Abstract
DNA polymerase β (Polβ) has been implicated in base excision repair in mammalian cells. However, the physiological significance of this enzyme in the body remains unclear. Here, we demonstrate that mice carrying a targeted disruption of the Polβ gene showed growth retardation and died of a respiratory failure immediately after the birth. Histological examination of the embryos revealed defective neurogenesis characterized by apoptotic cell death in the developing central and peripheral nervous systems. Extensive cell death occurred in newly generated post-mitotic neuronal cells and was closely associated with the period between onset and cessation of neurogenesis. These findings indicate that Polβ plays an essential role in neural development.
Keywords: apoptosis/base excision repair/DNA polymerase β/knockout mouse/neurogenesis
Introduction
Repair of DNA damage is essential for maintaining the integrity of the genetic information that is needed for normal development and physiological consequences. Spontaneous apurinic/apyrimidinic (AP) sites and DNA base modifications are induced by a variety of endogenous and exogenous agents such as reactive oxygen, ionizing radiation and alkylating agents. These forms of damage are corrected by the components of the base excision repair (BER) pathway. In mammalian cells, two types of BER have been found: a short-patch BER pathway involving replacement of one nucleotide and a long-patch BER pathway involving gap filling of several nucleotides.
DNA polymerase β (Polβ) is a 39 kDa protein made up of a single polypeptide and consists of two catalytic functional domains. The N–terminal 8 kDa domain carries a deoxyribose 5′–phosphatase activity as well as a single-stranded DNA binding activity. The C–terminal 31 kDa domain carries a DNA polymerase activity that fills short DNA gaps with a 5′–phosphate (Kumar et al., 1990; Singhal and Wilson, 1993; Matsumoto and Kim, 1995). The short-patch BER pathway generally is initiated by a specific DNA glycosylase that recognizes and removes a damaged base to generate an AP site in DNA; the AP site is corrected via incision by an AP endonuclease (APE), removal of a deoxyribose 5-phosphate and DNA synthesis to fill the gap by Polβ, and ligation by DNA ligase I or a complex of XRCC1–ligase III. Polβ has been shown to interact with APE, ligase I and XRCC1 (Caldecott et al., 1996; Prasad et al., 1996; Bennett et al., 1997), suggesting that these proteins must act coordinately in the BER pathway. On the other hand, the long-patch BER pathway requires proliferating cell nuclear antigen (PCNA) and FEN1 to excise a flap-like DNA structure produced by strand displacement. This pathway is divided further into two subpathways: a PCNA-stimulated, Polβ-directed pathway, and a PCNA-dependent, DNA polymerase δ/ɛ-directed pathway. Recent reconstitution studies with purified protein factors have confirmed a crucial role for Polβ in both of the pathways (Kubota et al., 1996; Klungland and Lindahl, 1997; Nicholl et al., 1997). A mutant cell line with no Polβ activity is viable and shows normal growth characteristics (Sobol et al., 1996). Although the mutant cells exhibit BER defects as shown by increased sensitivity to monofunctional DNA-alkylating agents, there still remains an activity in crude cell extracts to repair a damaged base residue in DNA substrate (Biade et al., 1998), indicating that there are both Polβ-dependent and -independent BER pathways in vivo.
The role of Polβ in the body is largely unknown. Recent work with Polβ-deficient mice generated by the Cre–loxP system has shown that they are not viable (Gu et al., 1994). Betz et al. (1996) have generated mosaic mice consisting of Polβ+/– and Polβ–/– cells by mating Polβ+/– and nestin–cre transgenic mice; some of the mosaic mice bypassed lethality and were viable but were often reduced in size and weight, indicating that Polβ is very important for development. However, in order to determine the essential function of the enzyme during development, detailed analysis should be carried out on the critical stage of death and the physiological defects in Polβ-deficient mice. In this study, we find that Polβ-deficient mice survive during the whole embryonic stage but die immediately after birth. We also find that the mutant embryos exhibit severe defects in the development of the nervous system.
