Abstract
The p38 mitogen-activated protein kinase (MAPK) group is represented by four isoforms in mammals (p38α, p38β2, p38γ and p38δ). These p38 MAPK isoforms appear to mediate distinct functions in vivo due, in part, to differences in substrate phosphorylation by individual p38 MAPKs and also to selective activation by MAPK kinases (MAPKKs). Here we report the identification of two factors that contribute to the specificity of p38 MAPK activation. One mechanism of specificity is the selective formation of functional complexes between MAPKK and different p38 MAPKs. The formation of these complexes requires the presence of a MAPK docking site in the N–terminus of the MAPKK. The second mechanism that confers signaling specificity is the selective recognition of the activation loop (T–loop) of p38 MAPK isoforms. Together, these processes provide a mechanism that enables the selective activation of p38 MAPK in response to activated MAPKK.
Keywords: MAP kinase/MKK3/MKK6/p38/signal transduction
Introduction
The p38 group of mitogen-activated protein kinases (MAPKs) is activated by treatment of cells with pro-inflammatory cytokines and by exposure to environmental stress (Cohen, 1997). One important function of the p38 signaling pathway appears to be the regulation of cytokine expression (Lee et al., 1999).
Molecular cloning studies have led to the identfication of four p38 isoforms: p38α (also known as SAPK2a), p38β2 (SAPK2b), p38γ (SAPK3) and p38δ (SAPK4). The p38α and p38β MAPK are 60% identical to p38γ and p38δ MAPK, indicating that these protein kinases represent related, but distinct, MAPK subgroups (Cohen, 1997). One subgroup (p38α and p38β) is inhibited by a class of pyridinyl imidazole drugs, while the other subgroup (p38γ and p38δ) is insensitive to these drugs (Lee et al., 1999). These p38 MAPKs phosphorylate both a common group of substrates and distinct substrates (Cohen, 1997) and they can be activated selectively by some extracellular stimuli (Wilk-Blaszczak et al., 1998; Conrad et al., 1999), suggesting that they may exert distinct biological actions. Indeed, studies of HeLa cells indicate that while p38α induces apoptosis, p38β2 promotes cell survival (Nemoto et al., 1998). These studies suggest that different p38 isoforms have overlapping, but also distinct physiological roles.
Specificity of p38 MAPK signaling has also been reported in studies of the MAPK kinases (MAPKKs) that activate the p38 MAPK isoforms. Two genes that encode p38-specific MAPKKs have been described: Mkk3 (Derijard et al., 1995) and Mkk6 (Han et al., 1996; Moriguchi et al., 1996a; Raingeaud et al., 1996; Stein et al., 1996). Targeted gene disruption studies in mice have demonstrated non-redundant functions of the Mkk3 and Mkk6 genes (Lu et al., 1999; Wysk et al., 1999). Furthermore, expression of activated forms of the MKK3 and MKK6 protein kinases causes different biological responses in cardiac myocytes (Wang et al., 1998). These actions are altered by co-expression of specific p38 isoforms; thus, p38α induces apoptosis while p38β2 promotes a hypertrophic response in cardiac myocytes (Wang et al., 1998). This signaling specificity is likely to be important for the generation of appropriate biological responses by the p38 MAPK pathway.
The mechanism that accounts for signaling specificity by the p38 MAPK pathway is not understood. Previously it has been demonstrated that MKK6 is a common activator of p38α, p38β2, p38γ and p38δ MAPK, while MKK3 activates only p38α, p38γ and p38δ MAPK (Jiang et al., 1996, 1997; Cuenda et al., 1997; Goedert et al., 1997; Wang et al., 1997; Enslen et al., 1998). This selectivity of p38 MAPK activation by MKK3 and MKK6 may contribute to the specificity of signal transduction by the p38 MAPK pathway.
The purpose of the study reported here was to examine the specificity of activation of p38 MAPK isoforms by MKK3 and MKK6. We show that two factors contribute to the specificity of p38 MAPK signaling: (i) selective docking interactions between the MAPKK and p38 MAPK; and (ii) sequences within the activation loop (T–loop) of individual p38 MAP isoforms. Together, these observations provide insight into the mechanism of p38 MAPK activation.
Results
Role of the N–terminal region of p38 MAPKK in the selective activation of p38β2 MAPK
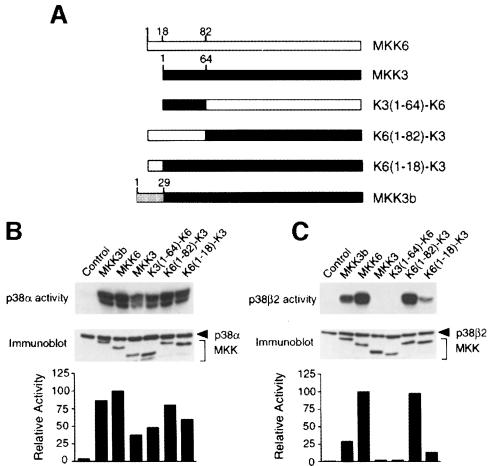
MKK3 and MKK6 selectively activate different p38 isoforms (Enslen et al., 1998). To identify the molecular determinants responsible for this selectivity, we constructed chimeric protein kinases using MKK3 and MKK6 sequences (Figure 1A). Constitutively active MAPKKs were constructed by replacing the sites of activating phosphorylation with glutamic acid. These chimeric kinases caused similar activation of p38α in co-transfection assays (Figure 1B). In contrast, differences were detected in assays using p38β2 (Figure 1C). As expected, MKK6, but not MKK3, caused p38β2 activation. However, replacement of the N–terminal region of MKK6 with sequences derived from MKK3 blocked the ability of MKK6 to activate p38β2 (Figure 1C). Conversely, chimeras of MKK3 with the N–terminal region of MKK6 were able to activate p38β2 (Figure 1C). Residues 1–18 of MKK6 were sufficient, but a larger activation of p38β2 was observed when MKK6 residues 1–82 were fused to the N–terminus of MKK3. These data indicated that the N–terminal region of MKK3 and MKK6 regulates substrate specificity (Figure 1C), but not activity (Figure 1B). This conclusion was confirmed by studies of an alternative form of MKK3 (MKK3b) (Moriguchi et al., 1996b; Han et al., 1997), which contains an additional 29 amino acids fused to the N–terminus of MKK3. We found that while MKK3 and MKK3b both activated p38α (Figure 1B), only MKK3b caused activation of p38β2 (Figure 1C). Together, these data identify a role for the N–terminal region of MKK3 and MKK6 in the determination of substrate specificity.
Fig. 1. Identification of a domain required for activation of p38β2 by MAP kinase kinases. (A) Schematic representation of MKK3, MKK3b, MKK6 and chimeras. MKK6 is shown in white, MKK3 in black and the N–terminal extension of MKK3b is gray. In the chimeras, domains from MKK3 are shown in black and domains from MKK6 in white. (B and C) Epitope-tagged p38α (B) or p38β2 (C) were immunoprecipitated from COS7 cells co-transfected with an empty vector (Control) or activated MAPKK. The activated MAPKKs were constructed by replacing the two sites of activating phosphorylation with glutamic acid residues. Immune complex kinase assays were performed to measure p38 MAP kinase activity using ATF2 as the substrate. The expression of MAPKK and p38 was examined by immunoblot analysis (lower panel). The rate of ATF2 phosphorylation was quantitated by PhosphorImager analysis and is presented as relative protein kinase activity.
