), CO and N decoupled CT-HSQC experiments (Grzesiek et al., 1993b; Vuister et al., 1993). Spectra were processed using NMRPipe (Delaglio et al., 1995) and analysed using NMRView (Johnson and Blevins, 1994) software packages.
Structure calculation
Approximate inter-proton distances were derived from the multidimensional NOE spectra (Clore and Gronenborn, 1991). The volumes of NOESY experiment non-overlapped cross peaks were integrated using the NMRView function Autocal2, which uses the ARIA calibration algorithm (Nilges, 1995). A constant value of 0.6 Å was added to the upper bounds of the strong and medium NOEs involving methyl groups, an extra allowance of 0.2 Å was added to the peaks involving amide protons. A lower limit of 1.8 Å was set for all the distance constraints. During the final stages of refinement H-bond constraints (dNH–O = 1.6–3.0 Å, dN–O = 2.4–3.6 Å) were included for those amide groups, within regular areas of secondary structure, that were still observable after 48 h of exchange with D2O at room temperature in a 1H–15N HSQC experiment. The zinc-binding site was constrained as previously described (Barbato et al., 1999).
The φ, ψ and χ1 angles were derived from homo- and heteronuclear three bond coupling constant data and consensus chemical shift index, employing as minimum range ±30, 45 and 25°, respectively.
The structures were calculated by simulated annealing with the program X–Plor 3.851 (Brunger, 1993) on an SGI O2 R10000 platform, following a previously published protocol (Omichinski et al., 1997). During the final steps a conformational database refinement was included in the protocol (Kuszewski et al., 1997).
The inhibitor was explicitly included in the calculations by modifying the topology files in X–Plor. The atom distances and valence angles used for the topology file at the ketoacid moiety were obtained by a search on the Chemical Structure Database (Allen et al., 1979, 1991). Structure calculations were performed in parallel, assuming alternatively both configurations (R,S) at the chiral centre. Surface evaluations, figures and statistical analysis were produced using the program InsightII and the modules Biopolymer and DelPhi (Molecular Simulations Inc.).
Accession number
The coordinates of the final 20 simulated annealing structures (S configuration) have been deposited in the Brookhaven Protein Data Bank, accession code 1dxw.
Acknowledgments
Acknowledgements
We thank P.Neddermann for her help in the preparation of the recombinant samples used throughout the project, and C.Steinkühler, A.Tramontano and A.Lahm for critically reading the manuscript. F.C. acknowledges the support of a Humboldt fellowship.
References
- Allen F.H., et al. (1979)The Cambridge crystallographic data centre: computer based search, retrieval, analysis and display of information. Acta Crystallogr. B, 35, 2331–2335. [Google Scholar]
- Allen F.H., et al. (1991)The development of versions 3 and 4 of the Cambridge structural database system. J. Chem. Inf. Comput. Sci., 331, 187–204. [Google Scholar]
- Avital J. (1998) Hepatitis C. Curr. Opin. Infect. Dis., 11, 293–299. [DOI] [PubMed] [Google Scholar]
- Bachovchin W.W. (1985) Confirmation of the assignment of the low field proton resonance of serine proteases by using specifically nitrogen-15 labeled enzyme. Proc. Natl Acad. Sci. USA, 82, 7948–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin W.W. (1986) 15N NMR spectroscopy of the hydrogen bonding interactions in the active site of serine proteases: evidence for a moving histidine mechanism. Biochemistry, 25, 7751–7759. [DOI] [PubMed] [Google Scholar]
- Barbato G., Cicero, D.O., Nardi, M.C., Steinkühler, C., Cortese, R., De Francesco, R. and Bazzo, R. (1999) The solution structure of the N–terminal protease domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism. J. Mol. Biol., 289, 370–384. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R. (1999) The NS3/4A protease of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepat., 6, 165–181. [DOI] [PubMed] [Google Scholar]
- Bax A. and Grzesiek, S. (1993) Methodological advances in protein NMR. Acc. Chem. Res., 26, 131–138. [Google Scholar]
