Abstract
Drosophila Staufen protein is required for the localization of oskar mRNA to the posterior of the oocyte, the anterior anchoring of bicoid mRNA and the basal localization of prospero mRNA in dividing neuroblasts. The only regions of Staufen that have been conserved throughout animal evolution are five double-stranded (ds)RNA-binding domains (dsRBDs) and a short region within an insertion that splits dsRBD2 into two halves. dsRBDs 1, 3 and 4 bind dsRNA in vitro, but dsRBDs 2 and 5 do not, although dsRBD2 does bind dsRNA when the insertion is removed. Full-length Staufen protein lacking this insertion is able to associate with oskar mRNA and activate its translation, but fails to localize the RNA to the posterior. In contrast, Staufen lacking dsRBD5 localizes oskar mRNA normally, but does not activate its translation. Thus, dsRBD2 is required for the microtubule-dependent localization of osk mRNA, and dsRBD5 for the derepression of oskar mRNA translation, once localized. Since dsRBD5 has been shown to direct the actin-dependent localization of prospero mRNA, distinct domains of Staufen mediate microtubule- and actin-based mRNA transport.
Keywords: bicoid mRNA/dsRNA-binding domain/mRNA localization/Staufen/translational control
Introduction
The establishment of cell polarity requires the targeting of specific proteins to the regions of a cell where they are required, and this is often achieved by localizing the mRNAs that encode them (St Johnston, 1995; Bashirullah et al., 1998). In many cases, mRNA localization is thought to be an active process that requires the cytoskeleton. For example, mating type switching in Saccharomyces cerevisiae is restricted to the mother cell by the myosin-dependent transport of ash1 mRNA into the emerging daughter cell, and the directed motility of cultured fibroblasts requires the actin-dependent localization of β–actin mRNA (Kislauskis et al., 1994; Bertrand et al., 1998). Other mRNAs are localized by microtubule-dependent mechanisms, such as Vg1 mRNA, which moves to the vegetal pole of the Xenopus oocyte, and bicoid (bcd) and oskar (osk) mRNAs, which localize to opposite poles of the Drosophila oocyte (Yisraeli et al., 1990; Pokrywka and Stephenson, 1991; Clark et al., 1994). The importance of microtubule-dependent mRNA localization has been most clearly demonstrated in the case of the latter two transcripts, since their positions define the anterior–posterior axis of the embryo. bcd mRNA localizes to the anterior of the egg, and is translated after fertilization to produce a morphogen gradient that patterns the head and thorax of the embryo; the localization of osk mRNA to the posterior of the oocyte directs the assembly of the pole plasm, which contains posterior and germline determinants (Ephrussi and Lehmann, 1992; Driever, 1993).
The cis–acting signals that direct mRNA localization have been mapped in several transcripts, and in a few cases biochemical approaches have led to the identification of RNA-binding proteins that interact with these signals (Bashirullah et al., 1998). However, the best characterized example of an RNA-binding protein required for mRNA localization is Drosophila Staufen protein (Stau), which was identified in a genetic screen (Schüpbach and Wieschaus, 1986). Subsequent work has shown that Stau plays an essential role in the localization of three different mRNAs during development. It is required for (i) the localization of osk mRNA to the posterior of the oocyte (Ephrussi et al., 1991; Kim-Ha et al., 1991, 1995); (ii) the anchoring of bcd mRNA at the anterior of the egg (St Johnston et al., 1989); and (iii) the basal localization of prospero mRNA during the asymmetric divisions of embryonic neuroblasts (Li et al., 1997; Broadus et al., 1998; Matsuzaki et al., 1998; Schuldt et al., 1998; Shen et al., 1998). Although it has not been possible to test whether Stau binds specifically to these mRNAs, it contains five copies of a double-stranded (ds)RNA-binding domain (dsRBD), and the third of these has been shown to bind to dsRNA in vitro (St Johnston et al., 1992). When mutations that abolish the RNA-binding activity of dsRBD3 are incorporated into a full-length Stau transgene, this construct no longer rescues the localization of either osk or bcd mRNAs (Ramos et al., 2000). Thus, the dsRNA-binding activity of dsRBD3 is required for bcd and osk mRNA localization, strongly suggesting that Stau binds these RNAs directly.
During stages 7–9 of Drosophila oogenesis, osk mRNA localizes transiently at the anterior of the oocyte, and then moves to the posterior pole at stage 9 in a microtubule-dependent manner (Ephrussi et al., 1991; Kim-Ha et al., 1991; Clark et al., 1994). In stau null mutants, however, osk mRNA fails to move to the posterior and remains at the anterior. Several lines of evidence indicate that Stau protein associates with osk mRNA to mediate its posterior localization. (i) Stau protein co-localizes with osk mRNA at the anterior of the oocyte and moves with the RNA to the posterior (St Johnston et al., 1991). (ii) Stau and osk RNA mislocalize to the same ectopic sites in mutants, such as gurken, which alter the polarity of the oocyte (González-Reyes et al., 1995; Roth et al., 1995). (iii) The posterior localization of Stau depends on osk mRNA (Ferrandon et al., 1994). In females carrying extra copies of an osk transgene, the increased quantity of osk mRNA produced induces a corresponding increase in the amount of Stau that localizes to the posterior pole. Thus, Stau is present in excess, and only the protein that is associated with osk RNA localizes to the posterior.
Translation of unlocalized osk mRNA is repressed by the binding of Bruno protein to Bruno-response elements (BRE) in the 3′UTR (Kim-Ha et al., 1995; Gunkel et al., 1998). An osk transgene lacking the BRE (oskBRE–) is therefore translated prior to its localization, leading to the production of ectopic Osk, which causes a range of patterning defects in the resulting embryos. Low levels of ectopic Osk result in a loss of head and thoracic segments, while higher levels induce the formation of bicaudal embryos, with abdomens at both ends. Although the translation of oskBRE– mRNA no longer depends on its localization, it still requires Stau protein, since the bicaudal phenotypes are suppressed in a stau null mutant background. Thus, Stau plays a role in the translation of oskBRE– mRNA that is independent of its role in posterior localization, suggesting that it may also be involved in the translational regulation of wild-type osk mRNA. Finally, Stau has also been implicated in the anchoring of osk mRNA at the posterior. When a temperature-sensitive stau allele is kept under semi-restrictive conditions, Stau and osk mRNA localize to the posterior of the oocyte at stage 10, but are not maintained there in the embryo (St Johnston et al., 1991; Rongo et al., 1995).
Although Stau is not involved in the initial anterior localization of bcd mRNA, it is required to anchor the mRNA during the final stages of oogenesis (St Johnston et al., 1989). bcd RNA is normally released from the cortex at some time between stage 12 of oogenesis and egg deposition, and remains tightly localized in a spherical region of cytoplasm at the anterior of the egg. In stau mutant eggs, however, the RNA forms a shallow anterior–posterior gradient, and the resulting embryos have head defects because there is insufficient Bicoid protein at the very anterior of the embryo. This function of Stau shows several parallels to its role in osk mRNA localization (Ferrandon et al., 1994). First, Stau protein co-localizes with bcd mRNA at the anterior of the egg, and this localization is bcd mRNA dependent. Second, when the bcd 3′UTR is injected into the egg, it recruits Stau into particles that localize to the poles of the mitotic spindles. Stau, therefore, mediates the microtubule-dependent localization of both bcd and osk mRNAs, but at two different stages of development. Furthermore, in each case the localization of Stau requires its interaction with the appropriate RNA, suggesting that Stau–RNA complexes are the substrate for localization.