The N–terminal region of MAPKK is required for binding to p38α and p38β2 MAPK
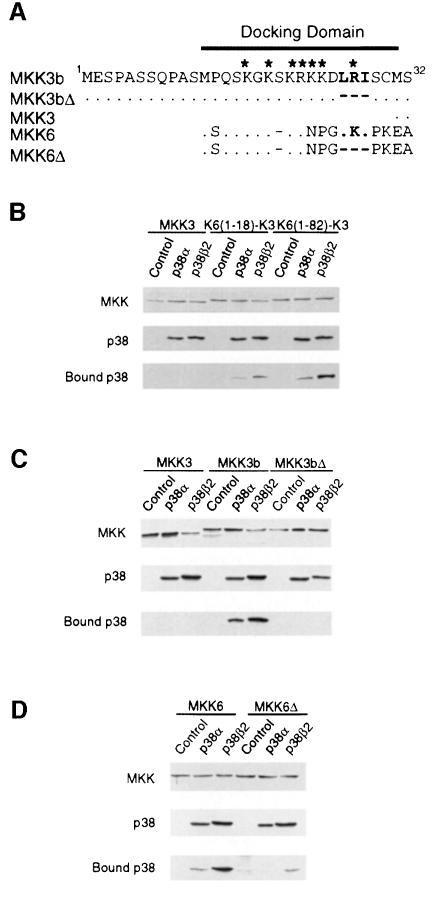
The analysis of MAPKK chimeras indicated that the first 18 amino acids of MKK6 contain a region that can confer on MKK3 the ability to activate p38β2 (Figure 1C). Alignment of the sequences of MKK3, MKK3b and MKK6 indicates that this 18 amino acid sequence present in the N–terminal region of MKK6 is absent in MKK3, but is conserved in MKK3b (Figure 2A). This region contains several basic amino acids and also a sequence motif that has previously been identified as a MAPK docking site (Holland and Cooper, 1999). The N–terminal region of MKK3b and MKK6 conforms to the consensus sequence for this type of MAPK docking site (-Lys/Arg-Xaa3-Leu/Ile-Xaa-Leu/Ile-). The N–terminal specificity-determining region of MKK3b and MKK6 may therefore function as a p38 docking site. To test this hypothesis, we examined the binding of p38α and p38β2 to MKK3 and MKK6 in co-precipitation assays using extracts prepared from transfected COS7 cells (Figure 2). We found that p38α and p38β2 co-precipitated with MKK6 and MKK3b, but not with MKK3. Interestingly, more p38β2 than p38α was observed to co-precipitate with MKK3b and MKK6. To test whether the putative docking site located in the N–terminal region of MKK6 and MKK3b was required for binding to p38, we examined the effect of mutations of the conserved Leu-Xaa-Ile motif on the interaction with p38. Mutational removal of the Leu-Xaa-Ile motif (MKK3Δ and MKK6Δ) reduced the co-precipitation of p38α and p38β2 with MKK6 and MKK3b. Furthermore, fusion of the N–terminal region of MKK6 (containing the putative MAPK docking site to MKK3) allowed co-precipitation of MKK3 with p38α and p38β2. Fusion of MKK6 residues 1–18 was sufficient for co-precipitation of p38α and p38β2, but a larger amount of co-precipitation was detected when MKK6 residues 1–82 were fused to MKK3. These data indicate that a p38 docking site is present in the N–terminal region of MKK3b and MKK6, but not MKK3.
Fig. 2. Binding of p38α and p38β2 to MAP kinase kinases. (A) Primary sequences of the N–terminal domain of MKK3, MKK3b, MKK6 and deletion mutants (Δ) are aligned. Residues that are identical to MKK3b are indicated with a dot (.). The residues of MKK3b (LRI) and MKK6 (LKI) deleted in MKK3bΔ and MKK6Δ are indicated in bold. The deleted residues are indicated with a dash (–). Basic residues are indicated by asterisks. (B) Activated GST-tagged MKK3, K6(1–18)–K3 or K6(1–82)–K3 were co-transfected with an empty vector (Control), Flag-tagged p38α or Flag-tagged p38β2 in COS7 cells. The activated MKKs were constructed by replacing the two sites of activating phosphorylation with glutamic acid residues. Protein expression was monitored by immunoblot analysis of cell extracts. The GST–MKK fusion proteins were isolated from the cell extracts by incubation with glutathione–Sepharose. The co-precipitation of p38α and p38β2 with the MKK was examined by immunoblot analysis with an antibody to the Flag epitope. (C and D) The interaction of Flag-tagged p38α and p38β2 with GST-tagged activated MKK3, MKK3b and MKK3bΔ (C) or MKK6 and MKK6Δ (D) co-expressed in COS7 cells was examined using the methods described in (B). The activated MKKs were constructed by replacing the two sites of activating phosphorylation with glutamic acid residues.
Binding to MKK3b and MKK6 potentiates p38 MAPK activation
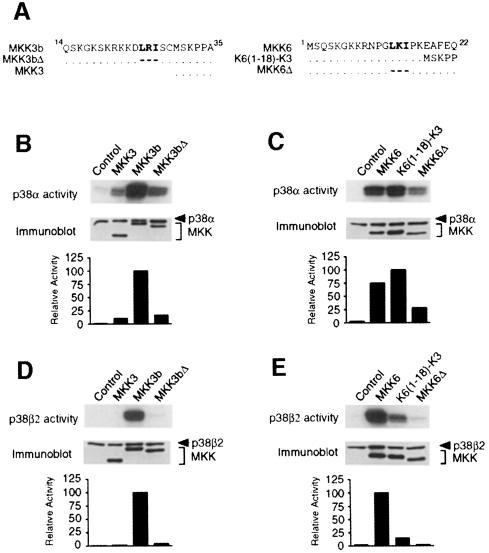
There is a strong correlation between MAPKK binding to p38 MAPK and the activation of p38α and p38β2. For example, MKK3b and MKK6 (but not MKK3) bind to and activate p38β2. Similarly, p38α (which is activated less potently by MKK3 than by MKK3b and MKK6) binds to MKK3b and MKK6, but not to MKK3. To examine the relationship between MAPKK binding and p38 activation, we examined the effect of mutations that inhibit MAPKK binding on p38 activation (Figure 3). Mutational removal of the MAPK docking site (Leu-Xaa-Ile) reduced the binding of MKK3b and MKK6 to p38α and p38β2 (Figure 2). The same mutation also reduced p38α activation caused by MKK3b and MKK6 (Figure 3B and C). Furthermore, like MKK3, these mutant MKK3b and MKK6 proteins were unable to activate p38β2 (Figure 3D and E). These data indicate that binding to MKK3b and MKK6 potentiates p38α activation and is required for p38β2 activation.
Fig. 3. Regulation of p38α and p38β2 by MAP kinase kinases. (A) The primary sequences of the N–terminal region of MKK3, MKK3b and MKK3bΔ (left panel) and of MKK6, K6(1–18)–K3 and MKK6Δ (right panel) are aligned. (B–E) Flag-tagged p38α (B and C) or p38β2 (D and E) were co-transfected together with activated MKK3, MKK3b and MKK3bΔ (B and D) or activated MKK6, K6(1–18)–K3 and MKK6Δ (C and E) in COS7 cells. The activated MKKs were constructed by replacing the two sites of activating phosphorylation with glutamic acid residues. The expression of p38 and MKK was examined by immunoblot analysis of cell lysates. The protein kinase activity of p38α and p38β2 was measured in immune complex kinase assays using ATF2 as the substrate. The phosphorylated ATF2 was detected after SDS–PAGE by auto- radiography and was quantitated by PhosphorImager analysis. The p38 activity is presented as relative protein kinase activity.
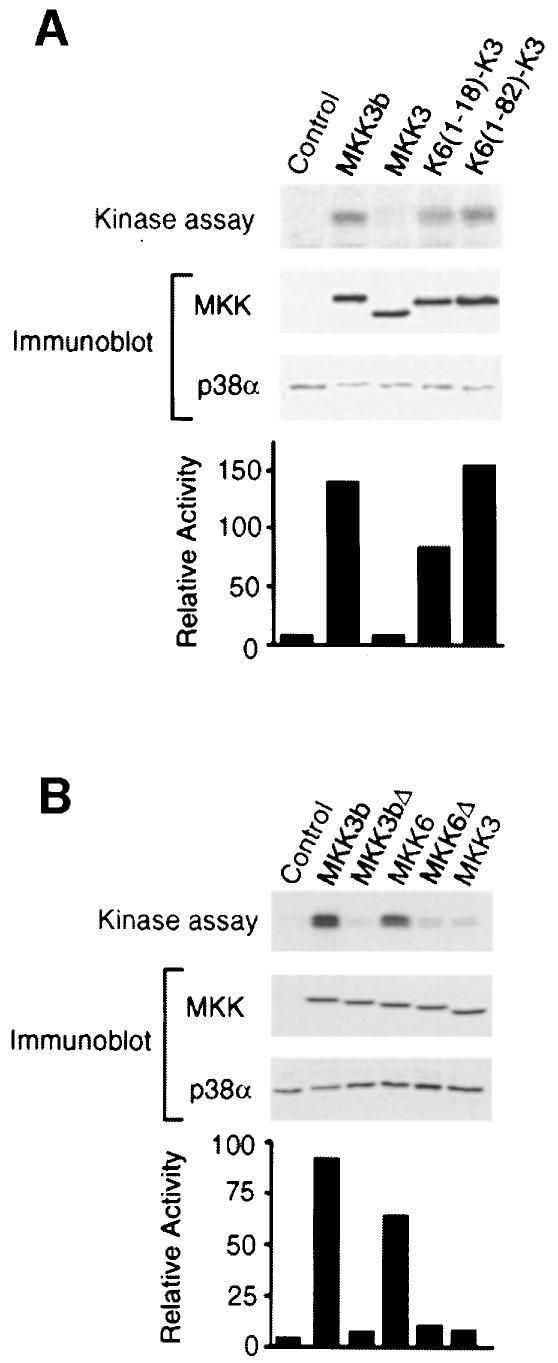
To confirm the conclusion that MAPKK binding increases p38 activation, we tested activated MKK3 and MKK6 on the activation of endogenous p38α in COS7 cells. We have reported previously that MKK6 (but not MKK3) potently activates endogenous p38α (Raingeaud et al., 1996). Here, we demonstrate that MKK3b, like MKK6, strongly activates endogenous p38α (Figure 4). Disruption of the MAPK docking site located in the N–terminal region of MKK3b and MKK6 reduced the activation of endogenous p38α. Furthermore, fusion of the N–terminal region of MKK6 (containing the putative MAPK docking site to MKK3) allowed MKK3 to activate endogenous p38α. These data suggest that the binding of MKK3b and MKK6 to p38 is a determinant of p38α activation in vivo. This requirement for a binding interaction can be compensated by overexpression of p38α, but not by overexpression of p38β2 (Figure 1).
