Abstract
Receptor binding to human poliovirus type 1 (PV1/M) and the major group of human rhinoviruses (HRV) was studied comparatively to uncover the evolution of receptor recognition in picornaviruses. Surface plas– mon resonance showed receptor binding to PV1/M with faster association and dissociation rates than to HRV3 and HRV16, two serotypes that have similar binding kinetics. The faster rate for receptor association to PV1/M suggested a relatively more accessible binding site. Thermodynamics for receptor binding to the viruses and assays for receptor-mediated virus uncoating showed a more disruptive receptor interaction with PV1/M than with HRV3 or HRV16. Cryo-electron microscopy and image reconstruction of receptor–PV1/M complexes revealed receptor binding to the ‘wall’ of surface protrusions surrounding the ‘canyon’, a depressive surface in the capsid where the rhinovirus receptor binds. These data reveal more exposed receptor-binding sites in poliovirus than rhinoviruses, which are less protected from immune surveillance but more suited for receptor-mediated virus uncoating and entry at the cell surface.
Keywords: cryo-EM and image reconstruction/picornavirus entry/poliovirus/rhinovirus/virus–receptor interaction
Introduction
Poliovirus (PV) and human rhinoviruses (HRV) are structurally related, non-enveloped RNA viruses (Rueckert, 1985). Because of their medical interest, they are among the most studied members of the picornavirus family. Up to 102 different serotypes of HRV have been described so far, in contrast to only three existing PV serotypes. Crystal structures of PV and HRV showed that the viral particles have icosahedral symmetry and are built up from 60 protomers arranged as 12 pentamers (Hogle et al., 1985; Rossmann et al., 1985). The PV and HRV capsids contain three external (VP1, 2 and 3) and one internal structural proteins (VP4). The crystal structure of the Mahoney strain of PV type 1 (PV1/M) revealed the presence of a fatty acid-related molecule (‘pocket factor’) in a cavity present in the protomers (Hogle et al., 1985). This molecule seems to stabilize the metastable virus particle. A similar ‘pocket factor’ molecule is present in structures of rhinovirus serotypes classified in antiviral group B (HRV1A and HR16), but absent in serotypes of group A (HRV3 and HRV14) (Zhao et al., 1996).
The cell surface receptors for PV (poliovirus receptor; PVR) and for 90% (major group) of the HRV serotypes (intercellular adhesion molecule-1; ICAM-1 or IC1) are related molecules that belong to the immunoglobulin superfamily (IgSF) (Greve et al., 1989; Mendelsohn et al., 1989; Staunton et al., 1989). PVR and IC1 contain three and five IgSF domains in their extracellular region, respectively, and a single transmembrane domain. IC1 belongs to a subfamily of related integrin ligands of the IgSF and binds to the leukocyte integrins LFA–1 and Mac–1, promoting a large range of cell–cell interactions related to inflammatory reactions (Springer, 1994). PVR is closely related in sequence to two other members of the IgSF known as poliovirus receptor related proteins 1 and 2 (PRR1 and PRR2) (Cocchi et al., 1998). These three molecules mediate entry of herpesviruses into the cell (Geraghty et al., 1998). Although N–terminal domains of PVR and IC1 contain the virus-binding epitopes, the presence of the second domain is important for proper folding and binding activity of the first domain (Staunton et al., 1990; Koike et al., 1991). Crystal structures of the N–terminal two domains of IC1 (Bella et al., 1998; Casasnovas et al., 1998b) showed that rhinovirus-binding residues sit in loops on the very ‘tip’ of the N–terminal domain of IC1. Mutants and models for domain 1 of PVR indicated that PV-binding residues reside on a side of the N–terminal domain of the receptor molecule (reviewed by Racaniello, 1996; Solecki et al., 1998).
Receptor binding to PV and major group of HRV induces conformational changes in the capsid leading to particle disruption and RNA release (Kaplan et al., 1990a; Hoover-Litty and Greve, 1993). These results suggested that PVR and IC1 not only function in virus attachment to the cell surface, but in the subsequent steps of RNA uncoating and virus entry into the cell (Racaniello, 1996). Upon binding to PVR, PV exposes hydrophobic residues, which mediate interaction of the particle with the cell surface (Fricks and Hogle, 1990). PV does not require endosomal acidification or clathrin-dependent endocytosis for entry and infection, and therefore uncoating might occur at the cell surface and be receptor driven (Perez and Carrasco, 1993; DeTuello and Kirchhausen, 1998). The entry pathway for the major group of HRV is not clearly defined, and both receptor- and pH-dependent uncoating could be required for cell entry (Hoover-Litty and Greve, 1993). A recent report showed that endosomal acidification was not required for cell infection by HRV14 (Schober et al., 1998), a serotype disrupted by receptor binding (Hoover-Litty and Greve, 1993). However, the inability of the receptor to mediate uncoating of major group serotypes containing ‘pocket factor’ suggests an alternative acid-dependent entry pathway.
Cryo-electron microscopy (cryo-EM) of HRV16 in complex with soluble IC1 mapped the receptor-binding site into a deep and narrow depression or ‘canyon’, around the 5–fold icosahedral vertices of the capsid (Olson et al., 1993). Kinetics for binding of soluble IC1 to HRV3 revealed a very low rate of association between monomeric receptor and virus, consistent with binding to a relatively inaccessible virus surface (Casasnovas and Springer, 1995). Docking of the IC1 structure into the electron density of the receptor in the complex showed that the tip of the receptor penetrates deep into the recessive virus-binding surface or ‘canyon’ on the virus capsid (Bella et al., 1998). Having the receptor-binding site in a quite inaccessible surface depression allows the major group rhinoviruses to escape immune surveillance (Rossmann, 1989).
Isolation of PV mutants resistant to the neutralization by solubilized receptor allowed the identification of putative receptor-binding residues on the virus (Kaplan et al., 1990b; Colston and Racaniello, 1994). The most critical and exposed receptor-binding residues defined by mutants were mapped to the ‘south wall’ of the ‘canyon’-like depression also present in PV. Some of these receptor-binding residues are part of neutralization antigenic sites (Solecki et al., 1998), suggesting that neutralizing antibodies can access conserved and critical residues for receptor recognition. While mutants defined a receptor-binding site close to the IC1-binding site in rhinovirus, the receptor-binding footprint in PV has not been mapped so far. Additionally, there is not any comparative information on receptor binding to the related PV and rhinoviruses, which can give new insights into the evolution of receptor recognition in picornaviruses and on virus uncoating processes mediated by the receptor. In this report, we present a comparative analysis on receptor binding to PV1/M and HRV3 and HRV16. Our analysis reveals a more accessible and disruptive receptor-binding site in PV1/M than HRV. Cryo-EM of PVR–PV1/M complexes shows receptor binding to surface protrusions and the junction of two capsid protomers and suggests how the PVR molecule triggers uncoating of the PV genome.
Results
Preparation of recombinant soluble receptor proteins
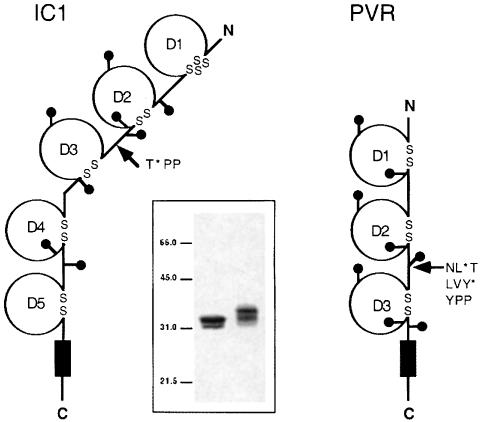
The extracellular regions of PVR and IC1 have three and five concatenated IgSF domains, respectively (Figure 1). Virus-binding epitopes in IC1 and PVR locate in the N–terminal domain (D1) of the receptors. D1 of IC1 belongs to the I1 set of IgSF domains and has an ‘extra’ disulfide bridge in the tip of the domain (Bella et al., 1998; Casasnovas et al., 1998b). In contrast, D1 of PVR is predicted to be part of the V set of IgSF domains and is, therefore, expected to be broader than the first domain of IC1 (Mendelsohn et al., 1989).
