Abstract
We recently identified intersectin, a protein containing two EH and five SH3 domains, as a component of the endocytic machinery. The N–terminal SH3 domain (SH3A), unlike other SH3 domains from intersectin or various endocytic proteins, specifically inhibits intermediate events leading to the formation of clathrin-coated pits. We have now identified a brain-enriched, 170 kDa protein (p170) that interacts specifically with SH3A. Screening of combinatorial peptides reveals the optimal ligand for SH3A as Pp(V/I)PPR, and the 170 kDa mammalian son-of-sevenless (mSos1) protein, a guanine-nucleotide exchange factor for Ras, con– tains two copies of the matching sequence, PPVPPR. Immunodepletion studies confirm that p170 is mSos1. Intersectin and mSos1 are co-enriched in nerve terminals and are co-immunoprecipitated from brain extracts. SH3A competes with the SH3 domains of Grb2 in binding to mSos1, and the intersectin–mSos1 complex can be separated from Grb2 by sucrose gradient centrifugation. Overexpression of the SH3 domains of intersectin blocks epidermal growth factor-mediated Ras activation. These results suggest that intersectin functions in cell signaling in addition to its role in endocytosis and may link these cellular processes.
Keywords: clathrin/EH domain/endocytosis/Ras/SH3 domain
Introduction
Src homology 3 (SH3) domains, 50–70 amino acid modules, mediate protein–protein interactions by binding specific proline-rich peptide sequences within cellular ligands. A well studied example of an SH3 domain-mediated interaction is the recruitment of the Ras guanine-nucleotide exchange factor, mammalian son-of-sevenless (mSos), to the plasma membrane by the adaptor Grb2 (Baltensperger et al., 1993; Buday and Downward, 1993; Chardin et al., 1993; Egan et al., 1993; Li et al., 1993; Rozakis-Adcock et al., 1993). mSos contains a proline-rich C-terminus that mediates its stable association with the SH3 domains of Grb2, a protein composed of two SH3 domains flanking a central SH2 domain (Lowenstein et al., 1992). Through its SH2 domain, Grb2 interacts with activated growth factor receptors either directly or through the adaptor protein SHC (Ceresa and Pessin, 1998), thereby recruiting mSos to the membrane where it, in turn, activates Ras (Aronheim et al., 1994; Quilliam et al., 1994). However, recent evidence suggests that mSos can be recruited to the membrane by additional mechanisms. Specifically, activation of Ras by mSos can occur in the absence of Grb–mSos interactions (Karlovich et al., 1995; McCollam et al., 1995; Wang et al., 1995), suggesting that other domains of mSos, such as its Dbl homology (DH) or pleckstrin homology (PH) domains, play important roles in mSos membrane targeting and activity (Chen et al., 1997; Qian et al., 1998), or that targeting of mSos to the membrane can be mediated through the actions of other adaptor proteins.
SH3 domain-mediated protein–protein interactions also function in vesicular trafficking, particularly in endocytosis (McPherson, 1999). For example, intersectin, a novel protein involved in clathrin-mediated endocytosis, contains two N–terminal Eps15 homology (EH) domains, a central helical region and five C–terminal SH3 domains (termed SH3A–E) (Guipponi et al., 1998; Yamabhai et al., 1998). Intersectin is homologous to Dap160 in Drosophila (Roos and Kelly, 1998), as well as to Ese1 in mouse (Sengar et al., 1999) and EHSH1 in rat (Okamoto et al., 1999). An alternatively spliced variant of intersectin, referred to as intersectin-long (intersectin–l), contains a C–terminal extension with DH, PH and C2 domains (Guipponi et al., 1998). Via its EH domains, intersectin interacts with epsin, a protein that binds to clathrin and the clathrin adaptor complex, AP–2 (Chen et al., 1998; Hussain et al., 1999), and through its SH3 domains (Yamabhai et al., 1998; Okamoto et al., 1999; Sengar et al., 1999), intersectin interacts with synaptojanin and dynamin, two proteins with critical enzymatic activities involved in the formation of clathrin-coated vesicles (Cremona et al., 1999; Sever et al., 1999). In addition, intersectin heterodimerizes through the formation of coiled-coils with Eps15 (Sengar et al., 1999), another accessory protein in endocytosis (Salcini et al., 1999). Thus, intersectin is believed to function as a scaffolding protein that assembles certain proteins at sites of clathrin-coated pit formation.
To study the role further of the SH3 domains of intersectin in endocytosis, we tested their ability to block transferrin receptor uptake using a permeabilized cell assay (Simpson et al., 1999). Intriguingly, while the SH3C and SH3E domains of intersectin, as well as the SH3 domains from the endocytic proteins endophilin I (de Heuvel et al., 1997; Ringstad et al., 1997), amphiphysin II (Ramjaun et al., 1997) and syndapin I (Qualmann et al., 1999), selectively inhibit late events of clathrin-coated vesicle formation involving membrane fission, intersectin SH3A was unique in its ability to block earlier stages (Simpson et al., 1999).
To follow up on this observation, we performed overlay assays with a glutathione S-transferase (GST)–SH3A fusion protein and identified an SH3A domain-specific binding partner of 170 kDa (p170). Analysis of the optimal ligand preference of the SH3A domain suggested that p170 was mSos1, a result confirmed by a variety of methods. Like intersectin, mSos1 is enriched in nerve terminals, and the two proteins strongly co-immuno– precipitate from embryonic brain extracts. Intersectin and Grb2 compete for binding to mSos1, and sucrose-density gradient analysis indicates that in brain, intersectin and mSos1 form a stable complex that primarily excludes Grb2. Interestingly, overexpression of the SH3 domains of intersectin strongly attenuates epidermal growth factor (EGF)-dependent GTP loading of Ras. Thus, it appears that intersectin may function as a scaffold molecule for protein components of both the endocytic machinery and signal transduction pathways.
Results
Identification of an intersectin SH3A domain-specific binding partner
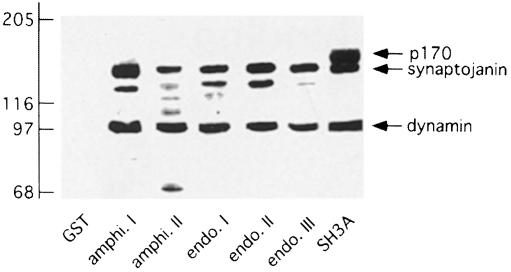
In previous collaborative studies, we determined that the SH3A domain of intersectin is unique among a wide variety of SH3 domains from endocytic proteins, including the other SH3 domains of intersectin, in its ability to block intermediate steps leading to the formation of a sequestered clathrin-coated pit (Simpson et al., 1999). We thus performed overlay assays using a GST–SH3A fusion protein in order to identify specific SH3A-binding partners. The intersectin SH3A domain, as well as the SH3 domains from amphiphysin I and II and endophilin I, II and III, each interact with bands at 100 and 145 kDa (Figure 1). The 100 and 145 kDa bands are most likely to be dynamin and synaptojanin, respectively, which have been observed to interact with the SH3 domains used here in several studies (Micheva et al., 1997; Ramjaun et al., 1997; Yamabhai et al., 1998). Interestingly, SH3A also interacted with an additional band at 170 kDa (Figure 1). The 170 kDa band (p170) was readily detectable at concentrations of fusion protein as low as 100 ng/ml, but was not detected in overlay assays with the other four SH3 domains of intersectin nor with the SH3 domains from the amphi– physins or the endophilins, at the same concentrations (Figure 1 and data not shown).
