Abstract
Background
Mercury-polluted environments are often contaminated with other heavy metals. Therefore, bacteria with resistance to several heavy metals may be useful for bioremediation. Cupriavidus metallidurans CH34 is a model heavy metal-resistant bacterium, but possesses a low resistance to mercury compounds.
Methodology/Principal Findings
To improve inorganic and organic mercury resistance of strain CH34, the IncP-1β plasmid pTP6 that provides novel merB, merG genes and additional other mer genes was introduced into the bacterium by biparental mating. The transconjugant Cupriavidus metallidurans strain MSR33 was genetically and biochemically characterized. Strain MSR33 maintained stably the plasmid pTP6 over 70 generations under non-selective conditions. The organomercurial lyase protein MerB and the mercuric reductase MerA of strain MSR33 were synthesized in presence of Hg2+. The minimum inhibitory concentrations (mM) for strain MSR33 were: Hg2+, 0.12 and CH3Hg+, 0.08. The addition of Hg2+ (0.04 mM) at exponential phase had not an effect on the growth rate of strain MSR33. In contrast, after Hg2+ addition at exponential phase the parental strain CH34 showed an immediate cessation of cell growth. During exposure to Hg2+ no effects in the morphology of MSR33 cells were observed, whereas CH34 cells exposed to Hg2+ showed a fuzzy outer membrane. Bioremediation with strain MSR33 of two mercury-contaminated aqueous solutions was evaluated. Hg2+ (0.10 and 0.15 mM) was completely volatilized by strain MSR33 from the polluted waters in presence of thioglycolate (5 mM) after 2 h.
Conclusions/Significance
A broad-spectrum mercury-resistant strain MSR33 was generated by incorporation of plasmid pTP6 that was directly isolated from the environment into C. metallidurans CH34. Strain MSR33 is capable to remove mercury from polluted waters. This is the first study to use an IncP-1β plasmid directly isolated from the environment, to generate a novel and stable bacterial strain useful for mercury bioremediation.
Introduction
Environmental decontamination of polluted sites is one of the main challenges for sustainable development. Bioremediation is an attractive technology for the clean-up of polluted waters and soils [1]–[6]. Mercury is one of the most toxic elements in the environment [7], [8]. Metal mining, fossil combustion and the chloralkali and acetaldehyde industries have raised mercury levels in water bodies and soils. Mercury enters from industrial sources mainly as Hg2+ into the environment [9]–[11]. Physicochemical and biological processes have been applied for mercury removal from contaminated environments. Physicochemical processes for heavy metal removal such as ion exchange and precipitation treatment procedures result in large volumes of mercury-contaminated sludge and are of high cost [12]–[13]. As an alternative to physicochemical processes, bacteria have been applied for the remediation of mercury pollution [1], [3], [10], [14]. The biological processes for mercury removal are of low cost, simple and environmentally friendly [10]. Mercury-polluted sites are often contaminated with other heavy metals [15]. Therefore, bacteria with resistance to several heavy metals may be useful for remediation.
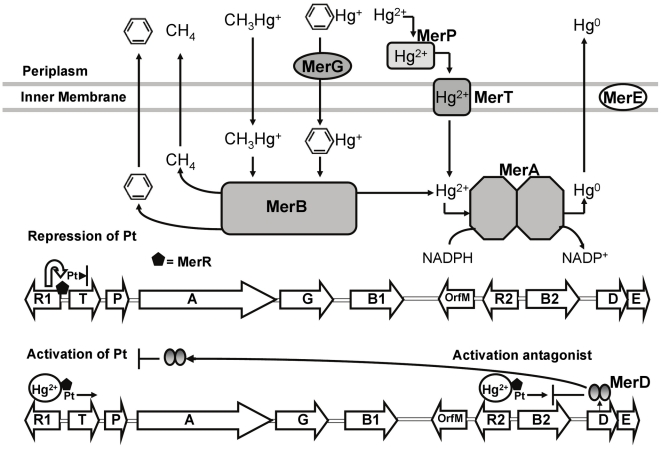
In bacteria, mercury resistance mer genes are generally organized in operons located on plasmids and transposons [16]–[17]. The narrow-spectrum mercury resistance merRTPADE operon confers resistance to inorganic mercury and the broad-spectrum mercury resistance merRTPAGBDE operon confers resistance to inorganic and organic mercury species (Fig. 1). The bacterial mechanism of mercury resistance includes the uptake and transport of Hg2+ by the periplasmic protein MerP and the inner membrane protein MerT. MerE is a membrane protein that probably acts as a broad mercury transporter mediating the transport of both methylmercury and Hg2+ [18]. The cytosolic mercuric reductase MerA reduces Hg2+ to less toxic elemental mercury [9]. The gene merB encodes an organomercurial lyase that catalyses the protonolytic cleavage of carbon-mercury bonds [19]–[20]. The merG gene product is involved in the reduction of cellular permeability to organomercurial compounds [21]. MerD probably acts as a distal co-repressor of transcriptional activation [9], [22]. MerR is the activator or repressor of the transcription of mer genes in presence or absence of mercury ions, respectively [23], [24]. At mercury stress condition the transcriptional activator MerR triggers the expression of the structural mer genes [25]. Sequencing of the native IncP-1β plasmid pTP6 that was originally isolated from mercury-polluted river sediment in a triparental mating showed that all these genes are part of transposon Tn50580 [26].
Figure 1. Bacterial mechanisms for organic and inorganic mercury detoxification and regulation of the mer genes.
MerA, mercuric reductase; MerB, organomercurial lyase; MerP, periplasmic mercury-binding protein; MerT, membrane mercury transport protein; MerG, periplasmic protein involved in cell permeability to phenylmercury; MerE membrane protein that probably acts as a broad mercury transporter; MerR, transcriptional activator or repressor of the transcription of mer genes (black pentagon); MerD, co-represor of transcriptional activation; Pt, promoter of merT and merB2 genes.
The heavy metal-resistant model bacterium Cupriavidus metallidurans strain CH34 harbors two large plasmids, pMOL28 and pMOL30, which carry genetic determinants for heavy metal resistance [27]. Each plasmid contains a merRTPADE operon that confers a narrow-spectrum mercury resistance. To improve inorganic and organic mercury resistance of strain C. metallidurans CH34, the IncP-1β plasmid pTP6 was introduced into strain CH34 in this study. The transconjugant strain Cupriavidus metallidurans MSR33 showed a broad-spectrum mercury resistance and was able to efficiently remove mercury from polluted water.
