Abstract
Low temperature is an important environmental factor that has effects on all living organisms. Various low-temperature-inducible genes encode products that are essential for acclimation to low temperature, but low-temperature sensors and signal transducers have not been identified. However, systematic disruption of putative genes for histidine kinases and random mutagenesis of almost all the genes in the genome of the cyanobacterium Synechocystis sp. PCC 6803 have allowed us to identify two histidine kinases and a response regulator as components of the pathway for perception and transduction of low-temperature signals. Inactivation, by targeted mutagenesis, of the gene for each of the two histidine kinases and inactivation of the gene for the response regulator depressed the transcription of several lowtemperature-inducible genes.
Keywords: histidine kinase/low-temperature-inducible gene/response regulator/signal perception and transduction/Synechocystis
Introduction
Poikilothermic organisms, such as prokaryotes, plants and fish, sense changes in ambient temperature and acclimate to such changes with greater or lesser efficiency (Kaye and Guy, 1995; Thieringer et al., 1998). Numerous low-temperature-inducible genes have been found in a wide range of organisms, e.g. csp genes for cold-shock proteins in Escherichia coli (Jones et al., 1987) and Bacillus subtilis (Willimsky et al., 1992), cor and cas genes in plants (Gilmour et al., 1992; Wolfraim et al., 1993), and genes for fatty acid desaturases in cyanobacteria (Wada et al., 1990; Murata and Wada, 1995; Murata and Los, 1997), plants (Gibson et al., 1994) and fish (Tiku et al., 1996). These low-temperature-inducible genes are thought to play an important role in acclimation to low temperature. To date, however, mechanisms for the perception and transduction of low-temperature signals remain to be characterized.
In a previous study (Vigh et al., 1993), we demonstrated that decreases in the degree of unsaturation of fatty acids in the plasma membrane of the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) by catalytic hydrogenation in vivo enhanced the expression of the desAgene for the Δ12 acyl-lipid desaturase, which is otherwise induced primarily by low temperature. Thus, a change in membrane fluidity appears to be important for the perception of temperature that results in induction of the synthesis of the desaturases (Los et al., 1997). However, it remains unclear how a change in membrane fluidity is perceived and how the signal is transduced from the membrane to chromosomes to induce the expression of low-temperature-inducible genes, such as genes for desaturases.
The physical state of the membrane also affects the high-temperature-induced expression of heat-shock genes (Vigh et al., 1998). Modification, by genetic manipulation, of the ratio of unsaturated to saturated fatty acids in Saccharomyces cerevisiae has a significant effect on the expression of the heat-shock genes hsp70 and hsp82 (Carratu et al., 1996). These findings support the hypothesis that changes in the state of the membrane might also be important in the regulation of the expression of heat-shock genes.
Physical and chemical stimuli that are generated extra- and intracellularly are perceived by a group of proteins that includes histidine kinases. These proteins are localized on the plasma membrane or in the cytosol in various prokaryotes (Appleby et al., 1996), yeast (Maeda et al., 1994) and plants (Chang et al., 1993; Kakimoto, 1996). It seems likely, therefore, that temperature-induced changes in membrane fluidity might be mediated by a membrane-bound histidine kinase. Kaneko et al. (1995, 1996) determined the sequence of the genome of Synechocystis and identified 43 putative genes for histidine kinases (Mizuno et al., 1996).
In this study we attempted to identify components of the pathway for perception and transduction of low-temperature signals. Among the various low-temperature-inducible genes that had been identified previously, we focused on the desB gene for ω3 fatty acid desaturase in Synechocystis because the low-temperature-induced expression of this gene has been characterized in detail (Los et al., 1997; Los and Murata, 1998). We monitored the promoter response of the desB gene to low temperature using a gene for bacterial luciferase as the reporter. Systematic mutagenesis of genes for histidine kinases and random mutagenesis of almost all the genes in the genome allowed us to identify genes for two histidine kinases and a response regulator as components of the pathway for perception and transduction of low-temperature signals.
Results
Systematic mutation of putative genes for histidine kinases in pdesB::lux cells
In order to monitor the inducibility by low temperature of the desB gene, we generated a strain of Synechocystis, designated pdesB::lux, in which the promoter region of the desB gene was ligated to the coding region of the luxAB gene for a bacterial luciferase (Los et al., 1997). Thus, luciferase activity, monitored in terms of luminescence, could be used as an indicator of low-temperature-inducible changes in the activity of the desB promoter.
The products of some of the 43 putative genes for histidine kinases in the genome of Synechocystis (Kaneko et al., 1995, 1996; Mizuno et al., 1996) might plausibly be expected to function as sensors or transducers of environmental or intracellular stimuli (Appleby et al., 1996; Mizuno et al., 1996). We designated the histidine kinases Hik1–Hik43 and their genes hik1–hik43. To investigate the contributions of the various histidine kinases to the induction by low temperature of transcription of the desB gene, we attempted to inactivate each of the genes for histidine kinase in pdesB::lux cells by inserting a spectinomycin-resistance gene (Spr) cassette (Prentki et al., 1991) into the coding region or by replacing part of the coding region with the cassette, creating a gene-knockout library.
