Abstract
The biogenesis of a number of RNA species in eukaryotic cells requires 3′ processing. To determine the enzymes responsible for these trimming events, we created yeast strains lacking specific 3′ to 5′ exonucleases. In this work, we describe the analysis of three members of the RNase D family of exonucleases (Rex1p, Rex2p and Rex3p). This work led to three important conclusions. First, each of these exonucleases is required for the processing of distinct RNAs. Specifically, Rex1p, Rex2p and Rex3p are required for 5S rRNA, U4 snRNA and MRP RNA trimming, respectively. Secondly, some 3′ exonucleases are redundant with other exonucleases. Specifically, Rex1p and Rex2p function redundantly in 5.8S rRNA maturation, Rex1p, Rex2p and Rex3p are redundant for the processing of U5 snRNA and RNase P RNA, and Rex1p and the exonuclease Rrp6p have an unknown redundant essential function. Thirdly, the demonstration that the Rex proteins can affect reactions that have been attributed previously to the exosome complex indicates that an apparently simple processing step can be surprisingly complex with multiple exonucleases working sequentially in the same pathway.
Keywords: exonuclease/maturation/processing/RNA
Introduction
The production of a wide variety of stable RNA species in eukaryotic cells requires specific 3′ exonucleolytic trimming reactions. This is in sharp contrast to how the end of most proteins is generated, where most of the C-termini of mature proteins correspond to the site of termination of synthesis. One possible explanation for the widespread use of 3′ RNA processing is that it allows for flexibility in the sequence of the 3′ end. This is due to the fact that the sequences that specify the 3′ end of the primary transcript restrict the possible sequences near its 3′ end. For example, the transcription termination signal for RNA polymerase III is a run of Us, such that the primary transcript necessarily ends in a run of Us. Similarly, the primary transcript of RNA polymerase II ends in a poly(A) tail. In addition, the information to cleave and polyadenylate is mostly located near the 3′ end of the primary transcript, putting further restrictions on the sequence of the primary transcript. Generating the 3′ end of stable RNA species by a processing reaction clearly allows for more flexibility in the sequence of the 3′ end of the mature transcripts. A key step in understanding the importance of RNA 3′-processing reactions is the identification and analysis of the 3′ exoribonucleases that process RNAs.
Examination of the yeast genome sequence predicted 17 3′ to 5′ exonucleases (Mian, 1996; Moser et al., 1997). In addition, at least one yeast protein (Rrp4p) not predicted to have 3′ exonuclease activity has been found to have such an activity (Mitchell et al., 1997). The presence of many 3′ exonucleases raises several questions, such as whether individual exonucleases are required for the processing of specific RNA species, and if so, what the specific roles of each exonuclease are. Alternatively, most of the exonucleases could be redundant with each other, with any one nuclease able to process a specific RNA species.
Some information is available on the function of selected yeast exonucleases. Nine of the (putative) 3′ exonucleases (Rrp4p, Rrp6p, Rrp41p, Rrp42p, Rrp43p, Rrp44p, Rrp45p, Rrp46p and Mtr3p) are present in one protein complex named the exosome. This complex is found in both the cytoplasm and the nucleus, and is thought to function in the processing of rRNA, snRNAs and snoRNAs and in the degradation of mRNAs and the external transcribed spacer of the rRNA. (Mitchell et al., 1997; Jacobs Anderson and Parker, 1998; Allmang et al., 1999a, b; van Hoof et al., 2000).
A second class of interesting 3′ exonucleases consists of five related yeast proteins that are part of a large family of proteins that includes known 3′ exoribonucleases [i.e. RNase D, RNase T and oligoribonuclease from Escherichia coli, Rrp6p from yeast and PARN from human and Xenopus (Moser et al., 1997; Korner et al., 1998)]. One of these five yeast proteins, Pan2p, has been shown to play a role in initial shortening of the poly(A) tails of mRNA (Boeck et al., 1996; Brown and Sachs, 1998). We have named the other four proteins Rex1p [RNA exonuclease 1; open reading frame (ORF) yGR276], Rex2p (ORF yLR059), Rex3p (ORF yLR107) and Rex4p (ORF yOL080). REX2 has recently been identified as YNT20 (Hanekamp and Thorsness, 1999), a suppressor of yme1-mediated escape of DNA from the mitochondrion. It was also shown that epitope-tagged Ynt20p when overexpressed sediments in a 10 min 10 000 g centrifugation step (Hanekamp and Thorsness, 1999). This, together with a cold-sensitive respiratory growth defect, was interpreted as evidence for a mitochondrial localization of Rex2p. REX1 has recently been identified as RNH70, a 70 kDa protein that copurifies with RNase H activity. However, Rex1p/Rnh70p does not show any sequence similarity to known RNase H proteins and rex1/rnh70 mutants do not show reduced RNase H activity, or the expected phenotype for an RNase H mutant (Frank et al., 1999; Qiu et al., 1999). It is therefore not clear whether Rex1p/Rnh70p indeed functions as an RNase H in vivo, and if so, whether it has exonuclease functions in addition to the RNase H function.
The Rex1p, Rex2p, Rex3p, Rex4p and Pan2p proteins are well conserved, with at least four of them having homologs in the human genome. The proteins encoded by the uncharacterized human ORFs BAA31685, AAC31668 and CAB53690 are homologs of Pan2p, Rex1p and Rex2p, respectively. Rex4p homologs have been studied to some extent in human and Xenopus. The human gene is named either ISG20 or HEM45, and encodes a protein whose expression is induced by interferon or estrogen and localizes to the nucleus (Gongora et al., 1997; Pentecost, 1998). The Xenopus homolog (XPMC2) is also a nuclear protein that, by an unknown mechanism, can rescue a cell cycle defect when expressed in a mutant fission yeast (Su and Maller, 1995). Given the conservation of this family of 3′ to 5′ exonucleases, we analyzed their possible role in RNA-processing reactions. Here we report specific RNA-processing defects for three of these conserved proteins. In addition, this work clarifies the role of the exosome in some RNA-processing reactions.
Results
To identify what role the Rex1p, Rex2p Rex3p and Rex4p putative exonucleases play in the cell we created null mutations in the REX1, REX2, REX3 and REX4 genes by precise deletion of each of the coding regions from the yeast genome. Each of these deletion mutants is alive, indicating that these proteins are not required for viability. In addition, we did not observe any growth defects on a variety of carbon sources and at a variety of temperatures in any of these mutants (data not shown). In our strains, none of the mutations in rex genes results in a cold-sensitive growth defect on glycerol. This is different from previous reports of rex2/ynt20 mutations (Hanekamp and Thorsness, 1999). Since we anticipated that there might be functional overlap between some of these proteins, we also carried out similar analyses in strains deleted for various combinations of these genes, and also failed to find an obvious growth defect in any of these multiple mutants (including the rex1Δ, rex2Δ, rex3Δ, rex4Δ, pan2Δ pentuple mutant; data not shown).