Materials and Methods
Chemicals
HgCl2 (analytical grade), CuSO4·5H2O, K2CrO4, NaBH4, NaOH, HCl (Suprapur) and standard Titrisol solution were obtained from Merck (Darmstadt, Germany). CH3HgCl (analytical grade) were obtained from Sigma Aldrich (Saint Louis, MO, USA). Stock solutions of Cu2+ (5,000 µg ml−1); CrO4 2− (2,500 µg ml−1); Hg2+ (1,000 µg ml−1) and CH3Hg+ (100 µg ml−1) were prepared. NaBH4 solution (0.25%) was prepared in NaOH (0.4%) solution. High purity hydrochloric acid was used for mercury dilutions before quantification by inductively coupled plasma optical emission spectrometer (ICP-OES). Sodium succinate and salts for media preparation were obtained from Merck (Darmstadt, Germany). Taq DNA polymerase and bovine serum albumin for PCR were obtained from Invitrogen (Carlsbad, CA, USA). RNA was extracted using an RNeasy Protect Bacteria Mini kit from Qiagen (Hilden, Germany). For RNA quantification the Quant-iT™ RNA Assay kit from Invitrogen (Carlsbad, CA, USA) was used. RT-PCR was performed using SuperScriptTM III One-Step RT-PCR System and Taq DNA Polymerase Platinum® from Invitrogen (Carlsbad, CA, USA).
Bacteria, plasmids, and culture conditions
Cupriavidus metallidurans CH34 [28], [29], Escherichia coli JM109 (pTP6) [26] and Cupriavidus necator JMP134 (pJP4) [30] were grown in Luria Bertani (LB) medium (contained per l: 10 g tryptone, 5 g yeast extract and 10 g NaCl) [31] in absence or presence of Hg2+. Cupriavidus metallidurans strains CH34 and MSR33 were cultivated also in low-phosphate Tris-buffered mineral salts (LPTMS) medium. The LPTMS medium contained (per l): 6.06 g Tris, 4.68 g NaCl, 1.49 g KCI, 1.07 g NH4Cl, 0.43 g Na2SO4, 0.2 g MgCl2·6H20, 0.03 g CaC12·2H20, 0.23 g Na2HPO4·12H20, 0.005 g Fe(III)(NH4) citrate, and 1 ml of the trace element solution SL 7 of Biebl and Pfennig [28]. Succinate (0.3%) was used as sole carbon source. To study the effect of inorganic mercury on growth, Hg2+ (0.04 mM) was added at time 0 or at exponential phase. Bacterial growth was determined by measuring turbidity at 600 nm.
The IncP-1β plasmid pTP6 that was originally isolated in a triparental mating from mercury-polluted river sediments contains merR1TPAGB1 and merR2B2D2E gene clusters conferring a broad-range mercury resistance [26].
Generation of transconjugant bacterial strains
Plasmid pTP6 was transferred from E. coli JM109 to C. metallidurans CH34 by biparental mating. Donor and recipient cells were mixed (1∶4) and placed in a sterile filter onto plate count agar (PCA) (Merck, Darmstadt, Germany). After overnight incubation at 28°C, transconjugants were selected on LPTMS agar plates containing succinate (0.3%) as sole carbon source and supplemented with Hg2+ (0.04 mM). The transconjugants were further tested for growth in liquid LPTMS medium containing succinate (0.3%) as sole carbon source and supplemented with Hg2+ (0.09 mM).
Genomic DNA purification was performed by standard procedures [31]. Plasmid extraction from C. metallidurans transconjugants, C. metallidurans CH34, C. necator JMP134 (pJP4) and E. coli JM109 (pTP6) was performed using the method of Kado and Liu with minor modifications [32], [33]. The presence of heavy metal resistance genes in genomic and plasmid DNA was analyzed by polymerase chain reaction (PCR) with specific primers. Primers used for merB gene amplification were the forward 5′-TCGCCCCATATATTTTAGAAC-3′ and the reverse 5′-GTCGGGACAGATGCAAAGAAA-3′ [34]. Primers used for amplification of chrB gene were the forward 5′-GTCGTTAGCTTGCCAACATC-3′ and the reverse 5′-CGGAAAGCAAGATGTCGAATCG-3′ [35], [36]. Primers used for copA gene amplification were the degenerated forward Coprun F2, 5′-GGSASBTACTGGTRBCAC-3′ and the degenerated reverse Coprun R1, 5′-TGNGHCATCATSGTRTCRTT-3′ [37]. Plasmids and PCR products were visualized in agarose gel electrophoresis (0.7%) in TAE buffer (0.04 M Tris, 0.04 M acetate, 0.001 M EDTA, pH 8.0).
The stability of the pTP6 plasmid in strain C. metallidurans MSR33 was evaluated through repeated cultivation in LB medium under non-selective conditions. A single colony of C. metallidurans MSR33 was used to inoculate 25 ml LB broth. After growth during 24 h at 28°C, serial dilutions were plated on PCA and 25 µl was used to inoculate fresh LB broth. The procedure was repeated six times. Forty-eight colonies randomly selected were picked from PCA plates of appropriate dilutions obtained when grown in LB broth for 1, 2, 3, 4, 5, 6 and 7 d and streaked on PCA supplemented with Hg2+ (0.5 mM). Plasmids were isolated and analyzed from 5 randomly selected mercury-resistant colonies from cultures grown during more than 70 generations under non-selective conditions.
Sequence analyses of MerA and MerB
The amino acid sequences of MerA and MerB proteins were obtained from GenPept database (http://www.ncbi.nlm.nih.gov). The multiple MerA and MerB sequences were aligned using Clustal W software [38].
