Abstract
Background
Japanese encephalitis virus (JEV) is a mosquito-borne flavivirus, leading to an acute encephalitis and damage to the central nervous system (CNS). The mechanism of JEV pathogenesis is still unclear. DNA microarray analyses have been recently employed to detect changes in host gene expression, which is helpful to reveal molecular pathways that govern viral pathogenesis. In order to globally identify candidate host genes associated with JEV pathogenesis, a systematic mRNA profiling was performed in spleens and brains of JEV-infected mice.
Results
The results of microarray analysis showed that 437 genes in spleen and 1119 genes in brain were differentially expressed in response to JEV infection, with obviously upregulated genes like pro-inflammatory chemokines and cytokines, apoptosis-related proteases and IFN inducible transcription factors. And the significant pathways of differentially expressed genes are involved in cytokine-cytokine receptor interaction, natural killer cell mediated cytotoxicity, antigen processing and presentation, MAPK signaling, and toll-like receptor signaling, etc. The differential expression of these genes suggests a strong antiviral response of host but may also contribute to the pathogenesis of JEV resulting in encephalitis. Quantitative RT-PCR (RT-qPCR) assay of some selected genes further confirmed the results of microarray assay.
Conclusions
Data obtained from mRNA microarray suggests that JEV infection causes significant changes of mRNA expression profiles in mouse spleen and brain. Most of differentially expression genes are associated with antiviral response of host, which may provide important information for investigation of JEV pathogenesis and therapeutic method.
Background
Japanese encephalitis virus (JEV), a mosquito-borne flavivirus belonging to family Flaviviridae, is responsible for an acute encephalitis and damage to the central nervous system (CNS) in wide areas of southern and eastern Asia. And recently, it has been isolated from previously non-affected areas, such as Australia [1]. Japanese encephalitis (JE) has a high fatality rate of 30% and around half of the JE survivors have severe neurological sequelae [2]. Approximately 50,000 JE cases with 10,000 deaths are reported annually [3]. Following entry into the host system through a mosquito bite, JEV may replicates in various organs such as liver and spleen, and then reaches the central nervous system, resulting in a rapid inflammatory response [4]. According to the observations from studies of other flaviviruses, specifically dengue virus, it has been proposed that JEV traverse through a lymphatic route that also involves cells of the monocyte/macrophage lineage. Recently, JEV has been shown to effectively replicate within lymphocytes and macrophages, thereby making these cell types possible carriers of the virus from the periphery to the CNS [5,6]. However, it remains to be elucidated how JEV infects the CNS via these peripheral cells. In addition, although neurological disorders caused by JEV are often characterized by evidence of immune system recognition and the presence of inflammatory components among the neuropathological changes, the mechanisms by which this virus causes neurological disease are not fully understood [7].
Recently, multiple DNA microarray analyses have been employed to detect changes in host gene expression after viral infection, which makes it possible to reveal molecular pathways that govern viral pathogenesis. Genechip analysis of human umbilical vein endothelial cells infected with Dengue Virus (DV) detected the upregulation of 269 genes and downregulation of 126 genes [8]. Gene profiling study of West Nile Virus (WNV) infected human embryonic kidney cells, human glioma cells and mice tissues were also performed [9,10]. Furthermore, identification of gene profiles in JEV-infected neuroblastoma cells and brain tissue have been reported recently, suggesting an increased expression of pro-inflammatory cytokines, chemokines, and anti-viral response genes after JEV infection [11,12]. However, both of studies on JEV were restricted to CNS, and few gene profiling researches about response in peripheral immune system has been carried out.
In present study, to globally identify candidate host genes associated with JEV pathogenesis, DNA microarray technology was utilized to investigate mRNA profile in spleen and brain tissues of mice infected with JEV wild strain P3, and some of the selected genes were further confirmed by quantitative RT-PCR. It was demonstrated that JEV infection resulted in significant changes in the expression of numerous genes in spleen and brain tissues, which could be crucial messages for revealing of JEV pathogenesis.
Results
mRNA expression profile of JEV-infected mice
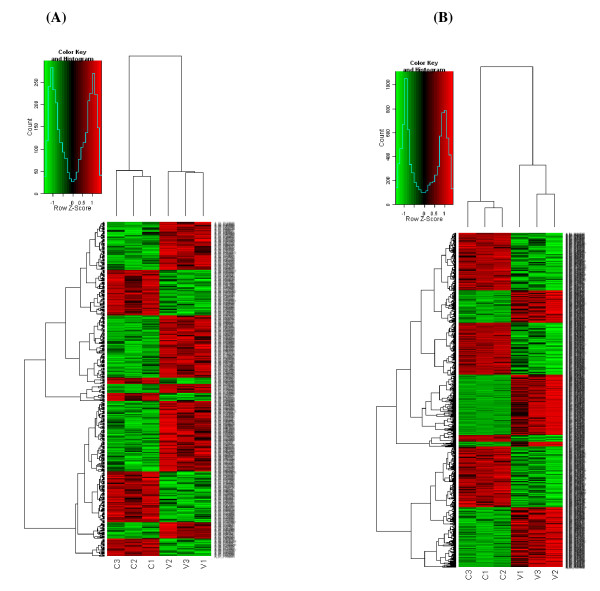
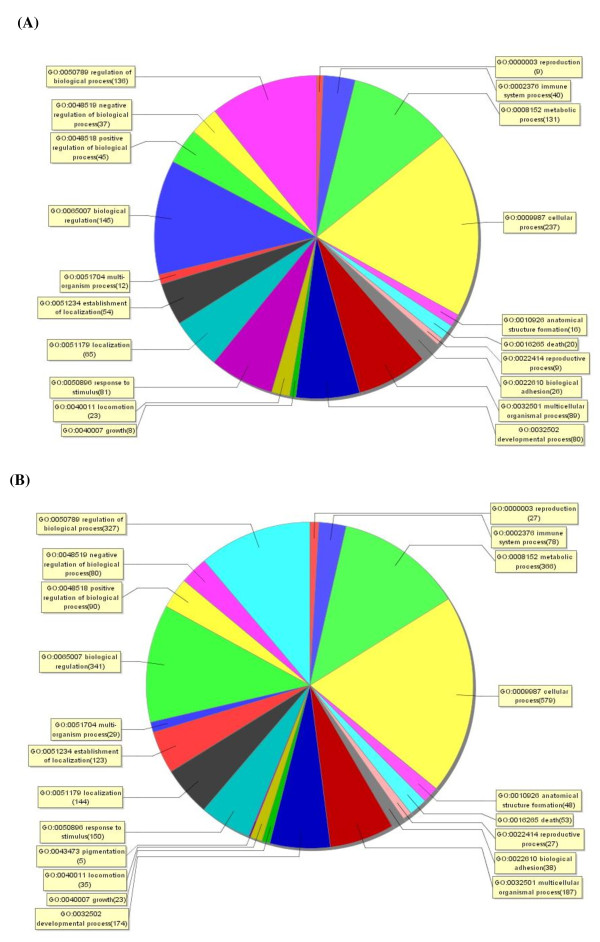
A mouse whole gene array was used to perform a systematic analysis of mRNA expression profile of spleen and brain tissues of JEV P3-infected mice which were sacrificed at day 3 and day 6 post-inoculation respectively. Genes that had ≥ |2|-fold change were identified as significantly differential expression. Of 41174 genes represented on the chip, 437 genes were differentially expressed in mouse spleens and 1119 genes were differentially regulated in brains in response to JEV infection (change fold ≥ 2.0, p value < 0.05). Unsupervised clustering (Figure 1) analysis of the expression profiles showed a distinct mRNA signature in both spleens and brains during JEV infection. To elucidate the correlation between gene expression pattern and JEV infection-induced biological processes, functional classification of mRNA transcripts and pathway analysis were performed. Differentially regulated genes in spleens of JEV-infected mice are involved in the biological processes such as cellular process, biological regulation and immune system process, etc (Figure 2A). And the significant pathways of differentially expressed genes are known to be involved in cytokine-cytokine receptor interaction, natural killer cell mediated cytotoxicity, antigen processing and presentation, and chemokine signaling, etc (Table 1). While biological processes which showed differentially regulated genes in brains of JEV infected mice are cellular process, metabolic process, and biological regulation, etc (Figure 2B). And the significant pathways of differentially expressed genes in mouse brain are cytokine-cytokine receptor interaction, MAPK signaling, neuroactive ligand-receptor interaction, and toll-like receptor signaling, etc. (Table 2).