In order to determine if the Rex proteins were required for any 3′ trimming reactions we isolated RNA from each mutant and analyzed it by Northern blotting for aberrant processing of various RNA species. While each of the rex1Δ, rex2Δ and rex3Δ mutants does show specific RNA-processing defects (see below), none of the mutants tested showed an obvious defect in a number of other RNA species, including several tRNAs, 7S RNA, U6 snRNA and several snoRNAs (data not shown). These results serve as important controls and indicate that the defects described below reflect specific roles for these genes. As additional negative controls, we analyzed strains deleted for the putative exonuclease genes SSD1, yDR514 and yCL036. These latter mutants, as well as rex4Δ, did not have any obvious growth defects, nor did they show defects in the processing of any of the RNA species tested, and thus serve as additional negative controls (data not shown).
Rex1p is required for 5S and tRNA-Arg3 maturation
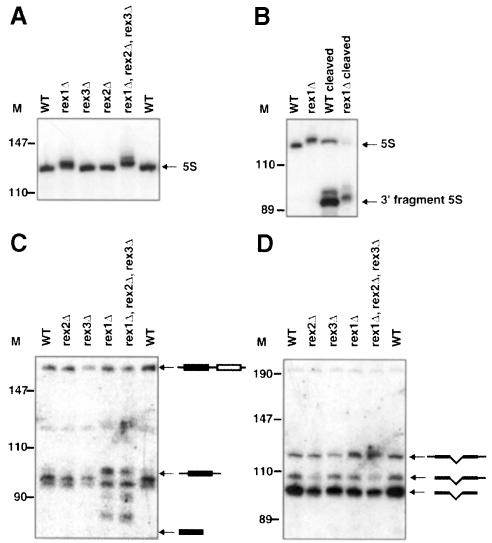
Strains deleted for REX1, but not REX2 or REX3, accumulate 5S rRNA that is longer than the corresponding RNA in wild type by ∼3 nt (Figure 1A). 5S rRNA is transcribed by RNA polymerase III as a precursor with 3′ extensions. Therefore, we hypothesized that Rex1p was required for the removal of these 3′ extensions. To test this possibility, we checked whether the longer form of 5S rRNA indeed carried 3′ extensions by cleaving the RNA with RNase H and probing for the 3′ cleavage product. As shown in Figure 1B, rex1Δ strains indeed accumulated 3′ extended forms of 5S rRNA. We interpret these observations to indicate that Rex1p is required for the proper maturation of the 3′ end of the 5S RNA (see Discussion).
Fig. 1. rex1Δ results in defects in 5S rRNA and tRNA-Arg3 processing, but not tRNA-Ser5. The strains indicated were grown to early- to mid-log phase in YPD at 30°C. RNA was extracted and analyzed by Northern blotting. Numbers to the left of each panel indicate the position of DNA molecular weight markers. (A) A Northern blot was probed for 5S rRNA. The migration of mature 5S rRNA from wild-type strains is indicated. (B) 5S rRNA was cleaved with oRP921 and RNase H, or treated with RNase H in the absence of any oligonucleotide before electrophoresis. The Northern blot was probed for 5S rRNA. The positions of mature 5S rRNA and the 3′ RNase H cleavage fragment are indicated. (C) A Northern blot was probed for 3′ extended forms of tRNA-Arg3. The migrations of a dicistronic precursor, as well as the monomeric processing intermediate with 5′ and 3′ extensions, are indicated. Also indicated is the migration of mature tRNA-Arg3, which does not hybridize to this probe. The faint band visible between 110 and 147 nt is a remaining signal from a previous probing for 5S rRNA that did not completely strip off. (D) The same Northern blot as in (C) was reprobed for intron-containing precursors to tRNA-Ser5. The positions of unspliced precursor, with and without 5′ and 3′ extensions, are indicated.
Strains lacking Rex1p were also defective in the processing of tRNA-Arg3. tRNA-Arg3 is encoded by four dicistronic genes and seven monocistronic genes (Hani and Feldmann, 1998). The precursor transcript from the dicistronic genes is processed into two monomeric intermediates. The 5′ cistron is then further processed, possibly by a 3′ exonuclease, to yield tRNA-Arg3 (Schmidt et al., 1980; Engelke et al., 1985). Figure 1C shows the altered pattern of accumulation of 3′ extended tRNA-Arg3 precursors in rex1Δ strains. This blot was probed with an oligonucleotide probe that is specific for 3′ extended forms of tRNA-Arg3 derived from the dicistronic tRNA-Arg3-Asp gene. We interpret these observations to indicate that Rex1p is required for the proper maturation of the 5′ cistron of this tRNA dicistronic unit.
No gross differences were seen when similar blots were probed for precursors to two other tRNAs (tRNA-Leu3 and tRNA-Ser5; Figure 1D and data not shown). The observation that Rex1p is not required for the processing of other tRNAs is not surprising. All other tRNA genes are transcribed as monomeric RNAs (Hani and Feldmann, 1998). The 3′ ends of these monomeric tRNAs are thought to be processed by endonucleolytic cleavage in an Lhp1p-dependent manner (Yoo and Wolin, 1997). Lhp1p is an RNA-binding protein associated with the 3′ end of nascent RNA polymerase III transcripts. The 5′ cistron of the dimeric transcript does not contain an RNA polymerase III transcription termination site and therefore is probably not bound by Lhp1p. Thus, this 5′ cistron can not be processed by an Lhp1p-dependent endonuclease.
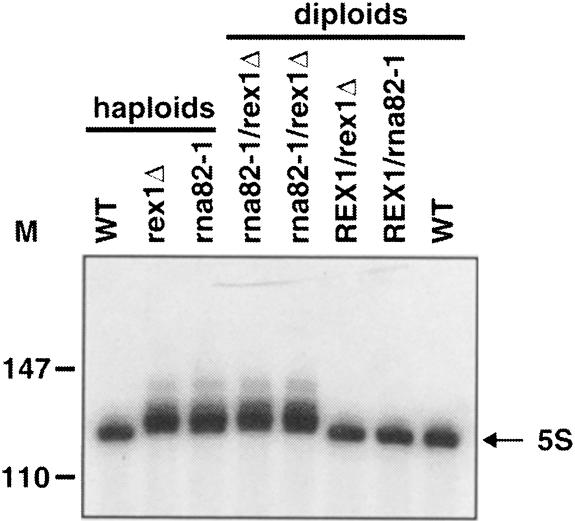
The phenotypes we described above for the rex1 deletion are similar to the phenotypes previously described for the rna82-1 mutation (Piper et al., 1983; Piper and Straby, 1989). Since the RNA82 gene has not been cloned it could either be a second gene required for the same processing steps, or RNA82 and REX1 could be the same gene. We tested whether rex1Δ and rna82-1 were in the same complementation group by crossing the two mutant strains to each other, and crossing each of them to a wild-type strain. Northern blot analyses revealed that indeed the 5S rRNA- and tRNA-processing defects were identical in the two mutants, and that they did fall in the same complementation group (Figure 2 and data not shown). To confirm that REX1 and RNA82 were the same gene we sequenced the REX1 gene from the rna82-1 strain, and found that this gene contained a mutation changing Trp433 to a stop codon. We thus conclude that REX1 and RNA82 are the same gene.
Fig. 2. rex1Δ and rna82-1 fail to complement each other. The strains indicated were grown to early- to mid-log phase in YPD at 30°C and analyzed by Northern blotting and probing for 5S rRNA. Numbers to the left indicate the position of DNA molecular weight markers.