Each cyanobacterial cell contains >10 copies of the chromosome (Mann and Carr, 1974). Therefore, replacement of the wild-type chromosomes by mutated chromosomes required a lengthy period of time under selective pressure due to spectinomycin in the medium. The 43 lines of transformed cells were cultured for 6 months on agar-solidified BG-11 medium supplemented with 20 μg/ml spectinomycin. If the gene that was the target of the mutation were not essential, we would expect all the copies of the wild-type chromosome eventually to disappear. If the gene were essential, some copies of the wild-type chromosome would be expected to remain. Therefore, using the polymerase chain reaction (PCR), we monitored the extent to which wild-type chromosomes had been replaced by mutated chromosomes (data not shown). The wild-type gene was completely absent from all copies of the chromosome in 33 of our mutant strains. It appeared, therefore, that these 33 hik genes were not essential for survival under our growth conditions. Eight hik genes were not completely removed. However, the number of copies of wild-type chromosomes relative to the total number of copies of the chromosome was quite small in each case (data not shown). It seemed likely that these eight hik genes were important for the growth and survival of Synechocystiscells under our culture conditions. We failed to obtain any spectinomycin-resistant cells in our attempts to mutate the hik13 (sll1003; this nomenclature refers to designations of open reading frames by Kaneko et al., 1995, 1996) and hik15 (sll1353) genes. These two hik genes might have been essential for growth under our conditions. Detailed information about these mutants can be found on the web page on which mutants of Synechocystis are listed: CyanoMutant, at http://www.kazusa.or.jp/cyano/mutants/.
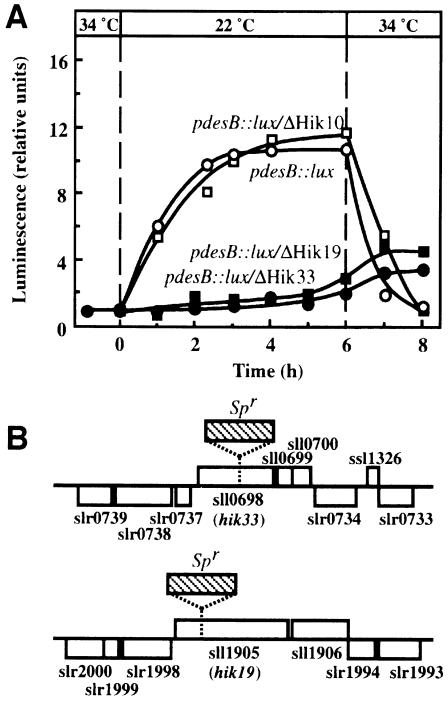
Inactivation of the hik33 and hik19 genes prevents the induction by low temperature of luciferase activity in pdesB::lux cells
We examined luciferase activity after a decrease in growth temperature in pdesB::lux cells and in each line of cells with a mutation in a gene for histidine kinase. Figure 1A shows that the shift in growth temperature from 34 to 22°C increased the luciferase activity ∼10-fold in pdesB::luxcells. By contrast, in the two mutants pdesB::lux/ΔHik33 and pdesB::lux/ΔHik19, in which, respectively, the hik33 gene and the hik19 gene had been inactivated by the insertion of the Spr cassette, no increase in luciferase activity was observed upon incubation of cells at 22°C (Figure 1A). This result indicated that mutations in the hik33 and hik19 genes eliminated the inducibility by low temperature of the desB promoter. By contrast, the luciferase activity in other lines with mutant genes for histidine kinase resembled that in pdesB::lux; for example, the response of luciferase activity to the shift in temperature in pdesB::lux/ΔHik10 cells, in which the hik10 gene (slr0533) had been inactivated, was the same as that in pdesB::lux cells (Figure 1A). The sites of insertion of the Spr cassette in hik33 and hik19 genes are shown in Figure 1B. Analysis by PCR revealed, however, that the native genes had not been completely eliminated in these lines, suggesting that the genes were essential under our growth conditions.
Fig. 1. Temperature-dependent changes in the activity of the desB promoter in pdesB::lux, pdesB::lux/ΔHik10, pdesB::lux/ΔHik33 and pdesB::lux/ΔHik19 cells, and sites of insertion of the Spr cassette in pdesB::lux/ΔHik33 and pdesB::lux/ΔHik19 cells. (A) Cells were grown on agar-solidified medium at 34°C and then transferred to 22°C. Luciferase activity was measured in terms of the intensity of luminescence, as described in Materials and methods. ○, pdesB::lux; □, pdesB::lux/ΔHik10; •, pdesB::lux/ΔHik33; ▪, pdesB::lux/ΔHik19. The results are the averages of results of three independent experiments. (B) The NcoI sites in the hik33 (sll0698) and hik19 (sll1905) genes at which the Spr cassette (hatched rectangle) was inserted are indicated by dashed vertical lines. Open rectangles indicate open reading frames.
Expression of low-temperature-inducible genes in wild-type, ΔHik33 and ΔHik19 cells
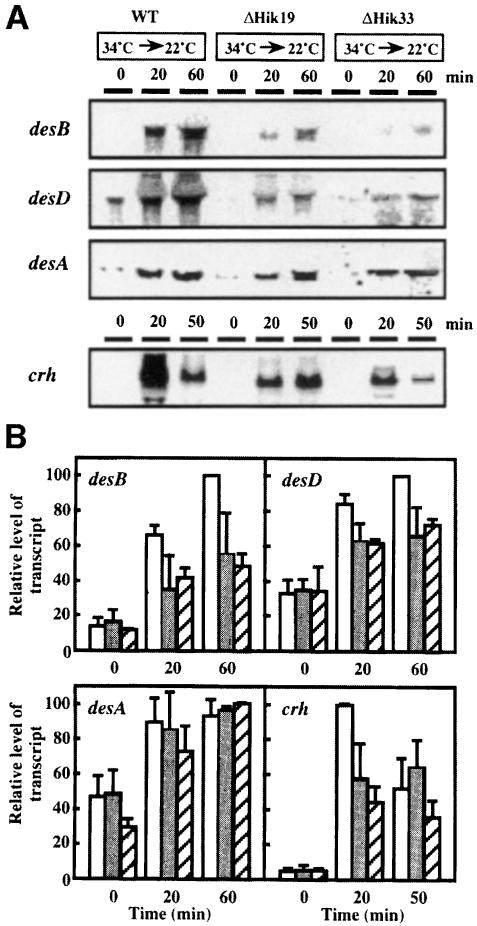
We next inactivated, separately, the hik33 and hik19 genes in wild-type cells to examine the effects of these genes on the expression of low-temperature-inducible genes. There are four genes for fatty acid desaturases in Synechocystis. The desC gene is expressed constitutively, while the desA, desB and desD genes are induced after a downward shift in temperature (Los et al., 1997). We performed Northern blotting analysis to examine the expression of the desB, desD and desA genes in wild-type, ΔHik19 and ΔHik33 cells before and after a shift in temperature from 34 to 22°C (Figure 2A). As observed previously in wild-type cells (Los et al., 1997), the increase in the level of the desB transcript was the most conspicuous. There was also a distinct increase in the level of the desD transcript. The level of the desA transcript increased least of all among the transcripts of the three genes for desaturases. Prior to exposure of cells to 22°C, the levels of the transcripts of the desB, desD and desA genes in the mutant cells were as low as those in wild-type cells (Figure 2A). However, the extent of induction at 22°C of the desB and desD genes, but not that of the desA gene, appeared to be reduced in both ΔHik19 and ΔHik33 mutant cells.