Figure 1.
Unsupervised hierarchical clustering of differentially expressed mRNAs. mRNA hybridization was performed with the use of 4 × 44 K Agilent Whole Mouse Genome Oligo Microarray. For each sample pair, the experiments were done with two independent hybridizations (Cy3 and Cy5 interchanging labeling). Genes that had ≥ |2|-fold change were identified as significantly differentially expressed. Differentially regulated genes were clustered using SAS (ShanghaiBio Analysis System) to identify significant gene expression patterns in spleens (A) and brains (B) of JEV-infected mice. Red indicates higher expression and green indicates lower expression in JEV-infected mice versus control. Black indicates no expression difference. The small figure represents color scales used in the cluster map.C indicates control group and V indicates viral-inoculated group. Each group contains 3 mice.
Figure 2.
Enriched Gene Ontology terms in the biological process category among differentially expressed genes. After mRNA microarray assay, significantly enriched Gene Ontology analysis in the biological process category among differentially expressed genes (fold change ≥ 2.0) in spleens (A) and brains (B) of JEV-infected mice was performed by using SAS (ShanghaiBio Analysis System). Each color section represents a different biological process and the gene number enriched in this section was shown following the biological process name.
Table 1.
Significant pathways of the differential expressed genes in spleens of JEV-infected mice.
| Pathway name | No. of genes | p-Value |
|---|---|---|
| Antigen processing and presentation | 9 | 0 |
| Chemokine signaling pathway | 13 | 0 |
| Complement and coagulation cascades | 7 | 0 |
| Cytokine-cytokine receptor interaction | 23 | 0 |
| Natural killer cell mediated cytotoxicity | 9 | 0 |
| Classical Complement Pathway | 3 | 1.00E-04 |
| Complement Pathway | 3 | 4.00E-04 |
| Alternative Complement Pathway | 2 | 0.0021 |
| Granzyme A mediated Apoptosis Pathway | 2 | 0.0036 |
| Toll-like receptor signaling pathway | 4 | 0.0071 |
| Intestinal immune network for IgA production | 3 | 0.009 |
| CCR3 signaling in Eosinophils | 2 | 0.0111 |
| Caspase Cascade in Apoptosis | 2 | 0.0121 |
| NOD-like receptor signaling pathway | 3 | 0.0121 |
| MAPK signaling pathway | 6 | 0.0168 |
| ECM-receptor interaction | 3 | 0.0232 |
| Jak-STAT signaling pathway | 4 | 0.0284 |
| IFN gamma signaling pathway | 1 | 0.0478 |
| B Lymphocyte Cell Surface Molecules | 1 | 0.0611 |
| T cell receptor signaling pathway | 3 | 0.069 |
Table 2.
Significant pathways of the differential expressed genes in brains of JEV-infected mice.
| Pathway name | No. of genes | p-value |
|---|---|---|
| Chemokine signaling pathway | 19 | 0 |
| Cytokine-cytokine receptor interaction | 30 | 0 |
| Jak-STAT signaling pathway | 19 | 0 |
| MAPK signaling pathway | 19 | 0 |
| NOD-like receptor signaling pathway | 10 | 0 |
| Purine metabolism | 14 | 0 |
| Toll-like receptor signaling pathway | 13 | 0 |
| IL-2 Receptor Beta Chain in T cell Activation | 5 | 6.00E-04 |
| Neuroactive ligand-receptor interaction | 16 | 6.00E-04 |
| NF-kB activation by Nontypeable Hemophilus influenzae | 4 | 0.0012 |
| Apoptosis | 7 | 0.0023 |
| Antigen processing and presentation | 7 | 0.0037 |
| IFN gamma signaling pathway | 2 | 0.0082 |
| Natural killer cell mediated cytotoxicity | 8 | 0.011 |
| T cell receptor signaling pathway | 7 | 0.0123 |
| IFN alpha signaling pathway | 2 | 0.0156 |
| FAS signaling pathway (CD95) | 3 | 0.0173 |
Genes with differential expression in spleens of JEV-infected mice
In spleens of mice with JEV infection, 263 genes were detected to be significantly upregulated and 174 genes were downregulated (Table 3). Genes with increased expression in spleens of JEV infected mice mainly fell into the function of immune response to viral infections. These include pro-inflammatory cytokine IFN-γ, IFN response transcription factor IRF7, IFN-induced proteins like IFIT1, IFITM3 and IFITM7, protein degradation gene ubiquitin-specific protease Usp29 and Usp18, apoptosis related genes granzymA, granzymeB, Porferin, and IMNB2, killer cell lectin-like receptors KLRC1, KLRC2 and KLRC3, and chemokines such as CXCL10, CXCL11, CCL12, CCL2 and CCL9. The marked increase in expression of these genes implies the occurrence of a strong antiviral protective response to JEV infection. The significantly downregulated genes are mainly involved in cell adhesion molecules such as monocyte/macrophage-lineage cell marker CD163, transmembrane cell surface receptor of Langerhans cells CD207, and ligand for myeloid cells receptor CD200. Evidence was also found for decreased expression of interferon transcription factor IRF6 and interleukin 7 receptor, which may contribute to JEV pathogenesis as well.