More recently, Stau has been shown to mediate the localization of prospero mRNA during the asymmetric divisions of embryonic neuroblasts (Li et al., 1997; Broadus et al., 1998; Fuerstenberg et al., 1998; Matsuzaki et al., 1998; Schuldt et al., 1998; Shen et al., 1998). In contrast to the localization of bcd and osk mRNAs, the localization of prospero mRNA–Stau complexes is disrupted by actin-destabilizing drugs, but not by microtubule-depolymerizing drugs (Broadus and Doe, 1997).
The discovery that Stau can mediate both microtubule- and actin-dependent mRNA localization raises the question of how different Stau–mRNA complexes are coupled to distinct transport pathways. It has previously been shown that Miranda protein binds to the dsRBD5 of Stau to direct the basal localization of prospero mRNA (Fuerstenberg et al., 1998; Schuldt et al., 1998; Shen et al., 1998). However, it remains unclear how Stau links osk and bcd mRNAs to the microtubule-based transport machinery, or how the activation of osk mRNA translation at the posterior is achieved. In this paper, we address this question by analysing the domains of Stau to determine which regions of the protein are required for these functions.
Results
Conservation of Stau throughout the animal kingdom
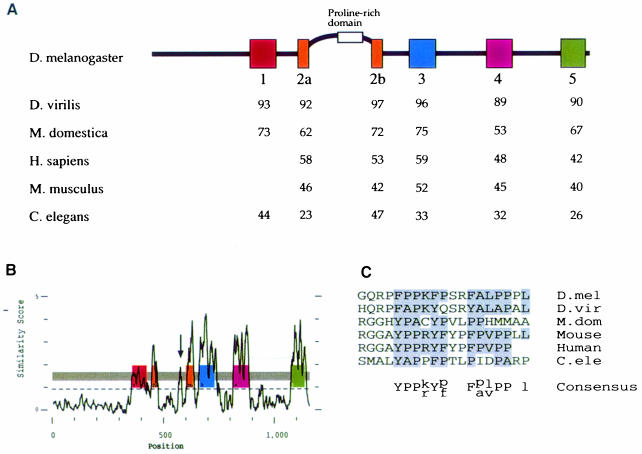
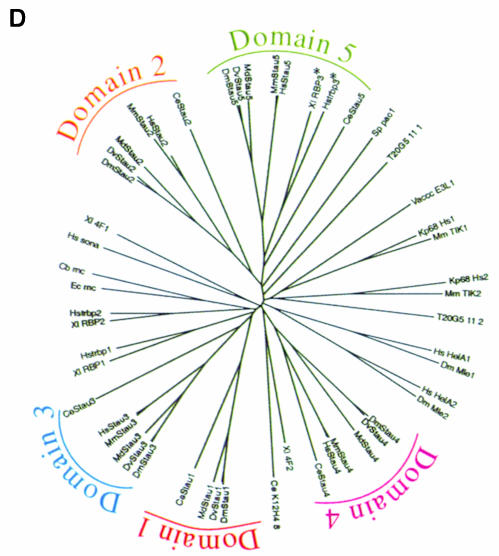
Since the functional domains of a protein can often be identified from their conservation during evolution, we cloned and sequenced homologues of stau from two other insect species, Drosophila virilis (Dvstau) and Musca domestica (Mdstau), which diverged from Drosophila melanogaster over 60 and 100 million years ago, respectively. In addition, Wickham et al. (1999) and Marión et al. (1999) have recently reported Stau homologues in Caenorhabditis elegans (Cestau), mouse (Mmstau) and human (Hsstau), and we also identified these on the basis of their homology to the insect genes and sequenced them in their entirety. The predicted amino acid sequences of the invertebrate homologues include five dsRBDs, corresponding to dsRBDs 1–5 of D.melanogaster Stau. In contrast, the human and mouse homologues include only four domains, which are most closely related to dsRBDs 2, 3, 4 and 5 of the invertebrate Stau (Figure 1D).
Fig. 1. A comparison of Staufen homologues from different species. (A) A diagram showing the positions of the conserved dsRBDs in D.melanogaster Staufen protein, and the percentage amino acid identity between these domains and the equivalent domains in other species. (B) A PlotSimilarity diagram of a ClustalW alignment of all six Staufen homologues. The positions of the dsRBDs are superimposed on the plot, and show that the similarity between homologues is almost entirely restricted to the dsRBDs. The arrow marks the only other region of similarity, which falls within the insertion in dsRBD2. Note that the degree of similarity shown for dsRBD1 appears lower than that for the other four dsRBDs because this domain is present in only 4/6 homologues. (C) Alignment of the conserved region in the dsRBD2 insertion. (D) An unrooted tree derived from a ClustalW alignment of dsRBDs from different proteins. Corresponding domains in the different Staufen homologues are more similar to each other than they are to any other dsRBDs, with the exception of CeStau dsRBD5, which is approximately similar to the other Stau dsRBD5s and to the third dsRBD of human TAR RNA-binding protein (Hstrbp) and Xenopus Xlrbpa (marked with asterisks). For simplicity, this diagram only includes dsRBDs described up to 1995.
Despite the absence of a dsRBD1 equivalent in the mouse and human sequences, we believe that they represent vertebrate homologues of Stau because of the high degree of similarity to DmStau within the remaining four domains (Figure 1A). A ClustalW analysis of dsRBDs from many proteins reveals that apart from one minor exception for dsRBD5, a given Stau domain is most similar to the equivalent domain in each of the other Stau homologues (Figure 1D). This suggests that evolution is not acting simply to maintain similarity to a dsRBD consensus sequence, but rather that each domain has unique features that have been conserved during evolution. It is particularly notable that in all six homologues, dsRBD2 is split into two parts by up to 118 aa of non-dsRBD sequence. While dsRBDs have been identified in many proteins, the distinctive split in dsRBD2 has only been observed in these six Stau homologues.
Analysis of an alignment of the Stau homologues reveals that the only portions of the protein to have been conserved during evolution are the five dsRBDs (Figure 1B). For example, the M.domestica and D.melanogaster proteins show an average of 67% amino acid identity within the dsRBDs, but <15% in the rest of the protein. dsRBD2 and dsRBD5 were originally described as ‘half domains’ showing similarity to the dsRBD consensus only over the C-terminal portion of the domain (St Johnston et al., 1992). However, the conservation extends over a region corresponding to the length of a whole domain, and these domains should therefore be considered as complete, albeit divergent, dsRBDs, in agreement with the results of Gibson and Thompson (1994). The only other obvious homology between these proteins is a short region within the insertion in the middle of dsRBD2 that is rich in proline and aromatic amino acids (Figure 1B and C). Since the regions of the protein essential for its activity are expected to be conserved during evolution, the dsRBDs and this proline-rich region are likely to mediate all of the functions of Stau, including its ability to bind both mRNA and the factors that localize Stau–mRNA complexes.
