Abstract
The transcription factor AP-1, composed of Jun and Fos proteins, is a major target of mitogen-activated signal transduction pathways. However, little is known about AP-1 function in normal cycling cells. Here we report that the quantity and the phosphorylation state of the c-Jun and JunB proteins vary at the M–G1 transition. Phosphorylation of JunB by the p34cdc2–cyclin B kinase is associated with lower JunB protein levels in mitotic and early G1 cells. In contrast, c-Jun levels remain constant while the protein undergoes N-terminal phosphorylation, increasing its transactivation potential. Since JunB represses and c-Jun activates the cyclin D1 promoter, these modifications of AP-1 activity during the M–G1 transition could provide an impetus for G1 progression by a temporal increase in cyclin D1 transcription. These findings constitute a novel example of a reciprocal connection between transcription factors and the cell cycle machinery.
Keywords: AP-1/cell cycle/cyclin D1/JunB/phosphorylation
Introduction
Progression through the cell cycle is controlled by the periodic, phase-specific regulation of the abundance and activity of a defined set of proteins. In eukaryotic cells, these include the different cyclins, cyclin-dependent kinases (cdks) and their inhibitors. This tight control also affects downstream genes that encode proteins required for nucleotide metabolism and DNA replication, such as thymidine kinase, DNA polymerase α, the DNA polymerase δ subunit, histones, etc. Regulation of these key components of the cell cycle machinery occurs via several interdependent mechanisms: post-translational modification, stage-specific association with different regulatory proteins, periodic degradation, cyclic induction of gene expression, etc. While experiments in different organisms have helped to dissect the post-transcriptional mechanisms by which cell cycle proteins are regulated, less is known about the mechanisms that regulate cyclin gene transcription during cell cycle progression. Specific cyclin–cdk complexes affect transcription through phosphorylation of components of the basal transcription apparatus (RNA polymerases and basal transcription factors), transcriptional modulators such as the retinoblastoma protein Rb, and transcription factors. The best studied example of the regulation of transcription factors by the cell cycle machinery is the modulation of the E2F protein family activity by cyclin–cdk complexes. Repression of E2F transactivation through binding to the Rb family of pocket proteins is relieved by cyclin–cdk-mediated pRb phosphorylation. Moreover, some but not all E2F proteins are direct substrates of specific cyclin–cdk kinase complexes and phosphorylation affects their binding to DNA. These proteins play a central role in regulating genes that promote cell cycle progression, including themselves and cyclins (Hiebert et al., 1992; Dynlacht, 1997).
Few other examples of cross-talk between transcription factors and cell cycle kinases are known. Other transcription factors have been proposed to be potential substrates for cyclin–cdk kinases (Boulikas, 1995; Dynlacht, 1997). Among them, only c-Myc has been shown to affect the expression of the cell cycle machinery proteins (Steiner et al., 1995; Grandori and Eisenman, 1997).
In the current study, we have extended this analysis to members of the AP-1 family of transcription factors. Although the expression of Jun and Fos proteins in various cellular contexts and their modulation after mitogenic stimulation or under stress conditions has been well documented (for a review, see Karin et al., 1997), less is known about AP-1 function in unstimulated cycling cells. While most cell types contain low basal levels of AP-1 activity and express only a subset of the Jun and Fos proteins (L.Bakiri and D.Lallemand, unpublished data), antibody microinjection and antisense experiments have suggested that these proteins are required for cell cycle progression (Holt et al., 1986; Kovary and Bravo, 1991, 1992; Cosenza et al., 1994). Further support for the role of c-Jun in normal cell growth came from the study of c-Jun-deficient mouse embryonic fibroblasts (Johnson et al., 1993; Schreiber et al., 1999). These findings indicate that the basal AP-1 activity is indispensible and that AP-1 not only mediates the response to environmental stimuli but exerts a specific role during the cell cycle. In this regard, it is noteworthy that numerous genes coding for components of both the cell cycle clock machinery (cyclins, cdks, etc.) and the DNA synthesis apparatus (PCNA, thymidine kinase, etc.) contain putative AP-1-binding sites in their promoters. Cyclin D1 is a prototype of such a gene that may directly link AP-1 to cell cycle progression; it is implicated in G1 progression and can be induced efficiently by treatments that increase AP-1 activity, e.g. mitogenic stimulation, ras transformation, ectopic c-Jun expression, etc. (Herber et al., 1994; Albanese et al., 1995).
Conversely, little is known about the effect of cell cycle progression on AP-1 activity. If such regulation occurs, it could affect AP-1 DNA binding, transactivation potential, localization or stability. Interestingly, like the E2F/DP transcription factor, AP-1 consists of different dimer combinations of Jun and Fos proteins that differ greatly in their abilities to transactivate a given TRE element (Nakabeppu et al., 1988; Smeal et al., 1989; Hirai et al., 1990; Hai and Curran, 1991; Ryseck and Bravo, 1991). In addition, Jun and Fos proteins are very sensitive to post-translational regulation of their activity, their stability and their binding to DNA (Binetruy et al., 1991; Musti et al., 1996). Like E2F and DP subfamilies, the specific modulation of one or several AP-1 components by a given cyclin–cdk complex could restrict target gene expression during the cell cycle and activate a phase-specific cellular response.
To clarify the potential role of AP-1 in cell cycle progression, we have monitored the quantitative and qualitative variations in c-Jun and JunB in exponentially cycling HeLa cells using two-dimensional flow cytometry, immunofluorescence and Western blotting. We observed that although the level of c-Jun protein remains constant, JunB decreases during mitosis. Moreover, the Jun proteins undergo specific post-translational modifications during M and early G1 phases. JunB is phosphorylated by the Cdc2–cyclin B complex on specific amino acids that are not conserved in c-Jun or JunD, triggering its degradation. c-Jun becomes phosphorylated on its N-terminal serines, an event that has been shown to increase both its transactivational potential and stability. To assess whether these stage-specific Jun modifications are important for cell cycle progression, we have used fibroblastic cell lines harbouring inducible Jun alleles together with transient transfection experiments, and demonstrated that changing the c-Jun to JunB ratio in the cell affects cyclin D1 gene transcription. Our results describe one mechanism by which AP-1 regulates G1 progression via its control of cyclin D1 gene expression. In return, the cell cycle machinery affects AP-1 activity during mitosis.
Results
Variation of Jun proteins during the cell cycle
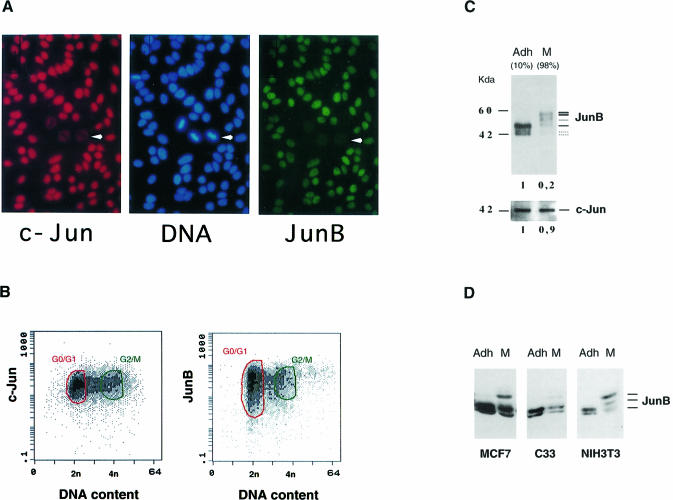
Exponentially growing HeLa cells express all three members of the Jun family, ATF-2 and barely detectable levels of Fos proteins (D.Lallemand, unpublished results). AP-1-responsive promoters in these cells would then be expected to bind mostly Jun–Jun or Jun–ATF dimers. The availability of polyclonal antibodies against c-Jun and monoclonal antibodies against JunB (Lallemand et al., 1997) enabled us to investigate, by indirect fluorescence microscopy, the co-expression of JunB and c-Jun within the same cell. Figure 1A shows the results of a representative experiment using cycling HeLa cells. While the c-Jun signal was relatively constant in all cells, more variation was observed for the JunB signal. Moreover, we noticed the almost complete absence of JunB signal from mitotic cells with disrupted nuclear membrane (indicated by an arrow). In the same mitotic cells, c-Jun is dispersed throughout the total cell volume, but its global level does not decrease drastically when corrected for the increase in the total cell volume.
Fig. 1. Variations of Jun protein levels during the cell cycle. (A) Co-immunofluorescence microscopy staining of exponentially growing HeLa cells for c-Jun (red), DNA (blue) and JunB (green). (B) Two-dimensional flow cytometry measurement of c-Jun and JunB levels in exponentially growing HeLa cells: FITC fluorescence (arbitrary units) of 10 000 individual cycling cells is plotted against propidium iodide fluorescence (DNA content). G0/G1 and G2/M cells are circled in green and red, respectively. (C) Analysis of c-Jun and JunB proteins in HeLa cells extracts from adherent (Adh) and nocodazole shake-off fractions (M). The percentage of 4n cells is indicated in parentheses. Equal amounts of total cell extracts were separated on SDS–PAGE and immunoprobed with the appropriate antibodies (Lallemand et al., 1997). Densitometry scan quantitation is presented below each gel. (D) Analysis of JunB proteins in different cell types. Equal amounts of extracts from adherent or M phase-enriched human breast cancer cells (MCF7), human cervical carcinoma cells (C33) and mouse fibroblasts (NIH 3T3) were probed for JunB.