Fig. 1. Design and expression of soluble receptor proteins. Scheme of intercellular adhesion molecule-1 (IC1) and poliovirus receptor (PVR). IgSF domains are labeled D1–D5 or D1–D3, and the transmembrane domain is represented by a black rectangle. Cysteine residues are indicated with ‘S’ and glycosylation sites with ‘lollipop’ representations. Arrows indicate positions in which the engineered translation stop codon (*) was introduced to generate the two-domain variants. Stop codons are located after residues Thr190 of the mature IC1 and after Leu238 and Tyr241 of the precursor PVR polypeptide (see Materials and methods). The inset shows silver-stained 10% SDS–PAGE of purified soluble IC1-2D (left) and PVR-2D (right) proteins. The position and size (kDa) of molecular weight markers are included.
Soluble fragments of receptors used in this study were expressed in a lectin-resistant CHO cell line as described in Materials and methods. The soluble IC1 fragment has the N–terminal two domains and 190 residues (IC1-2D/190), and it has proper folding and affinity for HRV (Casasnovas et al., 1998a). To generate this protein, the receptor was truncated prior to two consecutive proline residues (‘PP-motif’) present in the junction between domains 2 and 3 (Figure 1). A similar ‘PP-motif’ is also present between N–terminal domains 2 and 3 in PVR. Therefore, to generate a comparable soluble fragment to IC1-2D/190, PVR was also truncated prior to the PP-motif. Two recombinant cDNAs coding for the N–terminal two domains of the receptor, PVR-2D/238 and PVR-2D/241, were prepared (Figure 1). Only the longest PVR-2D construct was secreted to the supernatant of transfected cells. This protein contains the last N–linked glycosylation site of domain 2 that is missing in PVR-2D/238. A similar PVR variant with the two N–terminal domains has been reported to fold properly based on binding to antibodies and PV (Arita et al., 1998). Soluble PVR-2D/241 and IC1-2D/190 have similar mobility in SDS–PAGE (Figure 1, inset).
Specific binding of soluble receptors to immobilized viruses in BIAcore
Surface plasmon resonance (BIAcore 2000) was chosen to study virus–receptor interactions because of the high accuracy and reproducibility of this methodology (Casasnovas and Springer, 1995). Purified HRV3, HRV16 and PV1/M were immobilized through primary amino groups to the carboxy-methylated dextran attached to the gold film of the sensor chip used in BIAcore. Freshly purified and unfrozen viruses were used in all the experiments and similar immobilization conditions were used for PV1/M and HRV (see Materials and methods).
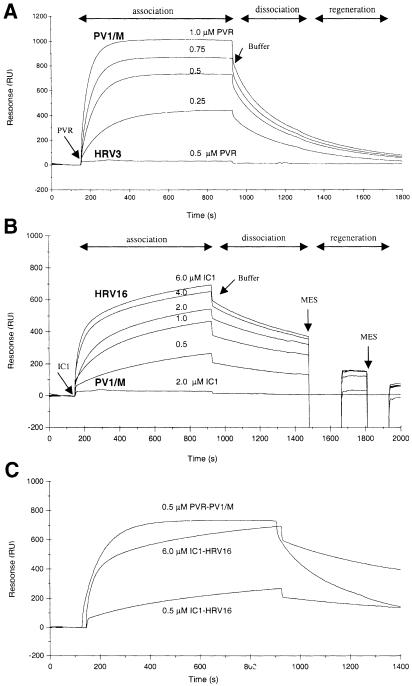
Binding specificity was examined by injecting soluble PVR (PVR-2D/241) and IC1 (IC1-2D/190) through surfaces coated with either PV1/M, HRV3 or HRV16 (Figure 2A and B). Binding, monitored as the increase in resonance units (RU), was dependent on receptor concentration and was detected only when the receptor protein was injected through a surface having its virus counterpart. About 10 times higher concentration of IC1 than PVR was needed to have similar binding units (Figure 2C). Successive cycles of association, dissociation and regeneration gave highly reproducible binding for all three receptor–virus interactions presented here (not shown). A range of receptor concentrations was usually injected through surfaces having 1000–15 000 RU of PV1/M or 5000–20 000 RU of HRV.
Fig. 2. Specific interaction of soluble receptor proteins with viruses in BIAcore. (A) Overlay plot of sensorgrams recording the response (RU) following injection of soluble PVR and running buffer (Buffer) through sensor chip surfaces containing immobilized PV1/M and HRV3 (∼12 000 RU of virus). Four successive cycles recorded at 20°C with a PV1/M surface and one with an HRV3 surface are shown. (B) Overlay plot of sensorgrams recording the response following injection of soluble IC1, running buffer (Buffer) and two pulses of 25 mM MES buffer (pH 6–6.25) through surfaces containing immobilized HRV16 and PV1/M (∼12 000 RU of virus). Four successive cycles recorded at 20°C with an HRV16 surface and one with a PV1/M surface are shown. The concentration of soluble receptors used in each cycle in (A) and (B) is shown on the top of the curves. Time ranges for the virus–receptor association and dissociation phases and surface regeneration are indicated. (C) Overlay plot of sensorgrams presented in (A) and (B) showing the association of PVR and IC1 to sensor chip surfaces with PV1/M and HRV and dissociation of the receptors from their respective virus surfaces. Receptor concentrations are indicated.
Sensorgrams recorded during the injection of soluble PVR through a PV1/M surface reached a plateau, consistent with steady-state binding (Figure 2A). Bound PVR at the end of the association phase dissociated quickly from the immobilized PV1/M and the response reached the starting baseline several minutes after buffer flow through the sensor chamber. However, IC1 dissociated slower than PVR and no steady-state situation was reached for binding of IC1 to HRV3 (not shown) or HRV16 (Figure 2B) during a 13 min association phase. These differences are consistent with faster kinetic rates for the PVR–PV1/M interaction, inferred from the overlay sensorgrams presented in Figure 2C. The slower dissociation of IC1 from HRV16 or HRV3 required short injections of mild acidic buffer (pH 6–6.25) for complete removal of the bound receptor and regeneration of the surface (Figure 2B).
Analysis of binding data and determination of kinetic rate constants
Sensorgram plots recorded for binding of soluble receptors to immobilized HRV and PV1/M at 20°C were analyzed using the linear transformation method and non-linear curve fitting (see Materials and methods). Binding curves recorded for binding of PVR to PV1/M fit quite well to a simple kinetic model (1:1 Langmuir interaction model, A+ B → AB) and the association phase was monophasic. The χ2 for fitting the sensorgram presented in Figure 2C to the Langmuir model was 2.1, equal to the χ2 for fitting to a binding model for a heterogeneous immobilized analyte (1:2 interaction model, A + B1 + B2 → AB1 + AB2). However, consistent with a previous report (Casasnovas and Springer, 1995), the association of IC1 to HRV3 and HRV16 was biphasic (not shown) and sensorgrams did not fit to a simple kinetic model. The sensorgram recorded with 6 μM of IC1 presented in Figure 2C gave χ2 of 928 and 1.7 when fitted to 1:1 and 1:2 interaction models, respectively. Two association rate constants were determined for binding of IC1 to rhinovirus.
Association rate constants (kass) determined using the linear transformation method and non-linear curve fitting were similar (not shown). In some experiments, however, non-linear curve fitting did not give consistent kass values for different receptor concentrations. We observed no significant differences in the rate of association with the amount of immobilized PV and rhinovirus (Table I), and similar association kinetic rates were determined with three different flows (not shown). Association of PVR to PV1/M was ∼4 times faster than association of IC1 to the first class of binding sites in HRV3 or HRV16. We believe that these differences reflect a receptor recognition site more accessible in PV1/M than HRV.