Table 3.
Differentially-regulated genes of our interest in spleens of mice with JEV Infection
| Genbank Accession | Gene symbol | Gene description | Fold change | p-Value |
|---|---|---|---|---|
| Up-regulated | ||||
| NM_177261 | Kndc1 | kinase non-catalytic C-lobe domain (KIND) containing 1 | 21.440 | 0.0252 |
| NM_019494 | Cxcl11 | chemokine (C-X-C motif) ligand 11 | 19.752 | 0.0084 |
| NM_013542 | Gzmb | granzyme B | 6.202 | 0.0286 |
| NM_031167 | Il1rn | interleukin 1 receptor antagonist (Il1rn), transcript variant 1 | 6.060 | 0.0015 |
| AK157531 | LOC629091 | activated spleen cDNA, RIKEN full-length enriched library, clone:F830221E13 | 5.500 | 0.0154 |
| NM_001033228 | Itga1 | integrin alpha 1 | 5.482 | 0.0308 |
| NM_013584 | Lifr | leukemia inhibitory factor receptor | 5.319 | 0.0109 |
| NM_177923 | H2-M10.2 | histocompatibility 2, M region locus 10.2 | 5.276 | 0.0009 |
| NM_008198 | Cfb | complement factor B | 5.188 | 0.0019 |
| NM_001005858 | LOC667370 | similar to interferon-induced protein with tetratricopeptide repeats 3 | 4.949 | 0.0459 |
| NM_145153 | Oas1f | 2'-5' oligoadenylate synthetase 1F | 4.619 | 0.0013 |
| NM_011331 | Ccl12 | chemokine (C-C motif) ligand 12 | 4.539 | 0.0308 |
| NM_027893 | Pvrl4 | poliovirus receptor-related 4 | 4.429 | 0.0352 |
| NM_010741 | Ly6c | lymphocyte antigen 6 complex, locus C | 4.287 | 0.0042 |
| NM_010370 | Gzma | granzyme A | 4.236 | 0.0010 |
| NM_009152 | Sema3a | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | 4.132 | 0.0358 |
| AF229257 | Usp29 | ubiquitin-specific processing protease 29 | 4.033 | 0.0093 |
| NM_133871 | Ifi44 | interferon-induced protein 44 | 3.843 | 0.0053 |
| NM_009912 | Ccr1 | chemokine (C-C motif) receptor 1 | 3.734 | 0.0013 |
| BC025535 | Fcgr1 | Fc receptor, IgG, high affinity I | 3.668 | 0.0096 |
| NM_145226 | Oas3 | 2'-5' oligoadenylate synthetase 3 | 3.531 | 0.0017 |
| NM_008331 | Ifit1 | interferon-induced protein with tetratricopeptide repeats 1 | 3.433 | 0.0460 |
| NM_021274 | Cxcl10 | chemokine (C-X-C motif) ligand 10 | 3.418 | 0.0307 |
| NM_016850 | Irf7 | interferon regulatory factor 7 | 3.378 | 0.0083 |
| NM_008530 | Ly6f | lymphocyte antigen 6 complex, locus F | 3.313 | 0.0027 |
| NM_011940 | Ifi202b | interferon activated gene 202B | 3.296 | 0.0266 |
| NM_145211 | Oas1a | 2'-5' oligoadenylate synthetase 1A | 3.283 | 0.0082 |
| NM_145227 | Oas2 | 2'-5' oligoadenylate synthetase 2 | 3.178 | 0.0083 |
| NM_144559 | Fcgr4 | Fc receptor, IgG, low affinity IV | 3.069 | 0.0067 |
| NM_027835 | Ifih1 | interferon induced with helicase C domain 1 | 2.940 | 0.0258 |
| NM_008462 | Klra2 | killer cell lectin-like receptor, subfamily A, member 2 | 2.906 | 0.0001 |
| NM_021792 | Iigp1 | interferon inducible GTPase 1 | 2.535 | 0.0232 |
| L38281 | Irg1 | immune-responsive gene 1 | 2.496 | 0.0329 |
| NM_008329 | Ifi204 | interferon activated gene 204 | 2.422 | 0.0290 |
| NM_025378 | Ifitm3 | interferon induced transmembrane protein 3 | 2.401 | 0.0002 |
| NM_021378 | Klrc3 | killer cell lectin-like receptor subfamily C, member 3 | 2.396 | 0.0063 |
| NM_011909 | Usp18 | ubiquitin specific peptidase 18 | 2.333 | 0.0473 |
| NM_011333 | Ccl2 | chemokine (C-C motif) ligand 2 | 2.314 | 0.0356 |
| NM_028968 | Ifitm7 | interferon induced transmembrane protein 7 | 2.246 | 0.0014 |
| NM_008337 | Ifng | interferon gamma | 2.229 | 0.0054 |
| NM_011338 | Ccl9 | chemokine (C-C motif) ligand 9 | 2.155 | 0.0065 |
| NM_010653 | Klrc2 | killer cell lectin-like receptor subfamily C, member 2 | 2.131 | 0.0076 |
| NM_009909 | Il8rb | interleukin 8 receptor, beta | 2.077 | 0.0036 |
| NM_010555 | Il1r2 | interleukin 1 receptor, type II | 2.069 | 0.0248 |
| NM_010652 | Klrc1 | killer cell lectin-like receptor subfamily C, member 1 | 2.025 | 0.0125 |
| NM_011073 | Prf1 | perforin 1 (pore forming protein) | 1.820 | 0.0326 |
| NM_010722 | Lmnb2 | lamin B2 | 1.381 | 0.1976 |
| Down-regulated | ||||
| NM_025989 | Gp2 | glycoprotein 2 (zymogen granule membrane) | 0.048 | 0.0005 |
| NM_053094 | Cd163 | CD163 antigen | 0.060 | 0.0171 |
| NM_177261 | Kndc1 | kinase non-catalytic C-lobe domain (KIND) containing 1 | 0.085 | 0.0070 |
| NM_144943 | Cd207 | CD 207 antigen | 0.159 | 0.0441 |
| NM_010378 | H2-Aa | histocompatibility 2, class II antigen A, alpha | 0.184 | 0.0055 |
| NM_028135 | Tmem163 | transmembrane protein 163 | 0.211 | 0.0196 |
| NM_007720 | Ccr8 | chemokine (C-C motif) receptor 8 | 0.242 | 0.0150 |