Fig. 4. Regulation of endogenous p38α activity by MAP kinase kinases. Activated epitope-tagged MKK3b, MKK3, K6(1–18)–K3 and K6(1–82)–K3 (A) or MKK3b, MKK3bΔ, MKK6, MKK6Δ and MKK3 (B) were expressed in COS7 cells. The activated MKKs were constructed by replacing the two sites of activating phosphorylation with glutamic acid residues. The epitope-tagged MKK expression and endogenous p38α were examined by immunoblot analysis of cell lysates. The activity of the endogenous p38α was measured in immune complex kinase assays using ATF2 as the substrate. The phosphorylated ATF2 was detected after SDS–PAGE by auto- radiography and was quantitated by PhosphorImager analysis. The p38 activity is presented as relative protein kinase activity.
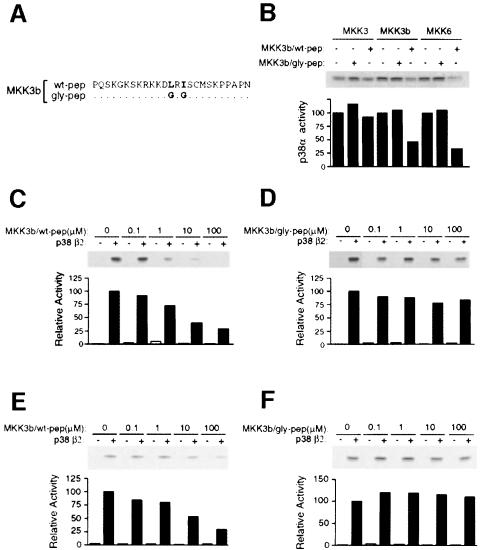
The N–terminal MAPK docking site is likely to contribute to p38 activation. To test this hypothesis, we performed competition analyses using synthetic peptides corresponding to the MAPK docking sites of MKK3b (Figure 5) and MKK6 (data not shown). The effect of wild-type peptides was compared with the effect of peptides in which the Leu-Xaa-Ile motif was replaced with Gly-Xaa-Gly (Figure 5A). If binding to the N–terminal docking site was required for p38 activation, we reasoned that a synthetic peptide containing this docking site should function as an inhibitor of p38 activation. Indeed, peptides derived from MKK3b (Figure 5B) and MKK6 (data not shown) caused inhibition of p38α activation by MKK3b and MKK6, but not by MKK3, which lacks the MAPK docking site (Figure 5B). In contrast, mutant synthetic peptides lacking the MAPK docking site caused no change in p38α activation. Inhibition of p38β2 activation by wild-type synthetic peptides, but not by mutated synthetic peptides, was detected in experiments using MKK3b (Figure 5C and D) and MKK6 (Figure 5E and F). However, the inhibitory effect of the peptides on p38β2 activation was larger than on p38α, confirming the previous observation (Figure 3) that binding is required for the activation of p38β2 in vitro, but serves only to potentiate the activation of p38α.
Fig. 5. Inhibition of p38β2 activity by peptide competition. (A) The primary sequence of synthetic peptides corresponding to the native (wt-pep) or mutated (gly-pep) N–terminal region of MKK3b is shown. The mutated peptide was prepared by replacing the residues of MKK3b (LRI), indicated in bold, with glycine. (B) Purified bacterially expressed GST–p38α was bound to glutathione–Sepharose and incubated with 100 μM wild-type or mutated MKK3b peptide. Purified activated MKK3, MKK3b or MKK6 were incubated with the immobilized GST–p38α in kinase buffer with ATP for 20 min. The GST–p38α was washed with kinase buffer and the p38α activity was measured using ATF2 and [γ-32P]ATP as the substrates. The phosphorylated ATF2 was detected after SDS–PAGE by auto- radiography and was quantitated by PhosphorImager analysis. The p38α activity is presented as relative protein kinase activity. (C–F) Purified bacterially expressed GST–p38β2 was bound to glutathione–Sepharose and incubated with increasing concentrations of the wild-type (C and E) or mutated (D and F) MKK3b peptide. Purified activated MKK3b (C and D) or MKK6 (E and F) were incubated with the immobilized GST–p38β2 in kinase buffer with ATP for 20 min. The GST–p38β2 activity was measured as described in (B).
Taken together, these data indicate that binding of p38 to a docking site in the N–terminal region of MAPKK contributes to p38 activation.
Docking of p38 MAPK to MAPKK selectively regulates phosphorylation on threonine and tyrosine
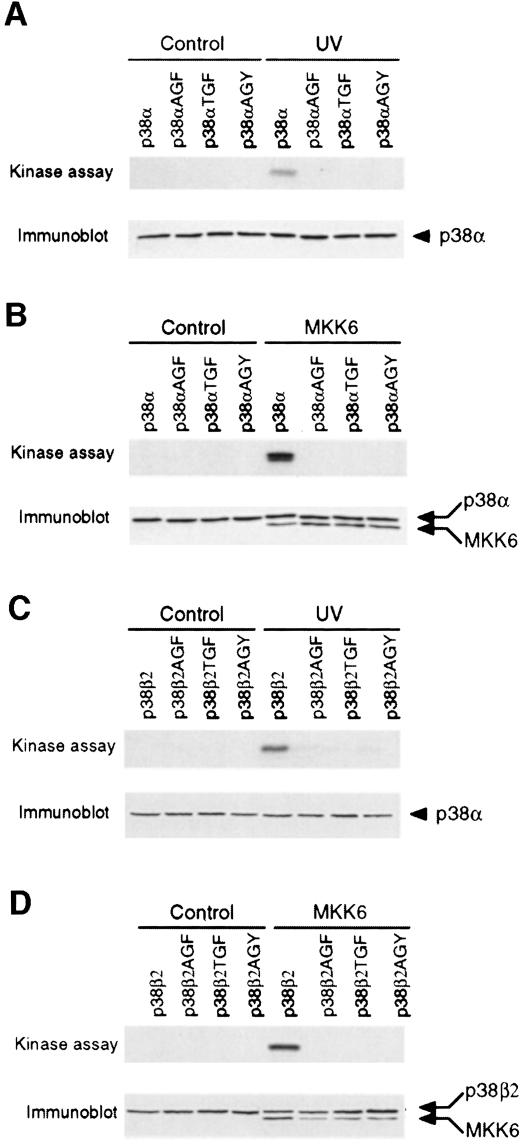
Activation of p38 is associated with increased threonine and tyrosine phosphorylation of the Thr-Gly-Tyr motif within the T–loop (Raingeaud et al., 1995). Simultaneous replacement of the Thr-Gly-Tyr dual phosphorylation motif by Ala-Gly-Phe blocks stress-induced p38 phosphorylation and activation (Raingeaud et al., 1995). To test whether phosphorylation of both threonine and tyrosine is required for p38α and p38β2 activation, we examined the effect of single point mutations (Figure 6). Each of these point mutations blocked p38 activation, indicating that dual phosphorylation on threonine and tyrosine was required for activation.