Determination of minimum inhibitory concentration (MIC) values
For testing the bacterial resistance to Hg2+, CH3Hg+, Cu2+ and CrO4 2−, LPTMS agar plates were used. Ten µl of cultures grown overnight in LPTMS were placed onto the agar plates supplemented with different concentrations of each heavy metal. LPTMS was supplemented with mercury (II) (chloride) in the concentration range from 0.01 to 0.16 mM (in increasing concentration of 0.01 mM steps), copper (sulphate) in the concentration range from 3.0 to 4.2 mM (in increasing concentration of 0.1 mM steps), and (potassium) chromate in the concentration range from 0.5 to 1.0 mM (in increasing concentration of 0.1 mM steps). The plates were incubated at 28°C for 5 d. The lowest concentration of metal salts that prevented growth was recorded as the MIC. MIC for methylmercury in the concentration range from 0.005 to 0.1 mM (in increasing concentration of 0.005 mM steps) was studied on paper discs on plates. The methylmercury in solution (10 µl) was added to sterile paper discs (diameter 6 mm) and the impregnated paper discs were placed on the bacterial culture in an agar plate. Methylmercury diffused in the area surrounding each paper disc. MIC for methylmercury was recorded as the lowest concentration at which growth inhibition was observed. MIC analyses were done in triplicate.
Transmission electron microscopy
To evaluate the morphology of cells exposed to Hg2+, cells were observed by transmission electron microscopy. Strain MSR33 and strain CH34 cells were grown in LPTMS medium and further incubated at exponential phase in presence of Hg2+ (0.04 mM) for 2 h. Cells were harvested by centrifugation and washed with 50 mM phosphate buffer (pH 7.0). Cells were fixed and treated for transmission electron microscopy as described [39]. Briefly, the cells were fixed with Karnowsky solution (2.5% glutaraldehyde, 3.0% formaldehyde) in 0.2 M cacodylate buffer, post fixed with 2% osmium tetroxide and dehydrated in ethanol and acetone series. Finally, cells were embedded in an epoxy resin (Eponate 812). Thin sections (500 nm) were obtained with diamond knife in an Ultracut E ultramicrotome (Reichert). Sections were contrasted with uranyl acetate and lead citrate and observed with a Zeiss EM900 electron microscope. Micrographs were obtained and enlarged for analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
SDS-PAGE was performed as previously reported [40]. Cells grown in LB medium supplemented with Hg2+ (0.075 mM) were harvested at exponential phase (Turbidity600 nm = 0.7) and washed twice with 50 mM phosphate buffer (pH 7.0). Cells were treated as previously described [40]. Briefly, Laemmli sample buffer was added (100 µl per 5 mg of wet weight of bacteria), cells were disrupted by boiling for 5 min and cell debris were removed by centrifugation at 10,733 g for 10 min at 4°C. Proteins were quantified using a Qubit fluorometer (Invitrogen). Proteins were separated by SDS-PAGE using 7 to 15% linear polyacrylamide gradient and visualized by staining with Coomassie brilliant blue R-250.
Bioremediation experiments in bioreactors and mercury quantification
A bioreactor was designed for mercury bioremediation assays. Each bioreactor contained 50 ml of Hg2+ (0.10 mM and 0.15 mM) spiked aqueous solution (50 mM phosphate buffer, pH 7.0) in a 250 ml sterile flask and was aerated by a blower at 300 ml min−1. Volatilized mercury (Hg0) was trapped on an external HNO3 (10%) solution in a 15 ml sterile tube. Bioreactors were bioaugmented by inoculating MSR33 or CH34 cells grown in LPTMS medium supplemented with Hg2+ (0.01 mM) for the induction of mer genes (cells were added to reach a final concentration of 2×108 cells ml−1). In control bioreactors, cells were not added. Thioglycolate (5 mM) was added to selected bioreactors. The bioreactors were incubated at 30°C for 5 h. All treatments were performed in triplicate. Samples (1 ml) were taken and centrifuged (10,733 g for 10 min) to remove the cells. The supernatants were used for mercury quantification. To quantify total mercury, samples were oxidized by adding 5 ml of HNO3 (analytical grade), heated at 90°C and then diluted to a final volume of 10 ml with HCl 10% (v/v). Samples were diluted to concentrations <100 µg of Hg per liter. Total mercury was quantified by cold vapor emission spectroscopy using a flow injection system Perkin Elmer (model FIAS 200) linked to an inductively coupled plasma optical emission spectrometer (ICP-OES) Perkin Elmer (model Optima 2000). Ionic mercury (II) from samples was reduced in the FIAS system (1.5 ml min−1) with NaBH4 (0.25% m/v) in NaOH (0.4% m/v) (1.4 ml min−1) to metallic mercury, which was volatilized using argon that carried atomic mercury to the plasma and detected at 253.652 nm. The standard Hg (Titrisol, Merck) concentration in the calibration solutions ranged from 10 to 80 µg l−1. Values were calculated as the mean ±SD of three independent experiments.
Results
Generation of a broad range mercury-resistant C. metallidurans strain
To generate a heavy metal-resistant strain with broad-range mercury species resistance, novel and additional mer genes were introduced into heavy metal-resistant C. metallidurans CH34. Plasmid pTP6 carrying a complex set of mer genes encoding a broad-spectrum mercury resistance was selected to transfer mer genes into strain CH34. Plasmid pTP6 is an IncP-1β plasmid that only carries mercury resistance genes including merB and merA genes and no additional resistance or catabolic genes [26]. MerB1 of pTP6 is closely related to MerB1 of Tn5058 (Pseudomonas sp. strain ED23–33) and MerB from pMR62 (Pseudomonas sp. strain K-62), whereas MerB2 of pTP6 is closely to MerB2 of Tn5058 and MerB of the isolated plasmid pQBR103. MerA from pTP6 is closely related to MerA of Tn5058 (Pseudomonas sp. strain ED23–33) and pMR26 (Pseudomonas sp. strain K-62), whereas MerA of the plasmids pMOL28 and pMOL30 are closely related to MerA of the catabolic plasmid pJP4 (C. necator strain JMP134) and the virulence plasmid pWR501 (S. flexneri strain 5a), respectively.
Plasmid pTP6 was transferred from E. coli JM109 (pTP6) to C. metallidurans strain CH34 by biparental conjugation. Transconjugants were selected by growth in LPTMS agar medium in presence of Hg2+ (0.04 mM). More than 100 mercury-resistant transconjugants were observed in the plate. Parental strain CH34 and E. coli strain JM109 (pTP6) did not grow in presence of Hg2+ (0.04 mM). All forty-four transconjugants that formed a large colony in presence of Hg2+ (0.04 mM) and that were picked were capable to grow in liquid LPTMS medium containing succinate (0.3%) as sole carbon source and Hg2+ (0.09 mM). One of these transconjugants, C. metallidurans strain MSR33, was selected for further characterization.