Fig. 1. Identification of a 170 kDa protein (p170) that binds specifically to the SH3A domain of intersectin. Strips of rat brain cytosolic fraction (100 μg/lane) were separated by SDS–PAGE, transferred to nitrocellulose, and overlaid with GST alone or GST fused to various SH3 domains of the proteins indicated on the blots (amphi., amphiphysin; endo., endophilin; SH3A, SH3A domain of intersectin). The positions of dynamin, synaptojanin and p170, which are recognized by various SH3 domains, are indicated by arrows on the right. The identities of the additional bands in the overlays are unknown.
GST–SH3A overlays of tissue extracts revealed that p170 was most enriched in brain, although significant levels were detected in all tissues tested, and in several tissues p170 was the only protein species identified (Figure 2A). Within the brain, subcellular fractionation revealed that p170 was present in both soluble and particulate fractions. Its overall distribution was similar to that of dynamin and synaptojanin (Figure 2B), proteins that are enriched in pre-synaptic nerve terminals (McPherson et al., 1994a), although a small pool of the protein was found in the P1 fraction, possibly in association with nuclei.
Fig. 2. Tissue and subcellular distribution of p170. (A) Proteins of cytosolic fractions from various adult rat tissues (100 μg/tissue) were separated by SDS–PAGE, transferred to nitrocellulose and overlaid with a GST fusion protein encoding the SH3A domain of intersectin. (B) Proteins of brain subcellular fractions (100 μg/fraction) were separated by SDS–PAGE, transferred to nitrocellulose and overlaid with a GST fusion protein encoding the SH3A domain of intersectin. Subcellular fractions were prepared as described (McPherson et al., 1994a). H, homogenate; P, pellet; S, supernatant. For both figures, the migratory positions of dynamin, synaptojanin and p170 are indicated by arrows on the right.
Identification of p170 as mSos1
SH3 domains interact with proline-rich peptides with the core sequence PXXP (where X is any amino acid). To define the ligand specificity of the intersectin SH3A domain, peptides were affinity selected from a phage-displayed X6PXXPX6 peptide library (Sparks et al., 1996a). The peptides that were selected from the library (Figure 3A) share a six amino acid motif with the consensus Pp(V/I)PPR, where p is typically proline. This motif represents a class II ligand for SH3 domains (Feng et al., 1994; Lim et al., 1994) and overlaps with the specificity of the Src SH3 domain (Sparks et al., 1996a). A computer search of DDBJ/EMBL/GenBank revealed two matching sequences (PPVPPR) in mSos1 (Figure 3B), a protein of 170 kDa (Chardin et al., 1993; Li et al., 1993). To determine if these two sites serve as peptide ligands for the SH3A domain of intersectin, the two sequences, along with a third highly related sequence from mSos1 (Figure 3B), were tested for binding to the five SH3 domains of intersectin expressed individually as GST fusion proteins. As seen in Figure 3C, only the proline-rich sequences within mSos1 that matched the defined consensus bound to the SH3A domain, whereas none of the peptides bound to the other four SH3 domains.
Fig. 3. Identification of consensus-binding sites for SH3A in mSos1. (A) The sequences of 12 peptides, affinity selected from a phage-displayed X6PXXPX6 peptide library (where X is any amino acid) using the SH3A domain from intersectin, are listed. The peptides define the SH3A-binding consensus sequence Pp(V/I)PPR, where p is typically proline. (B) Two putative ligand sites for the SH3A domain, PPVPPR, occur within human Sos1 at sequences between amino acids 1148 and 1161 and 1287 and 1300, and a third related site is found between amino acids 1208 and 1221, as indicated. (C) The segments of human Sos1 shown in (B) were fused to the N–terminus of secreted alkaline phosphatase and tested for binding to GST fusion protein encoding the individual SH3 domains of intersectin or to GST alone.
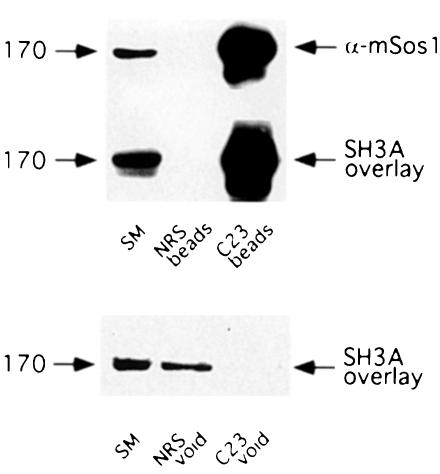
To determine if p170 is indeed mSos1, we performed GST–SH3A overlays on proteins immunoprecipitated from embryonic day 18 (E18) rat brain extracts with a polyclonal mSos1 antibody (C23). C23 efficiently immunoprecipitates mSos1 (Figure 4, top panel) and the immunoprecipitated protein is strongly reactive in a GST–SH3A domain overlay (Figure 4, middle panel). Furthermore, immuno– depletion of mSos1 from rat brain extracts depletes p170 (Figure 4, bottom panel), confirming that this interacting protein is mSos1. Moreover, the tissue distribution of mSos1 as determined by Western blotting (data not shown) closely parallels that of p170 (demonstrated in Figure 2A), suggesting that mSos1 is equivalent to p170 in all tissues tested.
Fig. 4. Conformation of p170 as mSos1. Aliquots of antiserum C23 against mSos1, as well as normal rabbit sera (NRS), were precoupled to protein A–Sepharose beads. Precoupled beads were washed, incubated overnight at 4°C with an E18 rat brain cytosolic fraction and extensively washed the next day. The material specifically bound to the beads (top two blots labeled beads) was eluted and processed, along with an aliquot of the cytosolic extract (starting material, SM), for Western blot analysis with the anti-mSos1 antibody (top blot), or for SH3A domain overlay (middle blot). The proteins that did not bind to the beads (void) were also subjected to an SH3A domain overlay assay (bottom blot).
Intersectin and mSos1 interact in neurons
To explore the potential interaction between mSos1 and intersectin in situ, we first performed immunoperoxidase staining to examine the regional distributions of the two proteins. Intersectin and mSos1 were expressed in discrete and highly overlapping neuronal populations in rat brain cortex, caudate and ventral pallidum (data not shown). Previously, it was demonstrated that Dap160, the Drosophila homologue of intersectin, was present in nerve terminals from third instar larvae (Roos and Kelly, 1998). Staining of 1–day-old neuron cultures from the CA3 region of hippocampus revealed that intersectin is broadly distributed in neurons including an enriched pool in growth cones (Hussain et al., 1999; data not shown). Interestingly, like intersectin, mSos1 is present throughout the hippocampal neurons and is enriched at the tip of their growth cones (Figure 5A). This enrichment is due to an increase in the density of mSos1 positive puncta in the growth cone area (Figure 5B) as well as an increase in the content of mSos1 per puncta (Figure 5C).