| NM_016851 | Irf6 | interferon regulatory factor 6 | 0.261 | 0.0037 |
| NM_013509 | Eno2 | enolase 2, gamma neuronal (Eno2) | 0.276 | 0.0234 |
| NM_011780 | Adam23 | a disintegrin and metallopeptidase domain 23 (Adam23) | 0.281 | 0.0009 |
| U96752 | H2-Q1 | major histocompability complex Q1b | 0.282 | 0.0247 |
| NM_008341 | Igfbp1 | insulin-like growth factor binding protein 1 | 0.289 | 0.0144 |
| NM_010565 | Inhbc | inhibin beta-C (Inhbc) | 0.304 | 0.0307 |
| AK089361 | AK089361 | B6-derived CD11 +ve dendritic cells cDNA | 0.307 | 0.0417 |
| NM_013517 | Fcer2a | Fc receptor, IgE, low affinity II, alpha polypeptide | 0.315 | 0.0318 |
| NM_016808 | Usp2 | ubiquitin specific peptidase 2 (Usp2), transcript variant 1 | 0.319 | 0.0266 |
| AK041838 | Il7r | interleukin 7 receptor | 0.319 | 0.0256 |
| NM_001033126 | Cd27 | CD antigen 27 (Cd27), transcript variant 1 | 0.326 | 0.0158 |
| NM_009142 | Cx3cl1 | chemokine (C-X3-C motif) ligand 1 | 0.343 | 0.0227 |
| NM_010215 | Il4i1 | interleukin 4 induced 1 | 0.387 | 0.0129 |
| NM_207105 | H2-Ab1 | histocompatibility 2, class II antigen A, beta 1 | 0.520 | 0.0255 |
| NM_033217 | Ngfr | nerve growth factor receptor (TNFR superfamily, member 16) | 0.440 | 0.0110 |
| AK041345 | Xlr4a | X-linked lymphocyte-regulated 4A | 0.460 | 0.0074 |
| NM_033042 | Tnfrsf25 | tumor necrosis factor receptor superfamily, member 25 | 0.465 | 0.0366 |
| NM_198297 | Trat1 | T cell receptor associated transmembrane adaptor 1 | 0.476 | 0.0334 |
| NM_011161 | Mapk11 | mitogen-activated protein kinase 11 | 0.478 | 0.0128 |
| NM_007549 | Blk | B lymphoid kinase | 0.483 | 0.0103 |
| NM_010818 | Cd200 | Cd200 antigen | 0.494 | 0.0156 |
Genes with differential expression in brains of JEV-infected mice
Compared to the expression after mock infection, 551 genes were detected to be significantly upregulated and 568 genes were downregulated in brains of JEV-infected mice (Table 4). Consistent to the results of spleen, chemokines CCL2, CCL3, CCL4 and CXCL10, IFN response transcription factor IRF7, IFN-induced proteins like Ifit1, Ifit2 and Ifit3, protein degradation gene Usp18 were obviously upregulated, and CD163 and IRF6 showed decreased expression upon JEV infection. In addition, increased expression of IFN-inducible transcription factors STAT1 and STAT2, TNF-α induced protein TNFAIP3, apoptosis-related proteins caspase3 and caspase4, suppressors of cytokine signaling Socs1 and Socs3, pro-inflammatory cytokines IL-1 and IL-6, TLR7, as well as IFN response antiviral genes of OAS family were also observed in microarray analysis. These results suggested the occurrence of a strong inflammatory response in mouse brain.
Table 4.
Differentially-regulated genes of our interest in brains of mice with JEV Infection.
| Genbank Accession | Gene symbol | Gene description | Fold change | p-Value |
|---|---|---|---|---|
| Up-regulated | ||||
| NM_021274 | Cxcl10 | chemokine (C-X-C motif) ligand 10 | 1760.024 | 0.0020 |
| NM_011333 | Ccl2 | chemokine (C-C motif) ligand 2 | 1387.794 | 0.0320 |
| NM_013652 | Ccl4 | chemokine (C-C motif) ligand 4 | 336.804 | 0.0151 |
| NM_010846 | Mx1 | myxovirus (influenza virus) resistance 1 | 229.188 | 0.0136 |
| NM_145209 | Oasl1 | 2'-5' oligoadenylate synthetase-like 1 | 209.863 | 0.0242 |
| NM_011940 | Ifi202b | interferon activated gene 202B | 201.477 | 0.0317 |
| NM_008329 | Ifi204 | interferon activated gene 204 | 165.395 | 0.0404 |
| NM_021792 | Iigp1 | interferon inducible GTPase 1 | 159.817 | 0.0168 |
| NM_011337 | Ccl3 | chemokine (C-C motif) ligand 3 | 111.943 | 0.0429 |
| NM_008176 | Cxcl1 | chemokine (C-X-C motif) ligand 1 | 107.659 | 0.0158 |
| AK085407 | Ifi44 | interferon gamma inducible protein 44 | 91.009 | 0.0062 |
| NM_010555 | Il1r2 | interleukin 1 receptor, type II | 89.273 | 0.0297 |
| NM_021893 | Cd274 | CD274 antigen | 87.250 | 0.0337 |
| NM_008330 | Ifi47 | interferon gamma inducible protein 47 | 83.552 | 0.0286 |
| NM_172648 | Ifi205 | interferon activated gene 205 | 82.479 | 0.0416 |
| NM_194336 | Mpa2l | macrophage activation 2 like | 77.279 | 0.0464 |
| NM_007609 | Casp4 | caspase 4, apoptosis-related cysteine peptidase | 77.077 | 0.0451 |
| NM_017466 | Ccrl2 | chemokine (C-C motif) receptor-like 2 | 74.783 | 0.0071 |
| NM_008332 | Ifit2 | interferon-induced protein with tetratricopeptide repeats 2 | 65.247 | 0.0222 |
| NM_011331 | Ccl12 | chemokine (C-C motif) ligand 12 | 65.192 | 0.0079 |
| NM_033601 | Bcl3 | B-cell leukemia/lymphoma 3 | 53.003 | 0.0138 |