Genetic characterization of the transconjugant strain MSR33
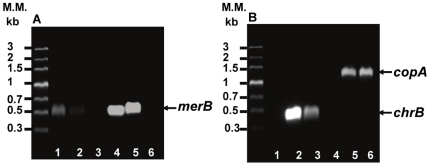
Strain MSR33 was genetically characterized. The presence of plasmid pTP6 in strain MSR33 was confirmed by plasmid isolation and visualization by agarose gel electrophoresis. The presence of merB gene in the isolated plasmid, pTP6, and in genomic DNA of strain MSR33 was confirmed by PCR (Fig. 2). Additionally, the C. metallidurans chromate-resistance chrB gene and the copper resistance copA gene were detected in the genome of transconjugant strain MSR33.
Figure 2. Detection by PCR of heavy metal resistance genes in C. metallidurans MSR33.
A, detection of merB gene in plasmid (lanes 1–3) and genomic DNA (lanes 4–6). B, detection of chrB (lanes 1–3) and copA (lanes 4–6) genes in genomic DNA. Lanes: 1 and 4, E. coli JM109 (pTP6); 2 and 5, C. metallidurans strain MSR33; 3 and 6, C. metallidurans strain CH34.
The genetic stability of a modified microorganism is critical to its release into the environment [6]. Therefore, the stability of the plasmid pTP6 in the transconjugant strain MSR33 was studied under non-selective conditions. All colonies of strain MSR33 grown under non-selective pressure after 70 generations, maintained their mercury resistance. All the colonies analyzed with improved mercury resistance after 70 generations, possessed the plasmid pTP6. These results indicated that additional mercury resistance mer genes were stable in strain MSR33. It has been reported that the IncP-1β plasmid pTP6 was stably maintained in different proteobacterial hosts under no-selective conditions [26].
Synthesis of MerB and MerA in C. metallidurans MSR33
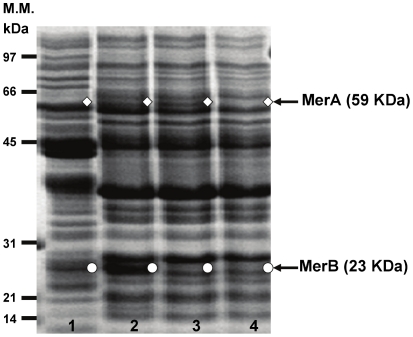
Plasmid pTP6 provided to the strain MSR33 novel genes that encode two organomercurial lyases MerB and an additional gene that encodes the mercuric reductase MerA. The synthesis of MerB and MerA in strain MSR33 grown in presence of Hg2+ was studied. The protein pattern of strain MSR33 in presence of Hg2+ was analyzed by SDS-PAGE (Fig. 3). The induction of proteins of approximately 23 kDa that probably are MerB enzymes was observed only in the transconjugant strain MSR33. Additionally, the induction of a protein of 50 KDa that probably is the mercuric reductase MerA was observed in strains MSR33 and CH34 during growth in presence of Hg2+.
Figure 3. Synthesis of MerB and MerA proteins in C. metallidurans strain MSR33.
Proteins of cells grown in LPTMS medium in presence (lanes 1–3) and absence of Hg2+ (lane 4) were separated by SDS-PAGE and stained with Coomassie blue. Lanes: 1, E. coli JM109 (pTP6) grown in presence of Hg2+; 2, C. metallidurans strain MSR33 grown in presence of Hg2+; 3, C. metallidurans strain CH34 grown in presence of Hg2+; 4, C. metallidurans strain CH34 grown in absence of mercury. White diamonds and circles indicate MerA and MerB proteins, respectively. Molecular mass of protein standards in kD are shown on the left side of gel.
Heavy metal resistance of strain MSR33
In order to study the inorganic mercury and organic mercury resistances of strain MSR33, cells were grown on LPTMS agar plates in presence of Hg2+ and CH3Hg+. Strain MSR33 showed a high resistance to Hg2+ (0.12 mM) whereas strain CH34 has a lower resistance to Hg2+ (0.05 mM). Therefore, the presence of plasmid pTP6 increased 2.4 fold Hg2+ resistance of strain MSR33. Noteworthy, strain MSR33 possesses a high resistance to methylmercury (0.08 mM). In contrast, strain CH34 was sensitive to methylmercury. Strain MSR33 maintained the Cu2+ and CrO4 2− resistances of the parental strain CH34 (Table 1).
Table 1. Minimum inhibitory concentrations of heavy metals for transconjugant C. metallidurans strain MSR33.
| Metal (mM) | Strain MSR33 | Strain CH34 | Increased resistance (fold) |
| Hg2+ | 0.12 | 0.05 | 2.4 |
| CH3Hg+ | 0.08 | <0.005 | >16 |
| Cu2+ | 3.80 | 3.80 | 0 |
| CrO4 2− | 0.70 | 0.70 | 0 |
Effect of Hg2+ on the growth and cell morphology
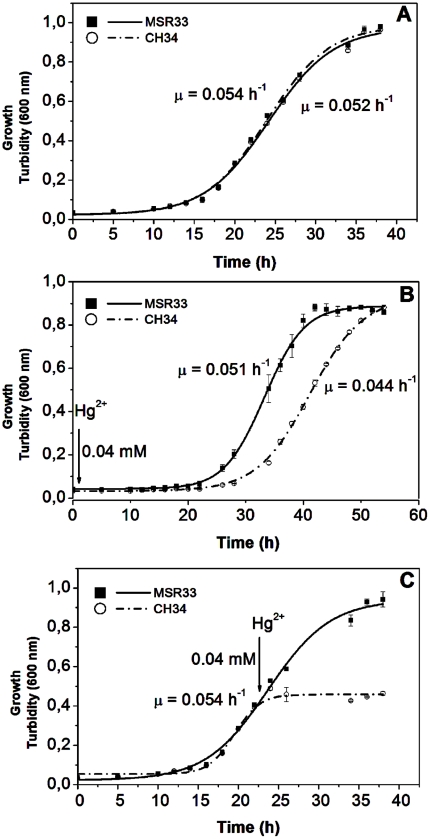
The effect of exposure to Hg2+ on the growth of strain MSR33 was studied (Fig. 4). Cells were grown in LPTMS medium with succinate as the sole carbon source in the absence or presence of Hg2+ (0.04 mM). Hg2+ did not affect the growth of MSR33 (µ = 0.051 h−1), whereas the growth of strain CH34 decreased slightly in presence of Hg2+ (µ = 0.044 h−1) (Fig. 4). Strains MSR33 and CH34 showed a similar growth rate in absence of Hg2+. In addition, the effect of Hg2+ (0.04 mM) exposure at exponential phase of growth was studied. Interestingly, strain MSR33 growth was not affected after addition of Hg2+ at exponential phase (Fig. 4). In contrast, no further growth of strain CH34 was observed when 0.04 mM of Hg2+ were added at exponential phase.