Fig. 5. Interaction of intersectin and mSos1 in situ. (A) mSos1 is found in puncta that are expressed throughout the cell body and neurites of hippocampal neurons maintained in culture for 1 day. The puncta are relatively homogeneous along the neurite but are enriched at growth cones (arrows). (B) A higher magnification image of the growth cone in (A) reveals that the density of mSos1 positive puncta is higher at the tip of the growth cone than along the neurite. (C) Color coding of fluorescent intensities of the area in (B) indicates that the intensity of individual mSos1 positive puncta is higher in the growth cone (red) than in other regions of the dendrite. Scale bar: (A), 10 μM; (B and C), 1.5 μM. (D) Aliquots of antisera 2173 and 2174 against intersectin, as well as pre-immune 2173 sera (NRS), precoupled to protein A–Sepharose beads, were incubated overnight at 4°C with an E18 rat brain cytosolic fraction. The beads were extensively washed and the material specifically bound to the beads was eluted and processed for Western blot analysis with polyclonal antibodies against mSos1 and intersectin. (E) As for (D) except that immuno– precipitations were performed from Triton X–100 solubilized membrane fractions. The antigens and their approximate molecular weights (kDa) are denoted with arrows on the right and left sides of the figure, respectively.
Next we performed co-immunoprecipitation experiments using a cytosolic fraction of E18 rat brain extracts. Two different antibodies against intersectin (2173, 2174) immunoprecipitate both the long (intersectin–l) and the short (intersectin–s) forms of intersectin and lead to a strong co-immunoprecipitation of mSos1 (Figure 5D; note the enrichment of mSos1 in the intersectin immunoprecipitated samples relative to the starting material). mSos1 also strongly co-immunoprecipitates with intersectin from a Triton X–100 solubilized particulate fraction, suggesting that the proteins also interact at the membrane (Figure 5E).
Intersectin competes with Grb2 for binding to mSos1
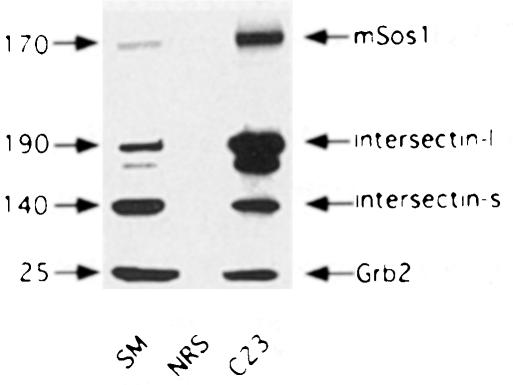
To explore the relationship between mSos1, intersectin and Grb2, we immunoprecipitated mSos1 from soluble E18 rat brain extracts and performed Western blots with antibodies against the various proteins. Intersectin–l, intersectin–s and Grb2 all co-immunoprecipitate with mSos1 (Figure 6). However, whereas intersectin–l was enriched in the immunoprecipitate relative to the starting material, Grb2 was not (Figure 6), even though in overlay assays, GST–Grb2 and GST–SH3A fusion proteins interact equally well with blotted mSos1 (data not shown). One plausible explanation is that intersectin and Grb2 compete for binding to mSos1, and the greater co-immunoprecipitation of intersectin versus Grb2 is due to intersectin being more abundant than Grb2 in the brain.
Fig. 6. Co-immunoprecipitation of intersectin and Grb2 with mSos1. Aliquots of antiserum C23 against mSos1, as well as normal rabbit sera (NRS), were precoupled to protein A–Sepharose beads. Precoupled beads were washed, incubated overnight at 4°C with an E18 rat brain cytosolic fraction, and extensively washed the next day. The material specifically bound to the beads was eluted and processed for Western blot analysis, along with an aliquot of the cytosolic extract (starting material, SM), with polyclonal antibodies against mSos1 and intersectin, and a monoclonal antibody against Grb2. The antigens and their approximate molecular weights (kDa) are denoted with arrows on the right and left sides of the figure, respectively.
mSos1 contains many SH3 domain-binding consensus sequences (i.e. PXXP). To determine if Grb2 and SH3A compete for binding to mSos1, GST–SH3A was used in overlay assays on E18 rat brain extracts in the presence of increasing concentrations of His6-tagged Grb2. Addition of Grb2 significantly reduced SH3A binding to mSos1 in a concentration-dependent manner, with binding completely eliminated at a molar ratio of 5:1 (Figure 7). Thus, Grb2 and SH3A may compete for binding to the same or overlapping site(s) in mSos1 although steric hindrance cannot be excluded. Previous mapping experiments have demonstrated that Grb2 binds to human Sos1 at three sites (PXXP cores at amino acids 1151–1154, 1212–1215 and 1290–1293) (Li et al., 1993; Rozakis-Adcock et al., 1993). The first and third of these sites are the same as those predicted by phage-display experiments to be ligand sites for the SH3A domain (Figure 3).
Fig. 7. In vitro competition binding assays of intersectin SH3A domain and Grb2 to mSos1. E18 rat brain cytosolic fractions were separated on SDS–PAGE, transferred to nitrocellulose membranes, and strips of the membrane were processed for overlay assays with GST–SH3A at 200 ng/ml. The overlay assays also contained His6-tagged Grb2 at increasing molar ratios of Grb2 to SH3A ranging from 0:1 (control) to 100:1 as indicated at the bottom of the figure. An example of the overlay results is shown at the top of the graph. The intensity of the stained mSos1 band was determined by densitometry of the autoradiographs and was normalized to control. The bars represent the mean ± SEM from three separate experiments.
To explore this issue further, we infected undifferentiated PC12 cells with an adenovirus construct encoding the five tandom SH3 domains of intersectin fused at their N–terminus to green fluorescent protein (GFP) (GFP–SH3A–E). Following expression of the construct, soluble cell extracts were prepared and processed for anti-GFP immunoprecipitation followed by blotting for co-immunoprecipitating mSos1. The anti-GFP antibody immuno– precipitates the GFP–SH3A–E fusion protein (which migrates with the expected molecular mass of 85 kDa) (Figure 8, middle panel) and leads to a strong co-immunoprecipitation of mSos1 (Figure 8, top panel). However, Grb2 is not detected in the immunoprecipitated samples (Figure 8, bottom panel), consistent with the observation that intersectin and Grb2 bind competitively to mSos1.