Results
Targeted disruption of the Polβ gene in mouse ES cells and generation of Polβ-deficient mice
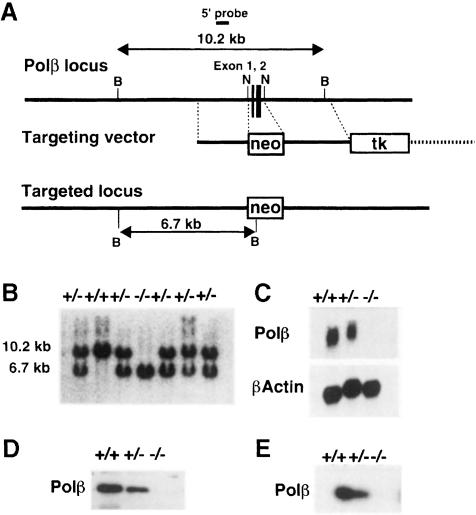
The targeting strategy used to disrupt the Polβ sequence in mouse embryonic stem (ES) cells is illustrated in Figure 1A. The targeting vector was constructed by replacing a 0.3 kb NaeI fragment containing exons 1 and 2 of the Polβ gene with the neo gene. In addition, a copy of the HSV-TK gene was placed at the 3′ end of the construct. Male chimeric mice generated using a correctly targeted ES clone transmitted the Polβ gene mutation through the germline when mated with C57BL/6 female mice. Mice homozygous for the targeted allele (Polβ–/–) were generated by intercrosses of heterozygous (Polβ+/–) mice, and the genotype of the resulting embryos was determined by Southern blot analysis with a 5′–flanking probe (Figure 1B). BamHI digestion of wild-type and targeted Polβ loci gave rise to 10.2 and 6.7 kb bands, respectively. It is evident that homozygous mutant embryos (–/–) at E18.5 (days post-coitum) carried two targeted loci as expected. Northern blot analysis of total RNA isolated from a Polβ–/– embryo at E18.5 showed no significant hybridization to a mouse Polβ cDNA probe, whereas a wild-type embryo (+/+) contained Polβ mRNA, indicating the absence of any detectable transcript for the gene in the mutant; a Polβ+/– littermate embryo (+/–) showed an expected 50% reduction in the level of mRNA (Figure 1C). Western blot analysis revealed that a Polβ–/– embryo at E10.5 synthesized no detectable Polβ protein (Figure 1D); DNA polymerase activity was not detected by an activity gel assay in cell-free extracts from an E10.5 Polβ–/– embryo (Figure 1E). Therefore, we conclude that Polβ–/– mutant embryos carry no enzyme activity.
Fig. 1. Targeted disruption of the mouse Polβ locus. (A) Structures of the Polβ genomic locus, a targeting vector and the targeted locus generated by gene targeting. Exons 1 and 2 are shown as black boxes and numbered. The targeting vector contains the neo gene (neo) in place of the 0.3 kb region spanning exons 1 and 2 of the Polβ gene. In addition, the HSV-TK gene (tk) is attached to the end of the region of homology; the broken line indicates the vector backbone sequence. The length of diagnostic BamHI restriction fragments and the location of a probe for Southern blot analysis are shown. B, BamHI; N, NaeI. Figures are not drawn to scale. (B) Southern blot analysis. Genomic DNA was isolated from embryos of a heterozygous cross, digested with BamHI and analyzed by Southern hybridization with the DNA probe indicated in (A). +/+, +/– and –/– represent genotypes of Polβ+/+, Polβ+/– and Polβ–/– embryos, respectively. (C) Northern blot analysis. Total RNA isolated from E18.5 embryos of each genotype as shown in (B) was electrophoresed, blotted and hybridized to a mouse Polβ cDNA probe including the entire sequence of the Polβ coding region. The amounts of RNA loaded are indicated by hybridization to a cDNA probe for β-actin. (D) Western blot analysis. Total homogenates were prepared from E10.5 embryos of each genotype as in (B), and equal amounts of protein were analyzed by SDS–PAGE followed by immunoblotting with anti-rat Polβ antibody. (E) Activity gel assay of Polβ. Total homogenates from E10.5 embryos of each genotype were subjected to SDS–PAGE containing activated calf thymus DNA; proteins in a gel were then renatured by removing SDS and assayed for DNA polymerase activity.
Null mutation of Polβ causes perinatal lethality
F1 Polβ+/– mice exhibited normal growth and development and were fertile. When they were intercrossed, however, no offspring homozygous for the targeted mutation were detected at 3 weeks after birth. Cumulative genotyping of liveborn mice indicated that Polβ+/– and Polβ+/+ mice were recovered at a ratio close to 2:1 (Table I). On the contrary, at E10.5, Polβ–/– embryos were recovered at the frequency predicted by Mendelian laws, consistent with a previous report (Gu et al., 1994). To define the critical stage of death for our mutant mice, we carried out genotyping of fetuses at different gestational days. We found that Polβ–/– embryos were alive at a Mendelian ratio up to E18.5, although they were significantly smaller than wild-type littermates throughout the stages E10.5–E18.5 (Figure 2A); and an ∼30% decrease in body weight was observed throughout the late developmental stage after E13.5 (Figure 2B). At post-natal day 1, however, no living Polβ–/– neonate was recovered (Table I).
Table I. Genotypic analysis of Polβ+/– intercrosses.
| Age (days) | Genotype |
Total number | ||
|---|---|---|---|---|
| +/+ | +/– | –/– | ||
| E10.5 | 5 | 11 | 9 | 25 |
| E13.5 | 9 | 20 | 7 | 36 |
| E16.5 | 4 | 15 | 6 | 25 |
| E18.5 | 76 | 118 | 58 | 252 |
| P1 | 32 | 54 | 0 (26) | 112 |
| 3 weeks | 48 | 82 | 0 | 130 |
The number of live embryos and mice of each genotype at the stages indicated is shown. The number of dead neonates is shown in parentheses. P1, post-natal day 1.