Figure 4. Effect of Hg2+ on the growth of C. metallidurans strains MSR33 and CH34.
Cells were grown in LPTMS medium in absence of Hg2+ A, presence of Hg2+ (0.04 mM) B, or Hg2+ (0.04 mM) were added to LPTMS medium at exponential phase C. The exposure to Hg2+ started at the point indicated with an arrow. Each value is the mean ± SD of three independent assays.
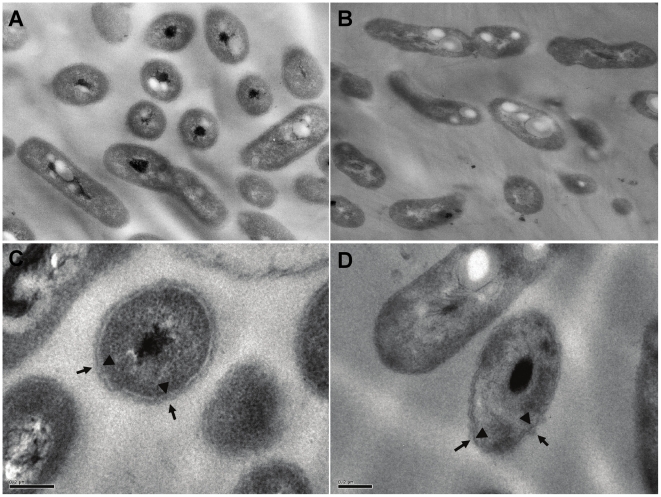
To evaluate changes in cell morphology by exposure to mercury, cells of MSR33 grown on LPTMS medium with succinate as sole carbon source and exposed to Hg2+ were analyzed by transmission electron microscopy (Fig. 5). Strain MSR33 cells exposed to Hg2+ showed no changes in cell morphology. In contrast, CH34 cells exposed to Hg2+ exhibited a fuzzy outer membrane. Electron dense granules in the cytoplasm of MSR33 and CH34 strains exposed to Hg2+ were also observed (Fig. 5).
Figure 5. Ultrathin section of C. metallidurans strain MSR33 exposed to Hg2+.
Cells grown in LPTMS medium with succinate until exponential phase were further incubated in presence of Hg2+ (0.04 mM) for 1 h. A and C, MSR33 cells; B and D, CH34 cells. Arrows and arrowheads indicate the outer and the inner membrane, respectively. The bars represent 0.2 µm.
Bioremediation of mercury-polluted water
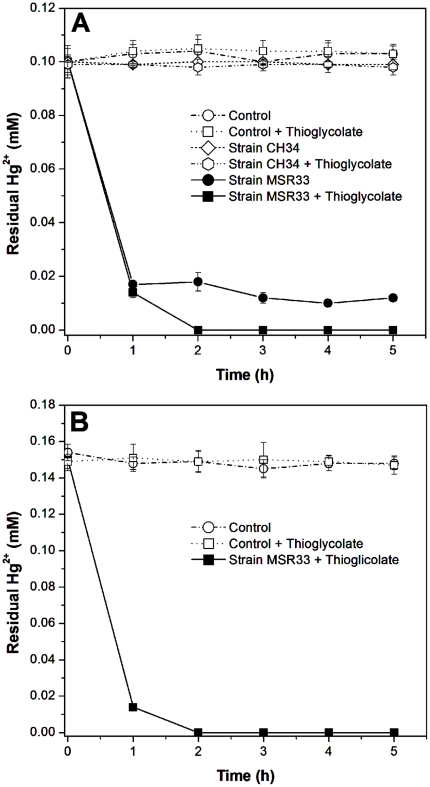
In this study, the effectiveness of C. metallidurans strain MSR33 as a biocatalyst for the bioremediation of two Hg2+-polluted aqueous solutions in a bioreactor was assessed. Bioaugmentation was studied in aqueous solutions containing Hg2+ (0.10 mM and 0.15 mM). The effect of addition of thioglycolate, a compound that provides an excess of exogenous thiol groups [41], [42] was evaluated on mercury bioremediation by strain MSR33. The effect of bioaugmentation by strain MSR33 on Hg2+ removal in two polluted waters is shown in Figure 6. In presence of thioglycolate, C. metallidurans strain MSR33 completely removed mercury (0.10 mM and 0.15 mM) from the two polluted waters after 2 h. In absence of thioglycolate, strain MSR33 removed 88% of Hg2+ (0.10 mM) after 3 h. In contrast, wild type C. metallidurans strain CH34 was not capable to remove mercury either in presence or absence of thioglycolate. No mercury removal was observed in the control bioreactors.
Figure 6. Effect of bioaugmentation with C. metallidurans strain MSR33 in Hg2+-polluted waters.
A, removal of Hg2+ from water polluted with Hg2+ (0.10 mM) by strain MSR33 or strain CH34 in presence or absence of thioglycolate (5 mM). B, removal by strain MSR33 of Hg2+ from water polluted with Hg2+ (0.15 mM) in presence of thioglycolate (5 mM). Control assays without cells were incubated in presence or absence of thioglycolate (5 mM).