Table I. Kinetic rate constants for receptor binding to viruses.
| Interaction | kass1 (M–1 s–1) | kass2 (M–1 s–1) | kdis×10–3 (s–1) | KD1 (nM) | KD2 (nM) | |
|---|---|---|---|---|---|---|
| PVR–PV1/M | H | 25 500 (6000) | 4.3 (0.1) | 180 (40) | ||
| L | 19 000 (1000) | 4.3 (0.1) | 230 (25) | |||
| Ave. | 23 000 (6000) | 4.3 (0.1) | 200 (40) | |||
| IC1–HRV3 | H | 5850 (50) | 1050 (140) | 1.7 (0.1) | 290 (10) | 1500 (160) |
| L | 6300 (500) | 900 (125) | 2.0 (0.1) | 310 (20) | 2420 (490) | |
| Ave. | 6100 (400) | 975 (150) | 1.8 (0.2) | 300 (20) | 2000 (600) | |
| IC1–HRV16 | H | 5400 (200) | 210 (50) | 1.4 (0.1) | 270 (01) | 5600 (1500) |
| L | 6300 (70) | 530 (10) | 1.3 (0.0) | 210 (00) | 2300 (100) | |
| Ave. | 5800 (500) | 340 (160) | 1.4 (0.1) | 240 (30) | 4300 (2000) |
Kinetic rate constants were determined from sensorgrams recording binding of PVR to PV1 and of IC1 to HRV3 and HRV16 in BIAcore at 20°C (see Figure 2). Determination of association and dissociation rate constants included in the table is described in Materials and methods. Rates for first (kass1) and second association phase (kass2) are shown for binding of IC1 to HRV. Experiments presented here were carried out with several surfaces containing high (H; >10 000 RU) or low amounts of immobilized viruses (L; <10 000 RU). Three experiments at 3, 10 and 20 μl/min were usually performed with each virus surface, and several receptor concentrations (0.1–2 μM for PVR and 0.5–8 μM for IC1) were injected in duplicate in each experiment. HEPES buffer (pH 8.0) with 150 mM NaCl was used as running buffer. kass1 and kdis were determined from cycles carried out at 10 and 20 μl/min. kass2 are the average of values determined at 3 and 10 μl/min. Affinity constants were calculated from the kinetic rate constants (KD = kdiss/kass). Averages for experiments carrried out with surfaces having high (H) and low (L) amount of viruses, and averages for all the experiments (Ave.) are shown. Standard deviations are in parentheses.
The first association rate (kass1) for IC1 binding to HRV3 or HRV16 did not differ significantly, but the second association rate (kass2) for binding of IC1 to HRV16 was lower than that to HRV3. Association rates for IC1 binding to HRV were higher than that reported previously (Casasnovas and Springer, 1995), most likely because we have used freshly purified viruses and a more sensitive BIAcore instrument here.
Faster dissociation of soluble PVR from immobilized PV1/M than of IC1 from HRV16 is clearly visualized in the overlay plot presented in Figure 2C. Consistently, the calculated kinetic dissociation rate (kdis) of PVR from PV1/M was ∼2.7 times higher than that for IC1–HRV (Table I). The faster dissociation of PVR from the virus indicates a weaker receptor binding to PV1/M than HRV.
In spite of the differences in rate of binding, soluble receptor molecules bound with similar affinity to PV1/M and HRV at 20°C (Table I). The affinity for soluble PVR binding to PV1/M determined in BIAcore is 2–4 times higher than that calculated using other methodologies (50–100 nM) (Arita et al., 1998). These authors noticed a fast dissociation of the receptor from PV1/M.
Effect of temperature on receptor binding to viruses and thermodynamics
Kinetics for receptor binding to PV1/M, HRV3 and HRV16 were analyzed at temperatures ranging from 10 to 25°C (Table II) to determine thermodynamics for these interactions. Cycles of receptor binding at 30°C gave a large decrease in baseline signal with surfaces having immobilized PV1/M and HRV3 (not shown), consistent with virus disruption. The behavior of these virus surfaces at 30°C made direct determination of kinetic constants at this temperature impossible in the BIAcore. Affinity constants (KD) decreased significantly for receptor binding to PV1/M and HRV3 with the increase of temperature (Table II). A small decrease was observed for binding of IC1 to HRV16. The decrease of KD with temperature indicates that the reaction is endothermic and, consequently, the calculated enthalpies were positive (Table II). The enthalpy for IC1–HRV3 is close to that reported previously (3.5 kcal/mol) (Casasnovas and Springer, 1995). The heat absorbed by receptor binding to the virus particles will contribute to decreasing the energy required for opening the viral particle and uncoating of the RNA (disruption). The enthalpy for PVR binding to PV1/M was the highest, which indicates that binding of PVR absorbed more heat than binding of IC1.
Table II. Effect of temperature on virus–receptor interactions and thermodynamics.
| Interaction | K10D | K15D | K20D | K25D | ΔHo (kcal/mol) |
|---|---|---|---|---|---|
| PVR–PV1/M | 430 (30) | 270 (20) | 200 (10) | 170 (20) | 10.6 |
| IC1–HRV3 | 380 (50) | 310 (25) | 300 (40) | 250 (15) | 4.3 |
| IC1–HRV16 | ND | 200 (6) | 180 (6) | 180 (10) | 1.7 |
Affinity constants (KD, nM) at four different temperatures (10–25°C) were determined from the kinetic rate constants as in Table I. KD for rhinovirus correspond to KD1. The average for two experiments for each temperature is shown, with errors in parentheses. Experiments were carried out using flows of 3 and 10 μl/min and with several concentrations of soluble receptors as described in Materials and methods and in Table I. Enthalpies (ΔH o) for the interactions were determined from the difference between the activation energy for the association and dissociation reaction as described in Materials and methods.
Efficiency of virus disruption by soluble receptors
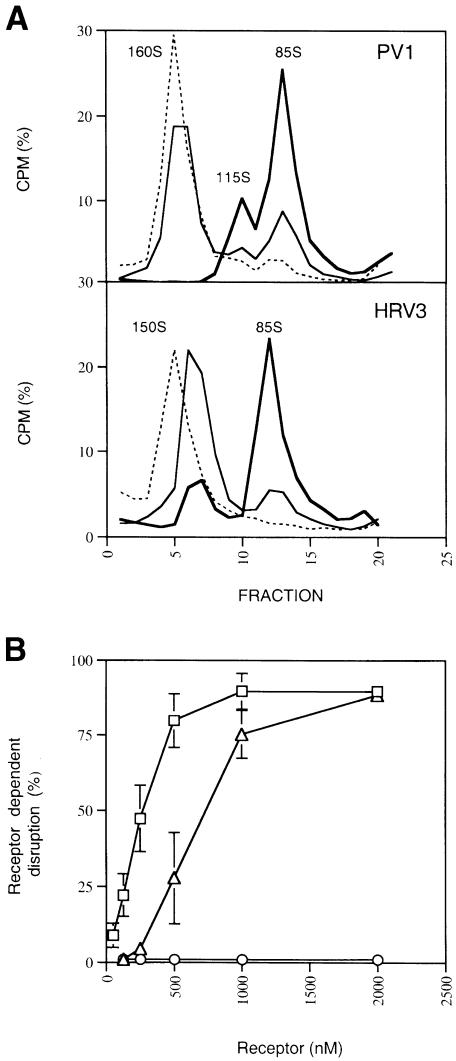
The positive enthalpies determined for the virus–receptor interactions studied here reveal virus destabilization by receptor binding, and are consistent with the reported receptor-mediated disruption of PV1/M and HRV3 (Kaplan et al., 1990a; Hoover-Litty and Greve, 1993; Arita et al., 1998). To analyze differences in the disruptive activity of PVR and IC1 further, we compared the effect of receptor binding on the conformation of PV1/M and HRV under similar experimental conditions. Incubation of either PV1/M or HRV3 with the two-domain receptor molecules at 37°C gave virus-derived particles with a sedimentation coefficient similar to empty capsids (∼80S) (Figure 3A). An intermediate particle with sedimentation of 115S was observed with PV1/M, but was absent in HRV3. This particle was not infective (not shown) and usually appeared together with empty capsids, but in a lower proportion. A similar particle reported as 135S-like has been shown to lack the internal VP4 protein (Arita et al., 1998). The temperature dependence of receptor-mediated disruption of PV1/M (not shown) was very similar to that reported in experiments with detergent-solubilized PVR receptor (Yafal et al., 1993) and for IC1–HRV3 (Casasnovas and Springer, 1994).