Fig. 6. Dual phosphorylation on threonine and tyrosine is required for p38 MAPK activation. Epitope-tagged wild-type and mutated p38α (A and B) or p38β2 (C and D) were expressed in COS7 cells. The mutated p38 contained point mutations in the Thr-Gly-Tyr (TGY) dual phosphorylation motif (replacement of threonine and tyrosine with alanine and phenylalanine, respectively). The effect of treatment with 80 J/m2 UV (A and C) or co-expression with activated MKK6 (B and D) was examined. Immune complex kinase assays were performed to measure p38 activity using ATF2 as the substrate. The expression of p38 and activated MKK6 was examined by immunoblot analysis (lower panel).
The observation that dual phosphorylation was required for p38 activation (Figure 6) has implications for the mechanism of p38 activation and the role of a docking interaction between p38 and upstream MAPKK. The requirement for a docking interaction for efficient p38 activation might reflect changes in overall phosphorylation on threonine and tyrosine. Alternatively, the docking interaction may selectively alter threonine or tyrosine phosphorylation. For example, the docking interaction might increase the processivity of the dual phosphorylation on threonine and tyrosine. Since dual phosphorylation is required for p38 activation, this mechanism could account for the effect of docking interactions on p38 activation. To test this hypothesis, we performed phosphoamino acid analysis of p38 phosphorylated by MKK3, MKK3b, MKK6 and chimeric protein kinases.
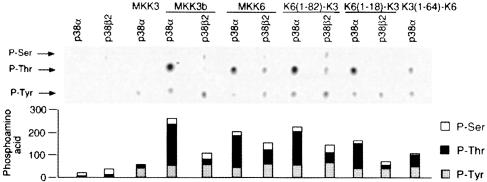
We found that p38α was phosphorylated preferentially on tyrosine by MKK3, which lacks the MAPK docking site (Figure 7). In contrast, increased p38α phosphorylation on threonine was observed in experiments using MAPKK with an N–terminal MAPK docking site, including MKK3b, K6(1–82)–K3 and K6(1–18)–K3 (Figure 7). These data suggest that the docking of MAPKK to p38α contributes to increased dual phosphorylation on threonine and tyrosine and therefore to p38α activation (Figure 6). However, K3(1–64)–K6 (which lacks a docking site) caused dual phosphorylation of p38α on threonine and tyrosine. Thus, MKK3 requires a docking domain for efficient dual phosphorylation of p38α, but MKK6 is able to phosphorylate p38α on both threonine and tyrosine in the absence of a docking domain. In contrast, p38β2 was not phosphorylated by MAPKK that lacked the MAPK docking site [e.g. MKK3 and K3(1–64)–K6], but was phosphorylated on threonine and tyrosine by all MAPKKs tested with a docking site [e.g. MKK3b, MKK6, K6(1–18)–K3 and K6(1–82)–K3].
Fig. 7. Phosphoamino acid analysis of p38α and p38β2 phosphorylated by MAP kinase kinases. Activated epitope-tagged MKKs (described in Figure 1A) were expressed in COS7 cells and isolated by immunoprecipitation. Immune complex kinase assays were performed using [γ-32P]ATP and purified bacterially expressed GST–p38α or GST–p38β2 as the substrates. The phosphorylated p38 was examined by phosphoamino acid analysis. The figure shows an autoradiograph of the phosphoamino acids separated by thin layer electrophoresis. The relative phosphorylation on serine, threonine and tyrosine was examined by PhosphorImager analysis.
These data demonstrate that the role of docking interactions differs between p38α and p38β2. Docking increases dual phosphorylation of p38α. In contrast, docking is required for p38β2 phosphorylation. Thus, p38α and p38β2 differ in their functional interactions with MAPKK.
The T–loop of p38α can direct activation by MKK3
Sequences within the N–terminal region of MAPKK appear to be important for the binding and activation of p38 (Figure 3). However, little is known concerning the p38 sequences that are required for activation. Differences in the sequences of p38 may contribute to the specificity of MAPKK-mediated activation of specific p38 isoforms. An example of isoform-specific p38 activation is provided by the observation that while p38α binds to and is activated similarly by MKK3b and MKK6, p38β2 is activated more potently by MKK6, although its binding to MKK3b and MKK6 is similar (Figures 1–3 ). These data indicate that factors other than binding can contribute to the activation of p38 isoforms. This conclusion was strongly confirmed by the observation that overexpressed MKK3 and MKK3bΔ, which do not bind p38α or p38β2 (Figure 2), caused activation of co-transfected p38α, but not p38β2 (Figures 1 and 3). Together, these data indicate that sequences within the structure of p38 contribute to the selectivity of the activation of different p38 isoforms.
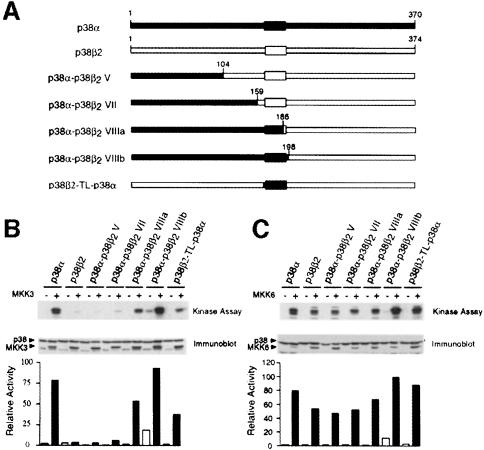
To test whether specific regions of p38α and p38β2 are important for selective activation, we created chimeric p38α/β2 molecules (Figure 8A) and examined the effect of activated MKK3 and MKK6 in co-transfection assays. We found that MKK6 activated the p38α, p38β2 and chimeric p38α/β2 similarly (Figure 8C). In contrast, marked differences were observed in experiments using MKK3 (Figure 8B). MKK3 activated p38α, but not p38β2. Examination of chimeric p38α/β2 indicated that molecules with an N–terminal region derived from p38α were activated by MKK3 if the fusion was created within subdomain VIII, but not if the fusion was created in subdomain V or VII (Figure 8C). These data suggested that the region located between subdomains VII and VIII of p38α is required for activation by MKK3. This region includes the Thr-Gly-Tyr motif located in the T–loop that is phosphorylated by MKK3 and MKK6. To test whether the T–loop of p38α can confer on p38β2 the ability to be activated by MKK3, we constructed a p38β2 molecule with the sequence of the T–loop of p38α (p38β2-TL-p38α). This chimeric p38α/β2 was activated by MKK3 (Figure 8B). Reciprocal studies of p38α with the T–loop of p38β2 were also performed. In contrast to p38α, p38α-TL-p38β2 was not activated by MKK3 and was poorly activated by MKK6 (data not shown). Together, these data indicate that the T–loop of p38 contributes to the specificity of activation of p38 isoforms.
Fig. 8. Regulation of chimeric p38 MAPK by MKK3 and MKK6. (A) Schematic representation of p38α, p38β2 and chimeric protein kinases. Regions of p38α are shown in black and regions of p38β2 in white. The p38β2-TL-p38α was constructed by replacing residues within the T–loop of p38β2 with those derived from p38α. (B and C) Epitope-tagged p38α, p38β2 or p38 chimeras were expressed in COS7 cells together with an empty vector (–) and activated (+) MKK3 (B) or MKK6 (C) The expression of MKK and p38 was examined by immunoblot analysis. The p38 activity was measured in immune complex kinase assays with ATF2 as the substrate. The rate of phosphorylation was quantitated by PhosphorImager analysis and is presented as relative p38 activity.
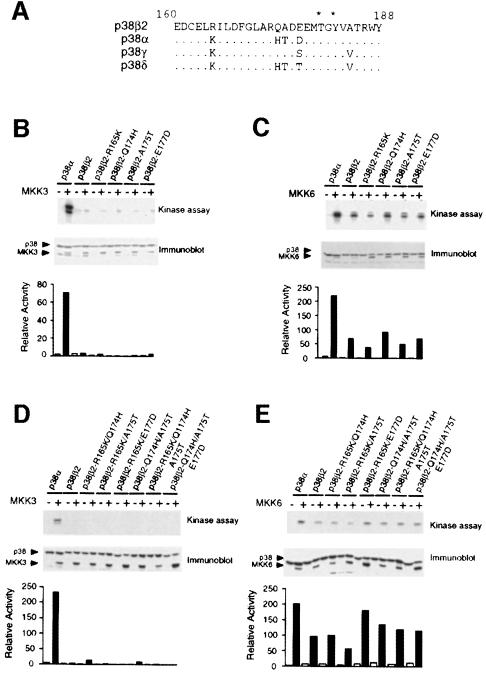
Inspection of the primary sequence of the T–loop of p38 isoforms indicates that this region is highly conserved within the p38 group (Figure 9A). Only four amino acids are different in this domain between p38α and p38β2. To test which of these amino acid substitutions is relevant to activation by MKK3, we examined the effect of point mutations at these four residues by substituting the corresponding amino acid found in p38α. Each of these mutated p38β2s was activated by MKK6 (Figure 9C), but none were activated by MKK3 (Figure 9B). We therefore constructed all 10 possible double and triple mutants of p38β2. The results obtained for six mutants are presented in Figure 9D and E, and similar data were obtained for the remaining four possible combinations of double and triple point mutants (data not shown). Each of these mutated p38β2s was activated by MKK6, but not by MKK3.