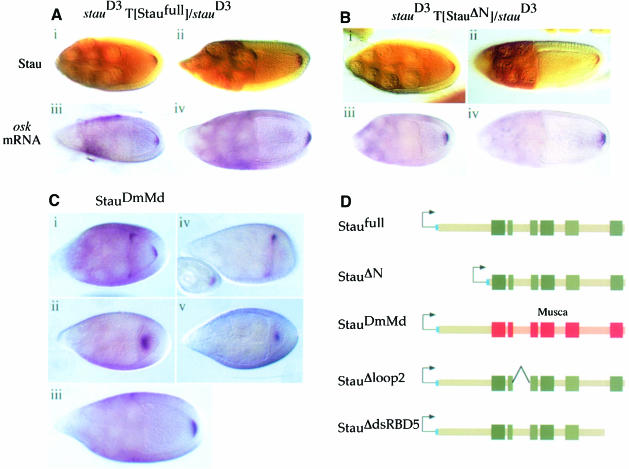
To determine whether the dsRBDs are indeed the only part of Stau necessary for its function, we generated a transgene in which the large non-conserved N-terminal region of DmStau was deleted, and crossed this construct into a stau null mutant background (stauD3). Like the full-length protein expressed from the same vector (Staufull), StauΔN localizes normally to the posterior of the oocyte, and completely rescues the posterior localization and anchoring of osk mRNA (Figure 2A and B). Furthermore, the eggs laid by StauΔN females show a wild-type localization of bcd mRNA at the anterior pole (data not shown). This rescue of the maternal function of stau is also reflected in the phenotype of the embryos produced by StauΔN females. Whereas the embryos laid by stauD3 mutant females die with head defects and no abdomen, almost all of the progeny of StauΔN females hatch into larvae, and have normal heads and almost wild-type abdominal segmentation (Table I). Furthermore, a similar proportion of the adult offspring of StauΔN females had gametic ovaries (85%) compared with those of females carrying Staufull (88%), indicating that this construct leads to the production of the high levels of Osk activity that are necessary to specify the germline (Table I). Thus, the portion of Stau that includes the dsRBDs is able to mediate all of Stau's functions during oogenesis, including localization of both osk and bcd mRNAs, activation of osk translation and maintenance of pole plasm at the posterior.
Fig. 2. The dsRBDs are the only conserved regions of Stau required for osk mRNA localization. (A) The localization of Staufen protein (i and ii) and osk mRNA (iii and iv) in stage 9 and 10 oocytes from stauD3 T[Staufull]/stauD3 females. Full-length Staufen protein expressed from the transgene localizes normally to the posterior of the oocyte, and rescues the osk mRNA posterior localization defect of a staufen null mutation. (B) The localization of Stau protein (i and ii) and osk mRNA (iii and iv) in stage 9 and 10 oocytes from stauD3 T[StauΔN]/stauD3 females. Stau protein lacking the non-conserved N-terminal 282 aa also localizes normally and rescues osk mRNA localization and anchoring. (C) (i) In wild-type egg chambers, osk mRNA shows a transient localization to the anterior of the oocyte during stage 9. (ii) In stauD3 T[StauDmMd]/stauD3 egg chambers, osk mRNA fails to accumulate at the anterior margin at stage 9, and localizes instead to the centre of the oocyte. (iii) However, the mRNA shows a normal localization at the posterior pole by stage 10. (iv) In stauD3 egg chambers, all osk mRNA remains anchored at the anterior of the oocyte. (v) osk mRNA shows a transient localization to a point in the centre of the oocyte, when StauDmMd is expressed in the presence of wild-type Drosophila Stau protein. (D) A diagram showing the structure of the Stau proteins encoded by the Staufull, StauΔN, StauDmMd, StauΔloop2 and StauΔdsRBD5 transgenes. The boxes indicate the positions of the dsRBDs. The short leader peptide containing the myc epitope tag is labelled in blue, Drosophila sequences in green and M.domestica sequences in red or pink.
Table I. Rescue of the stau null phenotype by stau transgenes.
| Genotype | Average No. of abdominal denticle belts | % adults with gametic ovaries | % normal heads |
|---|---|---|---|
| wild type | 8 ± 0 | 100 | 100 |
| stauD3 | 0.04 ± 0.002 | n/a | 0 |
| stauD3 T[Staufull] | 7.1 ± 0.21 | 88 | 100 |
| stauD3 T[StauΔN] | 6.9 ± 0.18 | 85 | 100 |
| stauD3 T[StauDmMd] | 7.4 ± 0.16 | 100 | 86 |
| stauD3 T[StauΔloop2] | 0.14 ± 0.06 | n/a | 74 |
| stauD3 T[StauΔdsRBD5] | 0.06 ± 0.03 | n/a | 68 |
As a more stringent test of whether the functional domains of Stau have been conserved during evolution, we generated transgenic lines in which the dsRBD-containing region of DmStau is replaced with the corresponding region from M.domestica (StauDmMd), and found that this transgene rescues all the phenotypes of a stau null mutation at least as well as the full-length Stau construct (Figure 2C; Table I). Since the only regions that are conserved between DmStau and MdStau are the five dsRBDs and the short sequence in the insertion in dsRBD2, it is likely that the most important functional domains of the protein reside in these regions.
Although the localization of osk mRNA to the posterior of the oocyte during stage 9 appears normal in stauD3 females carrying StauDmMd, osk mRNA localizes through an atypical intermediate stage. In wild-type flies, osk mRNA shows a transient association with the anterior pole of the oocyte before moving to the posterior, whereas all of the mRNA remains at the anterior in stauD3 homozygous females (Figure 2C, i and iv). In StauDmMd ovaries, however, osk mRNA does not accumulate at the anterior of the oocyte, and instead forms a ‘blob’ in the middle of the oocyte, which disperses as localization proceeds (Figure 2C, ii and iii). Furthermore, this effect of StauDmMd is dominant: in the absence of endogenous D.melanogaster Stau this ‘blob’ is diffuse, but in a wild-type background it forms a much sharper, well defined spot (Figure 2C, v). The nature of the cytoplasmic blob is unclear, but it is intriguing that D.virilis osk mRNA is localized through a similar intermediate when introduced into D.melanogaster (Webster et al., 1994).
dsRBD2 and dsRBD5 do not bind dsRNA
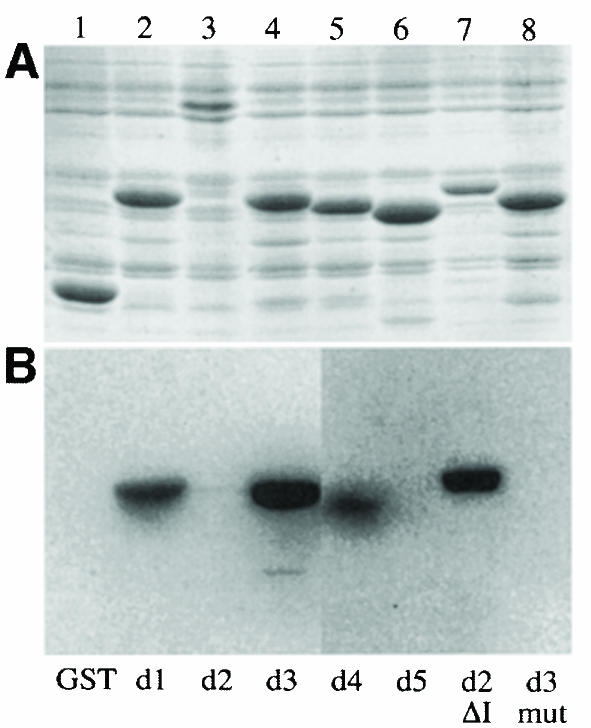
The discovery that the dsRBDs of Stau are the only conserved regions of the protein that are required for its function raises the question of whether all of these domains bind to dsRNA, and we therefore examined the ability of the five dsRBDs to bind to dsRNA on Northwestern blots (Figure 3A). As previously reported, dsRBD3 binds strongly to dsRNA, whereas a control domain in which 5 aa that contact the RNA have been mutated does not (Ramos et al., 2000). dsRBDs 1 and 4 also bind dsRNA, irrespective of its sequence, although this binding is weaker than that observed with dsRBD3. In contrast, dsRBD2 and dsRBD5 do not bind to any of the dsRNAs tested in this assay.