Discussion
Mercury-polluted sites often contain other heavy metals. Therefore, for the bioremediation of mercury-polluted sites broad range heavy metal-resistant bacteria are required. C. metallidurans strain CH34 is a model heavy metal-resistant bacterium [27], [28]. The main goal of this work was to generate a heavy metal-resistant bacterial strain with resistance to inorganic and organic mercury species. Therefore, the heavy metal-resistant strain C. metallidurans CH34 with low resistance to mercury (II) and sensitive to organic mercury was modified by introducing the plasmid pTP6 that has been directly isolated from the environment and that encodes Mer proteins that confer a broad-range mercury resistance [26]. The transconjugant C. metallidurans strain MSR33 was characterized and applied for remediation of mercury-polluted waters.
In the present study, improved mercury resistance of the transconjugant strain MSR33 was obtained by incorporation of broad-host range plasmid pTP6 with a complex mer system as sole accessory element into C. metallidurans CH34. To our knowledge, this is the first study to use an IncP-1β plasmid directly isolated from the environment to generate a novel bacterial strain with improved mercury resistance. Therefore, and in contrast to genetically engineered bacteria, environmental application of transconjugant strain MSR33 is not subjected to biosafety regulation, which is an important advantage for bioremediation of mercury-polluted sites. This study showed that pTP6 stably replicated even without selective pressure as other IncP-1 plasmids that are tightly controlled by korA and korB genes [26]. Interestingly, IncP-1 plasmids have an extremely broad-host range in Gram-negative bacteria and are efficiently transferred from the host to soil bacteria [43], [44]. The natural plasmid pTP6 provided novel merB and merG genes and conferred a broad-spectrum mercury resistance to C. metallidurans strain MSR33. The presence of plasmid pTP6 in strain MSR33 provided also additional mer gene copies (merR, merT, merP, merA, merD and merE genes) to the host strain, improving also inorganic mercury resistance. The genetic redundancy may increase the robustness of organisms to mercury stress [45]. The MerA protein encoded by plasmid pTP6 belonged to different subclasses than MerA proteins of pMOL28 and pMOL30. On the other hand, plasmid pTP6 provides two copies of merB genes encoding MerB1 and MerB2 enzymes.
The mechanisms for mercury detoxification in strain MSR33 are shown in Figure 1. The expression of mer genes of pTP6 in strain MSR33 support growth of strain MSR33 with Hg2+ at high concentrations and in presence of CH3Hg+. Strain MSR33 possesses a high organic mercury resistance (CH3Hg+, 0.08 mM) and a high inorganic mercury resistance (Hg2+, 0.12 mM) (Table 1). The novel resistance to methylmercury is conferred by the organomercurial lyase MerB. The organomercurial lyase catalyzes the protonolysis of the carbon mercury bond, removing the organic ligand and releasing Hg2+. Inorganic mercury is 100 times less toxic than the CH3Hg+ form [9], [24]. An additional MerA enzyme improved strain MSR33 detoxification of Hg2+, and additional MerT and MerP proteins in strain MSR33 probably allow a rapid uptake of mercury avoiding an extracellular accumulation. The mercury resistance of strain MSR33 is higher than that of other recombinant strains such as Pseudomonas putida F1::mer [46]. The addition of merTPAB genes in P. putida F1 increased broad-spectrum mercury resistance in the range of 14–86%.
Strain MSR33 showed a similar growth rate in absence and presence of Hg2+ (Fig. 4). As expected, the presence of Hg2+ decreased the growth rate of strain CH34. Noteworthy, the growth of MSR33 was not affected by the addition of mercury (II) at exponential phase (Fig. 4C). On the other hand, an immediate cessation of growth of strain CH34 was observed after the addition of Hg2+ at exponential phase. The additional mer genes incorporated into strain MSR33 probably improve the mercury detoxification system. The effects of Hg2+ on growth are in accordance with the effects on cell morphology (Fig. 5). Cell membrane of strain MSR33 was not affected by the presence of mercury. In contrast, after the exposure to Hg2+ strain CH34 showed a fuzzy outer membrane. Enterobacter exposed to mercury also showed a fuzzy cell membrane [47]. After exposure to mercury, both strains MSR33 and CH34 contained electron-dense granules in the cytoplasm that probably are polyhydroxybutyrate. C. metallidurans CH34 harbors the genes for polyhydroxybutyrate synthesis [48] and probably accumulates these intracellular storage granules during stress.
The bioremediation potential of strain MSR33 of mercury-polluted aqueous solutions was evaluated in a bioreactor. Strain MSR33 completely removed Hg2+ (0.10 mM and 0.15 mM) from polluted aqueous solutions. The presence of thioglycolate was required for complete mercury removal in water. The presence of thiol molecules ensures that Hg2+ will be present as the dimercaptide, RS-Hg-SR. The dimercaptide form of Hg2+ is the substrate of the NADPH-depending mercuric reductase MerA [42]. Thiol compounds such as 2-mercaptoethanol, cysteine and thioglycolate improved mercury volatilization by Escherichia coli, Pseudomonas sp. strain K-62, bacterial strain M-1 and Pseudomonas putida strain PpY101 (pSR134) [21], [41], [49]–[53].
In the present study, strain MSR33 showed a high mercury volatilization rate, 6.8×10−3 ng Hg2+ cell−1 h−1. Saouter et al. [54] studied mercury removal from freshwater pond by resting cells of mercury-resistant Aeromonas hydrophila strain KT20 and Pseudomonas aeruginosa PAO1 derivative containing a plasmid carrying mer genes. The mercury volatilization rates of A. hydrophila KT20 and P. aeruginosa PAO1 derivative were 1.0×10−3 ng Hg2+ cell−1 h−1 and 2.4×10−4 ng Hg2+ cell−1 h−1, respectively. Okino et al. [53] studied mercury removal from a mercury-polluted aqueous solution with the genetically engineered bacterium, P. putida strain PpY101 (pSR134) showing a volatilization rate of 6.7×10−3 ng Hg2+ cell−1 h−1.