Fig. 8. Co-immunoprecipitation of mSos1 with the SH3 domains of intersectin. Undifferentiated PC12 cells were infected with a recombinant adenovirus encoding the five tandom SH3 domains of intersectin fused to GFP (GFP–SH3A–E). Following protein expression, a soluble cell lysate was prepared and incubated overnight at 4°C with antisera against GFP or normal rabbit sera (NRS) precoupled to protein A–Sepharose beads. The bead samples were extensively washed the next day, and the material specifically bound to the beads was eluted and processed for Western blot analysis, along with an aliquot of the cell extract (starting material, SM), with polyclonal antibodies against mSos1 and GFP, and a monoclonal antibody against Grb2. The antigens and their approximate molecular weights (kDa) are denoted with arrows on the right and left.
We next investigated the association of intersectin with mSos1 by subjecting a soluble rat brain protein extract to sucrose-density gradient centrifugation and analyzing the distribution of the proteins by Western blotting (Figure 9, top panels; the gradient was fractionated from the bottom such that lower fraction numbers correspond to a greater density of sucrose). Intersectin–l was most abundant in fraction 9, but was detectable even in the densest gradient fractions. Interestingly, mSos1 migrated on the gradients with a molecular mass that was similar, but not identical, to that of intersectin–l. Both intersectin–l and mSos1 exhibited higher native molecular masses than either clathrin triskelia (∼650 kDa) or the AP–2 heterotetramer (∼300 kDa), the migratory positions of which were revealed by Western blots of the clathrin heavy chain or the α–subunit of AP–2, respectively. Intriguingly, Grb2 remained at the top of the gradient and was effectively separated from mSos1.
Fig. 9. Sucrose-density gradient analysis of mSos1 interactions. Proteins from a cytosolic extract of rat brain were separated on 5–20% linear sucrose-density gradients by centrifugation at 195 000 g for 2.5 h and the gradients were then collected into 19 fractions of 2 ml each from the bottom. Top panels: aliquots (100 μl) of odd gradient fractions (s.g. fraction) were analyzed by Western blotting using a variety of antibodies (the antigens and their approximate molecular masses in kDa are denoted with arrows on the right and left sides of the figure, respectively). Bottom panels: aliquots (750 μl) of the indicated gradient fractions were incubated with protein A–Sepharose beads precoupled to intersectin antibody (2173). Following an overnight incubation at 4°C, the samples were washed and the proteins specifically bound to the beads were eluted with gel sample buffer and processed for SDS–PAGE and Western blot analysis with intersectin and mSos1 antibodies. The antigens and their approximate molecular masses (kDa) are denoted with arrows on the right and left sides of the figure, respectively.
To confirm that the extremely large native molecular mass for mSos1 was due, at least in part, to its interactions with intersectin, we performed co-immunoprecipitation experiments from the gradient fractions (Figure 9, bottom panels). mSos1 could be co-immunoprecipitated with intersectin from each of the gradient fractions containing the two proteins. Overall, the distribution of mSos1 seemed to parallel more closely that of intersectin–l than intersectin–s. Whether this reflects a relatively higher affinity of intersectin–l versus intersectin–s for mSos1 or is due to the apparently greater abundance of intersectin–l in brain is not known. Regardless, these data suggest that in brain, a pool of the mSos1 protein is in a stable, large molecular weight complex that includes intersectin but that predominantly excludes Grb2.
Overexpression of the SH3 domains of intersectin blocks Ras activation
To determine if the SH3 domain-mediated interactions of intersectin with mSos1 might affect Ras activation, we measured the levels of Ras–GTP in cells with a GST fusion protein encoding the Ras-binding domain of Raf1. This fusion protein binds to Ras–GTP but has significantly weaker affinity for Ras in its GDP-bound form (de Rooij and Bos, 1997; Marais et al., 1998). In HEK-293 cells treated with EGF for 2 min, a substantial pool of the total cellular Ras binds to GST–Raf1 beads as detected by Western blots with a Ras monoclonal antibody, whereas little binding is detected in the absence of EGF stimulation (Figure 10, top panel). Overexpression of GFP has little effect on the ability of EGF to stimulate GTP loading on Ras (Figure 10, bottom panel; note that the ratio of Ras bound to the beads relative to that in the starting material is similar to that seen in the EGF-challenged non-infected cells in the top panel). In contrast, overexpression of the GFP–SH3A–E construct sharply attenuates EGF-activated GTP loading on Ras (Figure 10, bottom panel). There is no effect of the overexpression of either construct on the basal activity of Ras measured in the absence of EGF (data not shown).
Fig. 10. Inhibition of Ras activation by the SH3 domains of intersectin. (Top panel) HEK-293 cells were serum starved overnight and were then challenged for 2 min with serum-free media (–EGF) or with serum-free media containing 100 ng/ml EGF (+EGF). The cells were then washed, lysates prepared and incubated with a GST fusion protein encoding the Ras–GTP binding domain of Raf1 coupled to glutathione–Sepharose beads. Following incubation, the beads were washed and material bound to the beads (bead) was processed for SDS–PAGE, along with an aliquot of the cell lysate (starting material, SM) (equal to one-tenth the amount added to the beads) and an equal aliquot of the unbound material (void). The samples were processed for Western blotting with a monoclonal antibody against Ras as indicated on the figure. (Bottom panel) HEK-293 cells infected with recombinant adenovirus encoding GFP alone (GFP) or the five tandem SH3 domains of intersectin coupled to GFP (GFP–SH3A–E) were processed for Ras assays following a 2 min exposure to EGF.
Discussion
The identification and characterization of accessory proteins in endocytosis has received a great deal of attention in recent years. One of these proteins has been identified in Drosophila, Xenopus and mammals and is variously referred to as Dap160, intersectin, Ese and EHSH (Guipponi et al., 1998; Roos and Kelly, 1998; Yamabhai et al., 1998; Hussain et al., 1999; Okamoto et al., 1999; Sengar et al., 1999). This family of proteins contains two EH domains, followed by a putative helical region and four to five tandem SH3 domains. Intersectin is localized in part to clathrin-coated pits and interacts in vivo through its EH and SH3 domains with several proteins involved in clathrin-mediated endocytosis. In addition, intersectin interacts with another accessory component of the endocytic machinery, Eps15, through its coiled-coil region (Sengar et al., 1999). Thus, intersectin may function as a scaffolding protein in the assembly of clathrin-coated pits.
We have now identified mSos1 as a protein that interacts with the SH3A domain of intersectin. Previously, Leprince et al. (1997) had isolated amphiphysin II in a two-hybrid screen using the proline-rich C–terminus of human Sos1. In experiments reported here, we have found in a side-by-side comparison using overlay assays that the SH3A domain of intersectin can bind mSos1 better than the other four SH3 domains from intersectin or than the SH3 domains from amphiphysin I and II and endophilin I, II and III. These data suggest that intersectin can bind mSos1 with higher affinity than other SH3 domain-containing endocytic proteins but do not rule out a role for amphi– physin II in mSos1 function. Intersectin and mSos1 are both expressed at high levels in growth cones of developing neurons, and immunoprecipitation analysis from embryonic brain extracts confirms that the two proteins are associated in situ. In fact, sucrose-density gradients, coupled with co-immunoprecipitation analysis, suggest that in brain, intersectin and mSos1 are components of a large molecular mass protein complex that primarily excludes Grb2. Thus, the interaction of the SH3A domain with mSos1 appears specific and is likely to be biologically relevant.