Fig. 9. Effect of p38α T–loop mutations on p38β2 activation. (A) Comparison of the sequences of p38α, p38β2, p38γ and p38δ within the dual phosphorylation domain (T–loop). Residues in p38 isoforms that are identical to p38β2 are indicated with a dot (.). The sites of activating phosphorylation (threonine and tyrosine) are indicated with asterisks. (B–E) Flag-tagged p38α, p38β2 or T–loop mutants of p38β2 were expressed in COS7 cells and p38 activity was measured in immune complex kinase assays using ATF2 as the substrate. The effect of co-expression together with an empty vector (–) and activated (+) MKK3 (B and D) or activated MKK6 (C and E) was examined. The expression of MKK3, MKK6 and p38 was monitored by immunoblot analysis. The rate of phosphorylation was quantitated by PhosphorImager analysis and is presented as relative p38 activity.
Together, these data indicated a role for the T–loop in the recognition and activation of p38α by MKK3. The four amino acid differences in the primary sequence of the T–loops of p38α and p38β2 account for the observation that MKK3 activates p38α, but not p38β2. Replacement of these four residues in the T–loop of p38β2 with the corresponding residues of p38α allows the activation of p38β2 by MKK3 (Figure 8). All four residues in the T–loop contribute to the activation specificity of p38α and p38β2.
The T–loop of p38γ and p38δ can direct activation of p38β2 by MKK3
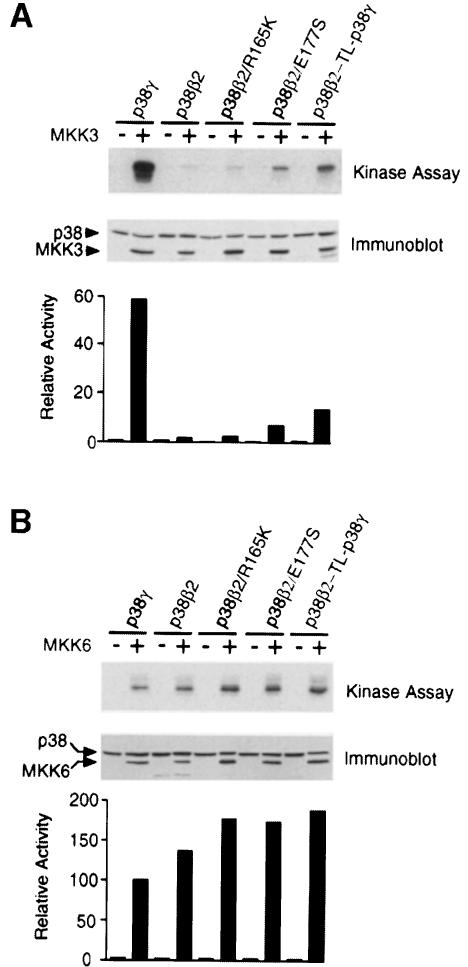
In addition to p38α, both p38γ and p38δ are activated by MKK3 (Enslen et al., 1998). This analysis suggested that the T–loops of p38α, p38γ and p38δ may allow activation by MKK3 and that the T–loop of p38β2 is distinct because it prevents activation by MKK3. Comparison of the primary sequence of the T–loops of p38γ and p38δ with that of p38β2 indicated three amino acid differences for p38γ and five for p38δ (Figure 9). To test the role of these amino acid differences, we examined the effect of mutations within the T–loop of p38β2.
Examination of the sequence of p38γ indicated that Arg165, Glu177 and Ala184 of p38β2 are replaced with lysine, serine and valine, respectively. To test the role of these amino acid differences in activation by MKK3, we examined the effect of point mutations. We focused our analysis on Arg165 and Glu177, which are located within the T–loop of p38β2. Control experiments demonstrated that MKK3 activated p38γ but not p38β2, while MKK6 caused similar activation of both p38γ and p38β2 (Figure 10). Simultaneous mutation of both Arg165 and Glu177 (p38β2-TL-p38γ) caused increased activation of p38β2 by MKK3, while a smaller increase was detected for a single point mutation at Glu177. These data suggest that the T–loop of p38γ contributes to the specificity of activation by MKK3.
Fig. 10. Effect of p38γ T–loop mutations on p38β2 activation. Flag-tagged p38γ, p38β2 or T–loop mutants of p38β2 were expressed in COS7 cells and p38 activity was measured in immune complex kinase assays using ATF2 as the substrate. The effect of co-expression together with an empty vector (–) and activated (+) MKK3 (A) or activated MKK6 (B) was examined. The expression of MKK3, MKK6 and p38 was monitored by immunoblot analysis. The rate of phosphorylation was quantitated by PhosphorImager analysis and is presented as relative p38 activity.
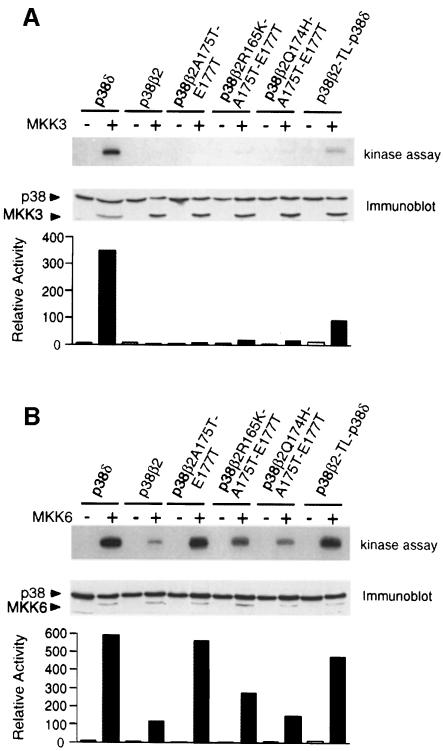
We performed a similar analysis of p38δ. Five residues differ between the T–loop of p38δ and p38β2. Four of these residues are also replaced in p38α (Figure 9A). MKK3 activated p38δ but not p38β2, and MKK6 activated both p38δ and p38β2 (Figure 11). Simultaneous replacement of the four residues in the T–loop of p38β2 (Arg165, Gln174, Ala175 and Glu177) (p38β2-TL-p38δ) increased the activation by MKK3 without altering activation by MKK6 (Figure 11). Single point mutations at these residues and Ala184 caused no change in p38β2 activation by MKK3. These data suggest that the T–loop of p38δ contributes to the specificity of activation by MKK3.
Fig. 11. Effect of p38δ T–loop mutations on p38β2 activation. Flag-tagged p38δ, p38β2 or T–loop mutants of p38β2 were expressed in COS7 cells, and p38 activity was measured in immune complex kinase assays using ATF2 as the substrate. The effect of co-expression together with an empty vector (–) and activated (+) MKK3 (A) or activated MKK6 (B) was examined. The expression of MKK3, MKK6 and p38 was monitored by immunoblot analysis. The rate of phosphorylation was quantitated by PhosphorImager analysis and is presented as relative p38 activity.
Discussion
MAPKs regulate a wide array of biological functions; therefore, mechanisms must exist to achieve signaling specificity and to ensure the correct biological response to extracellular stimulation. The complexity of each MAPK pathway provides multiple levels where specificity may be determined. Two simple mechanisms that can achieve signaling specificity are: (i) selective activation of different MAPKs; and (ii) distinct MAPK substrate specificities. Such differences in signaling specificity exist not only between the major groups of MAPK, but also between individual members of a single group of MAPKs.