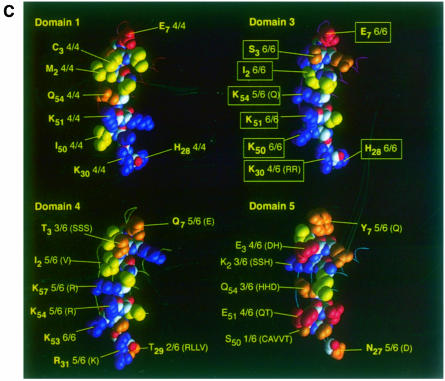
Fig. 3. dsRBDs 2 and 5 do not bind to dsRNA in vitro. (A) A Coomassie-stained SDS–PAGE gel showing the expression of the Staufen dsRBDs fused to glutathione S–transferase (GST). Lane 1, GST alone; lane 2, GST–dsRBD1; lane 3, GST–full-length dsRBD2; lane 4, GST–dsRBD3; lane 5, GST–dsRBD4; lane 6, GST–dsRBD5; lane 7, GST–dsRBD2 in which the large insertion has been replaced by the short loop 2 from dsRBD3; lane 8, GST–dsRBD3 containing five amino acid substitutions in residues that contact dsRNA (Ramos et al., 2000). (B) A Northwestern blot of the same samples as in (A) probed with [32P]dsRNA. The right hand side of this blot has been exposed approximately four times longer than the left to reveal the weak dsRNA-binding activity of dsRBD4. (C) A comparison of the RNA-binding faces of dsRBDs 1, 3, 4 and 5, showing the amino acids in domain 3 that are required for dsRNA binding (yellow boxes), and the identity and conservation of the amino acids in equivalent positions in the other domains (yellow). The structures of dsRBDs 1, 4 and 5 have been modelled by ‘threading’ them onto the known structure of dsRBD3 (Bycroft et al., 1995). Blue, basic residues; red, acidic; yellow, non-polar; orange, polar and uncharged. The amino acids are numbered from the first conserved residue of the domain (Ramos et al., 2000).
NMR and mutational analysis of Stau dsRBD3–dsRNA complexes have revealed that the dsRBD binds RNA through conserved amino acids that cluster on one face of the domain (Figure 3C) (Ramos et al., 2000). The corresponding positions in dsRBDs 1, 4 and 5 can be identified both by sequence alignment and by ‘threading’ their sequences onto the structure of dsRBD3. While dsRBDs 1 and 4 contain identical or similar conserved amino acids to dsRBD3, the amino acids on this face of dsRBD5 are much less well conserved, and are of a different type from those found in the other domains (Figure 3C). The lack of conservation of the dsRNA-contacting amino acids, coupled with the observed inability of dsRBD5 to bind dsRNA in vitro strongly suggest that this domain does not function as a dsRBD in vivo. However, the structural amino acids that comprise the hydrophobic core of the domain are highly conserved when compared with other dsRBDs, and amino acids on the other faces of dsRBD5 are also conserved across the different species. It is therefore likely that domain 5 folds into a typical dsRBD structure, but performs a distinct conserved function unrelated to dsRNA binding.
A highly conserved feature of all six Stau homologues is the presence of the large loop interrupting dsRBD2, and this most probably accounts for the inability of this domain to bind dsRNA in vitro. The NMR structure of the dsRBD3–dsRNA complex reveals that the dsRNA-binding regions of the domain span one turn of a dsRNA helix, and that their relative positions within the whole domain are crucial for RNA binding (Ramos et al., 2000). The insertion in loop 2 separates the two halves of dsRBD2, and the RNA-binding amino acids are therefore unlikely to have the correct spacing to contact RNA. Although the presence of the insertion in loop 2 makes it impossible to predict the structure of dsRBD2, the sequence of the domain suggests that it could bind dsRNA if it adopted a conformation in which these two halves were juxtaposed. To test this hypothesis, we constructed a version of dsRBD2 in which the extended loop 2 is replaced by the corresponding 8 aa loop of dsRBD3. When examined in the Northwestern assay, this dsRBD2Δloop2 binds dsRNA almost as efficiently as dsRBD3 (Figure 3). Thus, dsRBD2 can bind dsRNA when the removal of the large insertion allows the correct folding of the domain, suggesting that this domain binds dsRNA in vivo in the context of full-length protein.
The insertion in domain 2 is required for Stau–osk mRNA localization
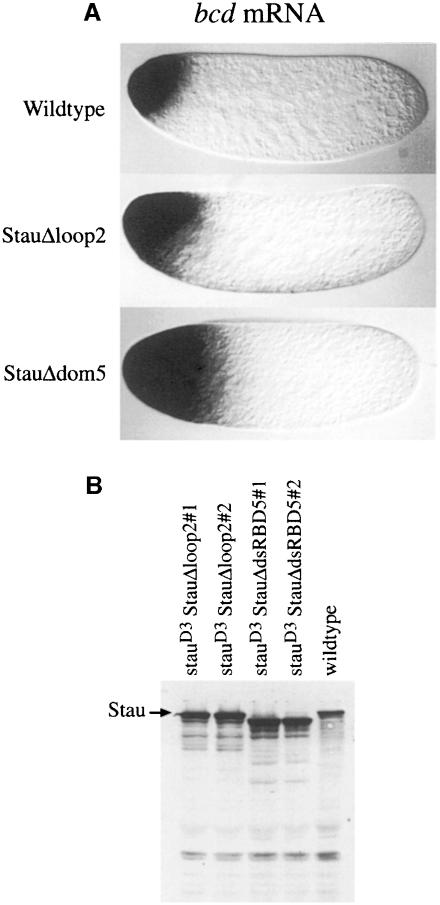
To determine what role, if any, the extended loop in dsRBD2 plays in Stau function, we constructed a transgene (StauΔloop2) in which the normal domain 2 is replaced by the truncated dsRBD2Δloop2 described above, and crossed this into a stauD3 mutant background. Although this transgene expresses high levels of a protein of the appropriate molecular weight (Figure 6B), it gives little or no rescue of the stau posterior phenotype; almost all osk mRNA and Stau protein fail to be transported to the posterior of the oocyte and remain trapped instead at the anterior margin (Figure 4A–E). However, a small amount of mRNA is occasionally seen at the posterior at stage 9. As a result of this defect in osk mRNA localization, none of the progeny of these flies hatch into larvae, and almost all develop less than one abdominal segment (Table I).
Fig. 6. dsRBD5 and the insertion in dsRBD2 are required for the anchoring of bcd mRNA. (A) bcd mRNA localization in freshly laid eggs from wild-type, stauD3 T[StauΔloop2]/stauD3 and stauD3 T[StauΔdsRBD5]/stauD3 females. (B) Western blot analysis of Stau expression in stau null mutant ovaries carrying the StauΔloop2 and StauΔdsRBD5 transgenes. Stau is expressed at higher levels than in the wild type in two independent lines of each transgene. Note that the StauΔdsRBD5 protein migrates slightly faster than the wild-type and StauΔloop2 proteins because it contains a larger deletion.