- Bazan J.F. and Fletterick, R.J. (1988) Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl Acad. Sci. USA, 85, 7872–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzo R., Cicero, D.O. and Barbato, G. (1995) A new 3D HCACO pulse sequence with optimised resolution and sensitivity. Application to the 21 kDa protein human interleukin-6. J. Magn. Reson. B, 107, 189–191. [DOI] [PubMed] [Google Scholar]
- Bianchi E., Orru, S., Dal Piaz, F., Ingenito, R., Casbarra, A., Biasiol, G., Koch, U., Pucci, P. and Pessi, A. (1999) Conformational changes in human hepatitis C virus NS3 protease upon binding of product based inhibitors. Biochemistry, 38, 13844–13852. [DOI] [PubMed] [Google Scholar]
- Bode W., Walter, J., Huber, R., Wenzel, H.R. and Tschesche, H. (1984) The refined 2.2 Å X–ray crystal structure of the ternary complex formed by bovine trypsinogen valine-valine and the Arg-15-analogue of bovine pancreatic trypsin inhibitor. Eur. J. Biochem., 144, 185–197. [DOI] [PubMed] [Google Scholar]
- Bode W., Turk, D. and Stuerzebecher, J. (1990) Geometry of binding of the benzamidine and arginine-based inhibitors NAPAP and MQPA to human α-thrombin. X–ray crystallographic determination of the NAPAP–trypsin complex and modeling of NAPAP–thrombin and MQPA–thrombin. Eur. J. Biochem., 193, 175–188. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. (1993) X-Plor Version 3.1: A System for X–ray Crystallography and NMR. Yale University Press, New Haven, CT. [Google Scholar]
- Cassidy C.S., Ling, J. and Frey, P. (1997) A new concept for the mechanism of action of chymotrypsin: the role of the low barrier hydrogen bond. Biochemistry, 36, 4576–4584. [DOI] [PubMed] [Google Scholar]
- Chen Z., Li, Y., Mulichak, A.M., Lewis, S.D. and Shafer, J.A. (1995) Crystal structure of human α-thrombin complexed with hirugen and p-amidinophenylpyruvate at 1.6Å resolution. Arch. Biochem. Biophys., 322, 198–203. [DOI] [PubMed] [Google Scholar]
- Choi H.K., Lu, G., Lee, S., Wengler, G. and Rossmann, M.G. (1997) Structure of Semliki Forest virus core protein. Proteins, 27, 345–359. [DOI] [PubMed] [Google Scholar]
- Cicero D.O., et al. (1999)Structural characterisation of the interactions of optimised product inhibitors with the protease domain of human hepatitis C virus by NMR and modelling studies. J. Mol. Biol., 289, 385–396. [DOI] [PubMed] [Google Scholar]
- Cleland W.W., Frey, P.A. and Gerlt, J.A. (1998) The low barrier hydrogen bond in enzymatic catalysis. J. Biol. Chem., 273, 25529–25532. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn, A.M. (1991) Structures of larger proteins in solution: three and four dimensional heteronuclear NMR spectroscopy. Science, 252, 1390–1399. [DOI] [PubMed] [Google Scholar]
- De Francesco R. and Steinkühler, C. (1999) Structure and function of the hepatitis C virus NS3–NS4A serine protease. Curr. Top. Microbiol. Immunol., 242, 149–169. [DOI] [PubMed] [Google Scholar]
- De Francesco R., Urbani, A., Nardi, M.C., Tomei, L., Steinkühler, C. and Tramontano, A. (1996) A zinc site in viral serine proteases. Biochemistry, 35, 13282–13287. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J. and Bax, A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Delbaere L.T.J. and Brayer, G.D. (1985) The 1.8Å structure of the complex between chymostatin and Streptomyces griseus protease A. A model for serine protease catalytic tetrahedral intermediates. J. Mol. Biol., 183, 89–103. [DOI] [PubMed] [Google Scholar]
- DeSantis G. and Jones, B. (1998) Chemical modifications at a single site can induce significant shifts in the pH profile of a serine protease. J. Am. Chem. Soc., 120, 8582–8586. [Google Scholar]
- Fersht A. (1985) Enzyme Structure and Mechanism. Freeman,W.H. and Co., New York, NY. [Google Scholar]
- Frey P.A., Whitt, S.A. and Tobin, J.B. (1994) A low-barrier hydrogen bond in the catalytic triad of serine proteases. Science, 82, 1927–1930. [DOI] [PubMed] [Google Scholar]
- Greer J. (1990) Comparative modeling methods: application to the family of the mammalian serine proteases. Proteins, 7, 317–334. [DOI] [PubMed] [Google Scholar]
- Grzesiek S., Anglister, J. and Bax, A. (1993a) Correlation of backbone amide and aliphatic sidechain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B, 101, 114–119. [Google Scholar]
- Grzesiek S., Vuister, G.W. and Bax, A. (1993b) A simple and sensitive experiment for measurement of JCC couplings between backbone carbonyl and methyl carbons in isotopically enriched proteins. J. Biomol. NMR, 3, 487–493. [DOI] [PubMed] [Google Scholar]