Fig. 2. Phenotypes of wild-type, Polβ+/– and Polβ–/– mice. (A) Comparison of the size of wild-type (+/+) and Polβ-null mutant (–/–) mice at different embryonic stages. Polβ–/– embryos were smaller than wild-type littermates at every stage. (B) Body weights of embryos with different genotypes. The average weights of 5–10 wild-type (filled bars), Polβ+/– (hatched bars) and Polβ–/– (white bars) embryos are plotted. (C) Survival curves of mice with different genotypes after Caesarean section at E18.5. All Polβ–/– neonates died within 15 min. (D) Hematoxylin/eosin staining of lungs in Caesarean-sectioned neonates. The lung of a wild-type (+/+) neonate is inflated with air, whereas that of a null mutant (–/–) neonate is not inflated with enough air.
To examine whether Polβ–/– mice die before or after birth, we dissected embryos at E18.5 from the uteri of Polβ+/– intercrosses (Caesarean section). The resulting 15 wild-type and 35 Polβ+/– neonates were breathing and acquired a normal pink coloration. On the contrary, all 14 Polβ–/– neonates were pale, failed to breath and showed cyanosis (Figure 2A); these neonates could respond to pain stimulus when their tails were pinched with forceps, but they rapidly lost the reflex action within 15 min (Figure 2C), demonstrating that these mutant mice were indeed alive for a very short period after the section. All wild-type and Polβ+/– neonates were alive for at least 40 min, except for one heterozygous neonate that died at 20 min.
Anatomical and histochemical examinations revealed that the lungs of a Polβ–/– neonate failed to inflate with air and that their alveolar air spaces were smaller than those of their wild-type littermate (Figure 2D). With any genotype, we could not find apparent histological abnormalities in various organs including the heart, liver and kidneys of neonates. These results demonstrate that Polβ–/– embryos survive the entire course of development but die at the perinatal stage, and strongly suggest that a respiratory defect is the cause of death for the null mutant neonates.
Extensive neuronal cell death in Polβ-deficient embryos
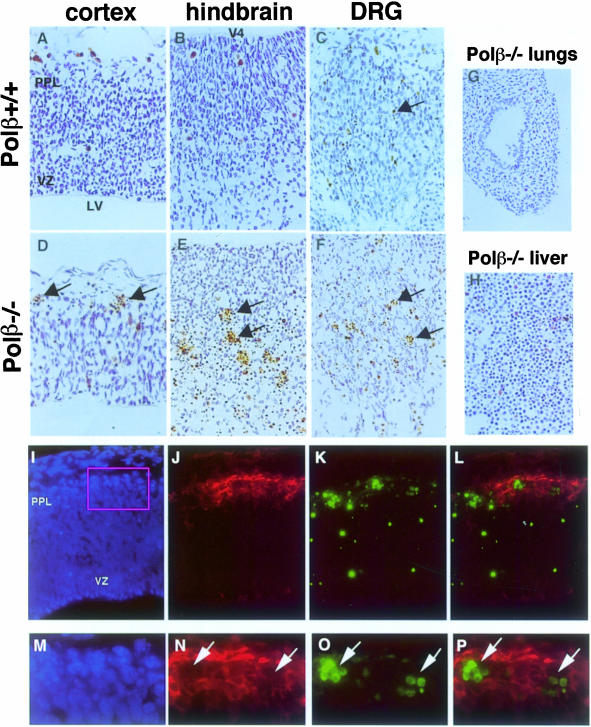
To identify specific pathological abnormalities in Polβ–/– mice that might cause neonatal death, we performed histological evaluations. Among all tissues examined, the only marked abnormality was severe cell death in the developing nervous system in Polβ–/– embryos at E12.5 (Figure 3D–F) compared with wild-type embryos (Figure 3A–C). In the mutant brain sections, we observed a significantly larger number of cells with pyknotic nuclei, which signifies the process of cell death in the cortex of forebrain, hindbrain and dorsal root ganglion (DRG). We next performed TUNEL (terminal deoxytransferase-mediated dUTP biotin nick end labeling) staining to detect apoptotic cells in wild-type and Polβ–/– embryo brains from two litters. A greatly increased number of TUNEL-positive cells was found in the E12.5 cortex, hindbrain and DRG in Polβ–/– embryos (Figure 3D–F), as compared with wild-type controls (Figure 3A–C). These TUNEL-positive cells were observed to be pyknotic. On the other hand, neither TUNEL-positive nor pyknotic cells were detected in other organs such as the lungs (Figure 3G) and liver (Figure 3H). These data indicate a severe induction of apoptotic cell death during the early development of the central and peripheral nervous systems in Polβ–/– embryo brains.