| NM_029803 | Ifi27 | interferon, alpha-inducible protein 27 | 47.264 | 0.0403 |
| NM_031168 | Il6 | interleukin 6 | 44.884 | 0.0106 |
| NM_016850 | Irf7 | interferon regulatory factor 7 | 42.752 | 0.0463 |
| NM_145211 | Oas1a | 2'-5' oligoadenylate synthetase 1A | 41.831 | 0.0497 |
| NM_011909 | Usp18 | ubiquitin specific peptidase 18 | 40.804 | 0.0052 |
| NM_010501 | Ifit3 | interferon-induced protein with tetratricopeptide repeats 3 | 39.282 | 0.0008 |
| NM_007707 | Socs3 | suppressor of cytokine signaling 3 | 36.912 | 0.0087 |
| NM_008331 | Ifit1 | interferon-induced protein with tetratricopeptide repeats 1 | 32.115 | 0.0013 |
| NM_013606 | Mx2 | myxovirus (influenza virus) resistance 2 | 31.018 | 0.0003 |
| NM_011854 | Oasl2 | 2'-5' oligoadenylate synthetase-like 2 | 29.655 | 0.0131 |
| NM_009896 | Socs1 | suppressor of cytokine signaling 1 | 28.609 | 0.0492 |
| NM_009140 | Cxcl2 | chemokine (C-X-C motif) ligand 2 | 24.678 | 0.0392 |
| NM_010397 | H2-T22 | histocompatibility 2, T region locus 22 | 21.010 | 0.0473 |
| NM_027835 | Ifih1 | interferon induced with helicase C domain 1 | 19.804 | 0.0233 |
| NM_009397 | Tnfaip3 | tumor necrosis factor, alpha-induced protein 3 | 19.385 | 0.0300 |
| NM_009283 | Stat1 | signal transducer and activator of transcription 1 | 18.134 | 0.0348 |
| NM_001083925 | Oas1b | 2'-5' oligoadenylate synthetase 1B | 17.574 | 0.0342 |
| NM_008361 | Il1b | interleukin 1 beta | 15.753 | 0.0043 |
| NM_133211 | Tlr7 | toll-like receptor 7 | 9.880 | 0.0343 |
| NM_027450 | Glipr2 | GLI pathogenesis-related 2 | 9.275 | 0.0405 |
| NM_008562 | Mcl1 | myeloid cell leukemia sequence 1 | 9.054 | 0.0336 |
| NM_029419 | Apol3 | apolipoprotein L 3 | 8.806 | 0.0165 |
| NM_001008700 | Il4ra | interleukin 4 receptor, alpha | 8.639 | 0.0360 |
| NM_013730 | Slamf1 | signaling lymphocytic activation molecule family member 1 | 8.274 | 0.0435 |
| NM_009841 | Cd14 | CD14 antigen | 6.666 | 0.0034 |
| NM_033541 | Oas1c | 2'-5' oligoadenylate synthetase 1C | 6.596 | 0.0120 |
| NM_009810 | Casp3 | caspase 3, apoptosis-related cysteine peptidase | 2.119 | 0.0117 |
| Down-regulated | ||||
| NM_053094 | Cd163 | CD163 antigen | 0.066 | 0.0007 |
| XM_898059 | Cd209f | CD209f antigen | 0.088 | 0.0271 |
| NM_026972 | Cd209b | CD209b antigen | 0.132 | 0.0149 |
| NM_033042 | Tnfrsf25 | tumor necrosis factor receptor superfamily, member 25 | 0.196 | 0.0288 |
| NM_016708 | Npy5r | neuropeptide Y receptor Y5 (Npy5r) | 0.242 | 0.0351 |
| NM_019577 | Ccl24 | chemokine (C-C motif) ligand 24 (Ccl24) | 0.302 | 0.0132 |
| NM_008409 | Itm2a | integral membrane protein 2A | 0.302 | 0.0075 |
| AK042749 | D230046B21Rik | 7 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:A730020N04 | 0.311 | 0.0358 |
| NM_030143 | Ddit4l | DNA-damage-inducible transcript 4-like | 0.318 | 0.0004 |
| NM_022723 | Scube1 | signal peptide, CUB domain, EGF-like 1 | 0.319 | 0.0400 |
| AK144387 | 4732444A12Rik | 21 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:G630051C23 | 0.319 | 0.0000 |
| AK082652 | Tmem44 | transmembrane protein 44 | 0.324 | 0.0108 |
| NM_175106 | Tmem177 | transmembrane protein 177 (Tmem177) | 0.330 | 0.0346 |
| NM_175564 | Tmem169 | transmembrane protein 169 | 0.336 | 0.0371 |
| NM_027016 | Tloc1 | translocation protein 1 | 0.336 | 0.0276 |
| NM_027163 | Il1f8 | interleukin 1 family, member 8 | 0.421 | 0.0267 |
| NM_022986 | Irak1bp1 | interleukin-1 receptor-associated kinase 1 binding protein 1 | 0.422 | 0.0169 |
| NM_016851 | Irf6 | interferon regulatory factor 6 | 0.447 | 0.0303 |
| NM_145826 | Il17re | interleukin 17 receptor E (Il17re), transcript variant 1 | 0.477 | 0.0403 |
Confirmation of microarray data by RT-qPCR
To confirm the microarray hybridization results, quantitative RT-PCR was performed on eight selected differentially expressed mRNAs in mouse spleen and brain respectively. As shown in the RT-qPCR result of spleen, granzymA, granzymeB, Porferin, IRF7, IFN-γ, CXCL10, and ILIR2 were significantly upregulated, while CD163 was downregulated (Table 5). Out of eight tested mRNAs in brains of mice infected with JEV, CCL2, CCL4, CXCL10, Casp3, Casp4, SOCS1 and SOCS3 showed increased expression, and CD163 was also found to have an obviously decreased expression (Table 5). Although absolute values are not identical due to the different sensitivity between the techniques, all genes showed a well comparative expression pattern with microarray data.
Table 5.