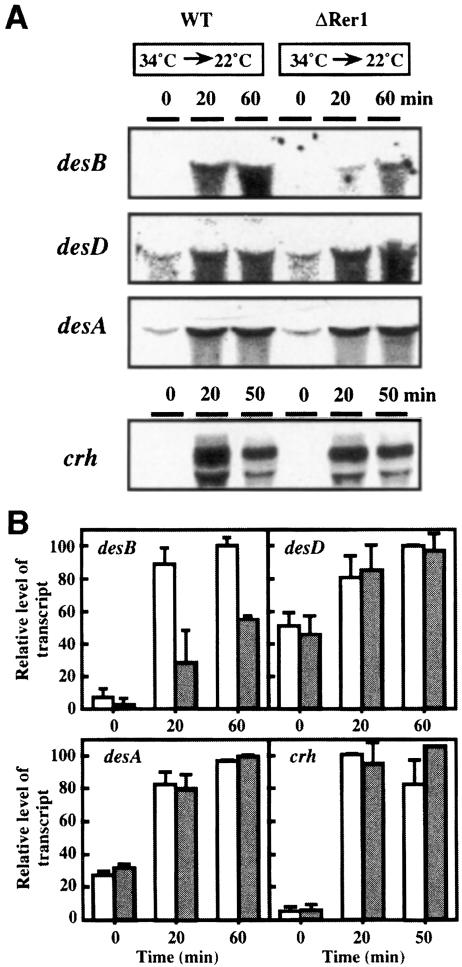
Fig. 2. Induction of desB, desD, desA and crh genes in wild-type, ΔHik33 and ΔHik19 cells after a downward shift in temperature. Cells that had been grown at 34°C for 16 h were transferred to 22°C and incubated for the periods of time indicated. Northern blotting analysis of the expression of the desB, desD, desA and crh genes was performed as described by Los et al. (1997). Total RNA (30 μg for analysis of des mRNAs and 5 μg for analysis of crh mRNA) was loaded in each lane. (A) Northern blots. (B) Quantification of transcripts. Open bars, wild-type cells; filled bars, ΔHik19 cells; hatched bars, ΔHik33 cells. The results are the averages from three independent experiments with experimental deviations.
Figure 2B shows the quantitative changes in the levels of the transcripts. The level of the desB transcript in wild-type cells was 8–fold higher after incubation for 60 min at 22°C than before the incubation. However, inactivation of the hik33 gene significantly depressed the lowtemperature-induced increase in the level of the desB transcript. The low-temperature-induced increase in the level of the desDtranscript in ΔHik33 cells was also reduced to two-thirds of that in wild-type cells after incubation for 60 min at 22°C. In addition, inactivation of the hik19 gene depressed the low-temperature-induced enhancement of expression of the desB and desD genes (Figure 2B). However, the low-temperature-induced enhancement of the expression of the desA gene was unaffected by the inactivation of hik33 and hik19 (Figure 2B). These results indicated that inactivation of hik33 and hik19 suppressed the low-temperature-induced expression of the desB and desD genes, but not of the desAgene.
The crh gene for a homolog of RNA helicase is also a gene that is induced at low temperatures (Chamot et al., 1999). Figure 2 shows that the level of crhmRNA increased within 20 min after a shift in temperature from 34 to 22°C and then decreased during further incubation at 22°C. The pattern of expression of crhmRNA in wild-type cells differed from that of the mRNAs for desaturases. Nonetheless, the increases in the level of crh mRNA in 20 min in ΔHik33 and ΔHik19 cells were smaller than those in wild-type cells.
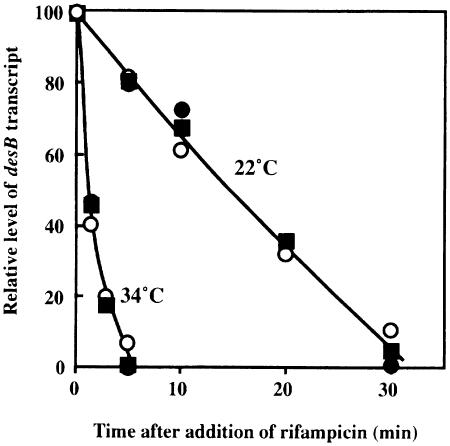
Degradation of desB mRNA in wild-type, ΔHik33 and ΔHik19 cells
In general, the level of an mRNA is regulated by the rate of transcription of the corresponding gene and the stability of the mRNA itself. These factors also control the accumulation of desB mRNA when Synechocystis cells are exposed to a low temperature (Los et al., 1997). Therefore, we compared the stability of desBmRNA at 34 and 22°C in wild-type, ΔHik33 and ΔHik19 cells in the presence of an inhibitor of transcription, rifampicin. Figure 3 shows that the rate of degradation of desB mRNA was the same in wild-type and mutant cells at both high and low temperatures. The half-life of the desB transcript was 15 min at 22°C and 1 min at 34°C. These results indicated that the reduction in the low-temperature-induced accumulation of the desB transcript in ΔHik33 and ΔHik19 cells was due to a decrease in the rate of transcription and not to a decrease in the stability of the mRNA, and they suggested, moreover, that both Hik33 and Hik19 might be involved in the low-temperature-induced regulation of transcription of the desB gene.