To quantify these variations, we performed flow cytometry analysis of immunostained exponentially growing cells. Two-dimensional flow cytometry allows the concomitant measurement of both DNA content and the specific fluorescence signal due to a given protein at the single-cell level. c-Jun- and JunB-specific fluorescence were plotted against the DNA content, measured using propidium iodide. Figure 1B shows that while c-Jun displayed limited variations in the relative fluorescence intensity during the cell cycle, JunB staining was more heterogeneous, especially during the G1 phase. The variation in signal intensity was of the order of two log units. It should be noted that M phase, which represents only 5–8% of the total cell number in a cycling cell population, is virtually indistinguishable from G2 by such conventional flow cytometry methods.
To investigate further the levels of c-Jun and JunB proteins at the G2–M boundary and to exclude that the variations observed were due to masking of the reactive epitopes, we performed Western blotting experiments with specific cell populations. We used nocodazole to enrich for mitotic cells (Rieder and Palazzo, 1992). M phase cells were collected by mechanical shaking, while G2 cells remained bound to the plate. The efficiency of cell synchronization was confirmed by flow cytometry (data not shown). Total cell extracts were prepared and analysed. No significant difference was observed in c-Jun protein level when exponentially growing and M phase cells were compared (Figure 1C). In contrast, JunB protein concentration decreased 5-fold in mitotic extracts and, most interestingly, its apparent molecular weight increased significantly. The mobility shift in JunB was estimated to be ∼15 kDa, was sensitive to alkaline phosphatase treatment (not shown) and is probably due to phosphorylation. The mobility shift and the quantitative decrease in the JunB protein levels in mitosis were also observed in extracts from mitotic HeLa cells obtained by mechanical shaking of exponentially growing cells, excluding that it was an artefact of the nocodazole treatment. Furthermore, similar results were obtained using other human and mouse cell lines (Figure 1D) and also primary mouse fibroblasts (not shown).
JunB binds to and is phosphorylated by Cdc2
We then attempted to identify the kinase(s) responsible for JunB phosphorylation during the G2–M transition. In contrast to c-Jun, JunB post-translational modifications have not yet been characterized extensively. The fact that the specific Jun N-terminal kinase phosphorylation sites present in c-Jun and JunD are not conserved in JunB, as well as the results of previous studies (Derijard et al., 1994; Kallunki et al., 1996) suggest that this protein is not a substrate for the JNKs. However, in a recent publication, it was claimed that the JNK MAP kinases can phosphorylate JunB on threonines 102 and 104, resulting in a slower migrating protein (Li et al., 1999). To check if the massive phosphorylation of JunB observed during mitosis is due to JNK activity, we irradiated HeLa cells with UV and followed the activation of JNK and the phosphorylation of JunB. While JNK was activated rapidly after irradiation, as demonstrated with an anti-phospho JNK-specific antibody, no shift in JunB mobility similar to that observed in mitotic cells could be seen (Figure 2A). Moreover, JNK activity was not detectable in mitotic cells using either anti-phospho JNK antibodies (Figure 2A) or in-gel kinase assays (not shown) under conditions where JunB was hyperphosphorylated. In conclusion, even though we cannot formally exclude that JNK can phosphorylate JunB, this phosphorylation does not induce the changes in the mobility of the protein seen during mitosis.
Fig. 2. Search for JunB phosphorylating kinase. (A) Total extracts from exponentially growing, mitotic (nocodazole shake-off) and UV-treated HeLa cells were separated by SDS–PAGE and probed with antibodies specific for JunB (top), phosphorylated Jun kinase (middle) and total JNK (bottom panel). (B) c-Jun and JunB were immunoprecipitated from total cell extracts of mitotic HeLa cells and the precipitated fractions were immunoblotted for Cdc2. A non-relevant antiserum (control) was also included. (C) The Cdc2–cyclin B kinase complex induced JunB mobility shift and degradation in vitro. Starfish oocyte Cdc2–cyclin B complex was incubated for the times indicated with equal amounts of in vitro synthesized JunB protein (Iv JunB) in the presence of ATP and protease inhibitors. As a control, in vitro JunB was also incubated in the absence of Cdc2 for 240 min. The reaction mixture was resolved by SDS–PAGE and immunoprobed for JunB. The position of the different forms of JunB is shown. A cross-reacting band in the reticulocyte lysate is indicated by an asterisk.
The cyclin B–Cdc2 complex is a major serine-threonine kinase active during mitosis, and is known to phosphorylate numerous proteins, triggering their degradation (Luscher et al., 1991; Boulikas, 1995). Therefore, we examined whether JunB could interact with this kinase complex. c-Jun and JunB were immunoprecipitated with specific antibodies from mitotic HeLa cell extracts, separated by SDS–PAGE and immunoprobed for Cdc2 by Western blotting. Cdc2 was undetectable after immunoprecipitation using anti c-Jun-specific antibodies, while it was clearly co-immunoprecipitated with JunB (Figure 2B). Similar results were obtained with extracts prepared from NIH 3T3 mouse fibroblasts. Cdc2 was also co-precipitated when we used an antibody reacting against a different domain of JunB, showing that this association is not due to a particular cross-reactivity of the antiserum. Quantitatively, only a minor fraction of total Cdc2 was associated with JunB (data not shown). This may explain why we were unable to co-immunoprecipitate JunB using Cdc2-specific antibodies. Alternatively, the JunB–Cdc2 complex could be disrupted by the Cdc2 antibodies that we used. Formally, the fact that we did not immunoprecipitate Cdc2 with c-Jun does not rule out a possible weak association.
To assess whether purified, active Cdc2–cyclin B can phosphorylate JunB in vitro, full-length JunB protein was synthesized in a reticulocyte lysate and incubated for increasing lengths of time with purified Cdc2–cyclin B complex from starfish oocytes (a gift from M.Dorée and J.C.Labbe) in the presence of ATP. The mixture was separated by SDS–PAGE and probed with an anti-JunB-specific antibody (Figure 2C). After 30 min, the JunB protein showed multiple hyperphosphorylated forms. The migration pattern obtained and the reduction in mobility were reminiscent of those seen in mitotic HeLa extracts (compare Figure 2A and C). Densitometry scanning showed that, despite the use of standard protease inhibitors, the overall amount of JunB protein decreases when compared with the same extracts incubated in the same conditions without Cdc2. Incubation of c-Jun or JunD with Cdc2–cyclin B did not produce any notable modification in their migration pattern (data not shown). Taken together, these results render plausible the hypothesis that the Cdc2–cyclin B complex phosphorylates JunB, an event that triggers its degradation.
Mutational analysis of mitosis-dependent JunB phosphorylation
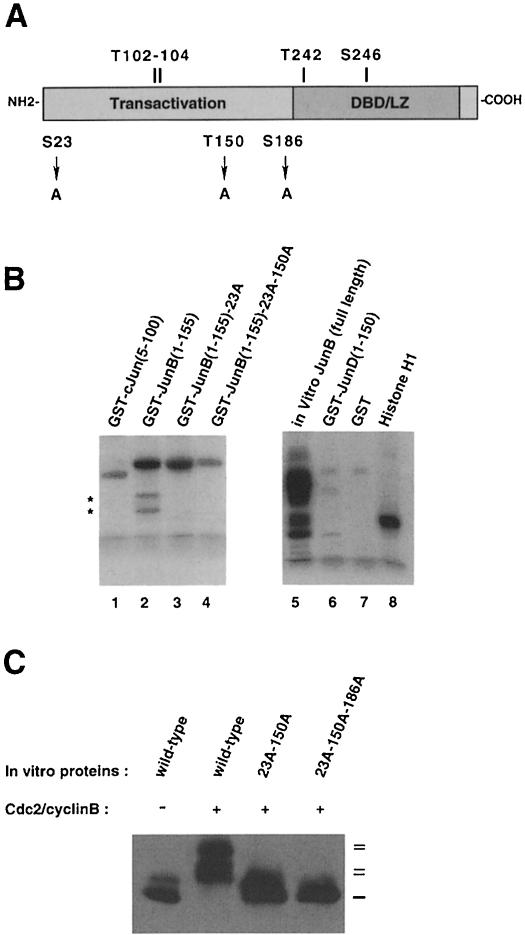
Examination of the JunB sequence reveals two Ser–Pro (residues at positions 23 and 186) and one Thr–Pro (position 150) motifs that are not conserved in c-Jun or JunD and that could be candidates for mitosis-specific phosphorylation (Figure 3A). To test if Ser28 and Thr150 could be phosphorylated, we used GST fusions containing the N-terminal part of JunB (amino acids 1–155) and performed in vitro phosphorylation assays with purified Cdc2–cyclin B and radiolabelled ATP. Figure 3B shows the results of a representative experiment. The Cdc2 kinase efficiently phosphorylated GST–JunB fragments (lane 2) compared with equivalent c-Jun (lane 1) or JunD (lane 6) fragments. Replacement of Ser23 (lane 3) or Thr150 (not shown) with alanine decreased 32P incorporation. Also, we noticed the disappearance of the two faster migrating bands (indicated by asterisks) that correspond to protein degradation products. Only mutation of both sites (lane 4), decreased 32P incorporation to practically background levels, showing that, in the N-terminal portion of JunB, Ser23 and Thr150 were indeed phosphorylated by Cdc2–cyclin B. Moreover, it is clear that threonines 102 and 104, identified as phosphorylation sites for JNK (Li et al., 1999), are not phosphorylated by Cdc2–cyclin B. We then mutated Ser23 and Thr150 to alanines in the full-length JunB and compared its migration profile with wild-type protein following Cdc2–cyclin B phosphorylation. As shown in Figure 3C, the mutation of these two sites did not completely abolish the JunB upshift. This was only achieved when we mutated the third putative site Ser186.