Given that intersectin is involved in the formation of clathrin-coated pits, and that the intersectin SH3A domain interacts specifically with cellular targets that function early in the formation of a clathrin-coated bud (Simpson et al., 1999), it is interesting to speculate that mSos1 may also play a role in clathrin-coated pit formation, possibly through activation of Ras. Many vesicular budding events that are mediated by coat proteins are initiated by the activation of small GTP-binding proteins through the actions of guanine-nucleotide exchange factors (Schekman and Orci, 1996). Very recently, Nakashima et al. (1999) have demonstrated that overexpression of mutant forms of Ras, as well as the small GTP-binding protein Ral, which plays a major role in mediating downstream Ras function (Feig et al., 1996), blocks the internalization of the EGF receptor. Furthermore, mSos1 can activate Rac (Nimnual et al., 1998; Scita et al., 1999), which has been implicated in transferrin receptor endocytosis (Lamaze et al., 1996). Finally, it should be noted that the long form of intersectin, which is generated by alternative splicing in neuronal tissues, contains DH, PH and C2 domains (Guipponi et al., 1998; Hussain et al., 1999; Okamoto et al., 1999; Sengar et al., 1999). Comparison of the primary structure of the DH and PH domains with other proteins suggests that the long form of intersectin may be a guanine-nucleotide exchange factor for Rho. Further work is necessary to clarify the involvement of GTP-binding proteins in clathrin-mediated endocytosis.
Another possible function of the intersectin–mSos1 complex is to couple the molecular machineries for endocytosis and signal transduction. For example, it has been demonstrated that dynamin-dependent endocytosis of the EGF receptor is necessary for EGF-dependent activation of the MAP-kinase pathway (Vieira et al., 1996). The ability of insulin-like growth factor-1 (IGF-1) to activate the SHC/MAP-kinase pathway, but not the insulin receptor substrate 1 pathway, is also dependent on clathrin-mediated endocytosis of the IGF receptor (Chow et al., 1998). Furthermore, endocytosis of the β2-adrenergic receptor is necessary for coupling to MAP-kinase activation (Daaka et al., 1998; Luttrell et al., 1999). Specifically, overexpression of a mutant form of β-arrestin, which prevents the β2-adrenergic receptor from targeting to clathrin-coated pits, blocks agonist activation of MAP kinase. Thus, it is possible that the clathrin-coated pit can function as a membrane microdomain, directing the assembly of signaling complexes, much as has been proposed for caveoli (Anderson, 1998). In fact, activation of the EGF receptor can lead to the formation of signaling complexes that include mSos1 and which are localized largely in endosomes (Di Guglielmo et al., 1994).
Given the evidence for a link between endocytosis and signaling, it is interesting to speculate that intersectin could play an important role in bringing together endocytic proteins such as dynamin, with signaling molecules such as mSos1. In fact, our data demonstrating that overexpression of the SH3 domains of intersectin functions in a dominant-negative manner to block EGF-dependent Ras activation strongly support a role for intersectin in cell signaling. Moreover, human intersectin has been found to interact by yeast two-hybrid screening with the proto-oncogene product, c-Cbl (Robertson et al., 1997), a tyrosine kinase substrate with ubiquitin ligase activity (Joazeiro et al., 1999), and transfection experiments have revealed that full-length intersectin functions in cell signaling pathways leading to activation of the Elk-1 transcription factor (J.O'Bryan, NIEHS, personal communication). Finally, in a genetic screen in Drosophila, Dap160, the Drosophila homologue of intersectin (Roos and Kelly, 1998), was selected as a negatively regulating component of the Sevenless receptor tyrosine kinase/MAP-kinase pathway (F.Rintelen and E.Hafen, University of Zurich, personal communication). Thus, intersectin appears to have a dual function in both endocytosis as well as signal transduction pathways, and it may play a role as an interface between these two important cellular processes.
Materials and methods
Antibodies
Polyclonal antibodies against intersectin were described previously (Hussain et al., 1999). Antibodies against clathrin (Simpson et al., 1996) and α–adaptin (Robinson, 1987) were a generous gift of Dr Margaret Robinson (Cambridge University). Polyclonal antibodies against mSos1 (C23) and GFP were purchased from Santa Cruz Biotechnology and Molecular Probes, respectively. Monoclonal antibodies against Grb2 and Ha-Ras were purchased from Transduction Laboratories.
Generation of fusion protein constructs
GST fusion protein constructs encoding the SH3 domains of amphi– physin I (David et al., 1994), amphiphysin II (Ramjaun et al., 1997), endophilin I (Micheva et al., 1997), the individual SH3 domains of intersectin (Yamabhai et al., 1998) and full length Grb2 (McPherson et al., 1994b) were described previously. The SH3 domains of endophilin II and III were generated by PCR using full length cDNAs (Sparks et al., 1996b) as templates in PCR with Vent DNA Polymerase (New England Biolabs) and the following primer pairs: endophilin II, forward primer 5′-GCGGGATCCGACCAGCCAAGCTGCAAG (nucleotides 919–936) and reverse primer 5′-GCGGAATTCTCACTGAGGCAGAGGCACCAG (nucleotides 1104–1087); endophilin III, forward primer 5′-GCGGGATCCGACCAGCCCTGCTGTCG (nucleotides 856–872) and reverse primer 5′-GCGCCCGGGTCACGGAGGTAAAGGCACAATG (nucleotides 1041–1023). The resulting PCR products were subcloned into the corresponding BamHI–EcoRI and BamHI–SmaI sites of pGEX-2T (Pharmacia Biotech Inc.), respectively. For His6-tagged Grb2, the GST–Grb2 construct in pGEX2T was digested with BamHI plus EcoRI enzymes and the liberated insert was subcloned into pTrcHisA (Invitrogen) using the same enzymes.
Generation of recombinant adenovirus
A recombinant adenovirus encoding the five tandem SH3 domains of intersectin with an N–terminal GFP tag was produced using the system of He et al. (1998). First, the appropriate region of intersectin was amplified by PCR using a full length cDNA (Yamabhai et al., 1998) with Vent DNA Polymerase (New England Biolabs) and with the forward primer 5′-GCGCTCGAGGGAGAAAGCCCCTCTAACG (nucleotides 2352–2370) and the reverse primer 5′-GCGGAATTCCTACGGTATTCCACCTGCTGG (nucleotides 3840–3823). The resulting PCR product was digested with XhoI and EcoRI and subcloned, in-frame, into the corresponding sites of pEGFP-C2 (Clontech) adding the N–terminal GFP tag. The resulting fusion protein construct was liberated with NheI and SmaI and subcloned into the compatible KpnI and EcoRV sites of pShuttle-CMV (He et al., 1998). The resulting plasmid was recombined with pAdEasy-1 in BJ5183 cells and recombinant plasmids were selected on kanamycin and identified by EcoRI and BstXI digests (He et al., 1998). Recombinant adenovirus was produced and amplified in HEK-293 cells and purified on CsCl2 gradients as described (He et al., 1998).