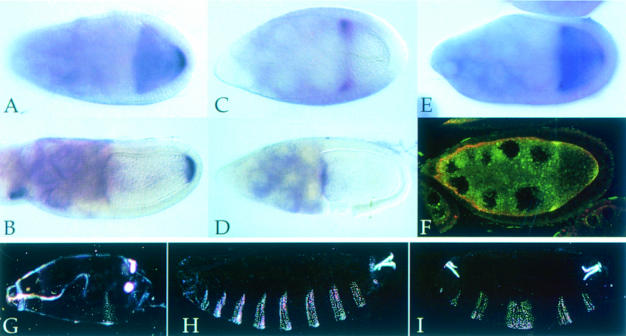
Fig. 4. The insertion in dsRBD2 is required for the posterior localization of osk mRNA. (A and B) Wild-type stage 9 and 10A egg chambers showing the normal localization of osk mRNA to the posterior of the oocyte. (C and D) stauD3 T[StauΔloop2]/stauD3 egg chambers, in which all osk mRNA remains anchored at the anterior of the oocyte. (E) A stauD3 T[StauΔloop2]/stauD3 stage 9 egg chamber, showing a small amount of osk mRNA at the posterior. (F) StauΔloop2 protein (yellow) co-localizes with osk mRNA to the anterior of the oocyte. (G) A cuticle preparation of a typical embryo from a stauD3;oskBRE– female, with a normal head and only one abdominal segment. (H and I) Typical embryos from StauΔloop2 stauD3;oskBRE– females, showing the loss of head structures and rescue of the abdomen (H), and the stronger symmetric bicaudal phenotype (I).
A trivial explanation for the inability of StauΔloop2 to localize osk mRNA is that the removal of the extended loop in dsRBD2 disrupts the folding of the protein and prevents it from binding to the RNA. While it is not possible to test the binding of full-length Stau to osk mRNA in vitro, three lines of evidence suggest that this is not the case. First, dsRBD2Δloop2 binds to RNA in vitro, whereas the wild-type domain does not, indicating that this deletion facilitates the folding of the domain into the RNA-binding configuration. Secondly, StauΔloop2 co-localizes with osk mRNA to the anterior of the oocyte (Figure 4F). The localization of Stau to the oocyte requires its association with osk mRNA, since the protein remains in the nurse cells in an osk mRNA null mutant (M.Weston and D.St Johnston, unpublished results). The normal co-localization of the mutant protein to the anterior of the oocyte with osk mRNA therefore indicates that it still binds to the RNA.
The third argument to suggest that StauΔloop2 interacts with osk mRNA makes use of the oskBRE– transgene to uncouple osk mRNA translation from localization. In wild-type ovaries, the ectopic Osk protein produced from oskBRE– mRNA suppresses head development in about half of the embryos, and causes a duplication of abdominal segments at the anterior (bicaudal phenotype) in over a third (Table I). Even though the translation of this mRNA does not require its localization to the posterior, it is not efficiently translated in the absence of Stau protein: none of the embryos laid by stauD3;oskBRE– mothers develop the bicaudal phenotype, and almost all form only one abdominal segment at the posterior (Figure 4G). In contrast, StauΔloop2 activates translation of oskBRE– mRNA almost as effectively as wild-type Stau: 36% of the embryos laid by StauΔloop2 stauD3;oskBRE– females develop a bicaudal phenotype, and another third show a suppression of head development and partial or complete rescue of the abdomen (Figure 4H and I). Thus, while StauΔloop2 is unable to localize osk mRNA, it is able to activate its translation, providing strong evidence that the protein is bound to the mRNA. This mutant protein therefore retains some of the functions of wild-type Stau, but has lost the ability to mediate the transport of Stau–osk mRNA complexes from the anterior to the posterior of the oocyte.
Domain 5 is required to activate the translation of osk mRNA at the posterior pole
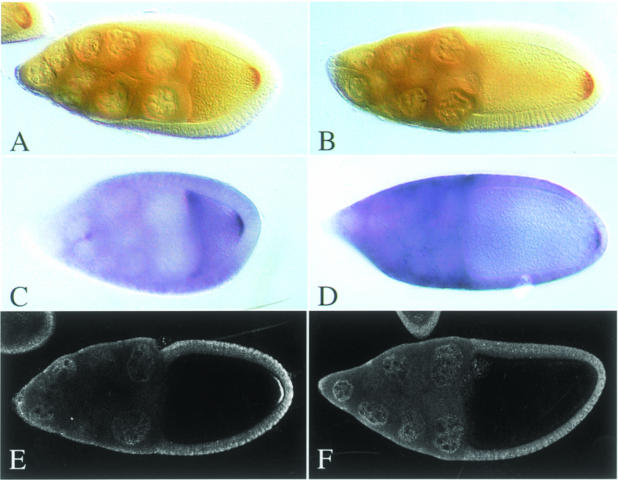
To determine which functions of Stau are mediated by dsRBD5, we constructed a transgene in which the C–terminus of the protein is deleted (StauΔdsRBD5). When introduced into flies lacking wild-type Stau, StauΔdsRBD5 completely rescues the posterior localization of osk mRNA, and co-localizes with the RNA to the posterior (Figure 5A–D). Thus, the mutant protein retains the ability both to bind osk mRNA and interact with the factors required for transport to the posterior. However, this construct shows almost no rescue of the stauD3 abdominal phenotype, and neither osk mRNA nor StauΔdsRBD5 protein are localized at the posterior by the time the egg is laid (Table I; data not shown). These observations show that dsRBD5 is required for a function of Stau that occurs after osk mRNA localization, suggesting that it may play a role in the translation of Osk protein. In wild-type ovaries, localized osk mRNA is translated to produce a tight posterior crescent of Osk protein at stage 10a (Figure 5E). In contrast, no detectable Osk is produced at the posterior of StauΔdsRBD5 oocytes, even though osk mRNA is correctly localized at this stage (Figure 5F). This requirement of Stau dsRBD5 for osk mRNA translation explains the failure of this transgene to rescue the development of the abdomen, and also accounts for the loss of osk mRNA and Stau from the posterior at later stages of oogenesis, since Osk protein has been shown to be necessary for the anchoring of Stau–osk mRNA complexes (Ephrussi et al., 1991; Kim-Ha et al., 1991).
Fig. 5. dsRBD5 is required for osk mRNA translation. (A–D) Both StauΔdsRBD5 protein (A and B) and osk mRNA (C and D) localize normally to the posterior of stage 9 (A and C) and 10 oocytes (B and D). (E and F) Osk antibody stainings of wild-type (E) and stauD3 T[StauΔdsRBD5]/stauD3 oocytes (F).
To examine the role of dsRBD5 in the regulation of osk mRNA translation further, we crossed the StauΔdsRBD5 transgene into the stauD3;oskBRE– background. Like StauΔloop2, StauΔdsRBD5 activates the translation of derepressed osk mRNA. The resulting embryos develop an average of 5.2 abdominal denticle belts, compared with 1.3 for stauD3;oskBRE– alone, and the majority show head defects caused by anterior Osk activity (Table II). This construct differs from StauΔloop2, however, in that it causes a lower frequency of bicaudal embryos, and this may be because osk mRNA is localized to the posterior rather than the anterior of the oocyte. Thus, StauΔdsRBD5 can activate the localization-independent translation of oskBRE– mRNA, but not the translation of wild-type osk mRNA at the posterior pole, suggesting that dsRBD5 is specifically required for the activation of translation once the mRNA has been localized.