Rex2p is required for proper U4 snRNA maturation
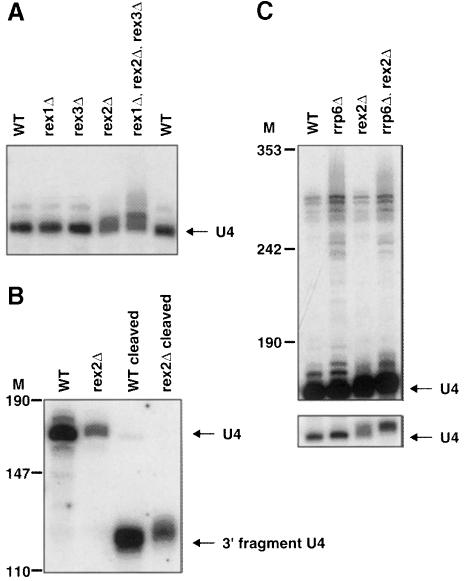
Strains deleted for REX2, but not REX1 or REX3, accumulate U4 snRNAs that were ∼1–4 nucleotides (nt) longer than the 160 nt RNA in wild type (Figure 3A). Moreover, cleavage of U4 snRNA with an oligonucleotide and probing for the 3′ fragment showed that the increase in size was due to a difference in the 3′ end of these molecules (Figure 3B). We interpret these observations to indicate that Rex2p is required for the final trimming of the 3′ end of U4 snRNA.
Fig. 3. rex2Δ results in a defect in U4 snRNA processing. The strains indicated were grown to early- to mid-log phase in YPD at 30°C. RNA was extracted and analyzed by Northern blotting. Numbers to the left of (B) and (C) indicate the position of DNA molecular weight markers. (A) A Northern blot was probed for U4 snRNA. The migration of mature U4 snRNA from wild-type strains is indicated. (B) U4 snRNA was cleaved with oRP756 and RNase H, or treated with RNase H in the absence of any oligonucleotide before electrophoresis. The Northern blot was probed for U4 snRNA. The positions of mature U4 snRNA and the 3′ RNase H cleavage fragment are indicated. (C) A dark (upper panel) and light (lower panel) exposure of the same Northern blot probed for U4 snRNA is shown. The migration of mature U4 snRNA from wild-type strains is indicated.
The 3′ trimming of the U4 transcript has been attributed previously to the exosome complex of 3′ exonucleases (Allmang et al., 1999b; van Hoof et al., 2000). Previous analyses have shown that U4 snRNA is initially transcribed as a species of >300 nt (and likely to be >500 nt). This precursor is cleaved by Rnt1p (an endonuclease) to a 295 nt product that contains U4 with a 135 nt 3′ extension. Mutants in RRP6 (and other exosome mutants) are defective in the processing of this Rnt1p cleavage product (Figure 3C; Allmang et al., 1999b; van Hoof et al., 2000). In contrast, the rex2Δ mutant showed a defect in removal of the last few nucleotides of the 3′ extension. This suggests that Rex2p acts on the product of trimming by the exosome. Interestingly, analysis of the rrp6Δ, rex2Δ double mutant showed that the rex2Δ phenotype was exacerbated. The rex2Δ, rrp6Δ double mutant contained little if any U4 snRNA of the normal size (Figure 3C, lower panel) indicating that in a rex2Δ mutant, Rrp6p (or perhaps the exosome) can partially take over the Rex2p role. These results suggest that the processing of the U4 snRNA either involves parallel pathways in which different exonucleases perform the trimming reactions or that the processing of this RNA requires the sequential action of various different exonucleases (see Discussion).
Rex3p is required for proper MRP RNA maturation
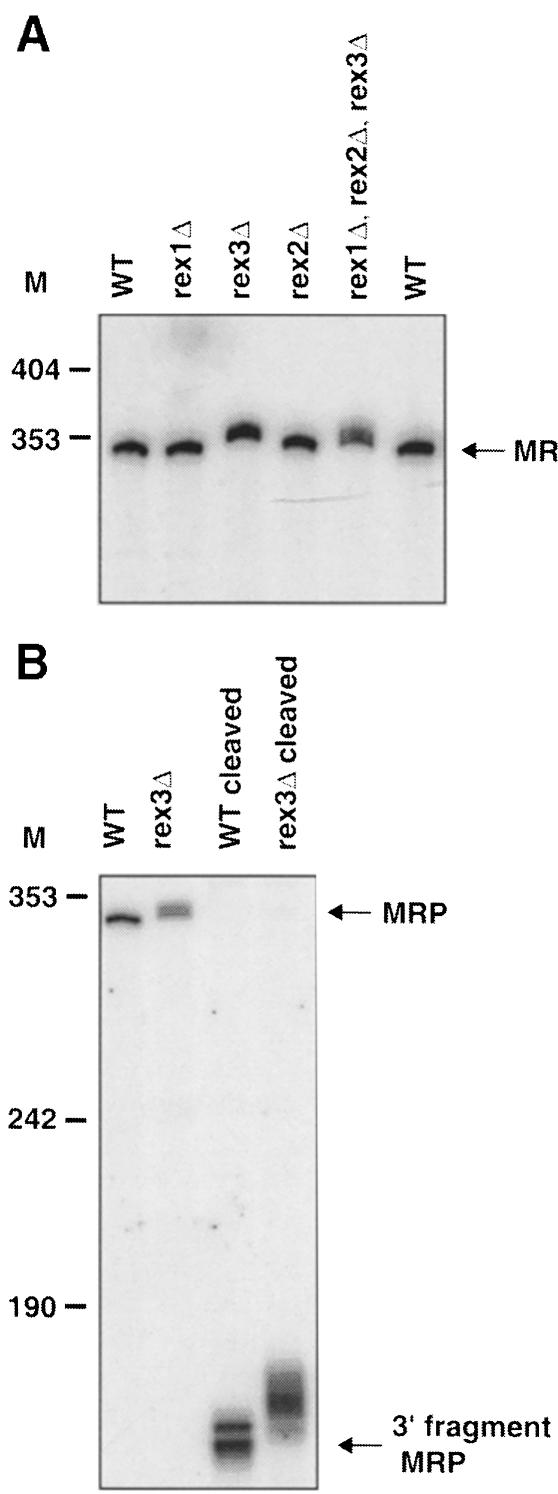
Strains deleted for REX3, but not REX1 or REX2, accumulated RNase MRP RNAs that were ∼7 nt longer than the corresponding RNA in wild type (Figure 4A). Moreover, cleavage of MRP RNA with an oligonucleotide and probing for the 3′ half showed that the increase in size was due to a difference in the 3′ end of these molecules (Figure 4B). We therefore conclude that Rex3p plays a role in 3′ end formation of the RNA subunit of RNase MRP.
Fig. 4. rex3Δ results in a defect in the processing of the RNA subunit of RNase MRP. The strains indicated were grown to early- to mid-log phase in YPD at 30°C. RNA was extracted and analyzed by Northern blotting. Numbers to the left of each panel indicate the position of DNA molecular weight markers. (A) A Northern blot was probed for the MRP RNA. The migration of mature MRP RNA from wild-type strains is indicated. (B) The MRP RNA was cleaved with oRP920 and RNase H, or treated with RNase H in the absence of any oligonucleotide before electrophoresis. The Northern blot was probed for MRP RNA. The positions of mature MRP RNA and the 3′ RNase H cleavage fragment are indicated.