The substrate specificity of MAPK depends, in part, on binding interactions between the MAPK and its substrates (Ip and Davis, 1998; Holland and Cooper, 1999). A docking interaction between MAPK and MAPKK has also been reported to contribute to the substrate specificity of MAPKK in yeast (Bardwell and Thorner, 1996). Here we report that a p38 docking domain in p38-specific MAPKK contributes to the selective activation of p38 isoforms. For example, p38β2 is not activated by MAPKK isoforms that lack a docking site (e.g. MKK3), but can be activated by MAPKK with a docking site (e.g. MKK6). We also demonstrate that differences in the primary sequence of the T–loop of the p38 isoforms contribute to signaling specificity. Together, these data indicate that the selective activation of p38 by MAPKK requires multiple molecular determinants present in both enzymes.
MAPK docking domains
Two genes encode proteins that act as specific activators of p38: Mkk3 and Mkk6 (Derijard et al., 1995; Han et al., 1996; Moriguchi et al., 1996a; Raingeaud et al., 1996; Stein et al., 1996). Targeted gene disruption studies in mice have demonstrated non-redundant functions of the Mkk3 and Mkk6 genes (Lu et al., 1999; Wysk et al., 1999), indicating that the MKK3 and MKK6 protein kinases have distinct biological functions.
MKK3 activates the isoforms p38α, p38γ and p38δ, but not p38β2. However, both MKK6 and MKK3b (a variant form of MKK3, which has an additional 29 amino acids fused to the N–terminus of MKK3) activate the four p38 isoforms p38α, p38β2, p38γ and p38δ. In this study, we have identified a p38 kinase docking site within the N–terminal region of MKK3b and MKK6. These sequences are highly conserved and are required for p38β2 activation by these two enzymes. MKK3, which lacks the docking site, does not activate p38β2. Fusion of the p38 docking site of MKK6 to the N–terminus of MKK3 allows activation of p38β2. Furthermore, synthetic peptides based on the primary sequence of the docking sites of MKK3b and MKK6 inhibit the activation of p38β2. These data indicate that the binding of p38β2 to an N–terminal region of the MAPKK is necessary for p38β2 activation. In contrast, binding to p38α is not a requirement for activation by MKK3b and MKK6 in vitro. However, the binding interaction does serve to potentiate p38α activation. These data provide an explanation for the selective activation of p38α, but not p38β2, by MKK3.
A similar role for a docking mechanism has been described in the yeast Saccharomyces cerevisiae where a high affinity interaction between a MAPKK (Ste7p) and MAPK (Kss1p and Fus3p) is required for MAPK activation (Bardwell et al., 1996). This interaction depends on a MAPK docking site present in the N–terminus of Ste7p. The sequence of the MAPK docking site of Ste7p is related to that identified in MKK3b and MKK6. These sequences are similar to the previously reported MAPK docking site consensus sequence found in MAPK substrates: -Arg/Lys-Xaa3-Leu/Ile-Xaa-Leu/Ile- (Yang et al., 1998a,b; Holland and Cooper, 1999). Interestingly, the N–terminal regions of MEK1 (32 amino acids), MKK4 (43 amino acids) and MKK7 (73 amino acids) have been demonstrated to be important for binding and activation of ERK and JNK (Fukuda et al., 1997; Xia et al., 1998; Tournier et al., 1999; Xu et al., 1999). Sequences similar to the MAPK docking sites of MKK3b and MKK6 are present in the N–terminal regions of MEK1, MKK4 and MKK7. Further studies are required to characterize the MAPK docking site with the N–terminus of MEK1, MKK4 and MKK7. In contrast, this study has established the presence of p38 docking sites on MKK3b and MKK6 by mutational analysis and peptide competition analysis.
The MAPK docking sites present in MAPKK appear to be targeted during infection by some pathogens. For example, the lethal factor (LF) of anthrax lethal toxin (the major cause of death in animals infected with anthrax) inhibits the ERK signal transduction pathway (Duesbery et al., 1998). LF is a protease that cleaves the N–terminus of MEK1 and MEK2, causing decreased activity towards ERK (Duesbery et al., 1998). The N–terminal cleavage of MEK1 and MEK2 destroys the putative ERK docking site. Loss of the ERK docking site may account for the ability of LF to block activation of ERK by MEK1 and MEK2 in vivo (Duesbery et al., 1998). Interestingly, sequences similar to the LF cleavage site in MEK1 and MEK2 are present in the N–terminus of MKK3b and MKK6. Cleavage of MKK3b and MKK6 by LF at these sites would remove the p38 docking domain and would therefore be predicted to prevent binding and activation of p38β2 and markedly decrease activation of p38α. Indeed, LF recently was reported to inhibit p38 signaling in macrophages (Pellizzari et al., 1999).
MAPK docking sites are present in MAPKK, MAPK substrates and MAPK phosphatases
Short peptide sequences that bind p38α and p38β2 have been identified in the transcription factors MEF2A and MEF2C (Yang et al., 1999). These domains are necessary for efficient phosphorylation and activation of MEF2A and MEF2C by p38α and p38β2 (Yang et al., 1999). Similar MAPK-binding motifs have been characterized in several MAPK substrates, including c-Jun (Hibi et al., 1993), JunB (Gupta et al., 1996; Kallunki et al., 1996), Elk-1 (Yang et al., 1998a,b), NFAT4 (Chow et al., 1997) and ATF-2 (Gupta et al., 1995; Livingstone et al., 1995). In general, these docking sites conform to the general consensus sequence -Arg/Lys-Xaa-Xaa-Xaa-Xaa-Leu/Ile-Xaa-Leu/Ile- (Yang et al., 1998a,b; Holland and Cooper, 1999). However, three additional classes of docking sites have been identified. These docking sites correspond to the Phe-Xaa-Phe-Pro motif identified in some Ets transcription factors that binds ERK (Jacobs et al., 1999), the -Leu-Ala-Gln-Arg-Arg-Xaa3-Leu/Ile- motif in Mnk2 and p90rsk that binds ERK (Gavin and Nebreda, 1999; Smith et al., 1999) and the Leu-hydrophobic-Lys/Arg-Lys/Arg-Lys/Arg-Lys/Arg- motif in PRAK and MAPKAP-K2 that binds p38 (Smith et al., 1999). Binding sites for MAPK have also been identified in phosphatases that regulate MAPK activation (Camps et al., 1998; Pulido et al., 1998; Oh-hora et al., 1999). These considerations indicate that MAPK docking sites are located on the kinases that activate MAPK (MAPKK), on phosphatases that inactivate MAPK and on MAPK substrates.
The observation that MAPKs can bind related sequences in both their substrates and enzymes that regulate MAPK activity (MAPKK and phosphatases) is intriguing. Is it possible that the regulatory enzymes and substrates compete for binding to MAPK? This hypothesis predicts that substrate phosphorylation by MAPK would only be observed following release of the activated MAPK from MAPKK. Indeed, evidence in favor of this hypothesis has been reported in studies of yeast MAPK signaling pathways. For example, the S.cerevisiae MAPK Kss1p is not able to phosphorylate exogenous substrates when bound to the MAPKK Kss1p (Bardwell et al., 1996). Similarly, the Schizosaccharomyces pombe MAPKK Pek1p binds to and inhibits the MAPK Pmk1p, but releases activated Pmk1p following stimulation (Sugiura et al., 1999). Similar models for binding and release of activated MAPK in mammals have been reported for JNK activation by MKK4 (Xia et al., 1998) and ERK activation by MEK1 (Fukuda et al., 1997).
In this study, we have identified protein regions that determine the specificity of p38 MAPK activation by MAPKK. Additional studies are required to identify the relationship of p38 MAPK docking to MAPKK with the interaction of MAPK with phosphatases and substrates. Differences in the interaction of substrates and phosphatases with individual p38 MAPK isoforms may contribute to the specificity of p38 MAPK signaling in vivo. For example, the C–terminus of p38γ functions as a docking site that interacts with PDZ domain-containing proteins (Hasegawa et al., 1999).
Docking interactions increase the dual phosphorylation of p38 MAPK on threonine and tyrosine
MAPKs are activated by dual phosphorylation on threonine and tyrosine. Analysis of p38 MAPK phosphorylation by MKK3 and MKK6 provides evidence for selective phosphorylation on these activating sites. The p38α MAPK was phosphorylated preferentially on tyrosine by MAPKK that lacked a MAPK docking site (e.g. MKK3). MAPKK with a MAPK docking site (MKK3b and MKK6) caused similar phosphorylation of p38α MAPK on both threonine and tyrosine. These data suggest that the docking interaction may increase the processivity of MAPK phosphorylation, leading to increased dual phosphorylation and, consequently, increased activation. The requirement for dual phosphorylation for MAPK activation is likely to account, in part, for the role of docking interactions between MAPK and MAPKK.