Fig. 3. Dependence on temperature of the stability of desB mRNA in wild-type, ΔHik33 and ΔHik19 cells. Cells were grown at 34°C for 16 h and then incubated for 2 h either at 22 or 34°C. An inhibitor of transcription, rifampicin, was added to the cultures at 50 μg/ml at time zero. Aliquots of cultures were withdrawn at the times indicated and levels of desB mRNA were determined by Northern blotting analysis. ○, wild-type cells; •, ΔHik33 cells; ▪, ΔHik19 cells. The results are averages from three independent experiments.
Random mutagenesis and screening of mutants with altered expression of the desB gene
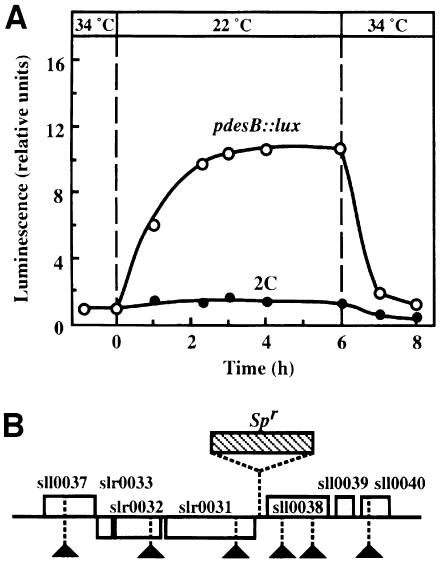
In order to find other components in the pathway for low-temperature signaling, we used random mutagenesis. We introduced the Spr cassette randomly into the chromosome of pdesB::lux cells by cassette mutagenesis (Hagemann et al., 1996). From among ∼20 000 spectinomycin-resistant mutants, we isolated 18 mutants in which the response of luciferase activity to a downward shift in temperature was different from that in parental pdesB::lux cells (Figure 4). We postulated that in cells of each of these mutant lines a component of the low-temperature signal-transduction pathway might have been mutated. We determined sequences on both sides of the sites of insertion of the cassette in these mutants. We found that in two of the 18 mutants the hik19 gene for Hik19 had been inactivated by insertion of the Spr cassette (data not shown).
Fig. 4. Changes in the activity of luciferase upon a change in temperature in pdesB::lux and 2C cells. (A) pdesB::lux and 2C cells were grown on agar-solidified medium at 34°C and then transferred to 22°C. Luciferase activity was measured in terms of the intensity of luminescence, as described in Materials and methods. ○, pdesB::lux; •, mutant 2C. The results are the averages of three independent experiments. (B) The site of insertion of the Spr cassette in the chromosome of mutant 2C cells (hatched rectangle) and the sites of insertion of the Sprcassette in wild-type cells (triangles) are indicated. The open reading frame sll0038 corresponds to the response regulator Rer1.
In another of these mutant lines, designated 2C, in which the desB promoter was not activated by low temperature (Figure 4A), the Spr cassette had been inserted in the upstream region of two operons, as shown in Figure 4B. The results suggested that one of the genes in the vicinity of the site of insertion encoded a component of the low-temperature signal-transduction pathway. In order to identify the gene responsible for the elimination of the transcriptional activity, we inactivated five putative genes separately (sll0037, slr0032, slr0031, sll0038, sll0040) in pdesB::lux by inserting an Spr cassette, as indicated by triangles in Figure 4B. Inactivation of gene sll0038, but not of any of the other genes, depressed the low-temperature-induced increase in luciferase activity in the same way as observed in 2C cells (data not shown). The amino acid sequence deduced from the nucleotide sequence of this gene indicated that its product was one of the 38 response regulators identified in Synechocystis (Mizuno et al., 1996). We designated the gene rer1 and its product Rer1.
Inactivation of the rer1 gene in wild-type cells inhibited the induction of the desB gene by low temperature
To examine the role of Rer1 in the regulation of expression of low-temperature-inducible genes, we inactivated the rer1 gene in wild-type cells by inserting the Spr cassette at the MscI site. The extent of the low-temperature-dependent induction of the desB transcript was reduced to half that in wild-type cells (Figure 5). By contrast, the inducibility by low temperature of the desD, desA and crh genes was unaffected by the mutation (Figure 5). These results indicated that Rer1 might specifically regulate the expression of the desBgene, but not that of the other genes examined.
Fig. 5. Induction of desB, desD, desA and crh genes in wild-type and ΔRer1 cells after a downward shift in temperature. Cells that had been grown at 34°C for 16 h were transferred to 22°C and incubated for the periods of time indicated. Northern blotting analysis of the expression of desB, desD and desA genes was performed as described by Los et al. (1997). Total RNA (30 μg for analysis of des mRNAs and 5 μg for analysis of crh mRNA) was loaded in each lane. (A) Northern blots. (B) Quantification of transcripts. Open bars, wild-type cells; filled bars, ΔRer1 cells. The results are the averages of results from three independent experiments with experimental deviations.
Discussion
Advantages of using Synechocystis for systematic inactivation of histidine kinases
There are 43 putative genes for histidine kinases in the chromosome of Synechocystis (Kaneko et al., 1995, 1996; Mizuno et al., 1996). Since histidine kinases have been shown, in many cases, to be sensors or components of signal-transducing systems (Appleby et al., 1996; Mizuno et al., 1996), some of the histidine kinases in Synechocystis might also be expected to have similar functions. However, it is impossible to predict the function of each individual histidine kinase on the basis of its primary structure. The functions of ∼30 histidine kinases have been identified in E.coli, but these enzymes have no obvious counterparts in Synechocystis (Mizuno et al., 1996).