Some combinations of 3′ exonucleases have redundant roles in the maturation of specific RNAs
We anticipated that some 3′ trimming reactions could be performed by multiple 3′ to 5′ exonucleases. Given this, we created and analyzed the phenotypes of a variety of multiple mutants that revealed a number of overlapping functions for these putative 3′ to 5′ exonucleases.
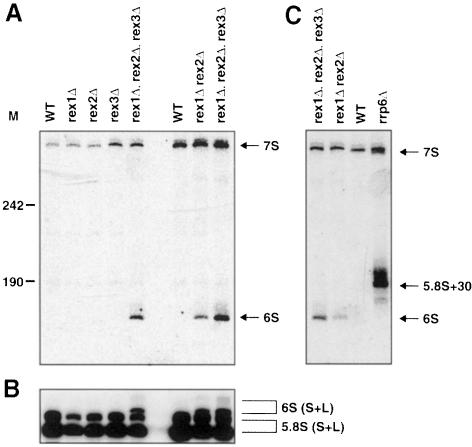
Rex1p and Rex2p are functionally redundant in the maturation of 5.8S rRNA. Analyses of various RNA-processing events in a rex1Δ, rex2Δ double mutant revealed that these two proteins have a redundant role in the processing of 5.8S rRNA. While each single mutant accumulated normal levels of 3′ extended precursors to 5.8S rRNA, the double mutant accumulated a species that we estimate to be ∼8 nt longer than the mature 5.8S rRNA (Figure 5A). This species is the correct size to correspond to the 6S pre-rRNA previously described in wild-type yeast strains (Mitchell et al., 1996). Elevated levels of 6S rRNA were not seen in any of the three single rexΔ strains, the rex1Δ, rex3Δ or rex2Δ, rex3Δ double mutants (Figure 5A and data not shown). Interestingly, the accumulation of the 6S rRNA species was higher in the rex1Δ, rex2Δ, rex3Δ triple mutant strain than in the rex1Δ, rex2Δ double mutant strain (Figure 5A), suggesting that Rex3p can also process 6S rRNA, albeit at a slower rate (see Discussion).
Fig. 5. Rex1p and Rex2p are redundant for 5.8S rRNA processing. The strains indicated were grown to early- to mid-log phase in YPD at 30°C. RNA was extracted and analyzed by Northern blotting. (A) A Northern blot was probed for 3′ extended forms of 5.8S rRNA. The positions of 7S pre-rRNA and 6S pre-rRNA are indicated. Numbers to the left of the panel indicate the position of DNA molecular weight markers. (B) The same Northern blot was reprobed with a probe for the mature 5.8S rRNA. The positions of both L and S isoforms of mature 5.8S rRNA and 6S pre-rRNA are indicated. These L and S isoforms differ from each other at their 5′ end. (C) A Northern blot was probed for 3′ extended forms of 5.8S rRNA. The positions of 7S pre-rRNA, 5.8S+30 pre-rRNA and 6S pre-rRNA are indicated.
To test to what extent rex mutants accumulate 3′ extended 5.8S rRNA we reprobed a blot containing RNA from the rex1Δ, rex2Δ, rex3Δ triple mutant with a probe for the mature 5.8S rRNA. This showed (Figure 5B) that, although most of the 5.8S rRNA in this strain is of the correct size, the rex1Δ, rex2Δ, rex3Δ triple mutant does accumulate a few percent of its 5.8S rRNA as a 6S species (see Discussion). Together these results suggest that Rex1p and Rex2p function redundantly in the 3′ end processing of 5.8S rRNA, but that Rex3p and an unidentified nuclease can to some extent substitute for Rex1p and Rex2p.
The 3′ trimming of pre-5.8S rRNA has been attributed previously to the exosome complex of 3′ exonucleases. Previous analyses indicate that the 156 nt 5.8S rRNA is initially co-transcribed with 18S and 25S rRNA as one large precursor. This precursor is processed by a combination of endonucleolytic cleavages and 5′ exonucleases to a 7S species of ∼300 nt. This 7S species is then thought to be processed by the exosome (Mitchell et al., 1997). Consequently, rrp6Δ mutants accumulate 5.8S rRNA precursors with ∼30 nt extensions (Briggs et al., 1998). Figure 5C shows that the 3′ extensions seen in rrp6Δ are longer than the ones seen in the rex1Δ, rex2Δ double mutant or the rex1Δ, rex2Δ, rex3Δ triple mutant. This suggests that the exosome acts on the 7S intermediate, and processes it to a 6S intermediate, which is subsequently processed by either Rex1p or Rex2p to yield ultimately the mature 5.8S rRNA.
In order to test the relationship between processing of 5.8S pre-rRNA by Rex1p, Rex2p and Rrp6p we attempted to create a strain lacking all three of these non-essential genes. Surprisingly, analysis of 21 tetrads from a cross between a rex1Δ strain and an rrp6Δ strain only yielded 77% viable spores, and no rex1Δ, rrp6Δ double mutants were recovered. In other crosses spore viability was 90–100% and all other combinations of double mutants of rex1Δ, rex2Δ, rex3Δ and rrp6Δ were recovered at the expected frequency in these other crosses. This indicates that rex1Δ is synthetically lethal with rrp6Δ (at least during spore germination), and therefore suggests that the Rex1p and Rrp6p proteins have a redundant role in the processing of one or more RNA species. The identity of this redundant role is not clear. One candidate is the processing of 5.8S rRNA precursors, since both Rex1p and Rrp6p are involved in this process. However, Rex2p can substitute for Rex1p in 5.8S processing, but apparently can not efficiently substitute for the Rex1p/Rrp6p redundant function.
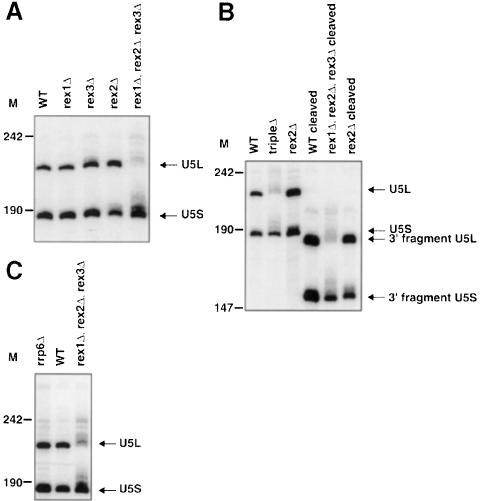
Rex1p, Rex2p and Rex3p are functionally redundant in the maturation of U5L snRNA. Analyses of the rex1Δ, rex2Δ, rex3Δ triple mutant revealed an additional role for these proteins in the processing of U5 snRNA. Wild-type yeast accumulates two forms of U5 snRNA, named U5S snRNA and U5L snRNA. U5L snRNA has been shown to be a distinct 3′ extended form of mature U5 snRNA, and is not a precursor to U5S snRNA (Chanfreau et al., 1997). The rex1Δ, rex2Δ, rex3Δ triple mutant accumulated drastically reduced levels of U5L snRNA (Figure 6A). All three single rexΔ strains, and all three possible double mutant strains accumulate normal levels of U5L snRNA (Figure 6A and data not shown), indicating that Rex1p, Rex2p and Rex3p have a redundant role in the processing of U5L snRNA. In the absence of this processing event U5L snRNA precursors are apparently degraded, or processed to U5S. This is similar to the proposed degradation of precursors of U5L snRNA and snR44, when their processing is blocked by rnt1 and exosome mutations, respectively (Chanfreau et al., 1997; van Hoof et al., 2000).