Fig. 3. Extensive cell death in the developing nervous system of Polβ–/– embryo brains. As indicated by arrows, extensive TUNEL-positive nuclei appeared in (D) cortex, (E) hindbrain and (F) dorsal root ganglion (DRG) in Polβ–/– brains at E12.5 (D–F), as compared with those in wild-type controls (A–C). On the other hand, no TUNEL-positive cells were observed in (G) lungs and (H) liver. Dorsolateral regions of E12.5 Polβ–/– coronal sections were stained with (I and M) Hoechst (blue), (J and N) anti-β3-tubulin antibody TuJ1 (red) and (K and O) TUNEL reagents (green). (L and P) Double staining with TuJ1 antibody and TUNEL reagents. TUNEL-positive cells are localized in the primordial plexiform layer (PPL) composed of newly generated neuronal cells. (M–P) Higher magnification of the boxed area in (I). Most cells are doubly stained with TUNEL reagents and TuJ1 as indicated by arrows. LV, lateral ventricle; V4, fourth ventricle; PPL, primordial plexiform layer; VZ, ventricular zone.
In the normal dorsolateral cortex, two layers, designated the ventricular zone (VZ) and the primordial plexiform layer (PPL), emerge in E12.5 mouse embryos (Takahashi et al., 1995). Mitotic neuronal progenitor cells reside in the VZ and give rise to neuronal cells, which then migrate to the PPL. Apoptotic cell death in Polβ–/– brains was obviously detectable in the PPL (Figure 3D), suggesting that this death occurs in post-mitotic neuronal cells. To prove the neuronal identity of these dead cells, we stained E12.5 Polβ–/– cortex sections with a TuJ1 antibody for β3-tubulin, an early marker of post-mitotic neurons (Lee et al., 1990), as well as with Hoechst and TUNEL reagents. TuJ1-positive (red) cells were detected in the PPL but not in the VZ (Figure 3J and N). Most of the cells with TUNEL-positive (green) nuclei (Figure 3K and O) resided within the TuJ1-positive cell population (Figure 3L and P). These results indicate that the extensive cell death observed in Polβ–/– embryos occurred in the post-mitotic neuronal cell population, while proliferating progenitors were spared. Furthermore, there existed some TUNEL-positive nuclei between the VZ and PPL (Figure 3L), suggesting that Polβ deficiency may lead a few early differentiating neuronal cells to apoptotic death.
Neuronal cell death observed in Polβ–/– embryo brains is associated with the temporal and spatial pattern of neurogenesis
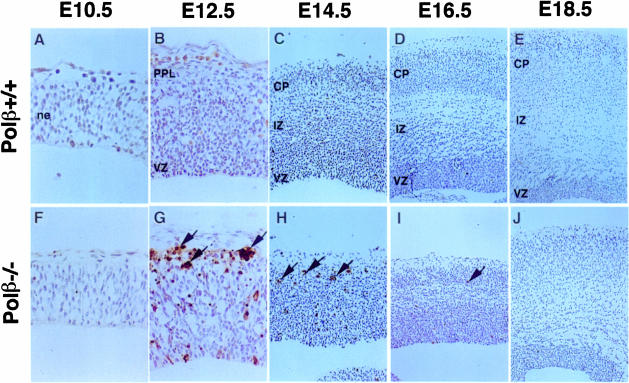
Neurogenesis in normal cortex is temporally and spatially controlled: it starts at E11.5 ventrolaterally and at E12.5 dorsomedially, peaks around E12.5–14.5, and continues at lower levels until E17.5 (Bayer and Altman, 1991; Takahashi et al., 1995). In Polβ–/– brains, abnormally increased dead cells in the neuroepithelium were not detected at E10.5 (Figure 4F) but first appeared at E12.5 in the dorsolateral region of the cortex (Figure 4G), following the onset of neurogenesis. Although the extensive cell death was still observed at E14.5 (Figure 4H), it decreased gradually thereafter (Figure 4I) and disappared almost completely at E18.5 (Figure 4J). Polβ+/+ wild-type brains exhibited no such abnormal pattern of neurogenesis (Figure 4A–E). These results indicate that the increased cell death in the dorsolateral region of the cortex of Polβ–/– brains was closely associated with the period between onset and cessation of neurogenesis. Also, gradually decreasing apoptosis found after neurogenesis (Figure 4H–J) supports the hypothesis that the cell death occurred in early post-mitotic, immature neurons but not in mature neurons. In the dorsolateral cortex of wild-type brains, three layers consisting of the cortical plate (CP), intermediate zone (IZ) and VZ emerged at E14.5 (Figure 4C). However, in Polβ–/– brain sections, the CP was not seen (Figure 4H). Furthermore, the mutant CP observed at E16.5 and E18.5 (Figure 4I and J) was thinner than the corresponding wild-type CP (Figure 4D and E), suggesting that the extensive cell death during neurogenesis results in an abnormal cortex structure composed of such thin CP.