Using Synechocystis, we were able to identify genes for histidine kinases that appear to be involved in the perception and transduction of low-temperature signals even though these genes are essential for survival and cannot be completely eliminated. Each cell of Synechocystis contains ∼10 identical copies of the chromosome and, thus, essential genes can be inactivated to some extent but not totally with detectable changes in phenotype. The hik33 and hik19 genes were not completely eliminated even under selective pressure due to spectinomycin and, therefore, they can be regarded as essential genes. It is impossible to identify genes that might correspond to hik33 and hik19 in experiments with E.coli or B.subtilis because each cell of these bacteria contains only a single copy of the chromosome.
Characteristics of Hik33
The amino acid sequence deduced from the hik33 gene (sll0698) indicates that Hik33 contains 663 amino acid residues. The strongly conserved histidine kinase domain is located near the C–terminus. An analysis using computer programs that predict the localization of proteins, such as PSORT (Nakai and Horton, 1999) and HMMTOP (Tusnady and Simon, 1998), indicated that Hik33 has two hydrophobic helices that might, in theory, span the membrane. A putative leucine zipper motif and a putative coiled-coil sequence are located between the second hydrophobic helix and the histidine kinase domain. These motifs are involved in the dimerization of a number of histidine kinases and are important for their activities (Lau et al., 1997; Yaku and Mizuno, 1997; Singh et al., 1998). Thus, we can predict that one or both of these sequences might be involved in the dimerization of Hik33 and the regulation of its activity.
A reduction in the fluidity of the plasma membrane of a cyanobacterium appears to be a primary signal for the low-temperature-induced expression of the genes for desaturases (Vigh et al., 1993; Murata and Los, 1997). The properties of Hik33 appear to be consistent with those of a sensor that can detect a decrease in membrane fluidity. A reduction in the fluidity of the membrane at sites at which Hik33 is located might alter the structure of Hik33, influencing the spatial relationship between monomers of the dimerized protein and thereby altering activity.
We searched for proteins homologous to Hik33 in standard databases. The kinase domain of Hik33, which includes an autophosphorylatable histidine residue and an ATP-binding motif (Park et al., 1998), was very similar to those of histidine kinases from bacteria, yeast, fungi and plants. However, the sequences outside the kinase domain, including the membrane-spanning domains, appeared to be unique. One relatively homologous gene was identified, namely ycf26, which was found in the chloroplast genome of the red alga Porphyra purpurea (Reith and Munholland, 1995). This gene encodes a homolog of histidine kinase of 656 amino acid residues. About 48% of the residues outside the histidine kinase domain are identical to those in Hik33 (data not shown). The hydropathy profiles of Hik33 and Ycf26 are also very similar (data not shown). However, it is unclear what kinds of signal might be perceived and what genes might be regulated by Ycf26.
Hik33 is also homologous to a Ycf26-like protein found in the chloroplast genome of Cyanidium caldarium (DDBJ/EMBL/GenBank accession No. AF022186) and to a protein encoded by the yycG gene in B.subtilis (Kunst et al., 1997). These proteins also contain putative membrane-spanning domains and coiled-coil motifs. This indicates that a pathway for the perception and transduction of cold signals identified in Synechocystis might be a common feature of the responses of many organisms to cold.
Characteristics of Hik19
The amino acid sequence deduced from the hik19 gene (sll1905) indicates that Hik19 contains 1014 amino acid residues and computer analysis suggests that it might be a soluble protein in the cytosol. A strongly conserved histidine kinase domain is located in a central region of the protein. One signal-receiver domain is localized at the N–terminus and another is near the C–terminal region. Furthermore, a histidine phospho-transfer (Hpt) domain is located at the C–terminus. Thus, Hik19 is a hybrid-type histidine kinase (Mizuno et al., 1996). The receiver domain at the N–terminus might accept a phosphate group from some other histidine kinase or protein that contains phosphorylated histidine and Hik19 might function downstream of the membrane-bound sensor Hik33. Hik19 is more likely than Hik33 to be a transducer of the low-temperature signal.
Characteristics of Rer1
The amino acid sequence deduced from the rer1 gene indicates that Rer1 contains 402 amino acid residues. Unlike most response regulators that have a signal-receiver domain at the N–terminus, Rer1 has a signal-receiver domain at the C–terminus. However, the N–terminal region is homologous to the DNA-binding domain, known as an HMG box, found in regulators of transcription in vertebrates, such as the Sox6 and Sox5 proteins (Connor et al., 1995). The central part of Rer1 is similar to the transcriptional activation domain of the aryl hydrocarbon (Ah) receptor nuclear translocator (Arnt; Li et al., 1994; Figure 6). Although Rer1 does not have a structure typical of response regulators, it is possible that it functions as a DNA-binding regulator of transcription.