Fig. 6. Rex1p, Rex2p and Rex3p are redundant for the processing of U5 snRNA. The strains indicated were grown to early- to mid-log phase in YPD at 30°C. RNA was extracted and analyzed by Northern blotting. Numbers to the left of each panel indicate the position of DNA molecular weight markers. (A) A Northern blot was probed for the U5 snRNA. The migration of mature U5S snRNA and U5L snRNA from wild-type strains is indicated. (B) The U5 snRNA was cleaved with oRP757 and RNase H, or treated with RNase H in the absence of any oligonucleotide before electrophoresis. The Northern blot was probed for U5 snRNA. The migration of mature U5S snRNA and U5L snRNA from wild-type strains and the 3′ RNase H cleavage fragment is indicated. (C) A Northern blot was probed for the U5 snRNA. The migration of mature U5S snRNA and U5L snRNA from wild-type strains is indicated.
The rex1Δ, rex2Δ, rex3Δ triple mutant also accumulated slightly larger forms of U5S snRNA (Figure 6A). These larger forms were shown to be 3′ extended by RNase H cleavage and probing for the 3′ fragment (Figure 6B), and were also seen to a lesser extent in the rex2Δ single mutant. We conclude that Rex2p is required for normal U5S snRNA processing, but that Rex1p and Rex3p can substitute reasonably effectively for Rex2p.
The 3′ trimming of this RNA has been attributed previously to the exosome complex of 3′ exonucleases (Allmang et al., 1999b). Comparison of the defects seen in U5L snRNA processing in rrp6Δ mutants with those seen in the rex1Δ, rex2Δ, rex3Δ triple mutants revealed that the rex triple phenotype was much more severe than the rrp6Δ phenotype (Figure 6C). We therefore conclude that the Rex proteins are primarily responsible for the 3′ processing of the U5L snRNA. The triple mutant may still accumulate low levels of U5L snRNA, suggesting that there is still another exonuclease that can process U5L snRNA precursors. We propose that this other exonuclease may be the exosome. Unfortunately we could not test this directly, since the rex1Δ, rex2Δ, rex3Δ, rrp6Δ quadruple mutant is inviable (data not shown), probably because of the synthetic lethality of rex1Δ and rrp6Δ described above.
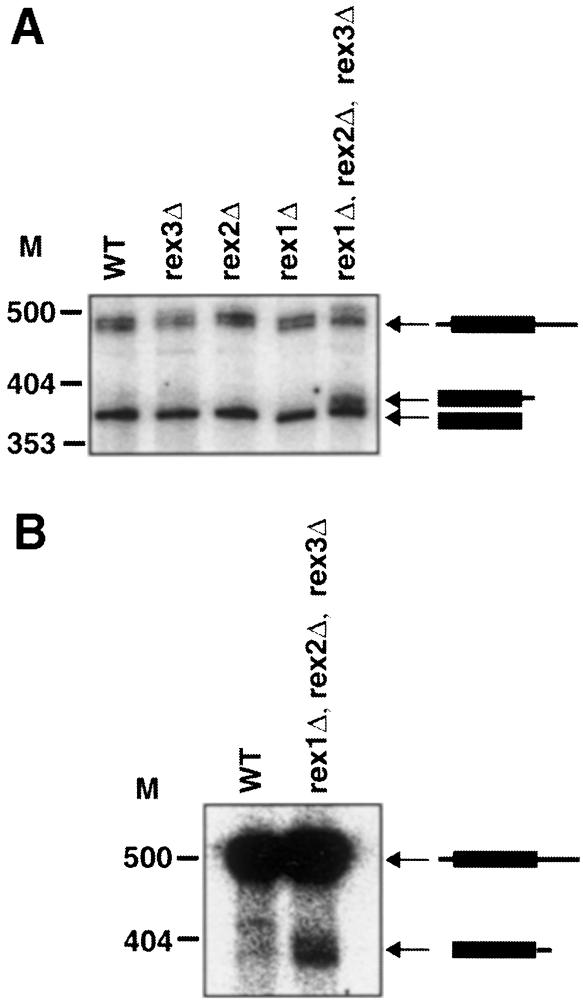
Rex1p, Rex2p and Rex3p are functionally redundant in the maturation of the RNA subunit of RNase P. The rex1Δ, rex2Δ, rex3Δ triple mutant showed a second defect in the processing of the RNA subunit of RNase P. The rex1Δ, rex2Δ, rex3Δ triple mutant accumulated a larger form of this RNA that was not present in any of the single or double mutants (Figure 7A and data not shown). This larger form was shown to be 3′ extended by probing a Northern blot with an oligonucleotide probe specific for 3′ extended forms of the RNA subunit of RNase P (Figure 7B). Wild-type yeast also accumulates two larger forms of RNase P, which are 5′ and 3′ extended and have been proposed to be precursors (Lee et al., 1991). The largest form of this putative precursor is also slightly larger in the rex1Δ, rex2Δ, rex3Δ triple mutant (Figure 7A). We interpret these results to suggest that the Rex1p, Rex2p and Rex3p proteins can all function redundantly to complete the proper maturation of the RNA subunit of RNase P.
Fig. 7. Rex1p, Rex2p and Rex3p are redundant for the processing of the RNA subunit of RNase P. The strains indicated were grown to early- to mid-log phase in YPD at 30°C. RNA was extracted and analyzed by Northern blotting. Numbers to the left of each panel indicate the position of DNA molecular weight markers. (A) A Northern blot was probed for the RNase P RNA. The migrations of mature RNase P RNA, a species with 5′ and 3′ extensions from wild-type strains and a 3′ extended species in the rex1Δ, rex2Δ, rex3Δ mutant are indicated. (B) A Northern blot was probed for 3′ extended forms of the RNA subunit of RNase P.