Previous studies of the JNK signaling pathway provide another example of selective phosphorylation of MAPK on threonine or tyrosine (Lawler et al., 1998). MKK4 was found to phosphorylate JNK1 preferentially on tyrosine, while MKK7 preferentially phosphorylated JNK1 on threonine. The mechanism that accounts for this selectivity has not been established. However, the different specificities of MKK4 and MKK7 in vitro suggest that these MAPKKs may collaborate to activate JNK1 in vivo (Lawler et al., 1998).
The T–loop of p38 MAPK is a specificity determinant for activation by MKK3, but not MKK6
MKK3 activates p38α MAPK, but not p38β2 MAPK. Since MKK3 does not bind to p38α or p38β2, the selectivity of MKK3 in activating p38α MAPK is not caused by a difference in docking interactions of MKK3 with p38α and p38β2. Instead, these data suggest that molecular determinants within p38α (that are absent in p38β2) contribute to recognition and activation by MKK3. Studies of chimeric p38α/β2 MAPK indicated that the T–loop was a critical element that determined the extent of activation by MKK3. In contrast, these changes in the T–loop did not alter activation by MKK6. Since the T–loop contains the sites of threonine and tyrosine phosphorylation by MKK3, the finding that the T–loop contributes to the specificity of substrate phosphorylation by MKK3 is intriguing because it suggests that it might play a direct role in substrate recognition by the active site of MKK3. Previous studies have not indicated that the sequence of the T–loop plays an important role in determining the specificity of MAPK activation by MAPKK. Although an important role for the T–loop in substrate recognition by MKK3 is established, our data do not exclude a role for other regions of the p38 MAPK in determining specificity (Brunet and Pouyssegur, 1996; Wilsbacher et al., 1999).
Materials and methods
Plasmids
The p38, MKK3 and MKK6 expression vectors have been described (Enslen et al., 1998). Point mutations and chimeric constructs were prepared using standard techniques.
Protein kinase isolation
Cells were solubilized in Tris lysis buffer (TLB) [20 mM Tris pH 7.5, 10% glycerol, 1% Triton X-100, 0.137 M NaCl, 25 mM β-glycerophosphate, 2 mM EDTA, 0.5 mM dithiothreitol (DTT), 1 mM sodium orthovanadate, 2 mM NaPPi, 10 μg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride (PMSF)] and centrifuged at 15 000 g for 15 min at 4°C. Epitope-tagged protein kinases were immunoprecipitated by incubation (4 h) with the M2 Flag monoclonal antibody (Sigma) bound to protein G–Sepharose (Amersham Pharmacia Biotech). The Sepharose beads were collected by centrifugation, washed twice with TLB and twice with kinase assay buffer (25 mM HEPES pH 7.4, 25 mM β-glycerophosphate, 25 mM MgCl2, 0.5 mM DTT, 0.1 mM sodium orthovanadate). The immunoprecipitation of endogenous p38α was performed similarly except that an anti-p38 rabbit polyclonal antibody bound to protein A–Sepharose (Sigma Chemical Co.) was employed (Raingeaud et al., 1995). In some assays, the immunoprecipitated epitope-tagged protein kinases were eluted by incubation with 0.1 mg/ml Flag synthetic peptide at 30°C (20 min).
Protein kinase assays
Protein kinase immunoprecipitates were used for kinase assays. The reactions were initiated by the addition of substrate protein (1 μg) and 50 μM [γ-32P]ATP (10 Ci/mmol), and the reactions were terminated after 20 min at 30°C by addition of Laemmli sample buffer. Assays of p38 kinase activity were performed using GST–ATF2 as the substrate (Raingeaud et al., 1995). Substrate phosphorylation was examined after SDS–PAGE by autoradiography and PhosphorImager analysis (Molecular Dynamics Inc.).
Peptide competition experiments were performed using bacterially expressed GST–p38 bound to glutathione–Sepharose and pre-incubated with synthetic peptides for 2 h at 4°C. The recombinant p38 was phosphorylated by MKK3b or MKK6 (isolated by elution from immunoprecipitates prepared from transfected COS7 cells) by incubation (20 min at 30°C) in kinase buffer with 50 μM ATP. The immobilized p38 was then washed four times with kinase buffer and the p38 activity was measured in kinase assay with GST–ATF2 (1 μg) and 50 μM [γ-32P]ATP (10 Ci/mmol). The reaction was terminated after 20 min at 30°C and the phosphorylated ATF2 examined by SDS–PAGE, detected by autoradiography and quantitated by PhosphorImager analysis.
Binding assays
GST-tagged MKK proteins were isolated from transfected COS7 cells using glutathione–Sepharose (Amersham Pharmacia Biotech) in TLB for 7 h at 4°C. The beads were washed five times with TLB, and the presence of bound co-transfected p38α or p38β2 was examined by immunoblot analysis.
Acknowledgments
Acknowledgements
We thank Drs J.Han and R.Ulevitch for providing reagents, Dr A.J. Whitmarsh for critical discussions, Dr C.Turck for peptide synthesis, T.Barrett and A.Quail for DNA sequencing, and K.Gemme for administrative assistance. R.J.D. is an investigator of the Howard Hughes Medical Instiute. Supported, in part, by grants CA65861 and CA72009 and by the Howard Hughes Medical Institute.
References
- Bardwell L. and Thorner, J. (1996) A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biochem. Sci., 21, 373–374. [PubMed] [Google Scholar]
- Bardwell L., Cook, J.G., Chang, E.C., Cairns, B.R. and Thorner, J. (1996) Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol., 16, 3637–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A. and Pouyssegur, J. (1996) Identification of MAP kinase domains by redirecting stress signals into growth factor responses. Science, 272, 1652–1655. [DOI] [PubMed] [Google Scholar]
- Camps M., Nichols, A., Gillieron, C., Antonsson, B., Muda, M., Chabert, C., Boschert, U. and Arkinstall, S. (1998) Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science, 280, 1262–1265. [DOI] [PubMed] [Google Scholar]
- Chow C.W., Rincon, M., Cavanagh, J., Dickens, M. and Davis, R.J. (1997) Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science, 278, 1638–1641. [DOI] [PubMed] [Google Scholar]
- Cohen P. (1997) The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell Biol., 7, 353–361. [DOI] [PubMed] [Google Scholar]
- Conrad P.W., Rust, R.T., Han, J., Millhorn, D.E. and Beitner-Johnson, D. (1999) Selective activation of p38α and p38γ by hypoxia. Role in regulation of cyclin D1 by hypoxia in PC12 cells. J. Biol. Chem., 274, 23570–23576. [DOI] [PubMed] [Google Scholar]
- Cuenda A., Cohen, P., Buee-Scherrer, V. and Goedert, M. (1997) Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6); comparison of the specificities of SAPK3 and SAPK2 (RK/p38). EMBO J., 16, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B., Raingeaud, J., Barrett, T., Wu, I.H., Han, J., Ulevitch, R.J. and Davis, R.J. (1995) Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science, 267, 682–685. [DOI] [PubMed] [Google Scholar]
- Duesbery N.S. et al. (1998)Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science, 280, 734–737. [DOI] [PubMed] [Google Scholar]
- Enslen H., Raingeaud, J. and Davis, R.J. (1998) Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J. Biol. Chem., 273, 1741–1748. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Gotoh, Y. and Nishida, E. (1997) Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J., 16, 1901–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C. and Nebreda, A.R. (1999) A MAP kinase docking site is required for phosphorylation and activation of p90 (rsk)/MAPKAP kinase-1. Curr. Biol., 9, 281–284. [DOI] [PubMed] [Google Scholar]
- Goedert M., Cuenda, A., Craxton, M., Jakes, R. and Cohen, P. (1997) Activation of the novel stress-activated protein kinase SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6); comparison of its substrate specificity with that of other SAP kinases. EMBO J., 16, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Campbell, D., Derijard, B. and Davis, R.J. (1995) Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science, 267, 389–393. [DOI] [PubMed] [Google Scholar]