Fig. 4. Abnormal neuronal cell death in Polβ–/– embryo brains is associated with neurogenesis. Dorsolateral regions of cortices of (A–E) wild-type and (F–J) Polβ–/– embryo brains at the developmental stages indicated were stained with TUNEL reagents. Arrows indicate TUNEL-positive apoptotic nuclei. CP, cortical plate; IZ, intermediate zone; ne, neuroepithelium; VZ, ventricular zone.
Discussion
Polβ-deficient mice die immediately after birth
The previous report with Polβ-knockout mice shows that Polβ–/– mice are detected at E10.5 but not at 4 weeks after birth (Gu et al., 1994). This observation has suggested that Polβ is essential for embryonic development since lack of this enzyme could cause embryonic death. In the present study, however, Polβ-null mutant mice were recovered at P1 at a Mendelian ratio, although all of them died soon afterwards (Table I). We found that these mutant embryos were alive at E18.5 but died within 15 min after Caesarian section (Figure 2A and C). Wild-type and Polβ+/– mice born under the same conditions were indistinguishable from those born by natural birth with respect to their survival and growth while suckling. Therefore, the phenotypes of mice born by Caesarian section should reflect those of mice born naturally. Our targeting construct disrupts exons 1 and 2 of the Polβ gene locus (Figure 1A). In this respect, the targeted allele is almost identical to that previously generated by Gu et al. (1994). Our mutation resulted in a complete elimination of the function of the Polβ gene, as shown by the absence of the mRNA, the protein and the enzyme activity in Polβ–/– embryos (Figure 1C–E). To assess the role of Polβ in DNA damage repair, we generated primary fibroblast cells from E12.5 embryos. Polβ–/– fibroblasts were clearly more sensitive than wild-type cells to the alkylating agent methyl methanesulfonate; similar results were obtained with other alkylating agents, N–methyl-N′-nitro-N–nitrosoguanidine and N–nitroso-N–methylurea. However, no difference in sensitivity to X-ray irradiation was found between the mutant and wild-type cells (data not shown). These data are consistent with those reported by Sobol et al. (1996), who studied cell lines established from the Polβ null mutant embryos of Gu et al. (1994). Therefore, we conclude that mice deficient in Polβ survive during embryonic development but die immediately after birth.
Polβ plays an essential role in neurogenesis
The pattern of neuronal cell death in Polβ–/– embryos (Figures 3 and 4) is strikingly similar to that observed in XRCC4- and ligase IV-deficient mouse embryos, which die at around E16.5 (Barnes et al., 1998; Gao et al., 1998). However, it seems that this cell death in Polβ–/– brains is slightly less severe than that in XRCC4- and ligase IV-deficient brains, as reflected by their later embryonic lethality. Both XRCC4 and ligase IV proteins are known to be components of V(D)J recombination and non-homologous end-joining (NHEJ) apparatus for DNA double strand break (DSB) repair (Li et al., 1995; Frank et al., 1998; Grawunder et al., 1998); therefore, the increased death of early post-mitotic neuronal cells in the mutant embryos may be due to increased occurrence of unrepaired DSBs (Barnes et al., 1998; Gao et al., 1998). However, the findings that cultured cells deficient in Polβ are highly sensitive to monofunctional DNA alkylating agents but resistant to X-ray irradiation (Sobol et al., 1996; data not shown) suggest its important role in the repair of DNA single-strand breaks (SSBs) but not DSBs. To date, the only known function of Polβ is to fill short DNA gaps through the BER pathway (Singhal and Wilson, 1993). Therefore, it is likely that extensive cell death in newly generated Polβ–/– neurons may result from defects in the repair of SSBs, which are not repaired in the absence of this enzyme, leading to apoptotic death in these particular cells. Although we do not know the actual source of SSBs, the post-mitotic neuronal cells might be suffering from higher levels of unrepaired SSBs compared with other cells. Alternatively, the neuronal cells may be hypersensitive to such unrepaired SSBs, even if their levels are similar in other cells. Another possibility is that the extensive neural cell death could be caused by a deficiency in a specific process, such as recombination, required for neural differentiation. The possible involvement of recombination in the CNS has been raised recently by the finding of the similarity of the genomic organization in 52 human neural cadherin-like cell adhesion genes and immunoglobulin/T-cell receptor gene clusters (Wu and Maniatis, 1999). Such a recombination would involve a NHEJ process that is initiated by a DSB and completed by filling short DNA gaps in its end and ligation. Since Polβ has been suggested to have an activity filling such gaps in NHEJ in Xenopus egg extracts (Pfeiffer et al., 1994), the enzyme could play a critical role in the recombination process in differentiating neural cells, and failure of this process in Polβ null mutants would result in abnormal cell death. If so, it may be possible that a deficiency in XRCC4, ligase IV and Polβ causes similar defects in neurogenesis. Polβ is ubiquitously expressed in the mouse but is especially active in the testis, thymus and brain (Hirose et al., 1989). Therefore, Polβ may participate not only in BER of the whole body but also in a recombination event(s) occurring in those active tissues.