Tissue and subcellular distribution experiments
Various rat tissues were homogenized in buffer A (10 mM HEPES–OH pH 7.4, 0.83 mM benzamidine, 0.23 mM phenylmethylsulfonylfluoride, 0.5 μg/ml aprotinin and 0.5 μg/ml leupeptin). Post-nuclear supernatants were obtained by centrifugation for 5 min at 800 gmax and were then separated into cytosolic and membrane fractions by ultracentrifugation at 205 000 gmax for 1 h at 4°C. Differential centrifugation of rat brain extracts leading to the defined subcellular fractions in Figure 2B was as described previously (McPherson et al., 1994a). Dissociated cell cultures were prepared from the CA3 and dentate regions of hippocampi from P1 rat pups as described (Hussain et al., 1999).
Overlay assays
Overlay assays with GST fusion proteins were performed as described (McPherson et al., 1994b). For competition experiments, GST–SH3A was mixed with increasing concentrations of His6-tagged Grb2 immediately prior to addition to the nitrocellulose transfers.
Immunoprecipitation analysis
Cytosolic extracts, prepared as described above, were made to 1% in Triton X–100 and precleared by incubation with protein A–Sepharose. The precleared samples were then incubated with various antibodies precoupled to protein A–Sepharose, and following an overnight incubation at 4°C, were washed three times in buffer A containing 1% Triton X–100 before the proteins specifically bound to the beads were eluted and processed for SDS–PAGE. In some cases, membrane fractions, generated as described above, were incubated for 30 min at 4°C in buffer A containing 1% Triton X–100, the samples were centrifuged at 205 000 gmax for 1 h at 4°C, and the soluble supernatants were used for immunoprecipitation. For immunoprecipitations from infected cells, PC12 cells were incubated with recombinant adenovirus encoding GFP–SH3A–E in 6-well plates at a multiplicity of infection of 100. The medium was changed the next day, and the cells were left for an additional 48 h. The cells were then washed, scraped off the plate in buffer B (buffer A with 5 mM EGTA, 5 mM EDTA, 50 mM sodium fluoride, 20 mM sodium pyrophosphate and 1 mM sodium vanadate), lysed, Triton X–100 was added to 1%, and the extracts were centrifuged in a Beckman TLA 100.2 rotor at 75 000 r.p.m. for 15 min. The solubilized extracts were precleared with protein A–Sepharose before the addition of anti-GFP antibody precoupled to protein A–Sepharose. Following a 2 h incubation, the samples were washed three times in buffer B containing 1% Triton X–100, and the proteins specifically bound to the beads were eluted and processed for SDS–PAGE.
Sucrose-density gradient centrifugation
Proteins from a rat brain cytosolic fraction were layered onto linear gradients of 5–20% sucrose prepared in buffer A. The gradients were centrifuged in a STEPSAVER 50 V39 vertical rotor (Sorvall) at 195 000 g for 2.5 h and were then collected into 19 fractions of 2 ml each from the bottom. Aliquots of each fraction were analyzed by SDS–PAGE and Western blotting. For immunoprecipitations, 750 μl of each fraction were made to 1% in Triton X–100 before the addition of protein A–Sepharose beads that had been precoupled to intersectin antibody. Following an overnight incubation at 4°C, the samples were washed three times in buffer A containing 1% Triton X–100, and the proteins specifically bound to the beads were eluted and processed for SDS–PAGE.
Determination of SH3A binding specificity by phage display
GST–SH3A was used to affinity select phage from a library of bacteriophage M13 displaying combinatorial peptides at the N–terminus of mature protein III. The library displayed peptides of the form X6PXXPX6, where X represents any of the 20 naturally occurring amino acids and P represents invariant proline residues (Sparks et al., 1996a). Three different 14mer peptides from mSos1 were fused to the N–terminus of secreted bacterial alkaline phosphatase (Yamabhai and Kay, 1997) and their binding to various GST–SH3 domains, which had been immobilized on the surface of microtiter dishes, was monitored with the substrate p-nitrophenyl phosphate. The optical absorbance (OD) of duplicate wells was determined with a plate spectrophotometer at 405 nm wavelength.
Ras assays
Recombinant adenoviruses encoding GFP–SH3A–E or GFP alone were added to HEK-293 cells plated on poly-l-lysine substrate in 10–cm2 dishes at a multiplicity of infection of 100. The medium was changed after 2 h and the cells were left for an additional 48 h. The cells were then transferred into serum-free medium (along with non-infected controls), and following an overnight incubation, were treated with 100 ng/ml EGF for 2 min at 37°C or were left untreated. The cells were then washed and immediately processed for Ras assays as described (de Rooij and Bos, 1997; Marais et al., 1998).
Acknowledgments
Acknowledgements
We would like to thank Dr Channing Der (University of North Carolina) for analysis of the intersectin–l-specific sequences, Dr Dengshun Miao (McGill University) for assistance with immunostaining of brain sections, Dr Margaret Robinson (Cambridge University) for the generous gift of antibodies to clathrin and AP–2, and Dr David Kaplan and Stephen Morris (Montreal Neurological Institute) for gifts of PC12 cells and recombinant GFP adenovirus, respectively. We would also like to thank Drs Felix Rintelen and Ernst Hafen (University of Zurich) and Dr John O'Bryan (National Institute for Environmental Health Sciences) for discussion and for sharing unpublished results. Finally, we thank Drs Wayne Sossin, John O'Bryan and Ted Fon for advice and discussion. This work was supported by Medical Research Council of Canada Operating Grants MT-13461 to P.S.M. and MOP-36413 to D.B., and by grants from The Leukemia Society of America (New York, NY) and Novalon Pharmaceutical Corporation (Research Triangle Park, NC) to B.K.K. The work was also supported by a research contract from BiochemPharma (Laval, QC) to P.S.M. C.C.Q. is supported by NINDS grant #NS22807 to Dr Susan Hockfield of Yale University. N.K.H. is supported by a McGill University Faculty of Graduate Studies and Research Fellowship and P.S.M. is a Scholar of the Medical Research Council of Canada.