C. metallidurans MSR33 is a stable transconjugant strain carrying plasmid pTP6, which conferred a novel organomercurial resistance and improved significantly the resistance to Hg2+. Strain MSR33 was able to cleave the organic moiety from methylmercury and to reduce Hg2+ into a less toxic Hg elemental form. Strain MSR33 efficiently removed Hg2+ (0.10 mM and 0.15 mM) through mercury volatilization from mercury-contaminated waters. This study suggests that strain MSR33 may be useful for mercury bioremediation of contaminated water bodies and industrial wastewater. In addition, strain MSR33 is an interesting biocatalyst for bioremediation of mercury-polluted soils.
Acknowledgments
The authors thank Max Mergeay for providing strain C. metallidurans CH34 and Gabriela Lobos for analytical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: M.S. gratefully acknowledges financial support from MILENIO P04/007-F (MIDEPLAN) (http://www.iniciativamilenio.cl), USM 130522, 130836, and 130948 (http://www.usm.cl), and FONDECYT 1070507 (CONICYT) (http://www.fondecyt.cl) grants. M.S. and K.S. gratefully acknowledge CONICYT-BMBF grant. L.A.R. gratefully acknowledges Nucleus Millennium EMBA, USM and PIIC-2009-USM fellowships. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.von Canstein H, Li Y, Timmis KN, Deckwer WD, Wagner-Döbler I. Removal of mercury from chloralkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl Environ Microbiol. 1999;65:5279–5284. doi: 10.1128/aem.65.12.5279-5284.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nealson KH, Belz A, McKee B. Breathing metals as a way of life: geobiology in action. Antonie Van Leeuwenhoek. 2002;81:215–222. doi: 10.1023/a:1020518818647. [DOI] [PubMed] [Google Scholar]
- 3.Valls M, de Lorenzo V. Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol Rev. 2002;26:327–338. doi: 10.1111/j.1574-6976.2002.tb00618.x. [DOI] [PubMed] [Google Scholar]
- 4.Pieper DH, Seeger M. Bacterial metabolism of polychlorinated biphenyls. J Mol Microbiol Biotechnol. 2008;15:121–138. doi: 10.1159/000121325. [DOI] [PubMed] [Google Scholar]
- 5.Morgante V, López-López A, Flores C, González M, González B, et al. Bioaugmentation with Pseudomonas sp. strain MHP41 promotes simazine attenuation and bacterial community changes in agricultural soils. FEMS Microbiol Ecol. 2010;71:114–126. doi: 10.1111/j.1574-6941.2009.00790.x. Erratum in FEMS Microbiol Ecol (2010) 72: 152. [DOI] [PubMed] [Google Scholar]
- 6.Saavedra JM, Acevedo F, González M, Seeger M. Mineralization of PCBs by the genetically modified strain Cupriavidus necator JMS34 and its application for bioremediation of PCBs in soil. Appl Microbiol Biotechnol. 2010;87:1543–1554. doi: 10.1007/s00253-010-2575-6. [DOI] [PubMed] [Google Scholar]
- 7.Nascimento AM, Chartone-Souze E. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet Mol Res. 2003;2:92–101. [PubMed] [Google Scholar]
- 8.Oehmen A, Fradinho J, Serra S, Carvalho G, Capelo JL, et al. The effect of carbon source on the biological reduction of ionic mercury. J Hazard Mater. 2009;165:1040–1048. doi: 10.1016/j.jhazmat.2008.10.094. [DOI] [PubMed] [Google Scholar]
- 9.Barkay T, Miller SM, Summers AO. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev. 2003;27:355–384. doi: 10.1016/S0168-6445(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 10.Wagner-Döbler I. Pilot plant for bioremediation of mercury-containing industrial wastewater. Appl Microbiol Biotechnol. 2003;62:124–133. doi: 10.1007/s00253-003-1322-7. [DOI] [PubMed] [Google Scholar]
- 11.Fatta D, Canna-Michaelidou S, Michael C, Demetriou Georgiou E, Christodoulidou M, et al. Organochlorine and organophosphoric insecticides, herbicides and heavy metals residue in industrial wastewaters in Cyprus. J Hazard Mater. 2007;145:169–179. doi: 10.1016/j.jhazmat.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Ritter JA, Bibler JP. Removal of mercury from wastewater: large-scale performance of an ion exchange process. Wat Sci Technol. 1992;25:165–172. [Google Scholar]
- 13.Chang JS, Hong J. Biosorption of mercury by the inactivated cells of Pseudomonas aeruginosa PU21 (Rip64). Biotechnol Bioeng. 1994;44:999–1006. doi: 10.1002/bit.260440817. [DOI] [PubMed] [Google Scholar]
- 14.Deckwer WD, Becker FU, Ledakowicz S, Wagner-Döbler I. Microbial removal of ionic mercury in a three-phase fluidized bed reactor. Environ Sci Technol. 2004;38:1858–1865. doi: 10.1021/es0300517. [DOI] [PubMed] [Google Scholar]
- 15.Baldrian P, in der Wiesche C, Gabriel J, Nerud F, Zadrazil F. Influence of cadmium and mercury on activities of ligninolytic enzymes and degradation of polycyclic aromatic hydrocarbons by Pleurotus ostreatus in soil. Appl Environ Microbiol. 2000;66:2471–2478. doi: 10.1128/aem.66.6.2471-2478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yurieva O, Kholodii G, Minakhin L, Gorlenko Z, Kalyaeva E, et al. Intercontinental spread of promiscuous mercury-resistance transposons in environmental bacteria. Mol Microbiol. 1997;24:321–329. doi: 10.1046/j.1365-2958.1997.3261688.x. [DOI] [PubMed] [Google Scholar]
- 17.Silver S, Phung LT. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J Ind Microbiol Biotechnol. 2005;32:587–605. doi: 10.1007/s10295-005-0019-6. [DOI] [PubMed] [Google Scholar]
- 18.Kiyono M, Sone Y, Nakamura R, Pan-Hou H, Sakabe K. The MerE protein encoded by transposon Tn21 is a broad mercury transporter in Escherichia coli. FEBS Lett. 2009;583:1127–1131. doi: 10.1016/j.febslet.2009.02.039. [DOI] [PubMed] [Google Scholar]
- 19.Moore MJ, Distefano MD, Zydowsky LD, Cummings RT, Walsh CT. Organomercurial lyase and mercuric ion reductase: nature's mercury detoxification catalysts. Acc Chem Res. 1990;23:301–308. [Google Scholar]
- 20.Misra TK. Bacterial resistance to inorganic mercury salts and organomercurials. Plasmid. 1992;27:4–16. doi: 10.1016/0147-619x(92)90002-r. [DOI] [PubMed] [Google Scholar]