Comparison of expression changes of some selected genes between microarray and qRT-PCR
| Gene name | Gene description | Fold change | |
|---|---|---|---|
| Microarray | qRT-PCR | ||
| Spleen | |||
| Cxcl10 | chemokine (C-X-C motif) ligand 10 | 3.418 | 3.375 (±1.065) |
| Ifng | interferon gamma | 2.229 | 3.062 (±0.507) |
| Gzmb | granzyme B | 6.202 | 7.285 (±0.311) |
| Gzma | granzyme A | 4.236 | 3.815 (±0.127) |
| Prf1 | Porferin 1 | 1.820 | 2.287 (±0.643) |
| Irf7 | interferon regulatory factor 7 | 3.378 | 4.926 (±0.309) |
| Il1r2 | Interleukin 1 receptor, type II | 2.069 | 2.078 (±0.105) |
| Cd163 | CD163 antigen | 0.060 | 0.083 (±0.017) |
| Brain | |||
| Cxcl10 | chemokine (C-X-C motif) ligand 10 | 1760.024 | 13.492 (±1.690) |
| Ccl2 | chemokine (C-C motif) ligand 2 | 1387.794 | 188.549 (±8.931) |
| Ccl4 | chemokine (C-C motif) ligand 4 | 336.804 | 61.007 (±3.735) |
| Casp3 | Caspase3, apoptosis-related cysteine peptidase | 2.119 | 1.207 (±0.073) |
| Casp4 | caspase4, apoptosis-related cysteine peptidase | 77.077 | 19.969 (±0.050) |
| Socs1 | suppressor of cytokine signaling 1 | 28.609 | 11.182 (±0.845) |
| Socs3 | suppressor of cytokine signaling 3 | 36.912 | 4.672 (±0.464) |
| Cd163 | CD163 antigen | 0.066 | 0.065 (±0.012) |
Discussion
Spleen is one of the major peripheral immunity organs and CNS is the ultimate infection target of JEV. Therefore, identification of the JEV-related host genes in spleens and brains of mice infected with JEV may be helpful for understanding of JEV pathogenesis. To this end, profiles of mRNA expression in both spleen and brain tissues of JEV-infected mice were analyzed systematically in this study.
In mRNA profiling assay, we found chemokines like CCL2 and CXCL10 were significantly up-regulated in both mouse spleen and brain in response to JEV-infection, suggesting a strong inflammatory response of host. Monocyte chemoattractant protein-1 (MCP-1/CCL2) is one of the key chemokines that regulate migration and infiltration of monocytes/macrophages. It was involved in neurological disorders such as encephalitis-related neuronal death, where its levels were elevated in astrocytes leading to neuronal death [13]. Previous study has demonstrated that the WNV-infection stimulated the expression of CCL2 in mice livers, suggesting a consistent result to our study of JEV [10]. CXCL10 is also found to play a crucial role in the host defense response against various viral infection of the CNS by enhancing innate immune responses [14,15], and our result of up-regulated CXCL10 mRNA has a good agreement with that was reported by Gupta et al. and Biswas et al. [11,12].
In addition, a strong IFN-pathway-related response was evident in mouse spleen and brain infected with JEV, with increased expression of IFN-γ, IFN response transcription factor IRF7, IFN-induced proteins IFIT1,IFITM3 and IFITM7 in spleen, and IRF7, IFN-inducible transcription factors STAT1 and STAT2, IFN-induced proteins IFIT1, IFIT2 and IFIT3 in brain, implying the occurrence of a protective response of host. Similar results were shown in previous reports as the upregulation of IFN-γ, STAT1 and STAT2 in JEV-infected mice brain [12], and increased expression of STAT1 and IFIT3 in JEV-infected N2A cells [11]. The reason why no increase in expression of IFN-γ was found in brains of JEV-infected mice in our study may be related to the difference of time points with Biswas' study [11]. IRF7 is activated in the presence of double stranded RNA following virus infection, which is functional as one of the regulators of the IFN-α/β gene promoter and the IFN-α/β responsive genes to create an antiviral state [16]. The increased expression of IRF7 has also been demonstrated in WNV infected mice, but wasn't found in JEV infected cells [17,18]. This may be due to the different signal pathways between intact host and cell culture.
Pore-forming protein perforin and the family of granzymes have been demonstrated to form an antiviral arsenal central to the function of cytotoxic T lymphocyte (CTL) and natural killer (NK) cells [19-21]. After JEV infection, pro-apoptotic genes found to have significant enhancement in mouse spleen, including porferin, granzyme A and granzymeB, which suggested a strong cytotoxic response against JEV infection. The consistent result as increased expression of granzyme A and B was also shown in the report about WNV infection [17]. Granzyme B is the most characterized granyzme which plays an important role in inducing apoptosis, and it is generally accepted that granzyme A can trigger a distinct nonapoptotic form of cell death [22]. The high expression of granzyme A and B in spleen could help clear virus infection, but may also involve lymphocyte injury. The upregulation of apoptosis-related proteins Caspase3 and 4 were also detected in brains of mice infected with JEV, indicating an inflammation-related neuronal apoptosis. Caspase 3 is an effector caspase that function as a central regulator of apoptosis. It has been reported that JEV infection triggers apoptosis in different cells, such as baby hamster kidney BHK-21 cells, mouse neuroblastoma N18 cells, human neuronal NT-2 cells, and human medulloblastoma cells, resulting in caspase 3 activation [23-25]. The function of caspase 4 is not fully known, but it is believed to be an inflammatory caspase, with a role in the immune system [26].
Pro-inflammatory cytokines like IL-1 and IL-6 were found to have increased expression in the brain, which was consistent with the results of the studies on DV and WNV [8,17]. IL-1α and IL-1β has been known to form an important part of the inflammatory response of the body against infection. IL-6 together with IL-1 and TNF-α acts as an endogenous pyrogen by causing fever following viral infections [27]. It's also associated with an unfavorable outcome following yellow fever virus infection [28]. Evidence suggests that circulating IL-6 can activate CNS mechanisms resulting in the development of the febrile response during disease [29]. Upregulation of IL-1 and IL-6 in brain may thus be protective against harmful JEV infection, but also have a pathogenic role in CNS.
The 2', 5'-oligoadenylate synthetase (OAS) and its downstream effector RNase L play important roles in host defense against virus infection. The OAS1b protein has been described as a flavivirus resistance factor, and OASl1 as a WNV-resistance factor in wild mice because a truncated version of the protein is expressed in laboratory mice which are susceptible to infection [30-32]. Human OAS1 p42/p46 and OAS3 p100 are likely to contribute to host defense against DEN infection and play a role in determining the outcomes of DEN disease severity [33]. Further, the activated expression of OAS2 has been demonstrated in mouse brain in response to JEV infection [34]. In present study, there was increased expression of various members of the OAS family in the brains of JEV-infected mice, including OAS1a, OAS1b, OAS1c, OAS1e, and OAS2. Therefore, the ability of these proteins to protect against JEV infection should also be further studied.
The significant downregulation of CD163 was detected both in mouse spleen and brain in response to JEV infection. CD163 is a novo identified marker for perivascular macrophages in humans, monkeys, and mice. And previously studies have found that perivascular CD163 expression is upregulated and the number of CD163-positive cells increases in HIV and SIV encephalitis (HIVE and SIVE) brains [35,36]. However, CD163 is not a "classical" activation marker, because peripheral blood monocytes and most tissues macrophages of normal uninfected controls all express it and because in vitro pro-inflammatory stimuli largely down-regulate its expression. These findings suggest that CD163 expression is regulated in association with a certain stage of differentiation. Here, our results showed a downregulated CD163 mRNA level in response to acute JEV infection, which probably suggested a decreased number of activated perivascular macrophages resulted by inflammatory disorder-related apoptosis response.