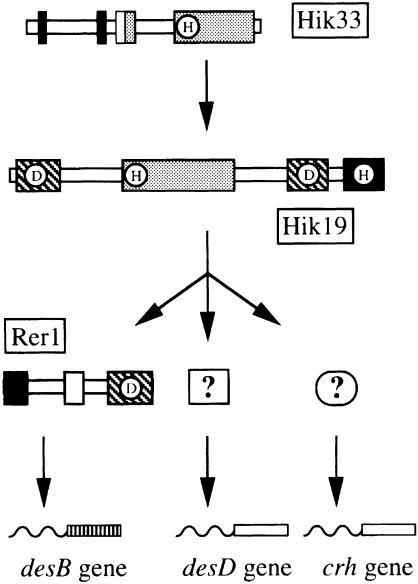
Fig. 6. A hypothetical scheme for the pathway for low-temperature signal transduction in Synechocystis. The histidine kinase domains, the receiver domains and the histidine phospho-transfer (Hpt) domain of Hik33, Hik19 and Rer1 are indicated by gray rectangles, hatched rectangles and a filled rectangle, respectively. The histidine and aspartate residues that might be involved in the phospho-relay reaction are indicated by H and D in circles, respectively. Filled rectangles, a gray rectangle and an open rectangle in Hik33 indicate the putative membrane-spanning domains, the coiled-coil domain and leucine-zipper domain, respectively. Closed and open rectangles in Rer1 indicate regions homologous to the HMG box and the Ah receptor nuclear translocator, respectively (see the text for details).
Characterization of the ΔHik33, ΔHik19 and ΔRer1 mutants
The extent of the low-temperature-inducible expression of the desB transcript in ΔHik33 cells was half that in wild-type cells (Figure 2). The low levels of Hik33 and Hik19 might be responsible for the low level of accumulation of the desB transcript in the mutant cells. However, we found that the low-temperature-induced activation of the desB promoter in pdesB::lux/ΔHik33 or pdesB::lux/ΔHik19 cells, monitored in terms of luciferase activity, was almost completely eliminated by the inactivation of the hik19 genes or the hik33 genes (Figure 1). The discrepancy between the extent of depression of the accumulation of the transcript and the extent of transcriptional activation might be explained by an increase in the stability of transcripts at low temperature, as shown in Figure 3 and in the previous report (Los et al., 1997). The half-life of the desB transcript increased 15-fold after a shift in growth temperature from 34 to 22°C in wild-type cells and in ΔHik33 and ΔHik19 cells. It is unclear how the transcripts of desB, desD and desA genes might be stabilized at low temperature.
Although inactivation of hik33 and hik19 reduced the low-temperature-induced accumulation of desBand desD transcripts (Figure 2), inactivation of the rer1 gene resulted in a reduction in the level of the desB transcript, while levels of desD, desA and crh transcripts were unaffected (Figure 5). These results indicate that Hik33 and Hik19 might be involved in a common mechanism that regulates the expression of desB, desD and crh genes, and that Rer1 might specifically regulate the expression of the desB gene (Figure 6). In E.coli, a hybrid-type histidine kinase, ArcB, transfers a phosphate group via its Hpt domain to several receivers, such as ArcA, OmpR and CheY (Perraud et al., 1999). Hik19 might also transfer phosphate groups to some, as yet unidentified, response regulators that, perhaps, contain a receiver domain and it might, thus, regulate the expression of desD, crh and certain other low-temperature-inducible genes.
A hypothetical pathway for perception and transduction of low-temperature signals
Figure 6 shows a hypothetical scheme for the transduction of low-temperature signals. Hik33 may span the plasma membrane twice and forms a dimer, whose structure may be influenced by the physical characteristics of lipids in the plasma membrane, such as their fluidity (or the extent of molecular motion), which is controlled by temperature and the extent of unsaturation of the fatty acids. When the temperature is decreased or the fatty acids are more saturated, the histidine residue in the histidine kinase domain may be phosphorylated. A phosphate group is then transferred to Hik19, and finally to Rer1, which regulates the expression of the desB gene. Hik19 and Hik33 are also involved in the regulation of expression of the crh and desD genes. However, we have not yet identified the response regulators (or transcriptional regulators) of these genes.
In E.coli, heat stress induces the expression of several genes, whose products are involved in the folding and degradation of denatured proteins. Some of these genes are regulated by the typical two-component system CpxA–CpxR. CpxA is a histidine kinase, which is bound to the plasma membrane and is autophosphorylated under heat stress. Phosphorylated CpxA transfers a phosphate group to the response regulator CpxR, which activates the transcription of several heat-inducible genes, such as degP, which encodes a protease, and dsbA, which encodes a disulfide isomerase (Mileykovskaya and Dowhan, 1997). The polypeptide deduced from the cpxA gene is different from Hik33 except in its histidine kinase domain. The pathway for low-temperature signal transduction in Synechocystis (Figure 6) appears to be more complex than the two-component system for high-temperature signal transduction in E.coli.
The expression of several sets of genes in response to low temperature occurs in all poikilothermic organisms examined to date. A pathway for the perception and transduction of low-temperature signals that includes two histidine kinases identified in Synechocystis might be a common feature of the responses of many organisms to low temperatures.
Materials and methods
Cells and culture conditions
A strain of Synechocystis sp. PCC 6803, which is tolerant to glucose (Williams, 1988), was obtained from Dr Williams at Dupont Co. Ltd. We generated strain pdesB::lux, in which the coding region of the desB gene was replaced by the luxAB gene for bacterial luciferase, as described previously (Los et al., 1997). In this construct, the luxAB gene was expressed under the control of the desB promoter. Wild-type cells were grown at 34°C in BG-11 medium (Stanier et al., 1971) buffered with 20 mM HEPES–NaOH pH 7.5 under continuous illumination from incandescent lamps, as described previously (Wada and Murata, 1989). Cells of mutants in which the Spr cassette had been inserted into the genome were grown under the same conditions as described above with the exception that the culture medium contained spectinomycin at 20 μg/ml up to the pre-cultures. Mutant cells were then transferred to medium without spectinomycin for the final cell culture used for the experiments.