Fig. 3. Mutational analysis of JunB degradation. (A) Schematic representation. Serine or threonine residues, conserved at similar positions in c-Jun, are indicated above, while serine and threonine residues specific for JunB and subject to mutational analysis are depicted below the scheme. (B) Cdc2–cyclin B phosphorylates Ser23 and Thr150 in vitro in JunB but not Thr102 and Thr104. Starfish oocyte Cdc2–cyclin B complex was incubated for 30 min with 5 µg of the indicated purified GST–Jun fusions in the presence of radioactive ATP. GST, histone H1 (0.1 µg) and reticulocyte lysate synthesized JunB were used as controls. The reactions were then resolved on SDS–PAGE and analysed with a phosphorimager. Asterisks on the left indicate degradation products. (C) Mutation of an additional residue (Ser186) in full-length JunB is necessary to abolish the Cdc2–cyclin B effect on JunB upshift. Equal amounts of in vitro sythesized JunB, JunB23A-150A and JunB23A-150A-186A were incubated with Cdc2–cyclin B. The mixture was resolved by SDS–PAGE and immunoprobed for JunB.
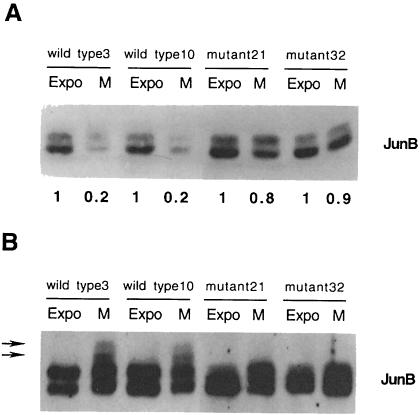
To assess whether the replacement of these three sites affects JunB degradation during mitosis, we established 293 clones stably expressing wild-type or mutated (triple) full-length JunB proteins. We chose 293 cells, a human kidney epithelial cell line transformed by adenoviral DNA, because they express very low levels of JunB. Equivalent amounts of total protein extracts from exponentially growing cells and M phase-enriched cultures were probed for JunB. Figure 4A shows the results for four independent representative clones. While ∼80% of the wild-type JunB protein was degraded during the G2–M transition, we observed a reduction of only 10–20% for the mutant protein. Moreover, when equivalent amounts of immunoreactive JunB protein were loaded and separated by SDS–PAGE and probed for JunB, the mutant protein was less retarded than wild-type JunB (Figure 4B). It should be noted that mitotic phosphorylation of JunB is less pronounced in 293 cells than in HeLa cells. This may be due to faster degradation of the phosphorylated JunB protein in these cells relative to HeLa cells.
Fig. 4. (A) Mutation of Ser23, Thr150 and Thr186 stabilizes JunB in mitotic cells. Equivalent amounts of total cell extracts from exponentially growing and mitotic 293 cells stably expressing wild-type (clones 3 and 10) or mutated JunB (clones 21 and 32) were resolved by Western blot and probed for JunB. Densitometry scan quantitation is presented. (B) Mutation of Ser23, Thr150 and Thr186 reduces JunB mitotic phosphorylation. Equivalent amounts of immunoreactive JunB proteins (based on the quantification in Figure 3B) were separated by Western blot and probed for JunB. Arrows indicate the position of slower migrating bands in mitotic wild-type JunB.
Analysis of cellular and molecular events at the G2–M–G1 transition
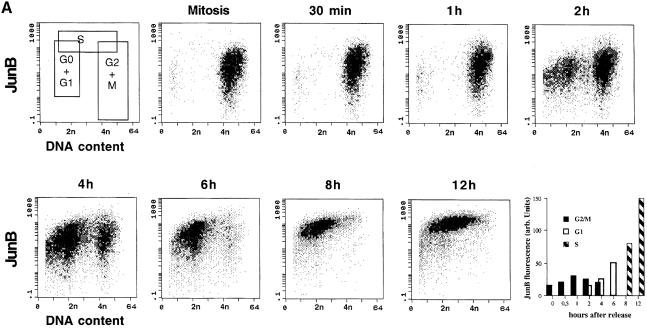
To follow more precisely the fate of JunB during the G2–M–G1 transition, nocodazole-arrested mitotic HeLa cells were collected by shaking, replated in the absence of nocodazole and followed for 12 h by both flow cytometry and Western blotting. Under such conditions (Figure 5A), mitotic and early G1 cells (until 2 h after release) showed a wide distribution of the JunB-specific fluorescence signal (>2 log units), with some G1 and mitotic cells containing very low amounts of JunB. As cells progressed through the G1 phase of the cycle (6–8 h) and entered S phase (8–12 h after release), JunB levels strongly increased (5- to 10-fold, see last panel), in agreement with our previous immunofluorescence and Western blot analysis, and cells exhibited a more homogeneous staining, especially in S phase. Under the same conditions, c-Jun showed little variation (data not shown), consistent with the results obtained with asynchronous cells.
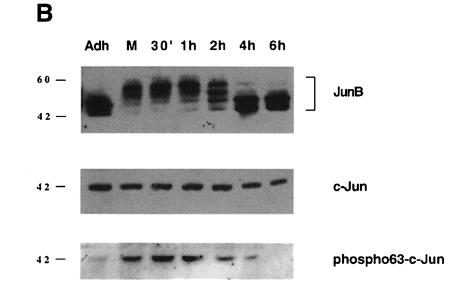
Fig. 5. Cellular and molecular events at the G2–M–G1 transition. Synchronized populations of HeLa cells were analysed by two-dimensional flow cytometry and Western blot. (A) Flow cytometry: variation of JunB during cell cycle progression of nocodazole-released cells. JunB-specific fluorescence is plotted against DNA content or (last panel) against time. The first panel (top left) indicates the limits of each subpopulation of cells. The positions of 2n and 4n cells are indicated. The last panel is based on JunB fluorescence peak values, obtained after gating the different populations. (B) Western blot analysis of JunB (the film was overexposed to reveal the slower migrating JunB bands), c-Jun and Ser63 phosphorylated c-Jun in cell extracts corresponding to the same time points.
Total cell extracts corresponding to the same time points were also analysed by Western blot. As expected, c-Jun showed neither quantitative nor qualitative modifications, while JunB was clearly shifted when floating M phase cells were compared with adherent cells blocked at the end of G2 (Figure 4B). This shift persisted for up to 2 h after replating the mitotic cells. Furthermore, JunB protein levels clearly decreased as cells proceeded through M and entered G1 phase. Finally, 4–6 h after release, JunB levels and the phosphorylation pattern became comparable with those of exponentially growing cells.
It was intriguing that, although c-Jun has been shown to be a key regulator of the G0–G1 transition (Kovary and Bravo, 1991; Johnson et al., 1993), its abundance did not change during the cell cycle. It is well established that the regulation of c-Jun activity occurs not only at the quantitative level, but also at the post-translational level. N-terminal phosphorylation of c-Jun has been shown to increase its transactivation potential strongly (Derijard et al., 1994). Therefore, we probed the same cell extracts with a specific monoclonal antibody directed against the phosphorylated Ser63 residue of the c-Jun protein (Lallemand et al., 1998). Figure 5B shows that indeed, c-Jun underwent a transient N-terminal phosphorylation as cells proceed from G2 to M and that this phosphorylation persists for up to 4 h after nocodazole release, i.e. until the majority of the cells have left the mitotic compartment (Figure 5A). Immunofluorescence and two-dimensional flow cytometry experiments on non-disturbed exponentially growing HeLa cell cultures confirmed that mitotic cells contained phosphorylated c-Jun (not shown). Since we could not detect specific JNK activation during mitosis (see Figure 2A), the nature of the kinase and the pathway responsible for c-Jun phosphorylation during mitosis remain to be determined.
The effects of c-Jun or JunB overexpression on cyclin D1 transcription
Previous studies have shown that JunB is a less potent activator of transcription than c-Jun (Chiu et al., 1989; Schutte et al., 1989; Castellazzi et al., 1991; Deng and Karin, 1993). The decrease in the concentration of JunB relative to c-Jun and the N-terminal phosphorylation of c-Jun at the beginning of the G1 phase could result in increased AP-1 activity and the induction of genes that respond better to c-Jun. A number of putative AP-1 target genes are known. Among them, cyclin D1 is interesting since it is synthesized early in the G1 phase and is required for progression to S phase (Quelle et al., 1993; Resnitzky and Reed, 1995). c-Jun has also been reported to induce cyclin D1 transcription in transient transfection assays (Herber et al., 1994; Albanese et al., 1995).