Discussion
The Rex1, Rex2 and Rex3 proteins are required for proper 3′ end maturation of several stable RNAs
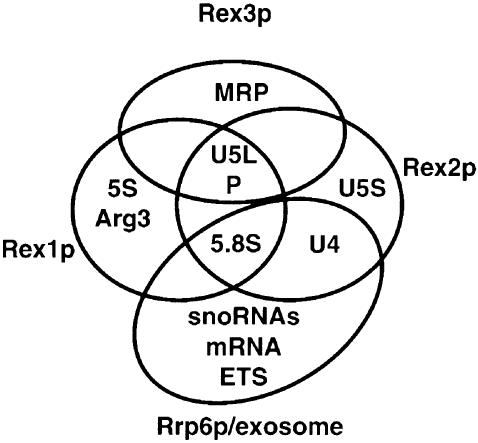
Our analyses identify the Rex proteins as being involved in a variety of RNA-processing reactions. The central observation is that strains lacking one or more of these proteins show the accumulation of 3′ extended forms of several RNAs. By this analysis, Rex1p is required for 5S rRNA and tRNA-Arg3 trimming, Rex2p is required for U4 snRNA trimming and Rex3p is required for trimming of the RNA subunit of RNase MRP. In addition, Rex1p and Rex2p function redundantly in 5.8S rRNA maturation, and Rex1p, Rex2p and Rex3p are redundant for the processing of U5 snRNA and the RNA subunit of RNase P. The identification of the Rex protein functions in RNA processing adds to a growing data set wherein various RNA 3′ to 5′ trimming reactions have been seen to be affected by lesions in specific (putative) 3′ to 5′ exonucleases (Figure 8). Preliminary evidence indicates that exonuclease activity copurifies with epitope-tagged versions of Rex1p, Rex2p and Rex3p (A.van Hoof and R.Parker, unpublished results). The simplest interpretation of these observations is that the protein lacking in each mutant is the actual 3′ exonuclease that performs the reaction. This assembly of mutant phenotypes for various (putative) exonuclease mutants allows several points to be made concerning the interaction of 3′ trimming enzymes and their substrates.
Fig. 8. Rex1p, Rex2p, Rex3p and the exosome function in a wide variety of RNA-processing reactions. A Venn diagram indicating which exonuclease is involved in the processing of various RNAs. RNA species indicated as needing two distinct proteins require both proteins at distinct steps, or require either protein for a redundant step. In cases where one protein is likely to have a major role and one has a minor role only the main role is indicated. MRP indicates processing of the RNA subunit of RNase MRP; P indicates processing of the RNA subunit of RNase P; U4, U5L and U5S indicate processing of the respective snRNA; 5S and 5.8S indicate processing of the respective rRNA; Arg3 indicates processing of tRNA-Arg3 from dicistronic precursors; snoRNAs indicates processing of many different snoRNAs; mRNA and ETS indicate degradation in the 3′ to 5′ direction of mRNA and the external transcribed spacer region of the pre-rRNA.
Some 3′ maturation events require a single specific nuclease
Our analyses indicate that in some cases 3′ processing is a simple reaction involving the removal of only a few nucleotides by a specific exonuclease. For example, 5S rRNA is transcribed by RNA polymerase III as a precursor with 7–13 nt 3′ extensions. While Rex1Δ and rna82-1 mutants accumulate mainly 5S rRNA with ∼3 nt extensions, it is important to keep in mind that this is a steady-state phenotype and may reflect a large reduction in the rate of processing. Indeed, previous analyses of the rna82–1 allele of REX1 have shown that at steady state rna82–1 mainly accumulates 5S rRNA that is extended by up to 3 nt. In contrast, pulse–chase experiments revealed a strong defect in processing of the primary transcript, which is extended by up to 13 nt (Piper et al., 1983). Therefore, Rex1p appears to be required for the efficient processing of the primary transcript to mature 5S rRNA, although in rex1 mutants an unknown nuclease can partially replace Rex1p. Similar to the defect in 5S rRNA processing, defects seen in rex mutants for other RNA-processing reactions might appear minor at steady state, but may reflect a major change in the rate of processing.
A second example of a single specific exonuclease being required for a processing step may be the RNA subunit of RNase MRP. Here, inactivation of Rex3p leads to the accumulation of a 3′ extended form. This 3′ extension maps close to an RNA polymerase III termination signal, which suggests that Rex3p is the only nuclease able to process the primary transcript.
Some 3′ maturation events can utilize multiple exonucleases for the same trimming reaction
Our analysis has also identified several 3′ trimming reactions that appear to be able to use multiple different exonucleases for the same apparent reaction. For example, efficient trimming of the 6S pre-rRNA occurs by either Rex1p or Rex2p, and therefore is defective in the double mutant. Similarly, either Rex1p, Rex2p or Rex3p can trim the U5L snRNA and RNase P precursors. It should be noted that although our analyses suggest multiple exonucleases can perform these 3′ end maturation events, it is likely that there is a preferred exonuclease under normal conditions and that the additional nucleases may be able to perform the reaction, but at a slower rate (but not detectable in the analysis of steady-state RNA). For example, Rex1p may normally process the 6S precursor to 5.8S rRNA and therefore may usually do all of this processing in vivo. However, Rex2p may substitute for Rex1p in a rex1Δ mutant, although at a slower rate. Similarly, Rex3p can substitute at a rate that is even slower, but does allow some RNA to be processed through this pathway. To resolve which of the redundant enzymes is the main activity in vivo, kinetic analyses of the various processing reactions in each mutant will be required.
3′ maturation events can utilize multiple exonucleases for different steps in the processing of a single precursor
Our analyses indicate that for some 3′ end-processing reactions, mutations in different exonucleases give rise to the accumulation of different 3′ extended forms. One example of this phenomenon is the case of U4 snRNA. Here mutations in the exosome or Rrp6p lead to the accumulation of long 3′ extended forms that can be polyadenylated (Allmang et al., 1999b; van Hoof et al., 2000). However, mutations in Rex2 lead to the accumulation of a slightly extended U4 snRNA molecule. A second example of this effect is the processing of the 5.8S rRNA precursor. Here, mutations in core exosome subunits lead to the accumulation of RNA species with long 3′ extensions, mutations in Rrp6p lead to the accumulation of a +30 species, and loss of Rex1p and Rex2p leads to the accumulation of a +8 species.
There are two possible interpretations of these observations. In one view these differences may reflect different parallel processing pathways. For example, in the case of U4 snRNA there could be one pathway that requires Rex2p, thus in the rex2Δ mutant intermediates specific for this pathway would accumulate, while other molecules that went through an exosome–Rrp6p-dependent pathway would be processed to the mature form. In essence, this is a variation on the redundancy discussed in the previous section, with the difference that in this case each pathway would proceed through distinct intermediates.
The more attractive explanation is that 3′ end-processing reactions proceed with the product of one nuclease being the substrate for another distinct nuclease. This has been suggested previously to be the case for the various exonucleases present in the exosome (Allmang et al., 1999b), but in that case it has not been resolved whether slightly different defects seen in various exosome mutants reflect the role of the mutated subunit, or reflect the altered activity of the exosome as a complex (reviewed in van Hoof and Parker, 1999). Based on both biochemical and genetic evidence, the Rex proteins do not appear to be part of the exosome complex. The Rex proteins are not present in purified exosomes (Allmang et al., 1999a), and the defect seen in rex mutants is clearly different from those seen in exosome mutants (Mitchell et al., 1997; Jacobs Anderson and Parker, 1998; Allmang et al., 1999a, b; van Hoof et al., 2000). Therefore, the roles of the Rex proteins can easily be separated from the roles of the exosome.
Experimental evidence for two exonucleases working sequentially in the same pathway can be found for the processing 5.8S rRNA. As described above, deletion of REX1 and REX2 leads to the accumulation of 6S pre-rRNA, suggesting that 6S RNA is a substrate for these proteins. In contrast, depletion of most of the exosome subunits leads to a reduction in the levels of 6S pre-rRNA (Allmang et al., 1999b), suggesting that 6S RNA is the product of the exosome. Together these two observations suggest that the product of the exosome is identical to the substrate of Rex1p and Rex2p, and thus indicate that in the processing of 5.8S rRNA precursors, the 3′ extension is removed sequentially by the exosome and the Rex proteins.