Fig. 3. Effect of soluble receptors on virus conformation. (A) Gradient sedimentation of [35S]methionine-labeled PV1/M (top) or HRV3 (bottom) particles incubated in the absence (dashed lines) or presence of soluble PVR at 37°C. The percentage of total CPM for each fraction is shown, together with the determined sedimentation of the particles. Thin lines shows sedimentation profiles of PV1/M and HRV3 particles treated with 125 and 500 nM concentrations of soluble PVR and IC1, respectively. Sedimentation profiles represented by thick lines come from treatments carried out with a 1000 nM receptor concentration. (B) Disruption of PV1/M (squares), HRV3 (triangles) and HRV16 (circles) at different receptor concentrations. Radiolabeled viruses were incubated with varying amounts of receptors as in (A). Receptor-dependent disruption was determined from the decrease in radioactivity for fractions corresponding to non-disrupted and infective virus particles (1–8 for PV1/M and 1–9 for HRV3 or HRV16) as described in Materials and methods. The average and standard deviation from three experiments are shown. No disruption was detected for HRV16 with 10, 25 and 50 μM IC1 concentrations (not included).
Receptor-dependent disruption of PV1/M, HRV3 and HRV16 was determined for a broad range of receptor concentrations (Figure 3B). The same amount of freshly purified viral particles was used for all viruses. Disruption of PV1/M by PVR was the most efficient, with a DC50 of ∼250 nM. Based on a calculated KD of 80 nM at 37°C from plots of rates with temperature (Arrhenius plots; see Materials and methods), we determined that ∼70% of the sites (3.5 sites/pentamer) would be occupied at 250 nM PVR. Significant disruption (25%) was observed at a receptor concentration of 150 nM, which gave a threshold disruptive occupancy of 3 sites/pentamer. A three times higher concentration of IC1 than PVR was needed to have 50% of the HRV3 particles disrupted and ∼500 nM to have 25% disruption. The determined KD at 37°C for IC1–HRV3 was 190 nM and the deduced occupancies for disruption of 50 and 25% of the HRV3 particles were 4.0 and 3.5 sites/pentamer, respectively. The lower DC50 found for PVR–PV1/M when compared with IC1–HRV3 is therefore related to the lower occupancy needed for PV1/M disruption and to the higher PV1/M-receptor affinity (lower KD) at 37°C.
Although IC1 binds to HRV3 and HRV16 with similar affinity, no disruption of HRV16 was observed even at 50 μM IC1 (Figure 3B). The low energy absorbed upon binding of IC1 to HRV16 (Table II) is, therefore, not sufficient to trigger uncoating of this rhinovirus serotype. The ‘pocket factor’ molecule found underneath the receptor-binding site in HRV16 could prevent capsid rearrangements and be responsible for the little destabilization of the virion upon receptor binding. Even though a related ‘pocket factor’ molecule is present in a similar location in the PV1/M capsid, these particles were disrupted by PVR quite efficiently.
Structure determination of PVR–PV1/M complexes by cryo-EM
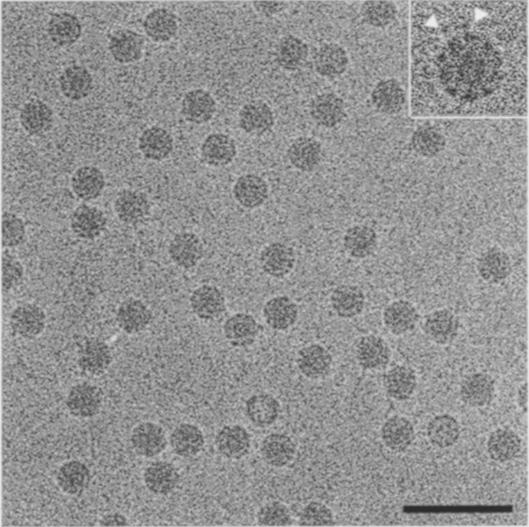
The biochemical data presented so far suggested significant differences in receptor recognition modes between PV1/M and HRV and a more disruptive receptor-binding interaction in PV. To understand further the differences in receptor binding and uncoating processes between rhinovirus and PV, we determined the structure of PVR–PV1/M complexes by cryo-EM. Complexes of PV1/M with bound PVR receptor were prepared in non-disruptive conditions (see Materials and methods). Images of unstained and purified virus–receptor were recorded as focal pairs with low and high defocus. The attached receptor molecule appeared as black dots on the virus particles in the cryo-EM images (Figure 4). Conditions for the preparation of PVR–PV1/M complexes gave few empty particles, easily detected by their low density in the projected center (Figure 4).
Fig. 4. Cryo-electron microscopy of PVR–PV1/M complexes. Image of unstained virus–receptor complexes recorded as focal pairs. Purified complexes were prepared as described in Materials and methods. The inset shows virus particles in complex with receptor at higher magnification (2×) and the arrows indicate the location of virion-bound receptor molecules. The bar corresponds to 100 nm.
Image reconstruction was performed first on the highly defocused image. A preliminary three-dimensional structure was obtained after a few cycles of orientation assignment with the Polar Fourier Transform (PFT) method. The data of the further defocused micrograph were used as a starting model for refinement of the data from the close-focus images. Particle selection for Fourier averaging was further sensitized with a scheme of interpolation and correlation of intermediate models within the redial bin of the bound PVR region. The coefficient of correlation was assessed to measure the occupancy of PVR binding and to exclude the image data with unsaturated receptor binding. The enantiomorphic information of the reconstruction was based on comparison with features on the PV1/M X–ray structure truncated at a resolution similar to cryo-EM data (Cheng, 1992). At the final stages of the reconstruction, 48 out of 198 selected individual particles were combined to calculate a map at 25 Å resolution.
The image reconstruction of virus–receptor complexes showed 60 spike-like projections on the virus particles with a similar density value to the capsid proteins (Figure 5, top). The number of spike projections (60) is consistent with the expected number of receptor-binding sites in PV1/M. The length of the spikes is 60–65 Å, similar to the length of the related two-domain CD4 molecule (65 Å) (Kwong et al., 1998). The narrower density in the middle of the spikes might correspond to the junction between the two IgSF domains of the receptor molecule. The electron density for the virus-binding first domain, proximal to the capsid, is very similar to the density level of the virus particle and ∼0.5 σ higher than for the second domain (not shown). This suggests that the location of the first domain is better defined in the complex and a certain degree of inter-domain flexibility. The density of the bound PVR molecule lies oblique to the particle radius, so that the C–terminal domains come close to the 3–fold axes (Figure 5, top). Density corresponding to the receptor molecule locates into a depressive region of the PV1/M particle, in between surface protrusions from neighboring protomer (Figure 5).