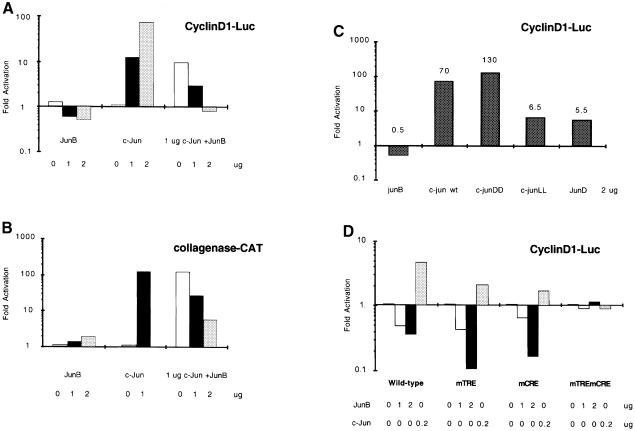
The human cyclin D1 transcriptional control sequences have been cloned and shown to contain different regulatory elements, including a TRE site and a CRE site, located 935 and 52 bp upstream of the initiation site, respectively. In exponentially cycling cells, transcriptional activation by c-Jun has been shown to be mediated essentially by these two sites (Herber et al., 1994; Albanese et al., 1995), and several groups have indicated that Jun, Fos and ATF proteins might be part of a complex bound to this site (Brown et al., 1998; Beier et al., 1999). Using cyclin D1 promoter constructs containing both sites for transient transfection in HeLa cells, we observed different responses to JunB and c-Jun (Figure 6A). While c-Jun strongly activated the cyclin D1 reporter, JunB repressed it in a dose-dependent manner. In contrast, JunB reproducibly activated a collagenase promoter construct containing the canonical TRE element, although this activation was weak compared with c-Jun. Furthermore, the strong activation by c-Jun was inhibited by co-expression of JunB. However, the inhibition of the cyclin D1 promoter was more pronounced than that of the collagenase promoter (Figure 6A and B). When we mimicked c-Jun N-terminal phosphorylation by using a c-Jun protein in which serines 63 and 73 were replaced by aspartic acid, the ability of c-Jun to activate cyclin D1 transcription increased (Figure 6C). In contrast, replacing these residues with leucines, which blocks c-Jun phosphorylation, strongly decreased its transcriptional activation of the cyclin D1 promoter. This c-JunLL protein exhibited activity similar to JunD which is a weak activator of cyclin D1 (Figure 6C). Finally, c-Jun and JunB seem to affect the same sites on the cyclin D1 promoter since only the mutation of both the proximal CRE and the distal TRE could abolish the effect of the two proteins (Figure 6D).
Fig. 6. Regulation of the human cyclin D1 promoter by Jun proteins. (A) JunB counteracts c-Jun activation of the cyclin D1 gene promoter. HeLa cells were transiently transfected with the Δ973cyclinD1LUC plasmid together with increasing amounts of expression constructs for c-Jun or JunB and a combination of 1 µg of c-Jun and increasing amounts of JunB. (B) JunB is a weak but positive transactivator on a canonical TRE-responsive promoter. In the same type of experiment, HeLa cells were transfected with a collagenase–CAT construct together with expression constructs for c-Jun and JunB. (C) Effect of each of the Jun proteins on the cyclin D1 gene promoter. HeLa cells were transiently transfected with the Δ973cyclinD1LUC plasmid together with expression constructs for c-Jun, JunB, JunD and mutants of c-Jun in the N-terminal phosphorylation sites. (D) JunB and c-Jun affect the same sites on the cyclin D1 promoter. HeLa cells were transiently transfected with Δ973cyclinD1LUC, Δ973cyclinD1mTRELUC or Δ973cyclinD1mCRELUC plasmids together with c-Jun and JunB expression vectors.The mean results of two representative experiments are presented.
Generation of NIH 3T3-derived cell lines expressing a functionally active JunB–ER protein
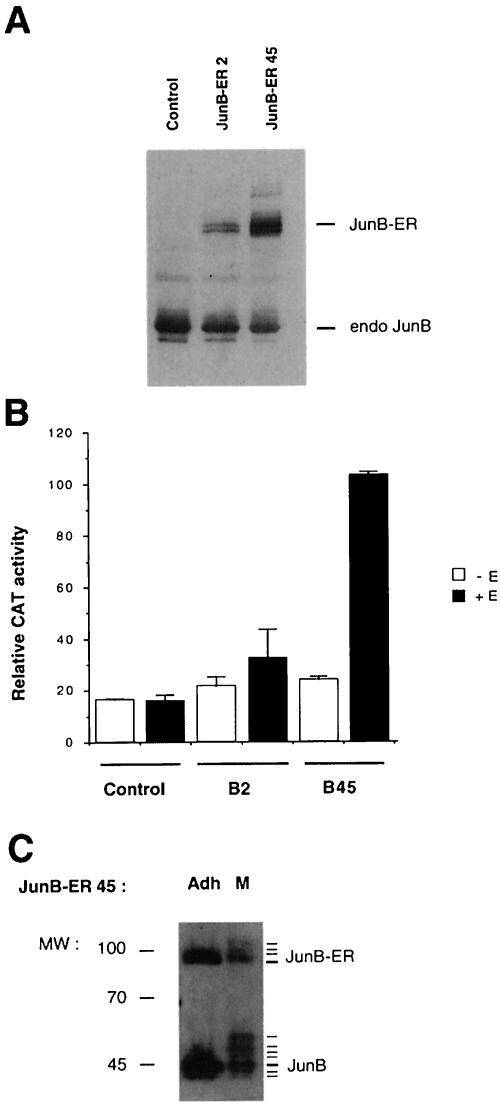
To investigate further the role of JunB during the cell cycle, a conditional allele for JunB was generated by fusing the full-length mouse junB gene with the hormone-binding domain of the human oestrogen receptor (ER). The fusion gene was inserted into a retroviral vector containing the neomycin resistance gene as a selectable marker. The principle of this inducible system is that, although the chimeric protein is constitutively expressed, it is inactive in the absence of hormone, and can be activated by addition of exogenous β-oestradiol (for a review, see Picard, 1994). Several independent NIH 3T3 clones with stable expression of the JunB–ER fusion protein were established and used for further studies. Control cells containing only the neomycin resistance cassette and two independent JunB–ER-expressing clones (B2 and B45) were analysed by Western blotting for the expression of the fusion protein and for the transactivation of a collagenase–CAT reporter construct in the absence or presence of oestradiol (Figure 7A and B). The level of fusion protein detected in the clones correlated well with the ability of the two JunB–ER-expressing clones to activate transcription from a reporter plasmid, containing a single consensus AP-1-binding site, in a hormone-dependent manner. It is noteworthy that irrespective of the amount of fusion protein produced, the transactivation of the collagenase–CAT construct was lower in the NIH 3T3/JunB-ER clones than in NIH 3T3/c-Jun-ER clones previously described (Bossy-Wetzel et al., 1997), confirming that JunB is a less potent transactivator than c-Jun (see also Figure 6B). The majority of the experiments using these inducible cell lines have been reproduced using hydroxytamoxifen, a ligand that does not potentiate the hormone-dependent transcriptional activation domain, in place of β-oestradiol. Finally, in mitotic cells, the JunB–ER fusion protein displayed a migration pattern similar to that of endogenous protein (Figure 7C), indicating that it is also targeted by Cdc2–cyclin B.
Fig. 7. (A) Immunoblotting analysis of JunB proteins in stable JunB–ER clones. Equal amounts of total extracts from β-oestradiol-stimulated NIH 3T3 control cells, NIH 3T3/JunB–ER clone 2 and clone 45 were separated by SDS–PAGE and probed for JunB. Labelled lanes indicate the position of the endogenous JunB protein and the ectopically expressed JunB–ER fusion protein of ∼80 kDa. (B) The JunB–ER protein activates transcription of a transiently transfected TRE reporter in a hormone-dependent fashion. Control and JunB–ER cells (clones B2 and B45) were transiently transfected with a collagenase–CAT construct and treated for 40 h with β-oestradiol or solvent. The values and standard deviation represent the results of three independent experiments. (C) The JunB–ER fusion protein is also subjected to Cdc2–cyclin B phosphorylation during mitosis. Equal amounts of total extracts from adherent (Adh) or mitotic (M) NIH 3T3/JunB–ER45 cells were separated by SDS–PAGE and probed for JunB.
c-Jun–ER and JunB–ER activation lead to opposite effects on cyclin D1 levels
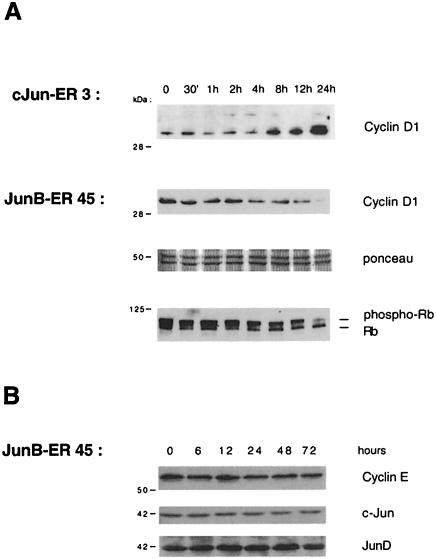
Considering the opposite effect of transient expression of JunB and c-Jun on cyclin D1 transcription, we examined the effects of c-Jun and JunB in the inducible ER cell system. We performed Western blot analysis using a cyclin D1-specific antibody and extracts from JunB–ER- and c-Jun–ER-expressing cells. As shown in Figure 8A, cyclin D1 protein levels increased significantly upon oestrogen stimulation of NIH 3T3/c-Jun-ER cells. In contrast, the activation of the JunB–ER fusion protein was followed by a significant decrease in cyclin D1 protein levels, consistent with the results previously obtained using transient transfections.