Systematic targeted mutagenesis of genes for histidine kinase
DNA fragments that contained complete or partial sequences of the 43 putative genes for histidine kinases of Synechocystis were amplified by PCR and cloned into pT7Blue (Novagen, Madison, WI), pBluescript II SK(+) (Stratagene, La Jolla, CA) or pUC18 (Toyobo, Osaka, Japan). Then the Spr cassette (Prentki et al., 1991) was inserted at suitable restriction sites in the Hik coding region in the same orientation as the open reading frame (Figure 1B). pdesB::lux cells were transformed with the resultant plasmids as described by Williams (1988). Transformed cells were cultured for at least 6 months on agar-solidified BG-11 medium (Stanier et al., 1971) that was buffered with 20 mM HEPES–NaOH pH 7.5 and contained 20 μg/ml spectinomycin. Genomic DNA was extracted from mutant cells and used as the template for analysis by PCR of the extent of replacement of copies of wild-type chromosomes by mutated chromosomes.
ΔHik33 and ΔHik19 were produced by inactivating the hik33 and hik19genes in wild-type cells in the same way as described above for pdesB::lux cells, with the Spr cassette being inserted at the NcoI site in each gene.
Random mutagenesis by insertion of the Spr cassette and identification of genes responsible for activation of the desB promoter
We modified pUC18 to generate pUCBam, in which all the multiple cloning sites were eliminated except the BamHI site. pUC18 was first treated with the restriction enzymes XbaI and EcoRI, and then sites of cleavage were blunted by T7 DNA polymerase (DNA Blunting Kit; Takara, Kyoto, Japan) and self-ligated. The resultant plasmid was digested with SalI and HindIII, blunted and self-ligated to construct pUCBam. The genomic DNA extracted from wild-type cells of Synechocystis was partially digested with Sau3AI and fractionated by electrophoresis on an agarose gel. DNA fragments of 1–5 kb were recovered from the gel and inserted into the BamHI site of pUCBam. The resultant genomic library was subjected to digestion with EcoRI, HindIII, XbaI, HincII, NcoI or NheI. The Spr cassette (Prentki et al., 1991) was ligated into the cleaved plasmids. Escherichia coli JM109 cells were transformed with the resultant plasmids. The transformed cells with Spr-tagged plasmids were selected on agar-solidified LB medium that contained 50 μg/ml ampicillin and 30 μg/ml spectinomycin. The Spr-tagged plasmids were recovered from selected E.coli cells and used for the transformation of Synechocystis cells as described by Williams (1988).
After the selection of mutants in which the low-temperature-induced activation of the desB promoter was depressed, we determined the DNA sequences on both sides of the sites of insertion of the Spr cassette in some of the mutants by inverse PCR (Ochman et al., 1988; Triglia et al., 1988) with DNA extracted from the mutant cells as template. Then we inactivated separately the genes located in the vicinity of the sites of insertion of the Spr cassette by targeted mutagenesis of pdesB::lux cells as described above. We identified genes responsible for the loss of inducibility by low temperature of the desB promoter by monitoring luminescence due to luciferase activity.
Determination of the activity of the desB promoter using a reporter gene for bacterial luciferase
Cells of pdesB::lux and mutants derived from it were grown on agar-solidified medium for 2 days at 34°C and then incubated at 22°C for appropriate periods of time. After n-decanal vapor had been applied to the cells for 1 min, the emission of photons from cells was monitored for 1 min with a photon-counting luminometer (ARGUS 50; Hamamatsu Photonics, Hamamatsu, Japan).
Analysis of transcripts
Northern blotting analysis was performed as described previously (Los et al., 1997). DNA fragments corresponding to the desA, desB, desD and crh genes were conjugated with alkaline phosphatase (Alkphos Direct kit; Amersham Pharmacia Biotech, Uppsala, Sweden) and the resultant conjugates were used as probes. After hybridization, the blots were soaked with CDP-star solution (Amersham Pharmacia Biotech) and signals from hybridized mRNA were detected with a luminescence image analyzer (LAS-1000; Fuji-Photo Film, Tokyo, Japan).
Acknowledgments
Acknowledgements
This work was supported by a Grant-in-Aid for Specially Promoted Research (No. 08102011) from the Ministry of Education, Science and Culture, Japan.
References
- Appleby J.L., Parkinson, J.S. and Bourret, R.B. (1996) Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell, 86, 845–848. [DOI] [PubMed] [Google Scholar]
- Carratu L., Franceschelli, S., Pardini, C.L., Kobayashi, G.S., Horvath, I., Vigh, L. and Maresca, B. (1996) Membrane lipid perturbation modifies the set point of the temperature of heat shock response in yeast. Proc. Natl Acad. Sci. USA, 93, 3870–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamot D., Magee, W.C., Yu, E. and Owttrim, G.W. (1999) A cold shock-induced cyanobacterial RNA helicase. J. Bacteriol., 181, 1728–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Kwok, S.F., Bleecker, A.B. and Meyerowitz, E.M. (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science, 262, 539–544. [DOI] [PubMed] [Google Scholar]
- Connor F., Wright, E., Denny, P., Koopman, P. and Ashworth, A. (1995) The Sry-related HMG box-containing gene Sox6 is expressed in the adult testis and developing nervous system of the mouse. Nucleic Acids Res., 23, 3365–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S., Arondel, V., Iba, K. and Somerville, C. (1994) Cloning of a temperature-regulated gene encoding a chloroplast ω-3 desaturase from Arabidopsis thaliana.Plant Physiol., 106, 1615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Artus, N.N. and Thomashow, M.F. (1992) cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana.Plant Mol. Biol., 18, 13–21. [DOI] [PubMed] [Google Scholar]
- Hagemann M., Richter, S., Zuther, E. and Schoor, A. (1996) Characterization of a glucosylglycerol-phosphate-accumulating, salt-sensitive mutant of the cyanobacterium Synechocystis sp. strain PCC 6803. Arch. Microbiol., 166, 83–91. [DOI] [PubMed] [Google Scholar]