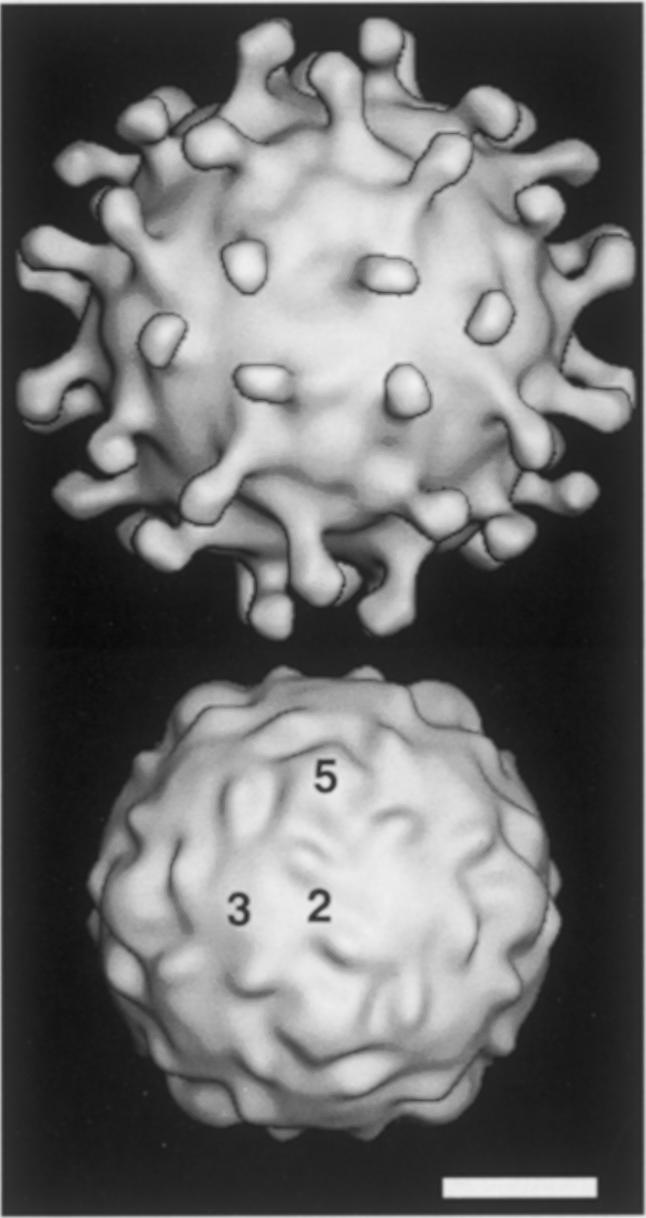
Fig. 5. Image reconstruction of PVR–PV1/M complexes. Surface representation generated from cryo-EM density maps of three-dimensional reconstructed images of the complexes (top) or from the crystal structure of PV1/M (bottom) (Hogle et al., 1985). Numbers indicate the location of the icosahedral 2-, 3- and 5–fold axes. The approximate location of the virus–receptor contact can be implied by comparison of complex (top) and virus (bottom) densities. The white bar corresponds to 100 Å.
Virus–receptor binding interfaces
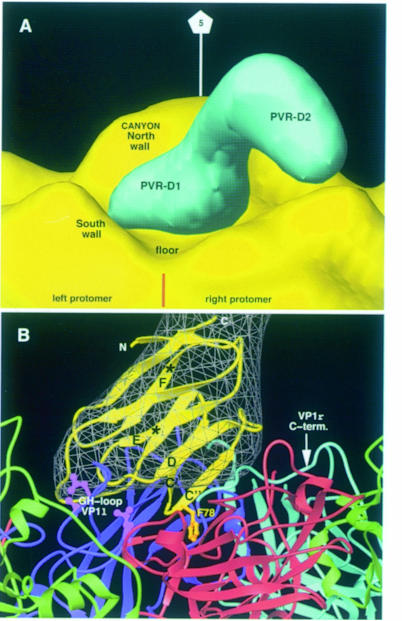
To define receptor–virus boundaries, electron density for the bound PVR molecule was computed by subtracting a 25 Å density map generated for the PV1/M structure from the cryo-EM density of the PVR–PV1/M complex. Electron density corresponding to the two-domain fragment has a ‘boot’-like shape (Figure 6A), with the virus-proximal density for domain 1 being the ‘foot’. Orientation is such that the ‘tip’ of the ‘foot’ contacts the so-called ‘south wall’ of a virus protomer (left protomer in Figure 6) and the ‘heel’ lays on a protrusion in the neighboring one (right protomer). No difference density appeared filling the bottom of the depressive virus surface or ‘canyon’.
Fig. 6. Poliovirus–receptor interactions. (A) Surface representation of electron density maps of a virion-bound PVR molecule (blue) and a PV1/M capsid pentamer (yellow). Density for the receptor was generated by subtracting the electron density of the virus particle (see Figure 5) from the cryo-EM density of the reconstructed PVR–PV1/M complex. Regions of the PVR density corresponding to domain 1 (D1) and domain 2 (D2) are indicated. The brown straight line marks the approximate location of an inter-protomer junction. The location is indicated for the components of the surface depression or ‘canyon’ surrounding the icosahedral 5–fold axis of the capsid pentamer. (B) View of virus–receptor-binding contacts. Ribbon diagrams of the modeled domain 1 of PVR (yellow) and interacting capsid proteins. The PVR domain 1 model is fitted into the corresponding electron density map (gray). Strands of the model referred to in the text are labeled. Asterisks mark the location of Cα atoms for the solvent-exposed Asn105 and Asn120 residues to which glycans attach. Two VP1 proteins from neighboring protomers are colored cyan and blue. VP2 is green and VP3 is red. Side chains of exposed receptor-binding residues in PV1/M (Asp1226, Leu1228, Leu1234, Ser3183) defined by soluble receptor-resistant PV mutants are colored magenta (Colston and Racaniello, 1994). The virus-binding Phe78 in PVR is in gold. This figure and Figure 7 were generated with Ribbons (Carson, 1987).
A model of PVR domain 1 fitted well into the ‘foot’-like electron density of the receptor molecule (Figure 6B), leaving the remaining density for domain 2 (compare Figure 6A and B). The shape of the difference density for domain 1 suggests that the long axis of the IgSF domain is quite oblique to the particle radius. Therefore, the PVR domain was oriented with the top in the ‘tip’ of the ‘foot’ and the C–terminal end located in the inter-domain junction. With this orientation, the N–linked sugars in strands E and F project toward the solvent and the C′–C″ ridge of the β–sandwich is proximal to the virus (Figure 6B). In the fitted domain 1, the N–linked glycan attached to Asn105 in the E–strand would project into a bulky density on the 5–fold distant side of the difference map (not shown), supporting accuracy of the domain orientation. The poor fitting for some of the loops may be related to the poor electron density for these more flexible regions or to structural differences between the model and the PVR structure.
The DE loop of the N–terminal domain of PVR locates in the ‘tip’ of the electron density and is in close contact with the GH loop of PV1 in the south wall of the ‘canyon’, where some of the defined receptor-binding residues locate in PV1/M (Figure 6B). Interaction of the DE loop with the virus is also consistent with results from PVR mutants (Racaniello, 1996; Solecki et al., 1998). The region between strands C′ and D has also been found to be important for virus binding in PVR. This region locates on the ‘heel’ of the domain density, in close contact with the C–terminal region of VP1 and with VP3 in the protomer on the right side of Figure 6B. The hydrophobic C′C″ loop of the domain contains the Phe78-binding residue (Figure 6B), homologous to Phe43 in CD4, a critical virus-binding residue that penetrates in a gp120 cavity (Kwong et al., 1998). Our fitting locates this loop in the depressive surface between two protomers (Figure 6B), with Phe78 close to the inter-protomer junction. We believe that the aromatic ring of the residue could be inserted in the junction upon receptor binding.
Discussion
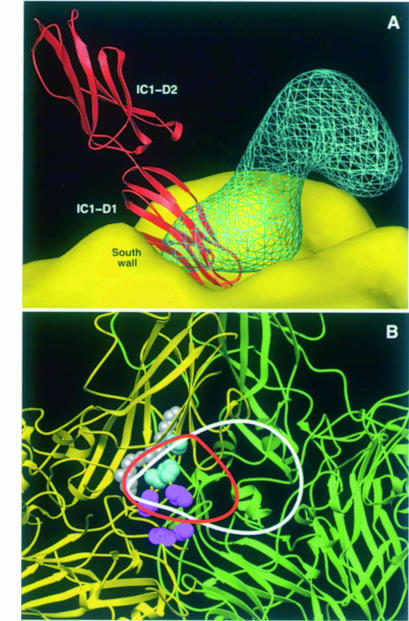
The orientation of the bound PVR to PV1/M presented here is strikingly different from that reported for bound IC1 to HRV16 (Figure 7A). The long axis of the IC1 molecule locates quite perpendicularly to the capsid, so that the top region of the N–terminal domain (BC and FG loops) reaches the bottom of the ‘canyon’ (Figure 7A), where receptor-binding residues defined by virus mutants reside (Figure 7B). A view of the virus–receptor complexes toward the particle center (Figure 7B) shows distinct but overlapping binding footprints around the depressive inter-protomer junction. PVR extends further in a second protomer, while the IC1 footprint is more circumscribed to the ‘canyon floor’ and the ‘pocket factor’ cavity. These structural data and binding kinetics reveal a receptor-binding site more accessible in PV1/M than HRV.