The observation that a simple 3′ trimming reaction is actually a series of discrete reactions may be a common property of 3′-processing reactions. Another example of this phenomenon is the 3′ to 5′ deadenylation and degradation of mRNA that also appears to require sequential action of diverse 3′ exonucleases. This degradation consists of initial deadenylation, poly(A) shortening, final deadenylation and digestion of the body of the mRNA. The initial deadenylation step is carried out by Pan2p (Brown and Sachs, 1998). The exonucleases carrying out poly(A) shortening and final deadenylation are not yet known. Finally, the body of an mRNA is degraded by the exosome (Jacobs Anderson and Parker, 1998). It therefore appears that in a growing number of cases 3′ processing and/or degradation requires sequential action of distinct 3′ exonucleases.
Importance of 3′ processing of stable RNAs
Our results suggest that the precise 3′ trimming of many stable RNAs may not be a critical process for cellular function. The central observation is that in a variety of mutant strains various stable RNAs are produced that are 3′ extended and in some cases no fully mature RNA is produced, yet those mutant strains are fully viable, at least under standard laboratory conditions. For example, the rex1Δ, rex2Δ, rex3Δ triple mutant is indistinguishable from wild type in growth, yet fails to accumulate substantial amounts of fully mature 5S, U4, U5L and RNase MRP RNA, and has additional defects in 5.8S rRNA and RNase P RNA processing. This suggests that these 3′ extended RNAs can carry out their normal function, as has been demonstrated previously for 3′ extended forms of 5.8S rRNA (Briggs et al., 1998). However, it should be noted that in some cases, such as tRNA processing, the precise 3′ end is essential.
Unique and overlapping specificity of exonucleases
The observation that the Rex proteins can have both unique and overlapping substrates raises an interesting question. How is the specificity of these nucleases controlled such that some substrates are uniquely processed by one exonuclease and other RNAs can be trimmed by any of the three exonucleases? Although the answer to this question is currently unclear, the requirement for a specific nuclease for a processing event could occur in at least three ways. First, it could be that there are specific features of the substrate RNA or RNP that actively recruit the required nuclease, and only that nuclease, to this substrate. For example, a Rex protein may have protein–protein interactions with the substrate RNP. Secondly, the substrate may have inhibitory regions that prevent any of the other exonucleases from acting on that substrate. For example, the exosome may not be able to remove the last few nucleotides from 5.8S rRNA precursors, because of steric hindrance by a secondary structure, or a bound protein. These two possibilities are not mutually exclusive and the proper match between substrate and enzyme may be a combination of both positive recruitment elements and negatively acting elements. Finally, it may be that the particular substrate and processing enzyme are co-compartmentalized such that there is only one exonuclease locally available for that event.
Materials and methods
Strains and plasmids
All yeast was grown in standard YEP media containing 2% glucose. The genotype of all strains can be found in Table I. All strains are completely isogenic to each other with the exception of strains containing the rna82-1 mutation.
Table I. Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| yRP840 | MATa leu2-3 112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | Hatfield et al. (1996) |
| yRP841 | MATα leu2-3 112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | Hatfield et al. (1996) |
| yRP1375 | MATa/MATα leu2-3 112/leu2-3 112 LYS2/lys2-201 HIS4/his4-539 trp1/trp1 ura3-52/ura3-52 cup1::LEU2/PGK1pG/MFA2pG/cup1::LEU2/PGK1pG/MFA2pG | van Hoof et al. (2000) |
| yRP1377 | MATα leu2-3 112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rrp6Δ::URA3 | van Hoof et al. (2000) |
| yRP1449 | MATa leu2-3 112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rex1Δ::TRP1 | this study |
| yRP1450 | MATα leu2-3 112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rex2Δ::LYS2 | this study |
| yRP1451 | MATa leu2-3 112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rex3Δ::HIS4 | this study |
| yRP1452 | MATa leu2-3 112 lys2-201 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rex1Δ::TRP1 rex2Δ::LYS2 | this study |
| yRP1453 | MATα leu2-3 112 lys2-201 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rex1Δ::TRP1 rex2Δ::LYS2 rex3Δ::TRP1 | this study |
| yRP1454 | MATα leu2-3 112 lys2-201 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rrp6Δ::URA3 rex2Δ::LYS2 | this study |
| pp1002 | MATa leu2 ade2 pep4-3 rna82-1 | Piper et al. (1983) |
| yRP1455 | MATa/MATα leu2-3 112/leu2 LYS2/lys2-201 HIS4/his4-539 TRP1/trp1 URA3/ura3-52 CUP1/cup1::LEU2/PGK1pG/MFA2pG ADE2/ade2 PEP4/pep4-3 rna82-1/rex1Δ::TRP1 | this study |
| yRP1456 | MATa/MATα leu2-3 112/leu2 LYS2/lys2-201 HIS4/his4-539 TRP1/trp1 URA3/ura3-52 CUP1/cup1::LEU2/PGK1pG/MFA2pG ADE2/ade2 PEP4/pep4-3 rna82-1/rex1Δ::TRP1 | this study |
| yRP1457 | MATa/MATα leu2-3 112/leu2-3 112 LYS2/lys2-201 HIS4/his4-539 trp1/trp1 ura3-52/ura3-52 cup1::LEU2/PGK1pG/MFA2pG/cup1::LEU2/PGK1pG/MFA2pG REX1/rex1Δ::TRP1 | this study |
| yRP1458 | MATa/MATα leu2/leu2-3 112 LYS2/lys2-201 TRP1/trp1 URA3/ura3-52 CUP1/cup1::LEU2/PGK1pG/MFA 2pG ADE2/ade2 PEP4/pep4-3 REX1/rna82-1 | this study |
Strain yRP1451 was created by transforming strain yRP840 with a fragment of pRP963. Strain yRP1450 was created by transforming strain yRP841 with a fragment of pRP964. Strain yRP1449 was created by transforming strain yRP841 with a fragment of pRP965. pRP963, pRP964 and pRP965 carry the HIS4 gene, the LYS2 gene and the TRP1 gene, respectively, in a pBluescript derivative flanked by 5′ and 3′ flanking sequence of the relevant REX gene. Plasmids pRP963, pRP964 and pRP965 were created by PCR amplifying two ∼500 nt fragments of the genome upstream and downstream of the gene of interest with oligonucleotides containing suitable restriction endonuclease sites, and cloning these PCR fragments into pBluescript derivatives already containing the relevant yeast marker. The resulting strains contain null alleles of the relevant ORF. Deletion of the gene of interest was confirmed by Southern blotting.
Strains yRP1452, yRP1453 and yRP1454 were created by crossing strains containing the relevant deletions. Strains yRP1455 and yRP1456 were created by crossing strain yRP1449 with strain pp1002. Strain yRP1457 was created by crossing strain yRP1449 to yRP840. Strain yRP1458 was created by crossing strain pp1002 to yRP841.