Table II. StauΔloop2 and StauΔdsRBD5 activate the translation of osk BRE–
| Genotype | % wild type | % anterior defects with full or partial abdomen | % bicaudal | % stauD3-like | Average No. of abdominal denticle belts |
|---|---|---|---|---|---|
| +;oskBRE– | 11 | 53 | 36 | 0 | 6.5 |
| stauD3;oskBRE– | 1 | 5a | 0 | 93 | 1.3 |
| stauD3 T[StauΔloop2];oskBRE– | 7 | 33a | 36 | 24 | 5.0 |
| stauD3 T[StauΔdsRBD5];oskBRE– | 5 | 52a | 9 | 34 | 5.2 |
aEmbryos were only assigned to this class if their head defects were stronger than those seen in stauD3 alone, to distinguish between the phenotype caused by the stau defect in bcd mRNA localization, and that caused by the ectopic anterior expression of Oskar protein.
bcd mRNA localization requires dsRBD2 and dsRBD5
Since Stau is also required for the anterior anchoring of bcd mRNA, we examined whether the StauΔdsRBD5 and StauΔloop2 transgenes could rescue the bcd mRNA localization defect of a stau null mutation. Although both mutant proteins are expressed at similar levels to endogenous Stau, neither anchors bcd mRNA at the anterior (Figure 6A and B). Surprisingly, both constructs almost completely rescue the stau head phenotype, even though they do not restore the wild-type localization of the bcd mRNA. Whereas 100% of the embryos laid by stauD3 homozygous females at 18°C lack all or part of the head skeleton, over two-thirds of the embryos laid by the transgenic stauD3 females have wild-type heads, and the rest have much milder head defects than in stauD3 alone (Table I). This suggests that Stau plays a second role in the regulation of bcd mRNA expression that is independent of its function in localization. In contrast to its role in anchoring, this activity does not require dsRBD5 or the insertion in dsRBD2.
Discussion
The only regions of Stau to have been maintained through evolution are the dsRBDs and a short region within the insertion in dsRBD2. Furthermore, the dsRBD-containing region from M.domestica retains all the essential functions of Stau during oogenesis. Thus, these conserved domains are likely to mediate all of the activities of Stau, such as binding to bcd and osk mRNAs, and its interactions with the factors that mediate mRNA transport and translational control. The alignment of the dsRBDs from all six species shows that natural selection is acting to maintain the unique characteristics of each. Consistent with this, only domains 1, 3 and 4 bind to dsRNA in vitro, while domains 2 and 5 are required for other activities of Stau.
The presence of a loop splitting dsRBD2 is the most pronounced conserved feature of all the Stau homologues. Replacement of this loop with the corresponding residues from dsRBD3 disrupts the posterior localization of almost all Stau and osk mRNA complexes, although a small amount is sometimes seen at the posterior of a few oocytes. A very similar variable localization of trace amounts of osk mRNA is also seen in all other mutants that specifically disrupt osk mRNA localization, such as mago nashi and Tropomyosin II, suggesting that this RNA may reach the posterior by a parallel translation-dependent pathway (Newmark and Boswell, 1994; Erdélyi et al., 1995). Wild-type osk mRNA is only translated once it is localized, and the resulting Osk protein anchors its own mRNA and Stau at the posterior pole. Thus, any RNA that diffuses to the posterior should be trapped by localized translation and anchoring, and this could localize a tiny fraction of the mRNA in the absence of the normal transport pathway.
Despite its inability to localize osk mRNA to the posterior, StauΔloop2 still associates with the RNA at the anterior of the oocyte, and activates the translation of oskBRE– RNA. These observations strongly suggest that StauΔloop2 interacts with osk mRNA, but that the removal of the insertion in dsRBD2 prevents the resulting complexes from associating with the transport machinery. This insertion probably disrupts the binding of the domain to dsRNA in vitro by changing the relative positions of the RNA-binding amino acids on either side. The domain binds dsRNA when the insertion is removed, however, and the domain contains several of the amino acids that contact RNA in other dsRBDs. Since these residues have been maintained by natural selection, it seems very likely that the domain interacts with RNA in vivo in the context of the full-length protein. Indeed, it is easy to envisage that the interaction of dsRBDs 1, 3 and 4 with RNA presents a very high local concentration of dsRNA to dsRBD2, which induces the domain to adopt the RNA-binding configuration.
Stau localization in both the oocyte and early embryo requires its association with the appropriate mRNA, suggesting that the protein undergoes a conformational change on binding RNA that allows it to associate with the factors that mediate RNA transport. Since the insertion in dsRBD2 is required for localization, it is attractive to propose that this conformational change occurs in dsRBD2. The two halves of dsRBD2 must come together for the domain to bind RNA, and this should loop out the insertion, which could then interact with the factors that transport Stau–RNA complexes. Thus, dsRBD2 could act as a conformational switch that senses the presence of bound RNA, and couples these complexes to the localization machinery. Alignment of the loops in dsRBD2 from different species reveals that they are highly divergent in both sequence and length, but there is a short block of amino acids that might represent a conserved motif with which these transport factors could interact (Figure 1B and C).
While the microtubule-dependent localization of osk mRNA requires the insertion in dsRBD2, but not dsRBD5, the converse is true for the actin-dependent localization of prospero mRNA in embryonic neuroblasts. In this case, localization is mediated by the binding of Miranda protein to dsRBD5 (Fuerstenberg et al., 1998; Matsuzaki et al., 1998; Schuldt et al., 1998; Shen et al., 1998). Thus, distinct domains of Stau mediate microtubule- and actin-dependent mRNA localization, presumably by recruiting different trans–acting factors. Although Stau is the first example of an RNA-binding protein that can direct localization along both actin and microtubules, it is likely that other proteins will also have this capacity. Chicken ZBP–1 protein, which binds to part of the β–actin mRNA localization sequence, is the homologue of VERA/Vg1RBP, which binds to the localization element that directs the microtubule-dependent localization of Xenopus Vg1 mRNA (Ross et al., 1997; Deshler et al., 1998; Havin et al., 1998). Thus, this protein is implicated in both microtubule- and actin-based localization, albeit in different organisms, but it remains to be seen whether distinct domains of the protein are required for each process.
While the localization of osk and prospero mRNAs requires either dsRBD5 or the insertion in dsRBD2, both domains are necessary for the Stau-dependent anchoring of bcd mRNA at the anterior of the egg. Very little is known about the steps in bcd mRNA localization that occur at the end of oogenesis, because it has been impossible to visualize the distribution of the RNA once the vitelline membrane is deposited around the egg. However, these results raise the possibility that Stau needs to interact with both the microtubule and actin cytoskeletons to anchor bcd mRNA.
Role of Stau in translational control
It has previously been difficult to investigate the role of Stau in osk mRNA translation for two reasons. First, stau null mutations disrupt the localization of osk mRNA, and it is not translated unless it is localized to the posterior pole (Markussen et al., 1995; Rongo et al., 1995). Second, it is difficult to distinguish between the effects of weak stau alleles on translation and anchoring, because Osk protein is required to anchor its own RNA, but the mRNA needs to be anchored at the posterior to be translated (Ephrussi et al., 1991; Kim-Ha et al., 1991). However, StauΔdsRBD5 seems to have a specific defect in osk mRNA translation, as osk mRNA is localized normally to the posterior at stage 10 in these ovaries, but no detectable Osk protein is produced. Furthermore, oskBRE– RNA produces significant amounts of Osk activity in these ovaries, indicating that StauΔdsRBD5 can function in the translation of derepressed osk mRNA. Taken together, these results strongly suggest that dsRBD5 is required to relieve Bruno repression once the mRNA has reached the posterior. This requirement cannot be absolute, however, since some Osk protein must be present early in oogenesis to anchor Stau–osk mRNA complexes.
Since dsRBD5 does not bind RNA, it presumably mediates its function in Osk translation through protein–protein interactions. Although Miranda binds to this domain, this interaction is unlikely to play any role during oogenesis, since miranda null germline clones have no phenotype (Matsuzaki et al., 1998). Thus, dsRBD5 presumably interacts with other proteins to regulate osk translation. A very similar translation defect is observed in osk transgenes that lack binding sites for 68 and 50 kDa proteins in the 5′UTR, whereas Stau is thought to associate with the localization signal in the 3′UTR (Gunkel et al., 1998). Thus, derepression is likely to involve cooperation between proteins bound to both ends of the RNA.