Polβ-deficient mice exhibit different phenotypes from other mice deficient in BER factors
The in vivo roles of several BER proteins have been investigated using knockout mice: APE and XRCC1 knockout mice die between E6.5 and E7.5 with abnormal morphogenesis of the embryonic body and apoptotic cell death (Xanthoudakis et al., 1996; Ludwig et al., 1998; Tebbs et al., 1999). Mice lacking ligase I are also embryonic lethal at E16.5 as a result of hypoxia secondary to anemia (Bentley et al., 1996). These results indicate that these BER factors are essential for early development. In contrast, knockout mice of two DNA glycosylases encoded by APNG (Engelward et al., 1997; Hang et al., 1997; Elder et al., 1998) and OGG1 (Klungland et al., 1999) are viable and fertile. Since there are six or more DNA glycosylases in mammalian cells, others could compensate for the defects in those mutant mice.
The effect of Polβ deficiency on survival is less severe compared with deficiency of APE, XRCC1 and ligase I, suggesting that there may be other pathways that are able to partially compensate for the Polβ defects. In fact, using a Polβ-deficient cell line, it has been reported that an AP site is repaired in vitro not only by the Polβ-dependent pathway but also by the PCNA-dependent BER pathway (Biade et al., 1998). As the latter pathway uses Polδ and/or Polɛ, it may function well in proliferating cells and tissues rather than in quiescent cells (Liu et al., 1995). As PCNA is not abundant in developing, post-mitotic neuronal cells compared with their mitotic progenitors, the PCNA-dependent pathway could be less active in the post-mitotic cells. The increased cell death observed in Polβ–/– neuronal cells (Figures 3 and 4) could result from the dysfunction of both Polβ- and PCNA-dependent pathways.
The respiratory control system is very complex and is not yet well understood. Respiration in mammals relies on a neural network located within the brain stem. The Krox-20 gene encodes a protein with three zinc fingers and is activated during hindbrain development before the onset of morphological segmentation (Wilkinson et al., 1989). Krox-20-deficient mice are defective in hindbrain segmentation, exhibit an abnormally slow respiratory and jaw opening rhythm, and die on the first day of post-natal life (Jacquin et al., 1996). Thus, abnormal hindbrain structures would cause a failure in the respiratory control system. The death of Polβ–/– neonates may result from abnormal hindbrain development due to increased death of post-mitotic neural cells.
Materials and methods
Construction of a targeting vector
A 7.1 kb EcoRI fragment of mouse genomic DNA, spanning from the promoter region to the second intron of the Polβ gene (Yamaguchi et al., 1987), was used to produce the targeting construct. A 0.3 kb NaeI region of the fragment containing exons 1 and 2 was replaced with the neo gene. The resulting construct possesses homologous arms of 2.3 and 3.4 kb flanking the neo gene. The HSV-TK gene was positioned downstream of the longer arm.
Disruption of the mouse Polβ gene
BK4 cells, a subclone of the ES cell line E14TG2a derived from strain 129/Ola mice, were cultured as described previously (Aratani et al., 1999). The targeting vector was linearized with NotI and introduced into ES cells by electroporation. Colonies doubly resistant to active G418 (250 μg/ml) and gancyclovir (2 μM) were isolated and screened for disruption of the Polβ gene by Southern blot analysis following complete digestion of genomic DNA with BamHI; a 485 bp SacI–SspI fragment containing the promoter region of the Polβ gene was used as a probe. An ES clone carrying one copy of the disrupted Polβ gene was injected into blastocysts to generate chimeras as described previously (Aratani et al., 1999). The chimeric mice obtained were mated with strain C57BL/6 mice, and F1 animals heterozygous for the Polβ gene were obtained. Polβ–/– mice were generated by Polβ+/– intercrosses; embryos and pups were genotyped by Southern blot analysis.
Northern and Western blot analysis
For Northern blot analysis, total RNA was extracted from the brain of E18.5 embryos with TRIzol reagent (Gibco-BRL) as recommended by the manufacturer. Each sample (20 μg) was electrophoresed in a 1% agarose–formaldehyde gel, blotted to Hybond-N+ (Amersham), and probed with a 1 kb fragment of a mouse Polβ cDNA. The probe including the entire sequence of the Polβ coding region was produced by RT–PCR using the following primers: 5′-TCCACCGGTAAGACCCAGGT-3′ and 5′-TAGGACTCCTAGTGGGCTGT-3′. Sequence analysis of the PCR product confirmed that the mouse Polβ cDNA fragment is 88.7% homologous to the human cDNA.
For Western blot analysis, cell-free extracts prepared from whole embryos at E10.5 were electrophoresed in a 12.5% SDS–polyacrylamide gel, transferred to an Immobilon membrane (Millipore), probed with a polyclonal rabbit anti-Polβ antibody (Date et al., 1990) and peroxidase-conjugated goat anti-rabbit IgG (BioRad), and stained with enhanced chemiluminescent detection reagents (ECL Kit, Amersham).