- Grzesiek S., Kuboniwa, H., Hinck, A.P. and Bax, A. (1995) Multiple quantum line narrowing for measurements of Hα–HβJ couplings in isotopically enriched proteins. J. Am. Chem. Soc., 117, 5312–5315. [Google Scholar]
- Hecht H.J., Szardenings, M., Collins, J. and Schomburg, D. (1991) Three dimensional structure of the complexes between bovine chymotrypsinogen A and two recombinant variants of human pancreatic secretory trypsin inhibitor. J. Mol. Biol., 220, 711–722. [DOI] [PubMed] [Google Scholar]
- Hijikata M., Mizushima, H., Akagi, T., Mori, S., Kakiuchi, N., Kato, N., Tanaka, T., Kimura, K. and Shimotono, K. (1993) Two distinct protease activities required for the processing of a putative non structural precursor protein of hepatitis C virus. J. Virol., 67, 4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingallinella P., Altamura, S., Bianchi, E., Taliani, M., Ingenito, R., Cortese, R., De Francesco, R., Steinkuehler, C. and Pessi, A. (1998) Potent inhibitors of human hepatitis C virus NS3 protease are obtained by optimising the cleavage products. Biochemistry, 37, 8906–8914. [DOI] [PubMed] [Google Scholar]
- Jackson S.E. and Fersht, A.R. (1993) Contribution of long range electrostatic interactions to the stabilisation of the transition state of the serine protease subtilisin BPN′. Biochemistry, 32, 13909–13916. [DOI] [PubMed] [Google Scholar]
- Johnson B. and Blevins, R.A. (1994) NMRView: a computer program for the visualisation and analysis of NMR data. J. Biomol. NMR, 4, 603–614. [DOI] [PubMed] [Google Scholar]
- Kim J.L., et al. (1996)Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell, 87, 343–355. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Kuboniwa H., Grzesiek, S., Delaglio, F. and Bax, A. (1994) Measurement of HN-HαJ coupling in calcium free calmodulin using new 2D and 3D water-flip-back methods. J. Biomol. NMR, 4, 871–878. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Gronenborn, A. and Clore, M. (1997) Improvements and extensions in the conformational database potential for the refinement of NMR and X–ray structures of proteins and nucleic acids. J. Magn. Reson., 125, 171–177. [DOI] [PubMed] [Google Scholar]
- Landro J.A., Raybuck, S.A., Luong, Y.P.C., O'Malley, E.T., Haberson, S.L., Morgenstern, K.A., Rao, G. and Livingston, D.J. (1997) Mechanistic role of an NS4A peptide cofactor with the truncated NS3 protease of hepatitis C virus: elucidation of the NS4A stimulatory effect via kinetic analysis and inhibitor mapping. Biochemistry, 36, 9340–9348. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lesk A. and Fordham, W.D. (1996) Conservation and variability in the structures of serine proteinases of the chymotrypsin family. J. Mol. Biol., 258, 501–537. [DOI] [PubMed] [Google Scholar]
- Love R.A., Parge, H.E., Wickersham, J.A., Hostomsky, Z., Habuka, N., Moomaw, E.W., Adachi, T. and Hostomska, Z. (1996) The crystal structure of hepatitis C virus NS3 protease reveals a trypsin-like fold and a structural zinc binding site. Cell, 87, 331–342. [DOI] [PubMed] [Google Scholar]
- Love R.A., et al. (1998)The conformation of the hepatitis C virus with and without NS4A: a structural basis for the activation of the enzyme by its cofactor. Clin. Diagn. Virol., 10, 151–156. [DOI] [PubMed] [Google Scholar]
- Markley J.L. (1978) Hydrogen bonds in serine proteases and their complexes with protein protease inhibitors. Biochemistry, 17, 4648–4656. [DOI] [PubMed] [Google Scholar]
- Markley J.L. and Westler, W. (1996) Protonation state dependence of hydrogen bond strengths and exchange rates in a serine protease catalytic triad: bovine chymotrypsinogen A. Biochemistry, 35, 11092–11097. [DOI] [PubMed] [Google Scholar]
- McGrath M.E., Vasquez, J.R., Craik, C.S., Yang, A.S., Honig, B. and Fletterick, R.J. (1992) Perturbing the polar environment of Asp 102 in trypsin: consequences of replacing conserved Ser 214. Biochemistry, 31, 3059–3064. [DOI] [PubMed] [Google Scholar]
- Narjes F., Brunetti,M., Colarusso,S., Gerlach,B., Koch,U., Biasiol,G., Fattori,D., De Francesco,R., Matassa,V.G. and Steinkühler,C. (2000) α-Ketoacids are potent slow binding inhibitors of the hepatitis C virus NS3 protease. Biochemistry, in press. [DOI] [PubMed] [Google Scholar]