Fig. 8. Western blotting analysis of NIH 3T3/JunB–ER-45 and NIH 3T3/c-Jun-ER cells. (A) Equivalent amounts of total cell extracts from stimulated cells were separated by SDS–PAGE, transferred to nitrocellulose membranes and probed with a mouse monoclonal anti-cyclin D1 antibody. In the case of JunB–ER, the filter was Ponceau stained to check the correct protein loading and reprobed with a monoclonal anti-Rb. (B) JunB–ER induction does not affect either cyclin E, c-Jun or JunD expression. Equivalent amounts of total cell extracts were subjected similarly to Western blot with the appropriate antibodies.
Cyclin D1 is a key regulator of the G1–S transition and promotes cell growth, in complex with Cdk4, by phosphorylating and inactivating the retinoblastoma tumour supressor protein (for a review, see Sherr, 1996). The inactivation of cyclin D1–cdk complexes by lowering the amounts of the cyclin partner should therefore result in a decrease in its Rb-phosphorylating activity and the accumulation of active, growth-suppressive, hypophosphorylated Rb protein. Figure 8A shows that this was indeed the case. An immunoblotting experiment using the same oestrogen-stimulated JunB–ER cell extracts and a specific anti-Rb antibody showed that the activation of the JunB–ER chimera correlated with an increase in the level of hypophosphorylated Rb. The decrease seen in total Rb protein levels remains to be investigated. In contrast, we did not detect any changes in either c-Jun or JunD protein levels, indicating that the cyclin D1 decrease was not due to the down-regulation of the other Jun proteins. Cyclin E levels also remain unaffected (Figure 8B).
Activation of the JunB–ER protein results in decreased cell proliferation
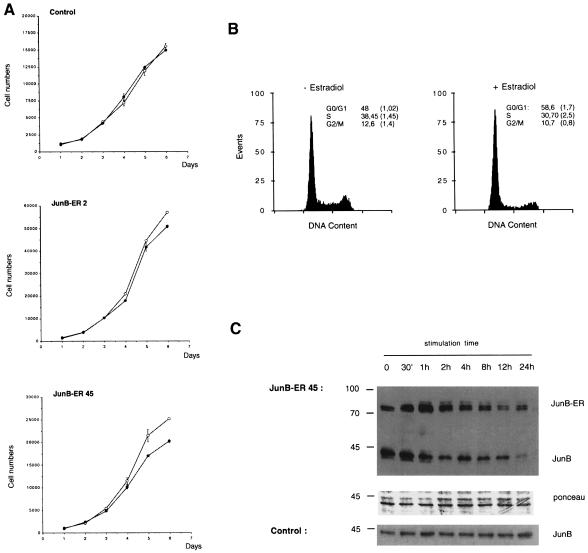
We next examined the effect of activation of the JunB–ER protein by β-oestradiol on cell growth. Control cells and the same clones (B2 and B45) were seeded at low density in the presence or absence of exogenous β-oestradiol, and were counted daily. The growth curves of β-oestradiol-treated or untreated control fibroblasts were virtually identical, demonstrating that β-oestradiol had no effect on the growth rate of these cells (Figure 9A). This was in marked contrast to NIH 3T3 cells expressing the JunB–ER protein. Activation of the JunB–ER protein by β-oestradiol resulted in a substantial decrease in cell numbers. The extent of growth retardation was dependent upon the expression levels of the JunB–ER protein found in these cells (Figure 7A). Cells expressing moderate levels of the JunB–ER protein (clone B2) showed a relatively mild inhibition of growth after β-oestradiol treatment, while cells with higher expression levels of the JunB–ER protein (clone B45) exhibited a stronger decrease in cell numbers in the presence of β-oestradiol (Figure 9A).
Fig. 9. (A) Effect of the JunB–ER protein on the growth of NIH 3T3 cells. Control NIH 3T3 and NIH 3T3/JunB–ER clones were cultured in the presence (closed circles) or absence (open circles) of 1 µM β-oestradiol. Cells were counted daily and the mean values of triplicate cultures have been plotted against time. (B) Cell cycle distribution of 48 h untreated and β-oestradiol-treated NIH 3T3/JunB–ER cells (clone B45). The DNA content is presented as relative fluorescence, and a quantitation of the results is presented. (C) Immunoblotting analysis of the JunB proteins during β-oestradiol stimulation of NIH 3T3/JunB–ER-45 or NIH 3T3 control cells. Equal amounts (documented for JunB–ER by Ponceau staining) of nuclear extracts from cells stimulated with β-oestradiol were probed for JunB. The endogenous JunB and the ectopically expressed JunB–ER fusion protein are indicated.
Activation of the c-Jun–ER fusion protein triggers an apoptotic cell death programme (Bossy-Wetzel et al., 1997). This appeared not to be the case with the JunB–ER-expressing cells. No visible morphological changes were observed in oestrogen-treated JunB–ER cells, while c-Jun–ER cell lines showed a marked apoptotic morphology upon oestrogen treatment (Bossy-Wetzel et al., 1997). To explore this issue further, cell cycle parameters were determined by flow cytometry. We have shown previously that oestrogen treatment had no effect on the cell cycle distribution of NIH 3T3 fibroblasts (Bossy-Wetzel et al., 1997). In contrast, the number of cells in the G0/G1 phase of the cell cycle was increased in β-oestradiol-treated JunB–ER fibroblasts, while S and G2/M phases decreased (Figure 8B). The extent of increase in the G0/G1 phase correlated with the expression levels of the JunB–ER protein found in these cells, since clone B45 showed a more marked arrest than clone B2 (not shown). No significant increase in cells with less than diploid DNA content, representative of apoptotic cells, was detected in JunB–ER cells, excluding the contribution of apoptosis to the growth retardation (data not shown). This accumulation of G1-arrested cells in exponentially growing, oestrogen-stimulated JunB–ER cultures presumably is due to the negative effect of JunB overexpression on cyclin D1 synthesis, slowing its accumulation early in G1.
We were surprised to observe that the increase in the G0/G1 compartment of the cell cycle observed upon JunB–ER induction reached a maximum after 36 h stimulation and remained constant, with a notable fraction of cells still cycling through S, G2 and M phases (not shown). Therefore, we decided to follow the nuclear accumulation of both the fusion protein and the endogenous JunB after oestrogen addition by Western blot using specific anti-JunB antibodies. As shown in Figure 9C, the level of the 80 kDa fusion protein reached a maximum 1 h after oestrogen addition. This accumulation was accompanied by a slightly decreased mobility of the JunB–ER protein probably representing the basal phosphorylation in JunB, analogous to the three migrating JunB species observed in growing NIH 3T3 cells (Lallemand et al., 1997). A similar mobility shift pattern was observed with a c-Fos–ER fusion protein by Preston and co-workers (Preston et al., 1996) and is probably due to the increased accessibility of the fusion protein to post-translational modifications in the presence of oestradiol when the protein is no longer complexed with heat-shock proteins.
Surprisingly, the initial increase in the JunB–ER protein was coupled with a decrease in the endogenous JunB protein (see also Figure 7A). Both the endogenous JunB and the JunB–ER fusion protein levels decreased further at later time points. The total amount of JunB was at least three times lower after 24 h of oestrogen treatment than in the untreated cells, and this was not observed in the control cell line (Figure 9C). Thus, while it accumulates rapidly in the nucleus, the JunB–ER fusion protein, together with the endogenous JunB protein, displays a substantial decrease at late times of induction, suggesting that JunB–ER accumulation may trigger a proteolysis cascade that decreases the concentration of total JunB protein in the cell. The link between this degradation and the Cdc2-associated JunB degradation remains to be clarified. We cannot formally exclude that the fusion protein also directly down-regulates JunB transcription or translation. However, the fact that the endogenous JunB and the fusion protein are transcribed from different promoters and their mRNAs differ in their 5′ and 3′ portions renders this possibility improbable. The decrease in the level of JunB and JunB–ER proteins may explain the fact that these cells are only partially arrested in the G0/G1 phase of the cycle. Similar instability of the activated fusion protein was shown for a c-Fos–ER chimera (Preston et al., 1996), but we did not observe it with the c-Jun–ER fusion protein (L.Bakiri, unpublished data).
Discussion
Members of the AP-1 transcription complex were shown to be important regulators of the G0–G1 transition of the cell cycle. In the present study, we investigated the role of some of these genes in normally cycling cells. Combining flow cytometry measurements, immunofluorescence staining and Western blotting experiments, we showed that c-Jun protein levels varied little during the cell cycle. In contrast, JunB displayed a highly heterogeneous distribution, especially in the G2/M and G1 phases. Immuno fluorescence microscopy revealed that the level of JunB protein was low in mitotic cells. Using synchronized cells, we were able to define more precisely the timing of the decrease in JunB abundance and showed that it was correlated with an increase in its apparent molecular weight, due to phosphorylation.