Fig. 7. PVR and IC1-binding modes and footprints. (A) View of the electron density for a PV-bound PVR molecule (blue lines), the PV1/M molecular surface (yellow) and an HRV16-bound IC1 molecule (red). A ribbon representation of the crystal structure of domains 1 (D1) and 2 (D2) of IC1 (Bella et al., 1998; Casasnovas et al., 1998b) fitted in the cryo-EM electron density of the reported HRV16–IC1 complex (Olson et al., 1993) is shown. The IC1 molecule has similar orientation to that previously reported (Bella et al., 1998). Structures of PV1/M and HRV16 were superimposed to generate the figure. (B) Lines defining the footprint of PVR (white) and IC1 (red) were drawn around the virus proximal cryo-EM density of the bound receptor molecules. The panel shows a view toward the center of the particle, with two PV1/M promoters colored in yellow and green. Defined receptor-binding residues in PV1 (see Figure 6) and HRV (Colonno et al., 1988) are colored magenta and cyan, respectively.
The kinetics for receptor binding to viruses provides valuable information on the conformation of binding sites and modes. Virus-binding residues in many virus receptors are highly exposed in loops, and therefore differences in association kinetics might reflect differences in the conformation of receptor-binding sites in viruses. Indeed, we determined a faster association kinetics for PVR than IC1 in BIAcore (Table I), and a receptor-binding site more exposed in PV1/M than HRV by cryo-EM (Figures 6 and 7). However, calculated kass values for receptor binding to PV1/M and HRV are 7 and 25 times lower than kass for binding of DAF (CD55) to echovirus 11 (∼1.5 × 105 M–1s–1) (Lea et al., 1998), which is close to the association of antibodies to rhinovirus-binding epitopes in IC1 (1–2 × 105 M–1s–1) (Casasnovas et al., 1998a). Therefore, receptor-binding sites in other picornaviruses might be even more exposed than in PV1/M, and lie outside of a surface depression.
The affinities reported for PV and HRV binding to the cell surface (KD between 10–10 and 10–11 M) (Colonno et al., 1988; Solecki et al., 1998) are several orders of magnitude higher than those for monomeric receptor binding to viruses. These differences may arise from multimeric attachment of viruses to receptors at the cell surface. The cryo-EM images showed that the second domains of three virus-bound PVR molecules approach each other around the icosahedral 3–fold axis of PV1/M (Figure 5), and we expect the third domain to be even closer. The organization of the PVR molecules on the virus particle suggests that a multimeric membrane-bound PVR receptor could interact around the 3–fold axis of the PV1/M capsid. There are no biochemical data supporting PVR oligomerization at the cell surface, but a third domain-dependent multimerization of PRR1 has been reported (Krummenacher et al., 1999).
Thermodynamic parameters and analysis of receptor-mediated virus disruption revealed a more disruptive receptor binding to PV1/M than HRV, which must be required for efficient PV uncoating and entry into the cell. Receptor-mediated entry of PV1/M at the cell surface is supported by reports showing no dependence of PV infection on endosomal acidification or clathrin-mediated endocytosis (Perez and Carrasco, 1993; DeTuello and Kirchhausen, 1998). The process by which PVR triggers opening of the capsid needs further structural analysis. However, the interaction of the PVR molecule with two neighboring capsid protomers (Figure 6) suggests that, upon binding, the receptor could move the two interacting protomers apart. This movement would allow fitting of the aromatic ring of Phe78 in the inter-protomer junction, which could move neighboring protomers further apart. This ‘wedge’-like role of the receptor could be critical for virus uncoating. Additionally, separation of protomers could facilitate release of the ‘pocket factor’ molecule and enhance capsid disruption.
Similarities in the affinity and kinetics for receptor binding to HRV3 and HRV16 show conservation in receptor-interacting residues among serotypes, and no influence of the ‘pocket factor’ molecule present in HRV16 on receptor binding. However, thermodynamics for IC1 binding to HRV3 and HRV16 differ significantly. Enthalpy for IC1–HRV16 was very low and, consequently, the virus was resistant to receptor-mediated uncoating (Figure 3) (Hoover-Litty and Greve, 1993). Receptor-sensitive HRV serotypes lacking ‘pocket factor’ molecules may follow a receptor-mediated entry pathway at the cell surface similar to that proposed for PV1/M entry. In good agreement, HRV14 infection occurs in the presence of agents that prevent endosomal acidification (Schober et al., 1998). Additional factors to the receptor will be required for uncoating and entry of receptor-resistant major group HRV serotypes. We have found cooperation of low pH and soluble receptor in HRV16 uncoating (unpublished observations), suggesting that both factors can cooperate in the entry of major group serotypes having ‘pocket factor’ molecules. Therefore, HRV16 must enter by receptor-mediated endocytosis, as described for the minor group of HRV (Schober et al., 1998).
Differences in receptor recognition modes and entry pathways between PV and HRV must be related to structural differences between these two viruses. The higher stability of PV than HRV in the low pH conditions found in endosomes must have forced PV to develop an efficient receptor-mediated uncoating process for entry at the cell surface. To fulfill these requirements, PV used receptor recognition sites more exposed and unprotected from immune surveillance than those of HRV, which most likely restricted PV evolution to a few serotypes. The major group of HRV use the most inaccessible receptor-binding epitope described so far in the picornavirus family, and therefore the most evolved to escape antibody neutralization (Rossmann, 1989). However, the receptor-binding mode in HRV is less suited to promoting virus uncoating than in PV. Structural alterations in the ‘pocket factor’ cavity (Zhao et al., 1996) may have facilitated HRV uncoating and entry. Diversity in the ‘pocket factor’ cavities among the major group of HRV correlates with differences in particle stability and entry pathways.
The comparative study on receptor binding to PV and HRV presented here has provided novel information on the evolution of receptor recognition and entry pathways in these related viruses. Comparative analysis on receptor binding to picornaviruses is, therefore, fundamental for understanding diversity in receptor recognition in this family of virus pathogens, and could guide the design of new strategies for controlling viral infections.
Materials and methods
Preparation of receptor proteins
The proteins were expressed in CHO Lec3.2.8.1 cells using the glutamine synthetase (GS) expression system (Casasnovas and Springer, 1995). Preparation of the recombinant IC1 cDNA has been described previously (Casasnovas et al., 1998a). Recombinant PVR cDNAs with a translation stop codon after positions 238 and 241 of the precursor polypeptide (Mendelsohn et al., 1989) were generated by PCR and subcloned in the pBJ5-GS expression vector for cell transfection. Fidelity of the PVR sequence in the constructs was determined by sequencing. The PVR protein was first purified from filtered cell supernatant using affinity chromatography with concanavalin A (Pharmacia) using standard procedures. Elution fractions containing the PVR protein were dialyzed against 20 mM HEPES pH 7.2, and run through an anion-exchange column (Q-Sepharose; Pharmacia). The flow through of this column contained ∼90% pure PVR protein, which was finally purified by size exclusion chromatography with Superdex-75 (Pharmacia) in 10 mM HEPES buffer pH 8.0, with 150 mM NaCl. The molar concentration of the purified protein was determined from the extinction coefficient at 280 nm of 1.2 ml/mg.cm and the molecular weight (23 600 kDa) of the deglycosylated and mature polypeptide. The soluble IC1 protein was purified as described (Casasnovas and Springer, 1995). The concentration of the protein was determined from the extinction coefficient of 0.8 ml/mg.cm and a molecular weight of 21 000 kDa.
Virus preparation
HRV3 and HRV16 stocks come from the American Type Culture Collection and PV1/M from the James Hogle laboratory. Viruses were propagated in HeLa-H1 cells at 35°C and purified from infected cells harvested 7, 8 and 9 h post-infection for PV1/M, HRV16 and HRV3, respectively. For radiolabeling of viruses, [35S]methionine (Amersham) was added to 1 × 108 infected cells in minimal essential medium lacking methionine. Viruses were first released from infected cells by freeze–thaw in the presence of protease inhibitors and purified from cell suspensions clarified by centrifugation at 12 000 g for 10 min. Purification was as described (Casasnovas and Springer, 1994), and included a final step of virus sedimentation through a 10–40% potassium tartrate gradient. Virus particles were sedimented from the gradient fractions by ultracentrifugation, and pellet was dissolved in 20–50 μl of HEPES buffer pH 7.4, with 10 mM MgCl2 and protease inhibitors. Virus concentration was determined from the OD at 260 nm (1 OD = 9.4 × 1012 virions/ml).