Assay for Polβ activity
An activity gel assay was performed as described previously (Hirose et al., 1989). Whole embryos at E10.5 were disrupted by sonication in 50 mM Tris–HCl at pH 7.8 containing 0.5 M KCl, 0.1 mM EDTA and 1 mM dithiothreitol (DTT), and supernatants (35 μg protein) obtained by centrifugation for 45 min at 10 000 g were loaded onto a 12.5% polyacrylamide gel including 0.1% SDS and 150 μg/ml of activated calf thymus DNA (Pharmacia). After electrophoresis, the proteins in the gel were renatured by removing SDS in 50 mM Tris–HCl pH 7.5, 0.1 mM EDTA, 1 mM DTT, and then incubated in a DNA synthesis mixture (50 mM Tris–HCl at pH 7.5, 7 mM MgCl2, 1 mM DTT, dATP, dGTP, dTTP and [α-32P]dCTP) for 17 h at 37°C. The gel was washed several times with 5% trichloroacetic acid, 1% sodium pyrophosphate, dried and autoradiographed.
Histological analysis
Embryos were fixed in 4% paraformaldehyde, embedded in OCT compound (Sakura Finetechinical Co.) and serially sectioned. TUNEL assay was performed on the fixed sections using an In situ Apoptosis Detection Kit (Takara) followed by counterstaining with cresyl violet (Sigma). Immunostaining was performed using monoclonal anti-β3-tubulin antibody TuJ1 (BAbCO), followed by rhodamine-conjugated goat anti-mouse IgG (Chemicon): nuclei were counterstained with Hoechst. Neonate lungs were fixed in 4% formalin, embedded in paraffin, serially sectioned and stained with hematoxylin and eosin.
Acknowledgments
Acknowledgements
We thank Dr A.Matsukage for the gift of the mouse Polβ gene and the anti-Polβ antibody, Dr N.Maeda for BK4 cells, and Dr N.Adachi for the neo gene. We also thank Dr K.Takiguchi-Hayashi for helpful discussions, Mr M.Kobayashi and Mrs T.Nishigaki for technical support, and Miss F.Oonuma for animal care. This work was supported in part by a Grant-in-Aid for Cancer Research from the Ministry of Education, Science, Sports and Culture, Japan, and by a Grant-in-Aid from the Ministry of Health and Welfare, Japan.
References
- Aratani Y., Koyama, H., Nyui, S., Suzuki, K., Kura, F. and Maeda, N. (1999) Severe impairment in early host defense against Candida albicans in mice deficient in myeloperoxidase. Infect. Immun., 67, 1828–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D.E., Stamp, G., Rosewell, I., Denzel, A. and Lindahl, T. (1998) Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol., 8, 1395–1398. [DOI] [PubMed] [Google Scholar]
- Bayer S.A. and Altman,J. (1991) Neocortical Development. Raven Press, New York, NY. [Google Scholar]
- Bennett R.A., Wilson, D., Wong, D. and Demple, B. (1997) Interaction of human apurinic endonuclease and DNA polymerase β in the base excision repair pathway. Proc. Natl Acad. Sci. USA, 94, 7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D., Selfridge, J., Millar, J.K., Samuel, K., Hole, N., Ansell, J.D. and Melton, D.W. (1996) DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nature Genet., 13, 489–491. [DOI] [PubMed] [Google Scholar]
- Betz U.A., Vosshenrich, C.A., Rajewsky, K. and Muller, W. (1996) Bypass of lethality with mosaic mice generated by Cre–loxP-mediated recombination. Curr. Biol., 6, 1307–1316. [DOI] [PubMed] [Google Scholar]
- Biade S., Sobol, R.W., Wilson, S.H. and Matsumoto, Y. (1998) Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J. Biol. Chem., 273, 898–902. [DOI] [PubMed] [Google Scholar]
- Caldecott K.W., Aoufouchi, S., Johnson, P. and Shall, S. (1996) XRCC1 polypeptide interacts with DNA polymerase β and possibly poly(ADP-ribose) polymerase and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro.Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date T., Yamamoto, S., Tanihara, K., Nishimoto, Y., Liu, N. and Matsukage, A. (1990) Site-directed mutagenesis of recombinant rat DNA polymerase β: involvement of arginine-183 in primer recognition. Biochemistry, 29, 5027–5034. [DOI] [PubMed] [Google Scholar]
- Elder R.H. et al. (1998)Alkylpurine-DNA-N–glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol. Cell. Biol., 18, 5828–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelward B.P. et al. (1997)Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 13087–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K.M., Sekiguchi, J.M., Seidl, K.J., Swat, W., Rathbun, G.A., Cheng, H.L., Davidson, L., Kangaloo, L. and Alt, F.W. (1998) Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature, 396, 173–177. [DOI] [PubMed] [Google Scholar]
- Gao Y. et al. (1998)A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell, 95, 891–902. [DOI] [PubMed] [Google Scholar]
- Grawunder U., Zimmer, D., Fugmann, S., Schwarz, K. and Lieber, M.R. (1998) DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell, 2, 477–484. [DOI] [PubMed] [Google Scholar]