References
- Anderson R.G.W. (1998) The caveolae membrane system. Annu. Rev. Biochem., 67, 199–225. [DOI] [PubMed] [Google Scholar]
- Aronheim A., Engelberg, D., Li, N., Al-Alawi, N., Schlessinger, J. and Karin, M. (1994) Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell, 78, 949–961. [DOI] [PubMed] [Google Scholar]
- Baltensperger K., Kozma, L.M., Cherniack, A.D., Klarlund, J.K., Chawla, A., Banerjee, U. and Czech, M.P. (1993) Binding of the Ras activator son of sevenless to insulin receptor substrate–1 signaling complexes. Science, 260, 1950–1952. [DOI] [PubMed] [Google Scholar]
- Buday L. and Downward, J. (1993) Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein and Sos nucleotide exchange factor. Cell, 73, 611–620. [DOI] [PubMed] [Google Scholar]
- Ceresa B.P. and Pessin, J.E. (1998) Insulin regulation of the Ras activation/inactivation cycle. Mol. Cell. Biochem., 182, 23–29. [PubMed] [Google Scholar]
- Chardin P., Camonis, J.H., Gale, N.W., van Aelst, L., Schlessinger, J., Wigler, M.H. and Bar-Sagi, D. (1993) Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science, 260, 1338–1343. [DOI] [PubMed] [Google Scholar]
- Chen H., Fre, S., Slepnev, V.I., Capua, M.R., Takei, K., Butler, M.H., Di Fiore, P.P. and De Camilli, P. (1998) Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature, 394, 793–797. [DOI] [PubMed] [Google Scholar]
- Chen R.-H., Corbalan-Garcia, S. and Bar-Sagi, D. (1997) The role of the PH domain in the signal-dependent membrane targeting of Sos. EMBO J., 16, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J.C., Condorelli, G. and Smith, R.J. (1998) Insulin-like growth factor-1 receptor internalization regulates signaling via the Shc/mitogen-activated protein kinase pathway, but not the insulin receptor substrate-1 pathway. J. Biol. Chem., 273, 4627–4680. [DOI] [PubMed] [Google Scholar]
- Cremona O. et al. (1999)Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell, 99, 179–188. [DOI] [PubMed] [Google Scholar]
- Daaka Y., Luttrell, L.M., Ahn, S., Della Roca, G.J., Ferguson, S.S., Caron, M.G. and Lefkowitz, R.J. (1998) Essential role for G protein-coupled receptor endocytosis in the activation of mitogen-activated protein kinase. J. Biol. Chem., 273, 685–688. [DOI] [PubMed] [Google Scholar]
- David C., Solimena, M. and De Camilli, P. (1994) Autoimmunity in Stiff-Man Syndrome with breast cancer is targeted to the C–terminal region of human amphiphysin, a protein similar to the yeast proteins Rvs167 and Rvs161. FEBS Lett., 351, 73–79. [DOI] [PubMed] [Google Scholar]
- de Heuvel E., Bell, A.W., Ramjaun, A.R., Wong, K., Sossin, W.S. and McPherson, P.S. (1997) Identification of the major synaptojanin-binding proteins in brain. J. Biol. Chem., 272, 8710–8716. [DOI] [PubMed] [Google Scholar]
- de Rooij J. and Bos, J. (1997). Minimal Ras-binding domain of Raf1 can be used as an activation-specific probe for Ras. Oncogene, 14, 623–625. [DOI] [PubMed] [Google Scholar]
- Di Guglielmo G.M., Baass, P.C., Ou, W.-J., Posner, B.I. and Bergeron, J.J.M. (1994) Compartmentalization of SHC, GRB2 and mSOS and hyperphosphorylation of Raf–1 by EGF but not insulin in liver parenchyma. EMBO J., 13, 4269–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S.E., Giddings, B.W., Brooks, M.W., Buday, L., Sizeland, A.M. and Weinberg, R.A. (1993) Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature, 363, 45–51. [DOI] [PubMed] [Google Scholar]
- Feig L.A., Urano, T. and Cantor, S. (1996) Evidence for a Ras/Ral signaling cascade. Trends Biochem. Sci., 21, 438–441. [DOI] [PubMed] [Google Scholar]
- Feng S., Chen, J., Yu, H., Simmon, J. and Schreiber, S. (1994) Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3–ligand interactions. Science, 266, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Guipponi M., Scott, H.S., Chen, H., Schebesta, A., Rossier, C. and Antonarakis, S.E. (1998) Two isoforms of a human intersectin (ITSN) protein are produced by brain-specific alternative splicing in a stop codon. Genomics, 53, 369–376. [DOI] [PubMed] [Google Scholar]
- He T.-C., Zhou, S., Da Costa, L.T., Yu, J., Kinzler, K.W. and Vogelstein, B. (1998) A simplified system for generating recombinant adenoviruses. Proc. Natl Acad. Sci. USA, 95, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain N.K., Yamabhai, M., Ramjaun, A.R., Guy, A.M., Baranes, D., O'Bryan, J.P., Der, C.J., Kay, B.K. and McPherson, P.S. (1999) Splice variants of intersectin are components of the endocytic machinery in neurons and nonneuronal cells. J. Biol. Chem., 274, 15671–15677. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A.P., Wing, S., Huang, H.-K., Leverson, J.D., Hunter, T. and Liu, Y.-C. (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science, 286, 309–312. [DOI] [PubMed] [Google Scholar]
- Karlovich C.A., Bonfini, L., McCollam, L., Rogge, R.D., Daga, A., Czech, M.P. and Banerjee, U. (1995) In vivo functional analysis of the Ras exchange factor son of sevenless. Science, 268, 576–579. [DOI] [PubMed] [Google Scholar]
- Lamaze C., Chuang, T.-H., Terlecky, L.J., Bokoch, G.M. and Schmid, S.L. (1996) Regulation of receptor-mediated endocytosis by Rho and Rac. Nature, 382, 177–179. [DOI] [PubMed] [Google Scholar]
- Leprince C., Romero, F., Cussac, D., Vayssiere, B., Berger, R., Tavitian, A. and Camois, J.H. (1997) A new member of the amphiphysin family connecting endocytosis and signal transduction pathways. J. Biol. Chem., 272, 15101–15105. [DOI] [PubMed] [Google Scholar]
- Li N., Batzer, A., Daly, R., Yajnik, V., Skolnik, E., Chardin, P., Bar-Sagi, D., Margolis, B. and Schlessinger, J. (1993) Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature, 363, 85–88. [DOI] [PubMed] [Google Scholar]
- Lim W.A., Richards, F.M. and Fox, R. (1994) Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature, 372, 375–379. [DOI] [PubMed] [Google Scholar]
- Lowenstein E.J. et al. (1992)The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell, 70, 431–442. [DOI] [PubMed] [Google Scholar]
- Luttrell L.M. et al. (1999)β-arrestin-dependent formation of β2 adrenergic receptor-Src protein kinase complexes. Science, 283, 665–661. [DOI] [PubMed] [Google Scholar]
- Marais R., Light, Y., Mason, C., Paterson, H., Olson, M. and Marshall, C.J. (1998) Requirement of Ras-GTP–Raf complexes for activation of Raf-1 by protein kinase C. Science, 280, 109–112. [DOI] [PubMed] [Google Scholar]