Virus immobilization in the BIAcore
Viruses were covalently immobilized to the dextran surface of certified CM5 sensor chips at 20°C via primary amino groups, using the amine coupling kit (Biacore AB, Uppsala, Sweden) as described previously for HRV3 (Casasnovas and Springer, 1995). About 1 μl of freshly purified virus solution (OD260 = 5–10) was diluted with 49 μl of 10 mM NaAc at pH 6.0 for PV1/M, pH 5.5–6.0 for HRV3 and pH 5.5 for HRV16 for immobilization. This mild acidification of rhinoviruses at room temperature was found to be non-disruptive, based on sucrose sedimentation experiments (not shown).
Determination of kinetic rate constants and thermodynamic parameters
Recorded sensorgrams for receptor binding to immobilized viruses were analyzed using the linear transformation method and non-linear curve fitting using the BIA-Evaluation 3.0 software (BIAcore). The linear transformation method uses the equation dRU/dt = ka[analyte]RUmax – RU(ka[analyte] + kd) to obtain the association kinetic constants from binding sensorgrams recorded with several receptor concentrations (Casasnovas and Springer, 1995). The linear transformation method was used for the determination of association rate constants (kass). Association of IC1 to HRV was biphasic and two kass values were calculated (Casasnovas and Springer, 1995). Dissociation rate constants were determined by non-linear curve fitting for different receptor concentrations and flows. Only sensorgrams with >100 RU of bound receptor were analyzed.
Activation energy for receptor–virus association and dissociation reactions was determined from Arrhenius plots as described (Casasnovas and Springer, 1995). Kinetic rates determined at different temperatures gave a very good linear fit in the plots, although values for kdis at 25°C were disregarded in some experiments due to deviation from linearity. Enthalpy (ΔHo) was determined from the difference between activation energy for association and dissociation reactions.
Analysis of virus conformation by sucrose gradient sedimentation
For analysis of virus conformation, radiolabeled viruses were run through linear 5–20% sucrose gradients in phosphate-buffered saline, which facilitate determination of the sedimentation coefficient for virus-derived particles based on the reported coefficients for PV1/M (160S) and HRV (150S) (Casasnovas and Springer, 1994). Freshly purified [35S]methionine-labeled viruses (2.8 × 1010 virions) were incubated in the presence or absence of soluble receptors at 37°C in 30 μl of HEPES buffer pH 8.0, with 150 mM NaCl and 5% fetal calf serum. After incubation, samples were chilled for 5 min on ice, loaded onto the 5 ml sucrose gradients and centrifuged at 40 000 r.p.m. and 4°C in a SW50.1 rotor for 50 min or 1 h, respectively, for PV1/M or HRV. Approximately 20 fractions were collected from the bottom and assayed using a scintillation counter. The percentage of the total counts per minute (%CPM) in the gradient was calculated for each fraction. Receptor-dependent disruption was determined from the decrease in the %CPM for the infectious fractions (%CPM-receptor – %CPM+receptor/%CPM-receptor) × 100.
Preparation of poliovirus–PVR complexes for cryo-EM
PV1/M virions and PVR (100 μM) were mixed at a molar ratio of 1:600 (10 receptor molecules/binding site) and incubated overnight at 4°C in 25 mM HEPES buffer pH 8.0, with 150 mM NaCl, 5 mM MgCl2 and 1% n-octyl-β-glucopyranoside. The sample (20 μl) was run through a 1 ml Superose-12 column in 25 mM HEPES buffer pH 8.0, with 50 mM NaCl and 5 mM MgCl2 to separate virus–receptor complex from unbound receptor molecules. Fractions containing the complex were determined from OD260 and used to prepare frozen specimens.
Cryo-sample preparation was performed essentially as described (Cheng et al., 1992). The specimen was observed and photographed at a temperature of –170°C using the Gatan 626 cryo-transfer system in a Philips CM120 operating at 120 kV. Images were recorded under low-dose conditions (7 e/Å2) with Kodak SO 163 film (Xing et al., 1999). Focus pair images were taken at 1.0 and 3.0 μm at a magnification of ×45 000.
Image reconstruction
Micrographs exhibiting minimal astigmatism and specimen drift were selected and both micrographs in the focal pair were digitized on a Zeiss Phodis scanner. The pixel size of 14 μm on the micrograph corresponds to 3.1 Å/pixel at the specimen. Individual particle images were extracted and analyzed with icosahedral symmetry processing procedures to reconstruct the three-dimensional structures (Crowther, 1971; Baker and Cheng, 1996; Fuller et al., 1996). Particle center and orientation were first estimated by the use of a cross-correlation and self-common-lines method, and were further refined with the PFT method (Baker and Cheng, 1996). Preliminary three-dimensional reconstruction was computed from the further defocus image. This reconstruction was subsequently used as the reference model to refine further the orientation and origin parameters of the close-defocus particle image. Radial filtering comparison of image data with the projected model map in real space was used to aid in particle selection.
The resolution of the final best density map was calculated to 25 Å and the reliability was indicated by the free R-factor. The PV1/M density map was computed at 25 Å with structure factors calculated from the capsid atomic coordinates (Hogle et al., 1985), where the scattering amplitudes were implemented to deconvolute the microscope contrast transfer function by least-square correlation (Cheng, 1992).
Modeling of the N–terminal domain of PVR and fitting
The model of the N–terminal domain of PVR was built with the program Modeller (Sali and Blundell, 1993), based on the structure of the homologous domain of CD4 and sequence alignment. Alignment was first carried out using MegAlign (DNAstart, Inc.). Manual modifications were included following reported aligments of predicted β–strands for PVR with homologous strands of CD4 (Racaniello, 1996; Solecki et al., 1998). The PVR model was fitted into the electron density map of the complex manually, together with PV1/M coordinates, and the adjustment was guided by the difference map. The difference map was computed after scaling the size and contrast of the cryo-EM map to the X–ray map on the basis of the capsid densities (Olson et al., 1993; Cheng et al., 1994).
Acknowledgments
Acknowledgements
The authors are especially thankful to Henrik Garoff for sharing infrastructure used to carry out the studies presented here. We also thank M.G.Rossmann for providing helpful protocols for virus preparation and the cryo-EM map for IC1–HRV16, and T.A.Springer for the antibody used in the purification of IC1 protein. This work has been sponsored by grants from the Swedish Medical Research Council (MFR-12175 and –12637) and the Natural Science Research Council (NFR-11691 and –11994) to R.H.C. and J.M.C. Support from the Karolinska Institute and a strategic fund for developing structural virology is also acknowledged.
Note added in proof
Cryo-EM structures of PV–PVR complexes were reported while this paper was under review (Belnap et al., 2000; He et al., 2000). Our structure shows better agreement with the second report.