We investigated which kinase may be responsible for JunB phoshorylation. JunB contains a JNK docking site, but lacks serine residues analogous to the phosphorylated sites in c-Jun. While initial studies indicated that it is not phosphorylated by JNK (Derijard et al., 1994; Kallunki et al., 1996), a recent report showed that two threonines are phosphorylated by excess JNK. In the present study, we were unable to induce a shift of JunB similar to that observed in mitotic cells, even after massive activtion of JNK by UV irradiation of different cell lines. In contrast, we were able to reproduce in vitro the mitotic migration pattern of JunB using purified Cdc2–cyclin B complex and, moreover, to co-immunoprecipitate JunB with the mitotic cyclin-dependent kinase p34cdc2. In comparison with mitosis-induced degradation of other proteins, it is reasonable to postulate that Cdc2-mediated phosphorylation of JunB may target the protein for a degradation pathway. Sequence comparison revealed only three proline-flanked serines or threonine residues, not conserved in c-Jun or JunD, that could be candidates for Cdc2–cyclin-mediated phosphorylation. Mutation of these sites, by replacement with alanines, abolished Cdc-2-mediated JunB phosphorylation in vitro and inhibited both JunB mitotic shift and degradation in vivo. Although, we have not ruled out the possibility that JunB might be a physiological substrate for other kinases during mitosis, it is likely that JunB, and not c-Jun or JunD, is a specific target for Cdc2–cyclin-dependent kinase activity, providing a link between AP-1 and the basal cell cycle machinery.
Although early microinjection and antisense oligonucleotide inhibition experiments have suggested that c-Jun is necessary for cell cycle progression (Kovary and Bravo, 1991; Cosenza et al., 1994), we did not detect any quantitative variations in c-Jun levels in cycling cells. However, we have observed that c-Jun is transiently phosphorylated in its N-terminal activation domain. This modification occurs in mitotic cells, remains until early G1 and is expected to increase both c-Jun transactivation potential and protein stability. The Jun N-terminal kinases are attractive candidate kinases for this phosphorylation. However, since phospho-specific antibodies and in-gel kinase assays failed to detect mitosis-specific JNK activation, we cannot exlude that other stress-activated kinases, such as p38 or the classical mitogen-activated protein kinases (ERK1 and ERK2), could phosphorylate c-Jun during mitosis. Indeed, the p38 kinase recently has been shown to be activated at the mitotic spindle assembly checkpoint (Takenaka et al., 1998).
In exponentially growing cells, JunB is detectable as two or three discrete Western blot bands. We have shown previously (Lallemand et al., 1997) that the slower migrating band is affected by phosphatase treatment, indicating, together with the results of previous reports (Kovary and Bravo, 1991), that this isoform is a phosphoprotein. The replacement of Ser23, Thr150 and Ser186 with alanines, while strongly decreasing the mitosis-specific upshift, did not alter this pattern (see Figures 3 and 4), suggesting that it is due to the phos phorylation of different residue(s).
As mentioned above, it was shown recently that JNK, and to a lesser extent p38, can phosphorylate JunB on threonines 102 and 104. This modification led to a slightly slower migrating protein and increased JunB transactivation potential on the interleukin-4 (IL-4) promoter (Li et al., 1999). We suggest that JunB can be phosphorylated by two different pathways: by MAP kinases on residues 102 and 104 in exponentially growing and growth factor-stimulated cells, and by Cdc2–cyclin B kinase on residues 23, 150 and 186 in mitotic cells. This last phosphorylation triggers its degradation. Even if c-Jun N-terminal phosphorylation provides only indirect evidence for MAP kinase activity during mitosis, we cannot exclude that the two JunB phosphorylation pathways are interdependent and that, for instance, the phosphorylation of threonines 102 and 104 is a prerequisite for Cdc2 kinase-mediated phosphorylation in vivo, even if this does not seem to be the case in vitro (see Figure 3B). Further studies with different combinations of JunB mutants are needed to clarify these issues.
The cell cycle-dependent decrease in JunB levels, together with the mitosis and early G1-specific phosphorylation and stabilization of c-Jun, led us to investigate whether the two proteins may have opposite roles in the regulation of genes that are important for early G1 progression. Among the cell cycle-regulated genes containing putative AP-1-responsive elements, cyclin D1 was an attractive candidate, as it displays induction kinetics during G1 compatible with the variations observed in AP-1 complexes (Sherr, 1996). Transient transfection experiments using a cyclin D1 reporter showed that c-Jun phosphorylation, mimicked by the use of aspartic acid N-terminal mutants (Papavassiliou et al., 1995), significantly increased the positive effect of c-Jun on cyclin D1 transcription. Interestingly, while JunD was a weaker transactivator, JunB clearly repressed the cyclin D1 construct and was able to counteract the effect of c-Jun. Moreover, we were able to show that the integrity of either the TRE or the CRE element in the cyclin D1 promoter is necessary for JunB repression. Curently, we cannot exclude the possibility that the mitotic phosphorylation of JunB also decreases its capacity to down-regulate target genes such as cyclin D1.
The c-Jun:JunB protein ratio appeared to be an important parameter for the control of cyclin D1 expression during early G1 phase. To test this hypothesis further, we used NIH 3T3 cells stably transfected with hormone-inducible c-Jun–ER (Bossy-Wetzel et al., 1997) or JunB–ER alleles and studied changes in cyclin D1 expression after induction of the fusion proteins. Indeed, JunB and c-Jun activation caused opposite effects on cyclin D1 protein levels. Moreover, JunB activation resulted in a slower cell division rate and a reproducible increase of the G0/G1 population. In fact, cells were accumulating in an early G1 compartment, as shown by the increase in unphosphorylated, growth-suppressive, retinoblastoma protein. It is of interest to note that ectopic expression of JunD also slows fibroblast growth (Pfarr et al., 1994), probably because JunD is a weaker inducer of the cyclin D1 gene than c-Jun.
It is notable that the JunB-induced cell cycle block was only partial. Western blotting analysis indicated that, while neither JunD nor c-Jun protein levels were affected, the induction of the JunB–ER protein is followed by a significant decrease in the quantity of both JunB and JunB–ER proteins at late induction time points, indicating that excess JunB switches on an inhibitory pathway. Treatment of JunB–ER cells with cell-permeant calpain inhibitors prevented this decrease and increased the G0/G1 cell arrest (data not shown). During mitosis, the JunB–ER fusion protein shows similar and concomitant modifications in its migration pattern and might be affected similarly by Cdc2-mediated degradation. The balance between induced synthesis and degradation may be sufficient for the partial inhibition of cell growth.
In contrast, despite a significant accumulation of cyclin D1 protein in c-Jun–ER cell lines, these cells did not show any shortening of their G1 phase, but activated a cell death programme. Therefore, cyclin D1 induction seems necessary, but not sufficient, to induce S phase entry (Bossy-Wetzel et al., 1997). Other events, such as subsequent activation of cyclin E-dependent kinases, are probably required, and the lack of these events might trigger apoptosis. The induction of cell death by cyclin D1 has been reported in other cell types, sometimes correlated with c-Jun induction (Freeman et al., 1994; Kranenburg et al., 1996; Sofer-Levi and Resnitzky, 1996). Furthermore, a direct link between JNK1 activation by Rac1 and cyclin D1 expression was shown recently (Westwick et al., 1997).
Although c-Jun and JunB are very homologous proteins, they seem to be modulated distinctly during the M–G1 transition. Moreover, they exert opposite biological effects on cell cycle progression. Consistent with these findings, fibroblasts derived from c-Jun–/– embryos display slower growth kinetics, an increase in G1 cells and reduced expression of cyclin D1 by a mechanism that involves at least the distal TRE element (Wisdom et al., 1999). Our data suport early findings showing that JunB inhibits c-Jun-mediated transactivation and transforming activity (Chiu et al., 1989; Schutte et al., 1989). In fact, numerous reports have associated JunB expression with differentiation (Melamed et al., 1993; Schlingensiepen et al., 1993; Li et al., 1999), which is often preceded by growth arrest, and others have described the induction of JunB by various growth inhibitors (Hashimoto et al., 1993; Nishina et al., 1993). On the other hand, c-Jun has been associated with ras transformation and mitogenic stimulation, and its overexpression can inhibit differentiation (Su et al., 1991). Our data indicate that the balance between JunB and c-Jun specifically modulated by the cell cycle machinery during mitosis is also important in regulating the cell cycle progression. This occurs at least through the transcriptional control of one component of the G1 cyclin–cdk complex, cyclin D1. Other cell cycle-implicated genes are probably also targeted by AP-1. For example, the absence of c-Jun has been shown to trigger an increase in p53 followed by the accumulation of the CDK inhibitor p21 (Schreiber et al., 1999).