In addition to its role in derepressing osk translation at the posterior, Stau is required for the efficient expression of derepressed oskBRE– RNA. Since neither the insert in dsRBD2 nor dsRBD5 are necessary for this activity, it presumably depends on the dsRBDs that bind RNA. It is possible that these dsRBDs also interact with other proteins, since only one face of the domain contacts RNA, and several amino acids on the other faces of these domains have been conserved during evolution. Alternatively, the binding of Stau may enhance osk mRNA translation indirectly, for example, by altering the folding of the RNA so that other factors can bind more efficiently.
Both StauΔloop2 and StauΔdsRBD5 partially rescue the stau head phenotype, even though they do not restore the wild-type localization of bcd mRNA. Thus, more Bcd activity must be produced from the mislocalized mRNA in the presence of these mutant proteins than in stauD3 alone, indicating that they provide a function of Stau that is independent of its role in anchoring. A comparison of the phenotypes produced by vasa exu and stau exu double mutants also indicates that Stau has a second function in the regulation of bcd mRNA. exu mutants block the localization of bcd mRNA early in oogenesis, and result in a uniform distribution of the RNA along the anterior–posterior axis of the embryo, while both vasa and stau mutants prevent the formation of the pole plasm, and therefore lack Nanos activity, which represses bcd mRNA translation (St Johnston et al., 1989; Wang and Lehmann, 1991; Wharton and Struhl, 1991; Wang et al., 1994). Despite the identical distributions of bcd RNA in these genotypes, vasa exu embryos develop anterior head structures everywhere, indicating that they contain high levels of Bcd activity, whereas stau exu form only thoracic structures (Schüpbach and Wieschaus, 1986). Thus, the removal of Stau reduces the level of Bcd expression, in the absence of any effect on mRNA localization. We can envisage two explanations for this localization-independent function of Stau. Stau binding could protect bcd RNA from degradation, and therefore increase the total amount of RNA. Alternatively, Stau could enhance the efficiency of bcd translation, in much the same way as it does for osk mRNA.
Since Stau has been conserved throughout animal evolution, it seems likely that the homologues will fulfil similar functions in mRNA localization and translational control in other organisms. In support of this view, recent evidence indicates that mammalian Stau mediates mRNA transport along microtubules in neurons (Köhrmann et al., 1999). The mouse and human Stau genes share an extra region of homology not found in the insect homologues, which resembles the microtubule-binding domain of MAP1B, and this region of HsStau binds to microtubules in vitro (Marión et al., 1999; Wickham et al., 1999). It will therefore be interesting to see whether this domain or the insertion in dsRBD2 is required for the microtubule-dependent movement of Stau in neurons.
Materials and methods
Cloning Stau homologues
Drosophila virilis and M.domestica homologues were obtained by low stringency screens of genomic libraries (Thummel, 1993; Curtis et al., 1995). These sequences have been submitted to the DDBJ/EMBL/GenBank under accession Nos AF225924 and AF225925. The human homologue (DDBJ/EMBL/GenBank accession No. T06248) was identified in the EST database (Adams et al., 1993). The corresponding clone (HFBDQ83) was obtained from the American Type Culture Collection and sequenced. The mouse homologue was obtained by low stringency screening of a 7.5 d.p.c. mouse embryonic cDNA library (L.-L.Li, unpublished) with HFBDQ83. Cestau was identified in cosmid F55A4 by BLAST searching, and corresponds to the predicted gene F55A4.5. ESTs directly confirm 6/10 of the predicted splice junctions. All but one of the remaining splice sites were confirmed by direct examination of the sequencing traces.
Drosophila mutants and transgenic lines
The oskBRE– stock was a kind gift from Paul MacDonald. stauD3 is a protein null (St Johnston et al., 1991).
Transgenes were expressed from a P-element transformation vector, pCaTubMycSTOP, which drives maternal germline expression from the α4 tubulin promoter (Micklem et al., 1997). The Staufull, StauΔdsRBD5 and StauΔloop2 constructs are identical to D288, StauΔdRBD5 and StauΔdRBD2 described previously (Schuldt et al., 1998). StauΔN was generated in a similar manner, but lacks the first 281 aa of Stau, upstream of the ClaI site at position 1140. StauDmMd is identical to Staufull from the start codon to 6 aa before the start of dsRBD1 (up to and including the sequence TSSSGRG). This portion is fused to the dsRBDs of MdStau, starting at an XmaI site 4 aa before the start of dsRBD1 (i.e. from REKTPMCLV). A single Ala residue replaces the Gly of DmStau (or the Ser of MdStau) 5 aa before the start of dsRBD1. Further details of these constructs are available on request.
Northwestern blots
Northwestern blots were performed using the procedure described in St Johnston et al. (1992), using a blocking buffer of 2.5% (v/v) Tween–20, 1% (w/v) milk, 25 mM NaCl, 10 mM MgCl2, 10 mM HEPES pH 8.0, 100 μM EDTA, 1 mM dithiothreitol (DTT), and a binding buffer of 2.5% Tween–20, 1% milk, 50 mM NaCl, 10 mM MgCl2, 10 mM HEPES pH 8.0, 100 μM EDTA and 1 mM DTT containing 500 000 c.p.m./ml of probe.
Phenotypic analysis
Antibody stainings and in situ hybridizations were performed as described in St Johnston et al. (1991). Antibodies were used at the following dilutions: rabbit α–Stau, 1/1000; rabbit α–Osk, 1/200.
Acknowledgments
Acknowledgements
We would like to thank Paul Macdonald for the oskBRE stocks, and Anne Ephrussi and Anne-Marie Michon for providing the Osk antibody. D.R.M. and D.St J. were supported by the Wellcome Trust; J.A. by the Boehringer Ingelheim Fonds; and S.G. by an EMBO Long Term Fellowship.