- 21.Kiyono M, Pan-Hou H. The merG gene product is involved in phenylmercury resistance in Pseudomonas strain K-62. J Bacteriol. 1999;181:762–730. doi: 10.1128/jb.181.3.726-730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Champier L, Duarte V, Michaud-Soret I, Covès J. Characterization of the MerD protein from Ralstonia metallidurans CH34: a possible role in bacterial mercury resistance by switching off the induction of the mer operon. Mol Microbiol. 2004;52:1475–1485. doi: 10.1111/j.1365-2958.2004.04071.x. [DOI] [PubMed] [Google Scholar]
- 23.Ni'Bhriain NN, Silver S, Foster TJ. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J Bacteriol. 1983;155:690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Permina EA, Kazakov AE, Kalinina OV, Gelfand MS. Comparative genomics of regulation of heavy metal resistance in Eubacteria. BMC Microbiol. 2006;6:49–60. doi: 10.1186/1471-2180-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 26.Smalla K, Haines AS, Jones K, Krögerrecklenfort E, Heuer H, et al. Increased abundance of IncP-1β plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of IncP-1β plasmids with a complex mer transposon as the sole accessory element. Appl Environ Microbiol. 2006;72:7253–7259. doi: 10.1128/AEM.00922-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mergeay M, Monchy S, Vallaeys T, Auquier V, Benotmane A, et al. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol Rev. 2003;27:385–410. doi: 10.1016/S0168-6445(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 28.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, et al. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monchy S, Benotmane MA, Janssen P, Vallaeys T, Taghavi S, et al. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal response to heavy metals. J Bacteriol. 2007;189:7417–7425. doi: 10.1128/JB.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Don RH, Pemberton JM. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd Ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Top E, Mergeay M, Springael D, Verstraete W. Gene escape model: transfer of heavy metal resistances genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liebert CA, Wireman J, Smith T, Summers AO. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl Environ Microbiol. 1997;63:1066–1076. doi: 10.1128/aem.63.3.1066-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nies A, Nies DH, Silver S. Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J Biol Chem. 1990;265:5648–5653. [PubMed] [Google Scholar]
- 36.Abou-Shanab RA, van Berkum P, Angle JS. Heavy metal resistance and genotypic analysis of metal resistances genes in gram-positive and gram-negative bacteria present in Ni-rich serpentine soil and in the rhizosphere of Alyssum murale. Chemosphere. 2007;68:360–367. doi: 10.1016/j.chemosphere.2006.12.051. [DOI] [PubMed] [Google Scholar]
- 37.Lejon DP, Nowak V, Bouko S, Pascault N, Mougel C, et al. Fingerprinting and diversity of bacterial copA genes in response to soil types, soil organic status and copper contamination. FEMS Microbiol Ecol. 2007;61:424–437. doi: 10.1111/j.1574-6941.2007.00365.x. [DOI] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Cámara B, Herrera C, González M, Couve E, Hofer B, et al. From PCBs to highly toxic metabolites by the biphenyl pathway. Environ Microbiol. 2004;6:842–850. doi: 10.1111/j.1462-2920.2004.00630.x. [DOI] [PubMed] [Google Scholar]
- 40.Seeger M, Jerez CA. Phosphate-starvation induced changes in Thiobacillus ferrooxidans. FEMS Microbiol Lett. 1993;108:35–42. doi: 10.1111/j.1574-6968.1993.tb06070.x. [DOI] [PubMed] [Google Scholar]
- 41.Summers AO, Sugarman LI. Cell-free mercury (II)-reducing activity in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1974;119:242–249. doi: 10.1128/jb.119.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fox B, Walsh CT. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem. 1982;257:2498–2503. [PubMed] [Google Scholar]
- 43.Pukall R, Tschäpe H, Smalla K. Monitoring the spread of broad host and narrow host range plasmids in soil microcosms. FEMS Microbiol Ecol. 1996;20:53–66. [Google Scholar]
- 44.Schlüter A, Szczepanowski R, Pühler A, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev. 2007;31:449–477. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 45.Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci USA. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horn JM, Brunke M, Deckwer WD, Timmis KN. Pseudomonas putida strains which constitutively overexpress mercury resistance for biodetoxification of organomercurial pollutants. Appl Environ Microbiol. 1994;60:357–362. doi: 10.1128/aem.60.1.357-362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaituzis Z, Nelson JD, Jr, Wan LW, Colwell RR. Effects of mercuric chloride on growth and morphology of selected strains of mercury-resistant bacteria. Appl Microbiol. 1975;29:275–286. doi: 10.1128/am.29.2.275-286.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janssen PJ, van Houdt R, Moors H, Monsieurs P, Morin N, et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE. 2010;5:e10433. doi: 10.1371/journal.pone.0010433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schottel JL. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978;253:4341–4349. [PubMed] [Google Scholar]
- 50.Nakamura K, Nakahara H. Simplified X-ray film method for detection of bacterial volatilization of mercury chloride by Escherichia coli. Appl Environ Microbiol. 1988;54:2871–2873. doi: 10.1128/aem.54.11.2871-2873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray S, Gachhui R, Pahan K, Chaudhury J, Mandal A. Detoxification of mercury and organomercurials by nitrogen-fixing soil bacteria. J Biosci. 1989;14:173–182. [Google Scholar]
- 52.Nakamura K, Hagimine M, Sakai M, Furukawa K. Removal of mercury from mercury-contaminated sediments using a combined method of chemical leaching and volatilization of mercury by bacteria. Biodegradation. 1999;10:443–447. doi: 10.1023/a:1008329511391. [DOI] [PubMed] [Google Scholar]
- 53.Okino S, Iwasaki K, Yagi O, Tanaka H. Development of a biological mercury removal-recovery system. Biotechnol Lett. 2000;22:783–788. [Google Scholar]
- 54.Saouter E, Gillman M, Barkay T. An evaluation of mer-specified reduction of ionic mercury as a remedial tool of mercury-contaminated freshwater pond. J Ind Microbiol. 1995;14:343–348. [Google Scholar]