Another important point with respect to the effect of AP-1 on transcription is that, in the same cellular system and depending on the promoter context, we have observed two opposite effects of JunB: JunB–ER activation reproducibly induced a collagenase reporter construct while it repressed the cyclin D1 reporter and RNA, and the same was observed in transient co-transfection experiments. This is an interesting example of differential regulation of two target genes by the same AP-1 component. JunB, like c-Jun, can form both active and inactive AP-1 dimers, depending on the nature of its partner (Fos, Jun or ATF) but also on the targeted promoter sequence (see, for example, JunB activation of the IL-4 promoter in Li et al., 1999). Since the AP-1-responsive elements in cyclin D1 and in the collagenase promoters are different, the opposing effects of JunB on these two genes could be explained by differences in cooperation with other transcription factors. The fact that JunB can activate or inhibit transcription depending on the promoter and partner context may extend to other genes and could contribute to the complexity of the genetic response to AP-1.
In conclusion, this work demonstrates that the antagonism between JunB and c-Jun is important not only in mediating the genetic response to extracellular stimuli, but also in regulating cell cycle progression through the transcriptional control of a key mediator of the cell cycle machinery, cyclin D1. In return, the cell cycle machinery seems to modulate this antagonism during mitosis by increasing c-Jun-activating phosphorylation and specifically triggering the phosphorylation-dependent degradation of JunB.
Materials and methods
Construction of expression vectors
To construct the pMV7junBER vector, a SalI–BspHI fragment from pBluescript-JunB was ligated with a BspHI–BamHI linker oligo into a SalI–BamHI-linearized pUC19-ER vector. An EcoRI fragment was subsequently excised and cloned into the pMV7 expression vector. To construct the JunB expression vectors, wild-type and mutated JunB were excised from pBluescript constructs using appropriate restriction sites and ligated dowstream of either a cytomegalovirus (pCG, a kind gift from Dr F.Thierry) or a Moloney murine leukaemia long terminal repeat (pVLMN1 a kind gift from Dr M.Piechaczyk). Mutation of JunB was performed using the Proto Quick change kit (Stratagene) and subsequently checked by sequencing.
Generation of cell lines and culture conditions
All cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco) containing antibiotics and 7% bovine calf serum (FBS). 17-β-Oestradiol (1 mM, Sigma) was stored at –20°C in ethanol. The NIH 3T3 PMV7 and NIH 3T3 PMV7-c-Jun-ER cells lines were described in Bossy-Wetzel et al. (1997). To generate the NIH 3T3/JunB-ER, 293JunB and 293JunBmut cell lines, NIH 3T3 and 293 cells were transfected similarly using the calcium phosphate precipitation method, and individual G418-resistant colonies were isolated.
For HeLa cell synchronization, 0.1 µg/ml nocodazole was added to subconfluent cultures. After 12 h, mitotic cells were collected by mechanical shaking and eventually released by replating in nocodazole-free medium. Synchronization efficiency was checked by both flow cytometry and microscopic observation of propidium iodide-stained cells.
Fluorescence microscopy
Fluorescence microscopy was described in Lallemand et al. (1997). Cells were co-stained using 4′,6-diamidino-2-phenylindole (DAPI) for DNA, mouse anti-JunB and rabbit anti-c-Jun, followed by fluorescein isothiocyanate (FITC)-coupled anti-mouse and Texas red-coupled anti-rabbit (Amersham). Cells were observed under a Zeiss Axiophot epifluorescence microscope and photographs recorded on Kodak films.
Flow cytometric analysis
Cells were trypsinized and pelleted at 2000 r.p.m. for 5 min. For two-dimensional flow cytometry, cells were fixed in 1% paraformaldehyde pH 7.2, post-fixed in ice-cold methanol and incubated overnight at 4°C in phosphate-buffered saline (PBS) containing 1% Tween, 10% FBS and anti-Jun antibodies. They were then washed and incubated for 1 h at room temperature with the appropriate FITC-coupled secondary antibody (Amersham), washed and resuspended for 15 min in 1 ml of 1% Na citrate, 0.1% Triton-X-100, 50 µg/ml propidium iodide and 50 µg/ml DNase-free RNase A (HSS). Analysis was carried out on an Epics XL Flow Cytometer (Coulter) and FITC fluorescence of individual cells was plotted against propidium iodide fluorescence.
For one-dimensional flow cytometry, the cells were resuspended directly in HSS and analysed. Cell numbers were plotted against propidium iodide fluorescence. Cell cycle phase distribution was estimated using the Multiplus Software (Phoenix Systems).
Preparation of protein extracts and immunoblotting
Whole-cell extracts were prepared in 50 mM Tris pH 7.6, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 0.5 µg/ml each of leupeptin, aprotinin and pepstatin A, and 1 mM phenylmethylsulfonyl fluoride (PMSF) (all from Sigma). Nuclear cell extracts preparation, Western blotting procedures and antibodies were described in Lallemand et al. (1997). The mouse monoclonal antibody specific for phosphorylated Ser63 in mouse c-Jun was described in Lallemand et al. (1998). Rb-, cyclin E-, JNK1- and phospho-JNK-specific antibodies were purchased from Pharmingen (Rb) and Santa-Cruz. Mouse cyclin D1 monoclonal antibody was a kind gift from Dr J.Bartek.
Co-immunoprecipitation and kinase assay
c-Jun and JunB were immunoprecipitated from total extracts using rabbit polyclonal antibodies. Immunocomplexes were collected using protein A–Sepharose beads (Pharmacia), washed several times with PBS-T, fractionated by SDS–PAGE and probed for the immunoprecipitated protein and for Cdc2 (Pharmingen).
JunB protein was prepared for in vitro studies using the TNT rabbit reticulocyte coupled transcription–translation system (Promega). Production and purification of GST fusions have been described elsewhere (Lallemand et al., 1997). Starfish oocyte cdc2–cyclin B complex (a kind gift from M.Dorée and J.C.Labbe) and ATP (eventually radioactive) were added and the reaction carried out at 30°C in the presence of protease inhibitors. The mixture was then separated by SDS–PAGE and exposed to a phosphoimager or probed for JunB.
Reporter assays
NIH 3T3, HeLa and JunB–ER cells were transfected by calcium phosphate co-precipitation with reporter plasmids (pCOL-517-CAT, provided by Dr P.Angel; Δ973CD1-Luc and the corresponding mutants in the TRE and CRE sites provided by Dr R.Muller) and Jun expression vectors (mutated c-Jun was a kind gift from Dr D.Bohmann). Oestrogen (1 µM) was added eventually. Luciferase assays were performed using the Promega kit following the manufacturer’s instructions. The CAT assay mixture included 50 µl of cell extract, 100 µl of 250 mM Tris–HCl pH 8, 8 µl of 20 µM acetyl co-enzyme A and 0.2 µCi of d-threo-[dichloroacetyl-1-14C]chloramphenicol (105 mCi/mmol, ICN). The reaction was separated by thin-layer chromatography and quantified using a phosphorimager.
Growth curves
Triplicate cultures of NIH 3T3 control or NIH 3T3/JunB-ER cells were performed in medium containing 10% serum in the presence or absence of β-oestradiol (1 µM) and cells were counted daily using a Coulter ZI cell counter.
Acknowledgments
Acknowledgements
We are grateful to F.Traincard and O.Jeannequin for help in the preparation of the monoclonal JunB antibody, M.Dorée and J.C.Labbe for the gift of purified Cdc2–cyclin B complex, D.Bohmann for the c-Jun mutant expression vectors, R.Muller for the cyclin D1 promoter constructs and J.Bartek for the cyclin D1 monoclonal antibody. We thank J.Weitzman, J.Ham and F. Mechta for critical reading of the manuscript, and E.Passegué and E.Wagner for fruitful discussions. This work was supported by grants from the ‘Association pour la Recherche contre le Cancer’ (ARC), the EEC Biomed and Training and Mobility Programs and from the ‘Ministry of Education and Research’ (MESR) and the Pasteur Weitzmann Council to L.B.