- Gupta S., Barrett, T., Whitmarsh, A.J., Cavanagh, J., Sluss, H.K., Derijard, B. and Davis, R.J. (1996) Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J., 15, 2760–2770. [PMC free article] [PubMed] [Google Scholar]
- Han J., Lee, J.D., Jiang, Y., Li, Z., Feng, L. and Ulevitch, R.J. (1996) Characterization of the structure and function of a novel MAP kinase kinase (MKK6) J. Biol. Chem., 271, 2886–2891. [DOI] [PubMed] [Google Scholar]
- Han J., Wang, X., Jiang, Y., Ulevitch, R.J. and Lin, S. (1997) Identification and characterization of a predominant isoform of human MKK3. FEBS Lett., 403, 19–22. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Cuenda, A., Spillantini, M.G., Thomas, G.M., Buee-Scherrer, V., Cohen, P. and Goedert, M. (1999) Stress-activated protein kinase-3 interacts with the PDZ domain of α1-syntrophin. A mechanism for specific substrate recognition. J. Biol. Chem., 274, 12626–12631. [DOI] [PubMed] [Google Scholar]
- Hibi M., Lin, A., Smeal, T., Minden, A. and Karin, M. (1993) Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev., 7, 2135–2148. [DOI] [PubMed] [Google Scholar]
- Holland P.M. and Cooper, J.A. (1999) Protein modification: docking sites for kinases. Curr. Biol., 9, R329–R331. [DOI] [PubMed] [Google Scholar]
- Ip Y.T. and Davis, R.J. (1998) Signal transduction by the c-Jun N–terminal kinase (JNK)—from inflammation to development. Curr. Opin. Cell Biol., 10, 205–219. [DOI] [PubMed] [Google Scholar]
- Jacobs D., Glossip, D., Xing, H., Muslin, A.J. and Kornfeld, K. (1999) Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev., 13, 163–175. [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Chen, C., Li, Z., Guo, W., Gegner, J.A., Lin, S. and Han, J. (1996) Characterization of the structure and function of a new mitogen-activated protein kinase (p38β) J. Biol. Chem., 271, 17920–17926. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Gram, H., Zhao, M., New, L., Gu, J., Feng, L., Di Padova, F., Ulevitch, R.J. and Han, J. (1997) Characterization of the structure and function of the fourth member of p38 group mitogen-activated protein kinases, p38δ. J. Biol. Chem., 272, 30122–30128. [DOI] [PubMed] [Google Scholar]
- Kallunki T., Deng, T., Hibi, M. and Karin, M. (1996) c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell, 87, 929–939. [DOI] [PubMed] [Google Scholar]
- Lawler S., Fleming, Y., Goedert, M. and Cohen, P. (1998) Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr. Biol., 8, 1387–1390. [DOI] [PubMed] [Google Scholar]
- Lee J.C., Kassis, S., Kumar, S., Badger, A. and Adams, J.L. (1999) p38 mitogen-activated protein kinase inhibitors—mechanisms and therapeutic potentials. Pharmacol. Ther., 82, 389–397. [DOI] [PubMed] [Google Scholar]
- Livingstone C., Patel, G. and Jones, N. (1995) ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J., 14, 1785–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.T., Yang, D.D., Wysk, M., Gatti, E., Mellman, I., Davis, R.J. and Flavell, R.A. (1999) Defective IL-12 production in mitogen-activated protein (MAP) kinase kinase 3 (Mkk3)-deficient mice. EMBO J., 18, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T. et al. (1996a)A novel kinase cascade mediated by mitogen-activated protein kinase kinase 6 and MKK3. J. Biol. Chem., 271, 13675–13679. [DOI] [PubMed] [Google Scholar]
- Moriguchi T. et al. (1996b)Purification and identification of a major activator for p38 from osmotically shocked cells. Activation of mitogen-activated protein kinase kinase 6 by osmotic shock, tumor necrosis factor-α and H2O2. J. Biol. Chem., 271, 26981–26988. [DOI] [PubMed] [Google Scholar]
- Nemoto S., Xiang, J., Huang, S. and Lin, A. (1998) Induction of apoptosis by SB202190 through inhibition of p38β mitogen-activated protein kinase. J. Biol. Chem., 273, 16415–16420. [DOI] [PubMed] [Google Scholar]
- Oh-hora M., Ogata, M., Mori, Y., Adachi, M., Imai, K., Kosugi, A. and Hamaoka, T. (1999) Direct suppression of TCR-mediated activation of extracellular signal-regulated kinase by leukocyte protein tyrosine phosphatase, a tyrosine-specific phosphatase. J. Immunol., 163, 1282–1288. [PubMed] [Google Scholar]
- Pellizzari R., Guidi-Rontani, C., Vitale, G., Mock, M. and Montecucco, C. (1999) Anthrax lethal factor cleaves MKK3 in macrophages and inhibits the LPS/IFNγ-induced release of NO and TNFα. FEBS Lett., 462, 199–204. [DOI] [PubMed] [Google Scholar]
- Pulido R., Zuniga, A. and Ullrich, A. (1998) PTP-SL and STEP protein tyrosine phosphatases regulate the activation of the extracellular signal-regulated kinases ERK1 and ERK2 by association through a kinase interaction motif. EMBO J., 17, 7337–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raingeaud J., Gupta, S., Rogers, J.S., Dickens, M., Han, J., Ulevitch, R.J. and Davis, R.J. (1995) Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem., 270, 7420–7426. [DOI] [PubMed] [Google Scholar]
- Raingeaud J., Whitmarsh, A.J., Barrett, T., Derijard, B. and Davis, R.J. (1996) MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol., 16, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.A., Poteet-Smith, C.E., Malarkey, K. and Sturgill, T.W. (1999) Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J. Biol. Chem., 274, 2893–2898. [DOI] [PubMed] [Google Scholar]
- Stein B., Brady, H., Yang, M.X., Young, D.B. and Barbosa, M.S. (1996) Cloning and characterization of MEK6, a novel member of the mitogen-activated protein kinase kinase cascade. J. Biol. Chem., 271, 11427–11433. [DOI] [PubMed] [Google Scholar]
- Sugiura R., Toda, T., Dhut, S., Shuntoh, H. and Kuno, T. (1999) The MAPK kinase Pek1 acts as a phosphorylation-dependent molecular switch. Nature, 399, 479–483. [DOI] [PubMed] [Google Scholar]
- Tournier C., Whitmarsh, A.J., Cavanagh, J., Barrett, T. and Davis, R.J. (1999) The MKK7 gene encodes a group of c-Jun NH2–terminal kinase kinases. Mol. Cell. Biol., 19, 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.S. et al. (1997)Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J. Biol. Chem., 272, 23668–23674. [DOI] [PubMed] [Google Scholar]
- Wang Y., Huang, S., Sah, V.P., Ross, J., Jr, Brown, J.H., Han, J. and Chien, K.R. (1998) Cardiac muscle cell hypertrophy and apoptosis induced by distinct members of the p38 mitogen-activated protein kinase family. J. Biol. Chem., 273, 2161–2168. [DOI] [PubMed] [Google Scholar]
- Wilk-Blaszczak M.A., Stein, B., Xu, S., Barbosa, M.S., Cobb, M.H. and Belardetti, F. (1998) The mitogen-activated protein kinase p38-2 is necessary for the inhibition of N-type calcium current by bradykinin. J. Neurosci., 18, 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbacher J.L., Goldsmith, E.J. and Cobb, M.H. (1999) Phosphorylation of MAP kinases by MAP/ERK involves multiple regions of MAP kinases. J. Biol. Chem., 274, 16988–16994. [DOI] [PubMed] [Google Scholar]
- Wysk M., Yang, D.D., Lu, H.T., Flavell, R.A. and Davis, R.J. (1999) Requirement of mitogen-activated protein kinase kinase 3 (MKK3) for tumor necrosis factor-induced cytokine expression. Proc. Natl Acad. Sci. USA, 96, 3763–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Wu, Z., Su, B., Murray, B. and Karin, M. (1998) JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino–terminal extension. Genes Dev., 12, 3369–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Wilsbacher, J.L., Collisson, T. and Cobb, M.H. (1999) The N–terminal ERK-binding site of MEK1 is required for efficient feedback phosphorylation by ERK2 in vitro and ERK activation in vivo. J. Biol. Chem., 274, 34029–34035. [DOI] [PubMed] [Google Scholar]
- Yang S.H., Whitmarsh, A.J., Davis, R.J. and Sharrocks, A.D. (1998a) Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J., 17, 1740–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Yates, P.R., Whitmarsh, A.J., Davis, R.J. and Sharrocks, A.D. (1998b) The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol. Cell. Biol., 18, 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Galanis, A. and Sharrocks, A.D. (1999) Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell. Biol., 19, 4028–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]