RNA extraction and Northern blotting
All yeast was grown in standard YEP containing 2% dextrose (YPD) at 30°C to an OD600 of 0.3 to 0.5. RNA was extracted, separated on polyacrylamide gels, transferred and probed as described by Jacobs Anderson and Parker (1998). Where indicated RNA was cleaved with RNase H. This was achieved by first annealing a specific oligo to 10 μg of RNA in 10 μl of 25 mM Tris–HCl pH 7.5, 1 mM EDTA, 50 mM NaCl for 10 min at 65°C, followed by addition of 10 μl of 40 mM Tris–HCl pH 7.5, 20 mM MgCl2, 100 mM NaCl, 2 mM dithiothreitol, 60 μg/ml bovine serum albumin, 1 U RNase H. The reaction was allowed to proceed for 1 h at 30°C. The RNA was then recovered by phenol/chloroform extraction and ethanol precipitation. The sequence of oligonucleotides used as probes or for RNase H cleavage can be found in Table II.
Table II. Oligonucleotides used in this study.
| RNA species | Name | Sequence |
|---|---|---|
| U4 snRNA | oRP756 | cggacgaatcctcactgata |
| U4 snRNA | oRP917 | gcgaacaccgaattgaccat |
| U5 snRNA | oRP757 | gttgacctccctccgcca |
| U5 snRNA | oRP918 | ctatggagacaacacccgg |
| 5S rRNA | oRP769 | gcgtatggtcacccactac |
| 5S rRNA | oRP921 | cgggaaacggtgctttctg |
| 5.8S rRNA | oRP924 | tttcgctgcgttcttcatc |
| 6S pre-rRNA | oJA003 | tgagaaggaaatgacgct |
| RNase P RNA | oRP766 | gttcgccactaatgacgtcc |
| pre-RNase P RNA | oRP940 | gttggatatgggctggaaca |
| RNase MRP RNA | oRP765 | cacgggaaagagcaatcgtc |
| RNase MRP RNA | oRP920 | tacccaagggcatcctcct |
| pre-tRNA-Ser5 | oRP779 | agccgaactttttattccattcg |
| pre-tRNA-Leu3 | oRP780 | cccacagttaactgcggtc |
| pre-tRNA-Arg3 | oRP781 | agaaacaaagcactcacgat |
Sequencing of rna82-1
The REX1 gene from strain pp1002 (including 345 nt 5′ of the coding region and 480 nt 3′ of the coding region) was PCR amplified using oligonucleotides oRP708 and oRP711. The PCR was done in duplicate, and each reaction was independently sequenced using oligonucleotides oRP708, oRP711, oRP822, oRP823 and oRP824 (sequence of oligonucleotides available on request). The sequence from both PCRs was identical and differed from the published genome sequence in SGD by only 1 nt; a TGG tryptophan codon was changed to a TGA nonsense codon.
Acknowledgments
Acknowledgements
We wish to thank Dr Peter W.Piper for the rna82-1 strain pp1002. We are grateful to members of the Parker laboratory for helpful comments on this work. This work was supported by the Howard Hughes Medical Institute and NIH grant number GM45443 to R.P.
References
- Allmang C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D. and Mitchell, P. (1999a) The yeast exosome and human PM-Scl are related complexes of 3′ to 5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E. and Tollervey, D. (1999b) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R., Tarun, S., Jr, Rieger, M., Deardorff, J.A., Muller-Auer, S. and Sachs, A.B. (1996) The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem., 271, 432–438. [DOI] [PubMed] [Google Scholar]
- Briggs M.W., Burkard, K.T. and Butler, J.S. (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem., 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- Brown C.E. and Sachs, A.B. (1998) Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol., 18, 6548–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Elela, S.A., Ares, M., Jr and Guthrie, C. (1997) Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev., 11, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke D.R., Gegenheimer, P. and Abelson, J. (1985) Nucleolytic processing of a tRNAArg–tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae.J. Biol. Chem., 260, 1271–1279. [PubMed] [Google Scholar]
- Frank P., Braunshofer-Reiter, C., Karwan, A., Grimm, R. and Wintersberger, U. (1999) Purification of Saccharomyces cerevisiae RNase H (70) and identification of the corresponding gene. FEBS Lett., 450, 251–256. [DOI] [PubMed] [Google Scholar]
- Gongora C., David, G., Pintard, L., Tissot, C., Hua, T.D., Dejean, A. and Mechti, N. (1997) Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem., 272, 19457–19463. [DOI] [PubMed] [Google Scholar]
- Hanekamp T. and Thorsness, P.E. (1999) YNT20, a bypass suppressor of yme1 yme2, encodes a putative 3′-5′ exonuclease localized in mitochondria of Saccharomyces cerevisiae.Curr. Genet., 34, 438–448. [DOI] [PubMed] [Google Scholar]
- Hani J. and Feldmann, H. (1998) tRNA genes and retro-elements in the yeast genome. Nucleic Acids Res., 26, 689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L., Beelman, C.A., Stevens, A. and Parker, R. (1996) Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae.Mol. Cell. Biol., 16, 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson J.S. and Parker, R. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner C.G., Wormington, M., Muckenthaler, M., Schneider, S., Dehlin, E. and Wahle, E. (1998) The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J., 17, 5427–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Rohlman, C.E., Molony, L.A. and Engelke, D.R. (1991) Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol. Cell. Biol., 11, 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian I.S. (1996) Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res., 25, 3187–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Petfalski, E. and Tollervey, D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski, E., Shevchenko, A., Mann, M. and Tollervey, D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Moser M.J., Holley, W.R., Chatterjee, A. and Mian, I.S. (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentecost B.T. (1998) Expression and estrogen regulation of the HEM45 mRNA in human tumor lines and in the rat uterus. J. Steroid Biochem. Mol. Biol., 64, 25–33. [DOI] [PubMed] [Google Scholar]
- Piper P.W. and Straby, K.B. (1989) Processing of transcripts of a dimeric tRNA gene in yeast uses the nuclease responsible for maturation of the 3′ termini upon 5 S and 37 S precursor rRNAs. FEBS Lett., 250, 311–316. [DOI] [PubMed] [Google Scholar]
- Piper P.W., Bellatin, J.A. and Lockheart, A. (1983) Altered maturation of sequences at the 3′ terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J., 2, 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Qian, Y., Frank, P., Wintersberger, U. and Shen, B. (1999) Saccharomyces cerevisiae RNase H (35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol., 19, 8361–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O., Mao, J., Ogden, R., Beckmann, J., Sakano, H., Abelson, J. and Soll, D. (1980) Dimeric tRNA precursors in yeast. Nature, 287, 750–752. [DOI] [PubMed] [Google Scholar]
- Su J.Y. and Maller, J.L. (1995) Cloning and expression of a Xenopus gene that prevents mitotic catastrophe in fission yeast. Mol. Gen. Genet., 246, 387–396. [DOI] [PubMed] [Google Scholar]
- van Hoof A. and Parker, R. (1999) The exosome: a proteasome for RNA?Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Lennertz, P. and Parker, R. (2000) Yeast exosome mutants accumulate 3′ extended polyadenylated forms of U4 snRNA and snoRNAs. Mol. Cell. Biol., 18, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.J. and Wolin, S.L. (1997) The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell, 89, 393–402. [DOI] [PubMed] [Google Scholar]