References
- Albanese C., Johnson,J., Watanabe,G., Eklund,N., Vu,D., Arnold,A. and Pestell,R.G. (1995) Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem., 270, 23589–23597. [DOI] [PubMed] [Google Scholar]
- Beier F., Lee,R.J., Taylor,A.C., Pestell,R.G. and LuValle,P. (1999) Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc. Natl Acad. Sci. USA, 96, 1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binetruy B., Smeal,T. and Karin,M. (1991) Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature, 351, 122–127. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E., Bakiri,L. and Yaniv,M. (1997) Induction of apoptosis by the transcription factor c-Jun. EMBO J., 16, 1695–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T. (1995) Phosphorylation of transcription factors and control of the cell cycle. Crit. Rev. Eukaryot. Gene Expr., 5, 1–77. [PubMed]
- Brown J.R., Nigh,E., Lee,R.J., Ye,H., Thompson,M.A., Saudou,F., Pestell,R.G. and Greenberg,M.E. (1998) Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol., 18, 5609–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellazzi M., Spyrou,G., La,V.N., Dangy,J.P. Piu,F., Yaniv,M. and Brun,G. (1991) Overexpression of c-Jun, junB, or junD affects cell growth differently. Proc. Natl Acad. Sci. USA, 88, 8890–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R., Angel,P. and Karin,M. (1989) Jun-B differs in its biological properties from and is a negative regulator of, c-Jun. Cell, 59, 979–986. [DOI] [PubMed] [Google Scholar]
- Cosenza S.C., Yumet,G., Soprano,D.R. and Soprano,K.J. (1994) Induction of c-fos and c-Jun mRNA at the M/G1 border is required for cell cycle progression. J. Cell. Biochem., 55, 503–512. [DOI] [PubMed] [Google Scholar]
- Deng T. and Karin,M. (1993) JunB differs from c-Jun in its DNA-binding and dimerization domains and represses c-Jun by formation of inactive heterodimers. Genes Dev., 7, 479–490. [DOI] [PubMed] [Google Scholar]
- Derijard B., Hibi,M., Wu,I.H., Barrett,T., Su,B., Deng,T., Karin,M. and Davis,R.J. (1994) JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell, 76, 1025–1037. [DOI] [PubMed] [Google Scholar]
- Dynlacht B.D. (1997) Regulation of transcription by proteins that control the cell cycle. Nature, 389, 149–152. [DOI] [PubMed] [Google Scholar]
- Freeman R., Estus,S. and Johnson,E.J. (1994) Analysis of cell cycle-related gene expression in postmitotic neurons: selective induction of cyclin D1 during programmed cell death. Neuron, 2, 343–355. [DOI] [PubMed] [Google Scholar]
- Grandori C. and Eisenman,R.N. (1997) Myc target genes. Trends Biochem. Sci., 22, 177–181. [DOI] [PubMed] [Google Scholar]
- Hai T. and Curran,T. (1991) Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl Acad. Sci. USA, 88, 3720–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Gaddy,K.D. and Vale,W. (1993) Protooncogene junB as a target for activin actions. Endocrinology, 133, 1934–1940. [DOI] [PubMed] [Google Scholar]
- Herber B., Truss,M., Beato,M. and Muller,R. (1994) Inducible regulatory elements in the human cyclin D1 promoter. Oncogene, 9, 2105–2107. [PubMed] [Google Scholar]
- Hiebert S.W., Chellappan,S.P., Horowitz,J.M. and Nevins,J.R. (1992) The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev., 6, 177–185. [DOI] [PubMed] [Google Scholar]
- Hirai S., Bourachot,B. and Yaniv,M. (1990) Both Jun and Fos contribute to transcription activation by the heterodimer. Oncogene, 5, 39–46. [PubMed] [Google Scholar]
- Holt J.T., Gopal,T.V., Moulton,A.D. and Nienhuis,A.W. (1986) Inducible production of c-fos antisense RNA inhibits 3T3 cell proliferation. Proc. Natl Acad. Sci. USA, 83, 4794–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.S., van,L.B., Papaioannou,V.E. and Spiegelman,B.M. (1993) A null mutation at the c-Jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev., 7, 1309–1317. [DOI] [PubMed] [Google Scholar]
- Kallunki T., Deng,T., Hibi,M. and Karin,M. (1996) c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell, 87, 929–939. [DOI] [PubMed] [Google Scholar]
- Karin M., Liu,Z.G. and Zandi,E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol., 9, 240–246. [DOI] [PubMed] [Google Scholar]
- Kovary K. and Bravo,R. (1991) The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol. Cell. Biol., 11, 4466–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovary K. and Bravo,R. (1992) Existence of different Fos/Jun complexes during the G0-to-G1 transition and during exponential growth in mouse fibroblasts: differential role of Fos proteins. Mol. Cell. Biol., 12, 5015–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranenburg O., van der Eb,A. and Zantema,A. (1996) Cyclin D1 is an essential mediator of apoptotic neuronal cell death. EMBO J., 1, 46–54. [PMC free article] [PubMed] [Google Scholar]
- Lallemand D., Spyrou,G., Yaniv,M. and Pfarr,C.M. (1997) Variations in Jun and Fos protein expression and AP1 activity in cycling, resting and stimulated fibroblasts. Oncogene, 14, 819–830. [DOI] [PubMed] [Google Scholar]
- Lallemand D., Ham,J., Garbay,S., Bakiri,L., Traincard,F., Jeannequin,O., Pfarr,C.M. and Yaniv,M. (1998) Stress-activated protein kinases are negatively regulated by cell density. EMBO J., 17, 5615–5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Tournier,C., Davis,R.J. and Flavell,R.A. (1999) Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J., 18, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B., Brizuela,L., Beach,D. and Eisenman,R.N. (1991) A role for the p34cdc2 kinase and phosphatases in the regulation of phosphorylation and disassembly of lamin B2 during the cell cycle. EMBO J., 10, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D., Resnitzky,D., Haimov,I., Levy,N., Pfarr,C.M., Yaniv,M. and Kimchi,A. (1993) Interleukin 6 induces DNA binding activity of AP1 in M1 myeloblastic cells but not in a growth resistant cell derivative. Cell Growth Differ., 4, 689–697. [PubMed] [Google Scholar]
- Musti A.M., Treier,M., Peverali,F.A. and Bohmann,D. (1996) Differential regulation of c-Jun and JunD by ubiquitin-dependent protein degradation. Biol. Chem., 377, 619–624. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Ryder,K. and Nathans,D. (1988) DNA binding activities of the three murine Jun proteins: stimulation by Fos. Cell, 55, 907–915. [DOI] [PubMed] [Google Scholar]
- Nishina Y., Sumi,T., Iwai,S.A., Kosaka,M. and Nishimune,Y. (1993) The induction of jun genes during the reversible changes induced with sodium butyrate on the differentiation of F9 cells. Exp. Cell Res., 208, 492–497. [DOI] [PubMed] [Google Scholar]
- Papavassiliou A.G., Treier,M. and Bohmann,D. (1995) Intramolecular signal transduction in c-Jun. EMBO J., 14, 2014–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr C.M., Mechta,F., Spyrou,G., Lallemand,D., Carillo,S. and Yaniv,M. (1994) Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell, 76, 747–760. [DOI] [PubMed] [Google Scholar]
- Picard D. (1994) Regulation of protein function through expression of chimaeric proteins. Curr. Opin. Biotechnol., 5, 511–515. [DOI] [PubMed] [Google Scholar]
- Preston G.A., Lyon,T.T., Yin,Y.,J. Lang,E., Solomon,G., Annab,L., Srinivasan,D.G., Alcorta,D.A. and Barrett,J.C. (1996) Induction of apoptosis by c-Fos protein. Mol. Cell. Biol., 16, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle D.E., Ashmun,R.A., Shurtleff,S.A., Kato,J.Y., Bar-Sagi,D., Roussel,M.F. and Sherr,C.J. (1993) Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev., 7, 1559–1571. [DOI] [PubMed] [Google Scholar]
- Resnitzky D. and Reed,S.I. (1995) Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol. Cell. Biol., 15, 3463–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L. and Palazzo,R.E. (1992) Colcemid and the mitotic cycle. J. Cell Sci., 102, 387–392. [DOI] [PubMed] [Google Scholar]
- Ryseck R.P. and Bravo,R. (1991) c-Jun, JUN B and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins. Oncogene, 6, 533–542. [PubMed] [Google Scholar]
- Schlingensiepen K., Schlingensiepen,R., Kunst,M., Klinger,I., Gerdes,W., Seifert,W. and Brysch,W. (1993) Opposite functions of jun-B and c-Jun in growth regulation and neuronal differentiation. Dev. Genet., 14, 305–312. [DOI] [PubMed] [Google Scholar]
- Schreiber M., Kolbus,A., Piu,F., Szabowski,A., Mohle-Steinlein,U., Tian,J., Karin,M., Angel,P. and Wagner,E.F. (1999) Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev., 13, 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte J., Viallet,J., Nau,M., Segal,S., Fedorko,J. and Minna,J. (1989) jun-B inhibits and c-fos stimulates the transforming and trans-activating activities of c-Jun. Cell, 59, 987–97. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (1996) Cancer cell cycles. Science, 274, 1672–7. [DOI] [PubMed] [Google Scholar]
- Smeal T., Angel,P., Meek,J. and Karin,M. (1989) Different requirements for formation of Jun:Jun and Jun:Fos complexes. Genes Dev., 2091–2100. [DOI] [PubMed] [Google Scholar]
- Sofer-Levi Y. and Resnitzky,D. (1996) Apoptosis induced by ectopic expression of cyclin D1 but not cyclin E. Oncogene, 13, 2431–2437. [PubMed] [Google Scholar]
- Steiner P., Philipp,A., Lukas,J., Godden-Kent,D., Pagano,M., Mittnacht,S., Bartek,J. and Eilers,M. (1995) Identification of a Myc-dependent step during the formation of active G1 cyclin–cdk complexes. EMBO J., 14, 4814–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H.Y., Bos,T.J., Monteclaro,F.S. and Vogt,P.K. (1991) Jun inhibits myogenic differentiation. Oncogene, 6, 1759–66. [PubMed] [Google Scholar]
- Takenaka K., Moriguchi,T. and Nishida,E. (1998) Activation of the protein kinase p38 in the spindle assembly checkpoint and mitotic arrest. Science, 280, 599–602. [DOI] [PubMed] [Google Scholar]
- Westwick J.K., Lambert,Q.T., Clark,G.J., Symons,M., Van Aelst,L., Pestell,R.G. and Der,C.J. (1997) Rac regulation of transformation, gene expression and actin organization by multiple, PAK-independent pathways. Mol. Cell. Biol., 17, 1324–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisdom R., Johnson,R.S. and Moore,C. (1999) c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J., 18, 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]