References
- Arita M., Koike, S., Aoki, J., Horie, H. and Nomoto, A. (1998) Interaction of poliovirus with its purified receptor and conformational alteration in the virion. J. Virol., 72, 3578–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T.S. and Cheng, R.H. (1996) A model-based approach for determining orientations of biological macromolecules imaged by cryo-electron microscopy. J. Struct. Biol., 116, 120–130. [DOI] [PubMed] [Google Scholar]
- Bella J., Kolatkar, P.R., Marlor, C.W., Greve, J.M. and Rossmann, M.G. (1998) The structure of the two amino–terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc. Natl Acad. Sci. USA, 95, 4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap D.M., McDermott, B.M., Jr, Filman, D.J., Cheng, N., Trus, B.L., Zuccola, H.J., Racaniello, V.R., Hogle, J.M. and Steven, A.C. (2000) Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl Acad. Sci. USA, 97, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M. (1987) Ribbon models of macromolecules. J. Mol. Graph., 5, 103–106. [Google Scholar]
- Casasnovas J.M. and Springer, T.A. (1994) The pathway of rhinovirus disruption by soluble intercellular adhesion molecule 1 (ICAM-1): An intermediate in which ICAM-1 is bound and RNA is released. J. Virol., 68, 5882–5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasnovas J.M. and Springer, T.A. (1995) Kinetics and thermodynamics of virus binding to receptor: Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1) and surface plasmon resonance. J. Biol. Chem., 270, 13216–13224. [DOI] [PubMed] [Google Scholar]
- Casasnovas J.M., Bickford, J.K. and Springer, T.A. (1998a) The domain structure of ICAM-1 and the kinetics of binding to rhinovirus. J. Virol., 72, 6244–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasnovas J.M., Stehle, T., Liu, J., Wang, J.-H. and Springer, T.A. (1998b) A dimeric crystal structure for the N–terminal two domains of intercellular adhesion molecule-1. Proc. Natl Acad. Sci. USA, 95, 4134–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R.H. (1992) Correlation of cryo-electron microscopic and X–ray data and compensation of the contrast transfer function. Proc. Electron Microsc. Soc. Am., 50, 996–997. [Google Scholar]
- Cheng R.H., Olson, N.H. and Baker, T.S. (1992) Cauliflower mosaic virus: a 420 subunit (T = 7) multilayer structure. Virology, 186, 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R.H., Reddy, V., Olson, N.H., Fisher, A., Baker, T.S. and Johnson, J. (1994) Functional implications of quasi-equivalence in a T = 3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure, 2, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F., Lopez, M., Menotti, L., Aoubala, M., Dubreuil, P. and Campadelli-Fiume, G. (1998) The V domain of herpesvirus Ig-like receptor (HIgR) contains a major functional region in herpes simplex virus-1 entry into cells and interacts physically with the viral glycoprotein D. Proc. Natl Acad. Sci. USA, 95, 15700–15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonno R.J., Condra, J.H., Mizutani, S., Callahan, P.L., Davies, M. and Murko, M.A. (1988) Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc. Natl Acad. Sci. USA, 85, 5449–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colston E. and Racaniello, V.R. (1994) Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J., 13, 5855–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther R.A. (1971) Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil. Trans. R. Soc. London B, 261, 221–230. [DOI] [PubMed] [Google Scholar]
- DeTuello L. and Kirchhausen, T. (1998) The clathrin endocytic pathway in viral infection. EMBO J., 17, 4585–4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricks C.E. and Hogle, J.M. (1990) Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol., 64, 1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S.D., Butcher, S.J., Cheng, R.H. and Baker, T.S. (1996) Three-dimensional reconstruction of icosahedral particles–the uncommon line. J. Struct. Biol., 116, 48–55. [DOI] [PubMed] [Google Scholar]
- Geraghty R.J., Krummenacher, C., Cohen, G.H., Eisenberg, R.J. and Spear, P.G. (1998) Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science, 280, 1618–1620. [DOI] [PubMed] [Google Scholar]
- Greve J.M., Davis, G., Meyer, A.M., Forte, C.P., Yost, S.C., Marlor, C.W., Kamarck, M.E. and McClelland, A. (1989) The major human rhinovirus receptor is ICAM-1. Cell, 56, 839–847. [DOI] [PubMed] [Google Scholar]
- He Y., et al. (2000)Interaction of the poliovirus receptor with poliovirus. Proc. Natl Acad. Sci. USA, 97, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle J.M., Chow, M. and Filman, D.J. (1985) Three-dimensional structure of poliovirus at 2.9 Å resolution. Science, 229, 1358–1365. [DOI] [PubMed] [Google Scholar]
- Hoover-Litty H. and Greve, J.M. (1993) Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J. Virol., 67, 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan G., Freistadt, M.S. and Racaniello, V.R. (1990a) Neutralization of poliovirus by cell receptors expressed in insect cells. J. Virol., 64, 4697–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan G., Peters, D. and Racaniello, V.R. (1990b) Poliovirus mutants resistant to neutralization with soluble cell receptors. Science, 250, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Koike S., Ise, I. and Namoto, A. (1991) Functional domains of the poliovirus receptor. Proc. Natl Acad. Sci. USA, 88, 4104–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummenacher C., et al. (1999)The first immunoglobulin-like domain of HveC is sufficient to bind herpex simplex virus gD with full activity, while the third domain is involved in oligomerization of HveC. J. Virol., 73, 8127–8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Wyatt, R., Robinson, J., Sweet, R.W., Sodroski, J. and Hendrickson, W.A. (1998) Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature, 393, 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea S.M., Powell, R.M., McKee, T., Evans, D.J., Brown, D., Stuart, D.I. and van der Merwe, P.A. (1998) Determination of the affinity and kinetic constants for the interaction between the human virus echovirus 11 and its cellular receptor, CD55. J. Biol. Chem., 273, 30443–30447. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C.L., Wimmer, E. and Racaniello, R. (1989) Cellular receptor for poliovirus: Molecular cloning, nucleotide sequence and expression of a new member of the immunoglobulin superfamily. Cell, 56, 855–865. [DOI] [PubMed] [Google Scholar]
- Olson N.H., Kolatkar, P.R., Oliveira, M.A., Cheng, R.H., Greve, J.M., McClelland, A., Baker, T.S. and Rossmann, M.G. (1993) Structure of human rhinovirus complexed with its receptor molecule. Proc. Natl Acad. Sci. USA, 90, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez L. and Carrasco, L. (1993) Entry of poliovirus into cells does not require a low-pH step. J. Virol., 67, 4543–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V.R. (1996) Early events in poliovirus infection: Virus–receptor interactions. Proc. Natl Acad. Sci. USA, 93, 11378–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. (1989) The Canyon Hypothesis: Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J. Biol. Chem., 264, 14587–14590. [PubMed] [Google Scholar]
- Rossmann M.G., et al. (1985)Structure of a human common cold virus and functional relationship to other picornaviruses. Nature, 317, 145–153. [DOI] [PubMed] [Google Scholar]
- Rueckert R.R. (1985) Picornaviruses and their replication. In Fields,B.N., Knipe,D.M., Melnick,J.L., Chanock,R.M., Roizman,B. and Shope,R.E. (eds), Virology. Raven Press, New York, NY, pp. 705–738. [Google Scholar]
- Sali A. and Blundell, T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Schober D., Kronenberger, P., Prchla, E., Blaas, D. and Fuchs, R. (1998) Major and minor receptor group human rhinoviruses penetrate from endosomes by different mechanisms. J. Virol., 72, 1354–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki D., Gromeier, M., Harber, J., Bernhardt, G. and Wimmer, E. (1998) Poliovirus and its cellular receptor: a molecular genetic dissection of a virus/receptor affinity interaction. J. Mol. Recogn., 11, 2–9. [DOI] [PubMed] [Google Scholar]
- Springer T.A. (1994) Traffic signals for lymphocyte recirculation and leukocyte emigration: The multi-step paradigm. Cell, 76, 301–314. [DOI] [PubMed] [Google Scholar]
- Staunton D.E., Merluzzi, V.J., Rothlein, R., Barton, R., Marlin, S.D. and Springer, T.A. (1989) A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell, 56, 849–853. [DOI] [PubMed] [Google Scholar]
- Staunton D.E., Dustin, M.L., Erickson, H.P. and Springer, T.A. (1990) The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell, 61, 243–254. [DOI] [PubMed] [Google Scholar]
- Xing L., Kato, K., Li, T., Takeda, N., Miyamura, T., Hammar, L. and Cheng, R.H. (1999) Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology, 265, 35–45. [DOI] [PubMed] [Google Scholar]
- Yafal A.G., Kaplan, G., Racaniello, V.R. and Hogle, J.M. (1993) Characterization of poliovirus conformational alteration mediated by soluble cell receptors. Virology, 197, 501–505. [DOI] [PubMed] [Google Scholar]
- Zhao R., Pevear, D.C., Kremer, M.J., Giranda, V.L., Kofron, J.A., Kuhn, R.J. and Rossmann, M.G. (1996) Human rhinovirus 3 at 3.0 Å resolution. Structure, 4, 1205–1220. [DOI] [PubMed] [Google Scholar]