- Jones P.G., Van Boegelen, R.A. and Neidhardt, F.C. (1987) Induction of proteins in response to low temperature in Escherichia coli.J. Bacteriol., 169, 2092–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science, 274, 982–985. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Tanaka, A., Sato, S., Kotani, H., Sazuka, T., Miyajima, N., Sugiura, M. and Tabata, S. (1995) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res., 2, 153–166. [DOI] [PubMed] [Google Scholar]
- Kaneko T. et al. (1996)Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res., 3, 109–136. [DOI] [PubMed] [Google Scholar]
- Kaye C. and Guy, C.L. (1995) Perspectives of plant cold tolerance: physiology and molecular responses. Sci. Prog., 78, 271–299. [PubMed] [Google Scholar]
- Kunst F. et al. (1997)The complete genome sequence of the gram-positive bacterium Bacillus subtilis.Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- Lau P.C., Wang, Y., Patel, A., Labbe, D., Bergeron, H., Brousseau, R., Konishi, Y. and Rawlings, M. (1997) A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc. Natl Acad. Sci. USA, 94, 1453–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Dong, L. and Whitlock, J.P., Jr (1994) Transcriptional activation function of the mouse Ah receptor nuclear translocator. J. Biol. Chem., 269, 28098–28105. [PubMed] [Google Scholar]
- Los D.A. and Murata, N. (1998) Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta, 1394, 3–15. [DOI] [PubMed] [Google Scholar]
- Los D.A., Ray, M.K. and Murata, N. (1997) Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis sp. PCC 6803. Mol. Microbiol., 25, 1167–1175. [DOI] [PubMed] [Google Scholar]
- Maeda T., Wurgler-Murphy, S.M. and Saito, H. (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature, 369, 242–245. [DOI] [PubMed] [Google Scholar]
- Mann N. and Carr, N.G. (1974) Control of macromolecular composition and cell division in the blue-green alga Anacystis nidulans.J. Gen. Microbiol., 83, 399–405. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E. and Dowhan, W. (1997) The Cpx two-component signal-transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J. Bacteriol., 179, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Kaneko, T. and Tabata, S. (1996) Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. DNA Res., 3, 407–414. [DOI] [PubMed] [Google Scholar]
- Murata N. and Los, D.A. (1997) Membrane fluidity and temperature perception. Plant Physiol., 115, 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N. and Wada, H. (1995) Acyl-lipid desaturases and their importance in the tolerance and acclimatization to cold of cyanobacteria. Biochem. J., 308, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K. and Horton, P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- Park H., Saha, S.K. and Inouye, M. (1998) Two-domain reconstitution of a functional protein histidine kinase. Proc. Natl Acad. Sci. USA, 95, 6728–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraud A.-L., Weiss, V. and Gross, R. (1999) Signalling pathways in two-component phosphorelay systems. Trends Microbiol., 7, 115–120. [DOI] [PubMed] [Google Scholar]
- Prentki P., Binda, A. and Epstein, A. (1991) Plasmid vectors for selecting IS1-promoted deletions in cloned DNA: sequence analysis of the ω interposon. Gene, 103, 17–23. [DOI] [PubMed] [Google Scholar]
- Reith M. and Munholland, J. (1995) Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep., 13, 333–335. [Google Scholar]
- Singh M., Berger, B., Kim, P.S., Berger, J.M. and Cochran, A.G. (1998) Computational learning reveals coiled coil-like motifs in histidine kinase linker domains. Proc. Natl Acad. Sci. USA, 95, 2738–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R.Y., Kunisawa, R., Mandel, M. and Cohen-Bazire, G. (1971) Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev., 35, 171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieringer H.A., Jones, P.G. and Inouye, M. (1998) Cold shock and adaptation. BioEssays, 20, 49–57. [DOI] [PubMed] [Google Scholar]
- Tiku P.E., Gracey, A.Y., Macartney, A.I., Beynon, R.J. and Cossins, A.R. (1996) Cold-induced expression of Δ9-desaturase in carp by transcriptional and posttranslational mechanisms. Science, 271, 815–818. [DOI] [PubMed] [Google Scholar]
- Tusnady G.E. and Simon, I. (1998) Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol., 283, 489–506. [DOI] [PubMed] [Google Scholar]
- Vigh L., Los, D.A., Horvath, I. and Murata, N. (1993) The primary signal in the biological perception of temperature: Pd-catalyzed hydrogenation of membrane lipids stimulated the expression of the desA gene in Synechocystis PCC6803. Proc. Natl Acad. Sci. USA, 90, 9090–9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigh L., Maresca, B. and Harwood, J.L. (1998) Does the membrane's physical state control the expression of heat shock and other genes?Trends Biochem. Sci., 23, 369–374. [DOI] [PubMed] [Google Scholar]
- Wada H. and Murata,N. (1989) Synechocystis PCC 6803 mutants defective in desaturation of fatty acids. Plant Cell Physiol., 30, 971–978. [Google Scholar]
- Wada H., Gombos, Z. and Murata, N. (1990) Enhancement of chilling tolerance of a cyanobacterium by genetic manipulation of fatty acid desaturation. Nature, 347, 200–203. [DOI] [PubMed] [Google Scholar]
- Williams J.G.K. (1988) Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol., 167, 766–778. [Google Scholar]
- Willimsky G., Bang, H., Fischer, G. and Marahiel, M.A. (1992) Characterization of cspB, a Bacillus subtilis inducible cold-shock gene affecting cell viability at low temperatures. J. Bacteriol., 174, 6326–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfraim L.A., Langis, R., Tyson, H. and Dhindsa, R.S. (1993) cDNA sequence, expression, and transcript stability of a cold acclimation-specific gene, cas18, of alfalfa (Medicago falcata) cells. Plant Physiol., 101, 1275–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaku H. and Mizuno, T. (1997) The membrane-located osmosensory kinase, EnvZ, that contains a leucine zipper-like motif functions as a dimer in Escherichia coli.FEBS Lett., 417, 409–413. [DOI] [PubMed] [Google Scholar]