Conclusions
In summary, our findings suggested that JEV infection resulted in significant changes in the expression of multiple genes in mouse spleen and brain, including inflammatory cytokines, chemokines, IFN inducible genes, IFN regulators, and apoptosis related genes, etc. These genes may play a critical role on antiviral response of host against JEV infection but could also contribute to the pathogenesis of JEV resulting in encephalitis. The mRNA profile obtained by microarray analysis in this study may provide a foundation for future investigation of JEV pathogenesis and therapeutic method.
Methods
Virus production
JEV wide type strain P3 was propagated in brains of suckling mice and titered in BHK-21 cells which was grown and maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% heated-inactivated fetal bovine serum (FBS, Hyclone, Logan, UT, USA), 100 g/ml streptomycin and 100 IU/ml penicillin (Sigma-Aldrich, MO, USA) at 37°C with 5% CO2.
Virus infection of mice
Four-weeks-old naive female BALB/c mice were purchased from Wuhan Institute of Biologic Products (Hubei Province, China) and inoculated subcutaneously (s.c.) with 5 × 106 PFU of JEV P3 strain. Dilutions were performed in serum-free DMED and experimental controls were mock injected with diluent. A part of JEV-infected mice were sacrificed at day 3 post-inoculation (the day before the neurological symptoms appeared), and spleens were harvested. The left mice were sacrificed at day 6 post-inoculation (the day before the mice started to die), and brains of mice were harvested. Spleen and brain homogenate was made in DMEM for RNA extraction.
Microarrays and bioinformatics
The total RNA was isolated from mouse spleens and brains respectively with trizol reagent (Invitrogen) for mRNA Microarray. mRNA hybridization was performed by shanghaiBio Corporation (shanghai, China) with the use of 4 × 44 K Agilent Whole Mouse Genome Oligo Microarray (total 41,174 oligo probes from 41,174 mouse genes). For each sample pair, the experiments were done with two independent hybridizations (Cy3 and Cy5 interchanging labeling). Hybridized arrays were scanned at 5 μm resolution on an Agilent DNA Microarray Scanner (Model G2565BA). Data extraction from images was done by using Agilent Feature Extraction software. Hierarchical cluster, gene ontology and pathway analysis were analyzed by using SAS (ShanghaiBio Analysis System).
Quantitative real-time RT-PCR (RT-qPCR)
For selected mRNA RT-qPCR, total RNA from the same samples used in microarray analysis was tested by using ABI 7500 FAST real-time PCR system. PCR primers were designed with Primer Express 2.0 software (Invitrogen). Results are shown as fold change. For mRNA RT-qPCR, experiments were carried out with the PrimerScript RT reagent Kit (TaKaRa) and SYBR Green Realtim PCR Master Mix (TaKaRa) according to manufacture's instruction. The housekeeping gene GHDAP was used for standardization of the initial RNA content of a sample. Relative changes of gene expression were calculated by the following formula, and the data are represented as fold upregulation/downregulation. fold change = 2-ΔΔCt, whereΔΔCt =(Ct of gene of interest, treated -Ct of HK gene, treated)-(Ct of gene of interest, control-Ct of HK gene, control), Ct is the threshold cycle number and HK is the house keeping gene.
Statistical analysis
Each gene in each infection group was subjected to a Student's t test to detect large expression differences relative to the mock-infected group. p-values <0.05 were considered to be statistically significant.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YY and JY carried out most of the experiments and wrote the manuscript. XY, RJ participated part of experiments. HC and SC conceived of the study, participated in its design and coordination, and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yang Yang, Email: yang55797961@163.com.
Jing Ye, Email: yeah_zaizai@hotmail.com.
Xiaohong Yang, Email: njyxh2006@sina.com.
Rong Jiang, Email: jrrabbit000@yahoo.com.cn.
Huanchun Chen, Email: chenhch@mail.hzau.edu.cn.
Shengbo Cao, Email: sbcao@mail.hzau.edu.cn.
Acknowledgements
This work was supported by the 973 Project of China (No. 2010CB530100), Transregional Collaborative Research Centre TRR 60 and PCSIRT (IRT0726).
References
- Hanna JN, Ritchie SA, Phillips DA, Shield J, Bailey MC, Mackenzie JS, Poidinger M, McCall BJ, Mills PJ. An outbreak of Japanese encephalitis in the Torres Strait, Australia, 1995. Med J Aus t. 1996;165:256–260. doi: 10.5694/j.1326-5377.1996.tb124960.x. [DOI] [PubMed] [Google Scholar]
- Solomon T. Japanese encephalitis. J Neurol Neurosurg Psychiatry. 2000;68:405–415. doi: 10.1136/jnnp.68.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Manual for the Laboratory Diagnosis of Japanese Encephalitis Virus Infection. 2007.