- McCollam L., Bonfini, L., Karlovich, C.A., Conway, B.R., Kozma, L.M., Banerjee, U. and Czech, M.P. (1995) Functional roles for the pleckstrin and Dbl homology regions in the Ras exchange factor Son-of-sevenless. J. Biol. Chem., 270, 15954–15957. [DOI] [PubMed] [Google Scholar]
- McPherson P.S. (1999) Regulatory role of SH3 domain-mediated protein–protein interactions in synaptic vesicle endocytosis. Cell Signal., 11, 229–238. [DOI] [PubMed] [Google Scholar]
- McPherson P.S., Takei, K., Schmid, S.L. and De Camilli, P. (1994a) p145, a major Grb2-binding protein in brain, is co-localized with dynamin in nerve terminals where it undergoes activity-dependent dephosphorylation. J. Biol. Chem., 269, 30132–30139. [PubMed] [Google Scholar]
- McPherson P.S., Czernik, A.J., Chilcote, T.J., Onofri, F., Benfenati, F., Greengard, P., Schlessinger, J. and De Camilli, P. (1994b) Interaction of Grb2 via its Src homology 3 domains with synaptic proteins including synapsin I. Proc. Natl Acad. Sci. USA, 91, 6486–6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva K.D., Kay, B.K. and McPherson, P.S. (1997) Synaptojanin forms two separate complexes in the nerve terminal. Interactions with endophilin and amphiphysin. J. Biol. Chem., 272, 27239–27245. [DOI] [PubMed] [Google Scholar]
- Nakashima S., Morinaka, K., Koyama, S., Ikeda, M., Kishida, M., Okawa, K., Iwamatsu, A., Kishida, S. and Kikuchi, A. (1999) Small G protein Ral and its downstream molecules regulate endocytosis of the EGF and insulin receptors. EMBO J., 13, 3629–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual A.S., Yatsula, B.A. and Bar-Sagi, D. (1998) Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science, 279, 560–563. [DOI] [PubMed] [Google Scholar]
- Okamoto M., Schoch, S. and Sudhof, T.C. (1999) EHSH1/intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25. A protein connection between exocytosis and endocytosis?J. Biol. Chem., 274, 18446–18454. [DOI] [PubMed] [Google Scholar]
- Qian X., Vass, W.C., Papageorge, A.G., Anborgh, P.H. and Lowy, D.R. (1998) N terminus of Sos1 Ras exchange factor: critical roles for the Dbl and pleckstrin homology domains. Mol. Cell. Biol., 18, 771–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B., Roos, J., DiGregorio, P.J. and Kelly, R.B. (1999) Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott–Aldrich syndrome protein. Mol. Biol. Cell, 10, 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam L.A., Huff, S.Y., Rabun, K.M., Wei, W., Park, W., Broek, D. and Der, C.J. (1994) Membrane-targeting potentiates guanine nucleotide exchange factor CDC25 and SOS1 activation of Ras transforming activity. Proc. Natl Acad. Sci. USA, 91, 8512–8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramjaun A.R., Micheva, K.D., Bouchelet, I.B. and McPherson, P.S. (1997) Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J. Biol. Chem., 272, 16700–16706. [DOI] [PubMed] [Google Scholar]
- Ringstad N., Nemoto, Y. and De Camilli, P. (1997) The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc. Natl Acad. Sci. USA, 94, 8569–8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H., Langdon, W.Y., Thien, C.B. and Bowtell, D.D.L. (1997) A c-cbl yeast two-hybrid screen reveals interactions with 14-3-3 isoforms and cytoskeletal components. Biochem. Biophys. Res. Commun., 240, 46–50. [DOI] [PubMed] [Google Scholar]
- Robinson M.S. (1987) 100-kD coated vesicle proteins: molecular heterogeneity and intracellular distribution studied with monoclonal antibodies. J. Cell Biol., 104, 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J. and Kelly, R.B. (1998) Dap160, a neural-specific Eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J. Biol. Chem., 273, 19108–19119. [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M., Fernley, R., Wade, J., Pawson, T. and Bowtell, D. (1993) The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature, 363, 83–85. [DOI] [PubMed] [Google Scholar]
- Salcini A.E., Chen, H., Iannolo, G., De Camilli, P. and Di Fiore, P.P. (1999) Epidermal growth factor pathway substrate, 15, Eps15. Int. J. Biochem. Cell Biol., 8, 805–809. [DOI] [PubMed] [Google Scholar]
- Schekman R. and Orci, L. (1996) Coat proteins and vesicle budding. Science, 271, 1526–1533. [DOI] [PubMed] [Google Scholar]
- Scita G., Nordstrom, J., Carbone, R., Tenca, P., Giardina, G., Gutkind, S., Bjarnegard, M., Betsholtz, C. and Di Fiore, P.P. (1999) EPS8 and E3B1 transduce signals from Ras to Rac. Nature, 401, 290–293. [DOI] [PubMed] [Google Scholar]
- Sengar A.S., Wang, W., Bishay, J., Cohen, S. and Egan, S.E. (1999) The EH and SH3 domain Ese proteins regulate endocytosis by linking to dynamin and Eps15. EMBO J., 18, 1159–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S., Muhlberg, A.B. and Schmid, S.L. (1999) Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature, 398, 481–486. [DOI] [PubMed] [Google Scholar]
- Simpson F., Bright, N.A., West, M.A., Newman, L.S., Darnell, R.B. and Robinson, M.S. (1996) A novel adaptor-related protein complex. J. Cell Biol., 133, 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F., Hussain, N.K., Qualmann, B., Kelly, R.B., Kay, B.K., McPherson, P.S. and Schmid, S.L. (1999) SH3 domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nature Cell Biol., 1, 119–124. [DOI] [PubMed] [Google Scholar]
- Sparks A., Rider, J., Hoffman, N., Fowlkes, D., Quilliam, L. and Kay, B.K. (1996a) Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCγ, Crk and Grb2. Proc. Natl Acad. Sci. USA, 93, 1540–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks A.B., Hoffman, N.G., McConnell, S.J., Fowlkes, D.M. and Kay, B.K. (1996b) Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nature Biotech., 14, 741–744. [DOI] [PubMed] [Google Scholar]
- Vieira A.V., Lamaze, C. and Schmid, S.L. (1996) Control of EGF receptor signaling by clathrin-mediated endocytosis. Science, 274, 2086–2089. [DOI] [PubMed] [Google Scholar]
- Wang W., Fisher, E.M., Jia, Q., Dunn, J.M., Porfiri, E., Downward, J. and Egan, S.E. (1995) The Grb2 binding domain of mSos1 is not required for downstream signal transduction. Nature Genet., 10, 294–300. [DOI] [PubMed] [Google Scholar]
- Yamabhai M. and Kay, B.K. (1997) Examining the specificity of Src homology 3 domain–ligand interactions with alkaline phosphatase fusion proteins. Anal. Biochem., 247, 143–151. [DOI] [PubMed] [Google Scholar]
- Yamabhai M., Hoffman, N.G., Hardison, N.L., McPherson, P.S., Castagnoli, L., Cesareni, G. and Kay, B.K. (1998) Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J. Biol. Chem., 273, 31401–31407. [DOI] [PubMed] [Google Scholar]