References
- Adams M.D., Kerlavage, A.R., Fields, C. and Venter, J.C. (1993) 3,400 new expressed sequence tags identify diversity of transcripts in human brain. Nature Genet., 4, 256–267. [DOI] [PubMed] [Google Scholar]
- Bashirullah A., Cooperstock, R.L. and Lipshitz, H.D. (1998) RNA localization in development. Annu. Rev. Biochem., 67, 335–394. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H. and Long, R.M. (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell, 2, 437–445. [DOI] [PubMed] [Google Scholar]
- Broadus J. and Doe, C. (1997) Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr. Biol., 7, 827–835. [DOI] [PubMed] [Google Scholar]
- Broadus J., Fuerstenberg, S. and Doe, C.Q. (1998) Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter cell fate. Nature, 391, 792–795. [DOI] [PubMed] [Google Scholar]
- Bycroft M., Grünert, S., Murzin, A.G., Proctor, M. and St Johnston, D. (1995) NMR solution structure of a dsRNA binding domain from Drosophila Staufen protein reveals homology to the N–terminal domain of ribosomal protein S5. EMBO J., 14, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I., Giniger, E., Ruohola-Baker, H., Jan, L. and Jan, Y. (1994) Transient posterior localisation of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol., 4, 289–300. [DOI] [PubMed] [Google Scholar]
- Curtis D., Apfeld, J. and Lehmann, R. (1995) nanos is an evolutionarily conserved organizer of anterior–posterior polarity. Development, 121, 1899–1910. [DOI] [PubMed] [Google Scholar]
- Deshler J.O., Highett, M.I., Abramson, T. and Schnapp, B.J. (1998) A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol., 8, 489–496. [DOI] [PubMed] [Google Scholar]
- Driever W. (1993) In Bate,M. and Martínez-Arias,A. (eds), Maternal Control of Anterior Development in the Drosophila Embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 1, pp. 301–324. [Google Scholar]
- Ephrussi A. and Lehmann, R. (1992) Induction of germ cell formation by oskar.Nature, 358, 387–392. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson, L.K. and Lehmann, R. (1991) oskar organizes the germ plasm and directs localization of the posterior determinant nanos.Cell, 66, 37–50. [DOI] [PubMed] [Google Scholar]
- Erdélyi M., Michon, A., Guichet, A., Bogucka Glotzer, J. and Ephrussi, A. (1995) A requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature, 377, 524–527. [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Elphick, L., Nüsslein-Volhard, C. and St Johnston, D. (1994) Staufen protein associates with the 3′UTR of bicoid mRNA to form particles which move in a microtubule-dependent manner. Cell, 79, 1221–1232. [DOI] [PubMed] [Google Scholar]
- Fuerstenberg S., Peng, C.-Y., Alvarez-Ortiz, P., Hor, T. and Doe, C.Q. (1998) Identification of Miranda protein domains regulating asymmetrical cortical localization, cargo binding and cortical release. Mol. Cell. Neurosci., 12, 325–339. [DOI] [PubMed] [Google Scholar]
- Gibson T.J. and Thompson, J. (1994) Detection of dsRNA-binding domains in RNA helicase A and Drosophila maleless: implications for monomeric RNA helicases. Nucleic Acids Res., 22, 2552–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Reyes A., Elliott, H. and St Johnston, D. (1995) Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature, 375, 654–658. [DOI] [PubMed] [Google Scholar]
- Gunkel N., Yano, T., Markussen, F.-H., Olsen, L.C. and Ephrussi, A. (1998) Localization-dependent translation requires a functional interaction between the 5′ and 3′ ends of oskar mRNA. Genes Dev., 12, 1652–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havin L., Git, A., Elisha, Z., Oberman, F., Yaniv, K., Schwartz, S.P., Standart, N. and Yisraeli, J.K. (1998) RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev., 12, 1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J., Smith, J.L. and Macdonald, P.M. (1991) oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell, 66, 23–35. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Kerr, K. and Macdonald, P.M. (1995) Translational regulation of oskar messenger RNA by Bruno, an ovarian RNA binding protein, is essential. Cell, 81, 403–412. [DOI] [PubMed] [Google Scholar]
- Kislauskis E.H., Zhu, X. and Singer, R.H. (1994) Sequences responsible for intracellular localization of β–actin messenger RNA also affect cell phenotype. J. Cell Biol., 127, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrmann M., Luo, M., Kaether, C., DesGroseillers, L., Dotti, C.G. and Kiebler, M.A. (1999) Microtubule-dependent recruitment of Staufen–green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Cell. Biol., 10, 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Yang, X., Wasser, M., Cai, Y. and Chia, W. (1997) Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell, 90, 437–447. [DOI] [PubMed] [Google Scholar]
- Marión R.M., Fortes, P., Belosco, A., Dotti, C. and Ortín, J. (1999) A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell. Biol., 19, 2212–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markussen F.H., Michon, A.M., Breitwieser, W. and Ephrussi, A. (1995) Translational control of oskar generates short Osk, the isoform that induces pole plasm assembly. Development, 121, 3723–3732. [DOI] [PubMed] [Google Scholar]
- Matsuzaki F., Ohshiro, T., Ikeshima-Kataoka, H. and Izumi, H. (1998) miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development, 125, 4089–4098. [DOI] [PubMed] [Google Scholar]
- Micklem D.R., Dasgupta, R., Elliott, H., Gergely, F., Davidson, C., Brand, A., González-Reyes, A. and St Johnston, D. (1997) The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila.Curr. Biol., 7, 468–478. [DOI] [PubMed] [Google Scholar]
- Newmark P.A. and Boswell, R.E. (1994) The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila.Development, 120, 1303–1313. [DOI] [PubMed] [Google Scholar]
- Pokrywka N.J. and Stephenson, E.C. (1991) Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development, 113, 55–66. [DOI] [PubMed] [Google Scholar]
- Ramos A., Grünert, S., Adams, J., Micklem, D.R., Proctor, M.R., Freund, S., Bycroft, M., St Johnston, D. and Varani, G. (2000) RNA recognition by a Staufen double-stranded RNA binding domain. EMBO J., 19, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C., Gavis, E.R. and Lehmann, R. (1995) Localization of oskar RNA regulates Oskar translation and requires Oskar protein. Development, 121, 2737–2746. [DOI] [PubMed] [Google Scholar]
- Ross A.F., Oleynikov, Y., Kislauskis, E.H., Taneja, K.L. and Singer, R.H. (1997) Characterization of a β–actin mRNA zipcode-binding protein. Mol. Cell. Biol., 17, 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S., Neuman-Silberberg, F.S., Barcelo, G. and Schüpbach, T. (1995) cornichon and the EGF receptor signaling process are necessary for both anterior–posterior and dorsal–ventral pattern formation in Drosophila.Cell, 81, 967–978. [DOI] [PubMed] [Google Scholar]
- Schuldt A.J., Adams, J.H.J., Davidson, C.M., Micklem, D.R., St Johnston, D. and Brand, A. (1998) Miranda mediates the asymmetric protein and RNA localisation in the developing nervous system. Genes Dev., 12, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T. and Wieschaus, E. (1986) Maternal-effect mutations altering the anterior–posterior pattern of the Drosophila embryo. Roux Arch. Dev. Biol., 195, 302–317. [DOI] [PubMed] [Google Scholar]
- Shen C.P., Knoblich, J.A., Chan, Y.M., Jiang, M.M., Jan, L.Y. and Jan, Y.N. (1998) Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila.Genes Dev., 12, 1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. (1995) The intracellular localization of messenger RNAs. Cell, 81, 161–170. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Driever,W., Berleth,T., Richstein,S. and Nüsslein-Volhard,C. (1989) Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development, (Suppl.), 107, 13–19. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Beuchle, D. and Nüsslein-Volhard, C. (1991) staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell, 66, 51–63. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Brown, N.H., Gall, J.G. and Jantsch, M. (1992) A conserved double-stranded RNA-binding domain. Proc. Natl Acad. Sci. USA, 89, 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C. (1993) Compilation of Drosophila cDNA and genomic libraries. Drosoph. Inf. Serv., 72, 180–183. [Google Scholar]
- Wang C. and Lehmann, R. (1991) Nanos is the localized posterior determinant in Drosophila.Cell, 66, 637–647. [DOI] [PubMed] [Google Scholar]
- Wang C., Dickinson, L.K. and Lehmann, R. (1994) Genetics of nanos localisation in Drosophila.Dev. Dyn., 199, 103–115. [DOI] [PubMed] [Google Scholar]
- Webster P., Suen, J. and Macdonald, P.M. (1994) Drosophila virilis oskar transgenes direct body patterning but not pole cell formation or maintenance of RNA localization in D.melanogaster.Development, 120, 2027–2037. [DOI] [PubMed] [Google Scholar]
- Wharton R.P. and Struhl, G. (1991) RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell, 67, 955–967. [DOI] [PubMed] [Google Scholar]
- Wickham L., Duchaîne, T., Luo, M., Nabi, I.R. and DesGroseillers, L. (1999) Mammalian Staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol., 19, 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli J., Sokol, S. and Melton, D. (1990) A two-step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development, 108, 289–298. [DOI] [PubMed] [Google Scholar]