- Gu H., Marth, J.D., Orban, P.C., Mossmann, H. and Rajewsky, K. (1994) Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- Hang B., Singer, B., Margison, G.P. and Elder, R.H. (1997) Targeted deletion of alkylpurine-DNA-N–glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc. Natl Acad. Sci. USA, 94, 12869–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F., Hotta, Y., Yamaguchi, M. and Matsukage, A. (1989) Difference in the expression level of DNA polymerase β among mouse tissues: high expression in the pachytene spermatocyte. Exp. Cell Res., 181, 169–180. [DOI] [PubMed] [Google Scholar]
- Jacquin T.D., Borday, V., Schneider-Maunoury, S., Topilko, P., Ghilini, G., Kato, F., Charnay, P. and Champagnat, J. (1996) Reorganization of pontine rhythmogenic neuronal networks in Krox-20 knockout mice. Neuron, 17, 747–758. [DOI] [PubMed] [Google Scholar]
- Klungland A. and Lindahl, T. (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J., 16, 3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland A., Rosewell, I., Hollenbach, S., Larsen, E., Daly, G., Epe, B., Seeberg, E., Lindahl, T. and Barnes, D.E. (1999) Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA, 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y., Nash, R.A., Klungland, A., Schar, P., Barnes, D.E. and Lindahl, T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Widen, S.G., Williams, K.R., Kedar, P., Karpel, R.L. and Wilson, S.H. (1990) Studies of the domain structure of mammalian DNA polymerase β. Identification of a discrete template binding domain. J. Biol. Chem., 265, 2124–2131. [PubMed] [Google Scholar]
- Lee M.K., Tuttle, J.B., Rebhun, L.I., Cleveland, D.W. and Frankfurter, A. (1990) The expression and posttranslational modification of a neuron-specific β-tubulin isotype during chick embryogenesis. Cell Motil. Cytoskel., 17, 118–132. [DOI] [PubMed] [Google Scholar]
- Li Z., Otevrel, T., Gao, Y., Cheng, H.L., Seed, B., Stamato, T.D., Taccioli, G.E. and Alt, F.W. (1995) The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell, 83, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Liu Y.C., Liu, W.L., Ding, S.T., Chen, H.M. and Chen, J.T. (1995) Serum responsiveness of the rat PCNA promoter. Exp. Cell Res., 218, 87–95. [DOI] [PubMed] [Google Scholar]
- Ludwig D.L., MacInnes, M.A., Takiguchi, Y., Purtymun, P.E., Henrie, M., Flannery, M., Meneses, J., Pedersen, R.A. and Chen, D.J. (1998) A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat. Res., 409, 17–29. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y. and Kim, K. (1995) Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science, 269, 699–702. [DOI] [PubMed] [Google Scholar]
- Nicholl I.D., Nealon, K. and Kenny, M.K. (1997) Reconstitution of human base excision repair with purified proteins. Biochemistry, 36, 7557–7566. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P., Thode, S., Hancke, J. and Vielmetter, W. (1994) Mechanisms of overlap formation in nonhomologous DNA end joining. Mol. Cell. Biol., 14, 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Singhal, R.K., Srivastava, D.K., Molina, J.T., Tomkinson, A.E. and Wilson, S.H. (1996) Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem., 271, 16000–16007. [DOI] [PubMed] [Google Scholar]
- Singhal R.K. and Wilson, S.H. (1993) Short gap-filling synthesis by DNA polymerase β is processive. J. Biol. Chem., 268, 15906–15911. [PubMed] [Google Scholar]
- Sobol R.W., Horton, J.K., Kuhn, R., Gu, H., Singhal, R.K., Prasad, R., Rajewsky, K. and Wilson, S.H. (1996) Requirement of mammalian DNA polymerase-β in base-excision repair. Nature, 379, 183–186. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Nowakowski, R.S. and Caviness, V., Jr (1995) The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J. Neurosci., 15, 6046–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbs R.S., Flannery, M.L., Meneses, J.J., Hartmann, A., Tucker, J.D., Thompson, L.H., Cleaver, J.E. and Pedersen, R.A. (1999) Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol., 208, 513–529. [DOI] [PubMed] [Google Scholar]
- Wilkinson D.G., Bhatt, S., Chavrier, P., Bravo, R. and Charnay, P. (1989) Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature, 337, 461–464. [DOI] [PubMed] [Google Scholar]
- Wu Q. and Maniatis, T. (1999) A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell, 97, 779–790. [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S., Smeyne, R.J., Wallace, J.D. and Curran, T. (1996) The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA, 93, 8919–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Hirose, F., Hayashi, Y., Nishimoto, Y. and Matsukage, A. (1987) Murine DNA polymerase β gene: mapping of transcription initiation sites and the nucleotide sequence of the putative promoter region. Mol. Cell. Biol., 7, 2012–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]