- Vaughn DW, Hoke CH. The epidemiology of Japanese encephalitis: prospects for prevention. Epidemiol Rev. 1992;14:197–221. doi: 10.1093/oxfordjournals.epirev.a036087. [DOI] [PubMed] [Google Scholar]
- Yang KD, Yeh WT, Chen RF. A model to study neurotropism and persistency of Japanese encephalitis virus infection in human neuroblastoma cells and leukocytes. J Gen Virol. 2004;85:635–642. doi: 10.1099/vir.0.19426-0. [DOI] [PubMed] [Google Scholar]
- Aleyas AG, George JA, Han YW. Functional modulation of dendritic cells and macrophages by Japanese encephalitis virus through MyD88 adaptor molecule-dependent and - independent pathways. J Immunol. 2009;183:2462–2474. doi: 10.4049/jimmunol.0801952. [DOI] [PubMed] [Google Scholar]
- Wolinsky JS. In: Immunological mechanisms in neurologic and psychiatric disease. Waksman BH, editor. New York: Raven Press; 1990. Subacute sclerosing panencephalitis progressive rubella panencephalitis, and multifocal leukoencephelapathy; pp. 259–268. [Google Scholar]
- Warke RV, Xhaja K, Martin KJ. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J Virol. 2003;77:11822–11832. doi: 10.1128/JVI.77.21.11822-11832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Smith M, Katze MG. The host response to West Nile Virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J Virol. 2004;78:7737–7747. doi: 10.1128/JVI.78.14.7737-7747.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh WL, Ng ML. Molecular mechanisms of West Nile Virus pathogenesis in brain cells. Emerg Infect Dis. 2005;11:629–631. doi: 10.3201/eid1104.041076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Santhosh SR, Babu JP, Parida MM, Rao PV. Chemokine profiling of Japanese encephalitis virus-infected mouse neuroblastoma cells by microarray and real-time RT-PCR: Implication in neuropathogenesis. Virus Research. 2010;147:107–112. doi: 10.1016/j.virusres.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SM, Kar S, Singh R, Chakraborty D, Vipat V, Raut CG, Mishra AC, Gore MM, Ghosh D. Immunomodulatory Cytokines Determine the Outcome of Japanese Encephalitis Virus Infection in Mice. Journal of Medical Virology. 2010;82:304–310. doi: 10.1002/jmv.21688. [DOI] [PubMed] [Google Scholar]
- Sakurai-Yamashita Y, Shigematsu K, Yamashita K. Expression of MCP-1 in the hippocampus of SHRSP with ischemia-related delayed neuronal death. Cell Mol Neurobiol. 2006;26:823–831. doi: 10.1007/s10571-006-9077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MF, Lai SL, Chen JP. Both CXCR3 and CXCL10/IFN-inducible protein 10 are required for resistance to primary infection by dengue virus. J Immunol. 2006;177:1855–1863. doi: 10.4049/jimmunol.177.3.1855. [DOI] [PubMed] [Google Scholar]
- Hardison JL, Wrightsman RA, Carpenter PM. The chemokines CXCL9 and CXCL10 promote a protective immune response but do not contribute to cardiac inflammation following infection with Trypanosoma cruzi. Infect Immun. 2006;74:125–134. doi: 10.1128/IAI.74.1.125-134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marietjie V, Timothy GM, Michael AW, Thomas JK, Janusz TP, Felicity JB, Patricia AL, Robert S. Gene expression in mice infected with West Nile virus strains of different neurovirulence. Virology. 2005. pp. 119–140. [DOI] [PubMed]
- Chang TH, Liao CL, Lin YL. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect. 2006;8:157–71. doi: 10.1016/j.micinf.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Park HL, Henkart PA. Synergistic roles of granzymes A and B in mediating target cell death by rat basophilic leukemia mast cell tumors also expressing cytolysin/perforin. J Exp Med. 1995;181:1037–1046. doi: 10.1084/jem.181.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne KA, Blink E, Sutton VR. Cytosolic delivery of granzyme B by bacterial toxins: evidence that endosomal disruption, in addition to transmembrane pore formation, is an important function of perforin. Mol Cell Biol. 1999;19:8604–8615. doi: 10.1128/mcb.19.12.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Hengartner H, Podack ER. Purification and characterization of a cytolytic pore-forming protein from granules of cloned lymphocytes with natural killer activity. Cell. 1986;44:849–859. doi: 10.1016/0092-8674(86)90007-3. [DOI] [PubMed] [Google Scholar]
- Joseph AT, Phillip IB. A renaissance in understanding the multiple and diverse functions of Granzymes? Immunity. 2008;29:665–667. doi: 10.1016/j.immuni.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Liao CL, Lin YL. Replication-incompetent virions of Japanese encephalitis virus trigger neuronal cell death by oxidative stress in a culture system. J Gen Virol. 2004;85:521–533. doi: 10.1099/vir.0.19496-0. [DOI] [PubMed] [Google Scholar]
- Tsao CH, Su HL, Lin YL. Japanese encephalitis virus infection activates caspase-8 and -9 in a FADD-independent and mitochondrion-dependent manner. J Gen Virol. 2008;89:1930–1941. doi: 10.1099/vir.0.2008/000182-0. [DOI] [PubMed] [Google Scholar]
- Yang TC, Shiu SL, Chuang PH, Lin YJ, Wan L, Lan YC, Lin CW. Japanese encephalitis virus NS2B-NS3 protease induces caspase 3 activation and mitochondria-mediated apoptosis in human medulloblastoma cells. Virus Research. 2009;143:77–85. doi: 10.1016/j.virusres.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Martinon F, Tschopp J. "Inflammatory caspases and inflammasomes: master switches of inflammation". Cell Death Differ. 2007;14:10–22. doi: 10.1038/sj.cdd.4402038. [DOI] [PubMed] [Google Scholar]
- Heinrich PC, Castell JV, Andus T. Interleukin-6 and the acute phase response. Biochem J. 1990;265:621–636. doi: 10.1042/bj2650621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulen J, Sakho M, Koulemou K. Activation of the cytokine network and unfavorable outcome in patients with yellow fever. J Infect Dis. 2004;190:1821–1827. doi: 10.1086/425016. [DOI] [PubMed] [Google Scholar]
- Cartmell T, Poole S, Turnbull AV. Circulating interleukin-6 mediates the febrile response to localized inflammation in rats. J Physiol. 2000;526:653–661. doi: 10.1111/j.1469-7793.2000.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Mashimo T, Frenkiel MP. Infection of mouse neurones by West Nile virus is modulated by the interferoninducible 2V-5V oligoadenylate synthetase 1b protein. Immunol Cell Biol. 2003;81:230–236. doi: 10.1046/j.1440-1711.2003.01166.x. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Lucas M, Simon-Chazottes D. A nonsense mutation in the gene encoding 2V-5V-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc Natl Acad Sci USA. 2002;99:11311–11316. doi: 10.1073/pnas.172195399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel CE. Host genetic variability and West Nile virus susceptibility. Proc Natl Acad Sci USA. 2002;99:11555–11557. doi: 10.1073/pnas.202448899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Yu HP, Chang BL. Distinct antiviral roles for human 2',5'-oligoadenylate synthetase family members a gainst dengue virus infection. J Immunol. 2009;183:8035–43. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- Saha S, Rangarajan PN. Common host genes are activated in mouse brain by Japanese encephalitis and rabies viruses. Journal of General Virology. 2003;84:1729–1735. doi: 10.1099/vir.0.18826-0. [DOI] [PubMed] [Google Scholar]
- Borda JT, Alvarez X, Mohan M, Hasegawa A, Bernardino A, Jean S, Aye P, Lackner AA. CD163, a marker of perivascular macrophages, is up-regulated by microglia in Simian immunodeficiency Virus Encephalitis after Haptoglobin-Hemoglobin Complex stimulation and is suggestive of breakdown of the Blood-Brain Barrier. The American Journal of Pathology. 2008;172:725–737. doi: 10.2353/ajpath.2008.070848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WK, Alvarez X, Fisher J, Bronfin B, Westmoreland S, McLaurin J, Williams K. CD163 identifies perivascular macrophages in normal and viral encephalitic brains and potential precursors to perivascular macrophages in blood. American Journal of Pathology. 2006;168:822–834. doi: 10.2353/ajpath.2006.050215. [DOI] [PMC free article] [PubMed] [Google Scholar]