- Navia M.A., McKeever, B.M., Springer, J.P., Lin, T.Y., Williams, H.R., Fluder, E.M., Dorn, C.P. and Hoogsteen, K. (1989) Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84 Å resolution. Proc. Natl Acad. Sci. USA, 86, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P., Tomei, L., Steinkühler, C., Gallinari, P., Tramontano, A. and De Francesco, R. (1997) The non structural proteins of the hepatitis C virus: structure and functions. Biol. Chem., 378, 469–476. [PubMed] [Google Scholar]
- Neri D., Szyperski, T., Otting, G., Senn, H. and Wüthrich, K. (1989) Specific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry, 28, 7510–7516. [DOI] [PubMed] [Google Scholar]
- Nilges M. (1995) Calculation of protein structures with ambiguous distance restraints. Automated assignment of ambiguous NOE crosspeaks and disulphide connectivities. J. Mol. Biol., 245, 645–660. [DOI] [PubMed] [Google Scholar]
- Omichinski J.G., Pedone, P.V., Felsenfeld, G., Gronenborn, A.M. and Clore, G.M. (1997) The solution structure of a specific GAGA factor–DNA complex reveals a modular binding mode. Nature Struct. Biol., 4, 122–132. [DOI] [PubMed] [Google Scholar]
- Ortiz C., Tellier, C., Williams, H., Stolowich, N.J. and Scott, A.I. (1991) Diastereotopic covalent binding of the natural inhibitor leupeptin to trypsin: detection of two interconverting hemiacetals by solution and solid state NMR. Biochemistry, 30, 10026–10034. [DOI] [PubMed] [Google Scholar]
- Pelton J.G., Torchia, D.A., Meadow, N.D. and Roseman, S. (1993) Tautomeric states of the active site histidines of phosphorylated and unphosphorylated IIIGlc, a signal transducing protein from Escherichia coli, using two dimensional heteronuclear NMR techniques. Protein Sci., 2, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.A. and Fletterick, R.J. (1992) Proteases. Curr. Opin. Struct. Biol., 2, 713–720. [Google Scholar]
- Polgar L. (1989) Mechanism of Protease Action. CRC Press, Boca Raton, FL. [Google Scholar]
- Russell A.J. and Fersht, A. (1987) Rational modification of enzyme catalysis by engineering surface charge. Nature, 328, 496–500. [DOI] [PubMed] [Google Scholar]
- Schechter I. and Berger, A. (1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun., 27, 157–162. [DOI] [PubMed] [Google Scholar]
- Shah D.O., Lai, K. and Gorenstein, D.G. (1984) 13C NMR spectroscopy of ‘transition state analogue’ complexes of N-acetyl-l-phenylalaninal and α-chymotrypsin. J. Am. Chem. Soc., 106, 4272–4273. [Google Scholar]
- Tong L., Wengler, G. and Rossmann, M.G. (1993) Refined structure of Sindbis virus core protein and comparison with other chymotrypsin-like serine protease structures. J. Mol. Biol., 230, 228–247. [DOI] [PubMed] [Google Scholar]
- Tong L., Qian, C., Massario, M.J., Deziel, R., Yoakim, C. and Lagace, L. (1998) Conserved mode of peptidomimetic inhibition and substrate recognition of human cytomegalovirus. Nature Struct. Biol., 5, 819–826. [DOI] [PubMed] [Google Scholar]
- Urbani A., Bazzo, R., Nardi, M.C., Cicero, D.O., De Francesco, R., Steinkühler, C. and Barbato, G. (1998) The metal binding site of the hepatitis C virus NS3 protease. J. Biol. Chem., 273, 18760–18769. [DOI] [PubMed] [Google Scholar]
- Vuister G.W., Wang, A.C. and Bax, A. (1993) Measurement of three bond nitrogen carbon J coupling in proteins uniformly enriched in 15N and 13C. J. Am. Chem. Soc., 115, 5334–5335. [Google Scholar]
- Walter J. and Bode, W. (1983) The X–ray crystal structure analysis of the refined complex formed by bovine trypsin and p-amidino– phenylpyruvate at 1.4Å resolution. Hoppe Seylers Z. Physiol. Chem., 364, 949–959. [DOI] [PubMed] [Google Scholar]
- Warshel A. (1998) Electrostatic origin of the catalytic power of enzymes and the role of preorganised active sites. J. Biol. Chem., 273, 27035–27038. [DOI] [PubMed] [Google Scholar]
- World Health Organization (1999) Global surveillance and control of hepatitis C. J. Viral Hepat., 6, 35–47. [PubMed]
- Yan Y., et al. (1998)Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 Å resolution structure in a hexagonal crystal form. Protein Sci., 7, 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A.S., Gunner, M.R., Sampogna, R., Sharp, K. and Honig, B. (1993) On the calculation of pKas in proteins. Proteins, 15, 252–265. [DOI] [PubMed] [Google Scholar]