Abstract
Different patterns of motor nerve activity drive distinctive programs of gene transcription in skeletal muscles, thereby establishing a high degree of metabolic and physiological specialization among myofiber subtypes. Recently, we proposed that the influence of motor nerve activity on skeletal muscle fiber type is transduced to the relevant genes by calcineurin, which controls the functional activity of NFAT (nuclear family of activated T cell) proteins. Here we demonstrate that calcineurin-dependent gene regulation in skeletal myocytes is mediated also by MEF2 transcription factors, and is integrated with additional calcium-regulated signaling inputs, specifically calmodulin-dependent protein kinase activity. In skeletal muscles of transgenic mice, both NFAT and MEF2 binding sites are necessary for properly regulated function of a slow fiber-specific enhancer, and either forced expression of activated calcineurin or motor nerve stimulation up-regulates a MEF2-dependent reporter gene. These results provide new insights into the molecular mechanisms by which specialized characteristics of skeletal myofiber subtypes are established and maintained.
Keywords: calcineurin/CaMKIV/MEF2/myogenic cells/skeletal muscle
Introduction
Skeletal muscle fibers of vertebrate organisms are heterogeneous with respect to size, metabolism and contractile function (Williams and Neufer, 1996). Specialized myofiber phenotypes are matched to environmental demands for different types of locomotor function, and are based on distinctive programs of gene expression (Schiaffino and Reggiani, 1996). Different myofiber subtypes can be detected during embryonic development (DiMario et al., 1993; Ontell et al., 1993; Stockdale, 1997), and genetically determined patterning of fiber types within major muscle groups becomes fully established in the early postnatal period (Garry et al., 1996). In skeletal muscles of adult animals, however, specialized myofiber phenotypes are highly plastic, and are subject to physiological control by variations in motor neuron activity (Vrbova, 1963; Williams et al., 1986; Pette and Staron, 1997). Specifically, a sustained, tonic pattern of motor nerve firing directs myofibers to the slow, oxidative myofiber phenotype, while fibers assume one of several fast fiber phenotypes based on other patterns of neural activity. Shorter durations of tonic motor nerve activation associated with endurance exercise training are sufficient to increase the proportion of oxidative fibers without promoting fast to slow fiber transformation (Saltin and Gollnick, 1983). The dominant influence of the motor neuron over fiber type-specific gene expression is demonstrated most clearly by experiments in which cross-innervation or electrical pacing can convert one myofiber subtype to another (Romanul and Van der Meulen, 1966; Williams et al., 1986; Pette and Vrbova, 1992).
Although our understanding of muscle development has advanced significantly during the past decade, molecular mechanisms regulating skeletal fiber diversity are only beginning to be elucidated. In zebrafish, myogenic precursor cells are programmed to differentiate into slow or fast fiber phenotypes in a manner that is influenced by the sonic hedgehog signaling pathway (Blagden et al., 1997), but there is no current evidence that this pathway is relevant to fiber type transitions in adult mammalian myofibers. The dominant role of motor nerve activity in directing the expression of fiber type-specific genes in adults led us to postulate that the relevant signaling pathways are triggered by changes in intracellular calcium (Chin et al., 1998). We proposed that, as a consequence of tonic neural stimulation, slow and oxidative fibers maintain a sustained elevation of intracellular free calcium in a pattern that activates calcineurin, a cyclosporin-sensitive, calcium/calmodulin-dependent serine/threonine phosphatase.
We reported previously that activated calcineurin selectively up-regulates slow or oxidative fiber-selective gene promoters, and that pharmacological inhibition of endogenous calcineurin in intact animals promotes slow to fast fiber transformation (Chin et al., 1998). The hypothesis that calcineurin activity directs myofibers towards the slow fiber phenotype has been supported subsequently by other investigators (Dunn et al., 1999). Calcineurin-dependent gene regulation in skeletal myocytes was shown to be transduced, at least in part, by nuclear translocation and DNA binding of proteins of the nuclear factor of activated T cells (NFAT) family. However, as noted in our initial experiments (Chin et al., 1998) and subsequently by others (Calvo et al., 1999), ablation of NFAT binding motifs within slow or oxidative fiber-specific promoter/enhancer regions does not completely abolish transcriptional regulation by calcineurin or fiber type-specific gene expression. This observation revealed that gene regulatory signals triggered by calcineurin can be transduced to target genes in the absence of DNA binding by NFAT proteins, and suggested that other transcription factors are capable of responding to calcineurin-dependent signals in skeletal muscles.
Here we demonstrate that MEF2 proteins function in this capacity, and that the functional activity of MEF2 as a transcriptional activator in skeletal myocytes is stimulated synergistically by calcineurin and other calcium-regulated signaling molecules. Functional activation of MEF2 correlates with its dephosphorylation by a calcium-activated, calcineurin-dependent mechanism. MEF2 binding is required for the correctly regulated function of a slow fiber-specific enhancer in intact animals. In addition, the transactivating function of MEF2 proteins is detectable selectively in slow and oxidative myofibers, and is induced either by motor nerve stimulation or expression of a constitutively active form of calcineurin.
Results
MEF2 transduces calcineurin-dependent activation of fiber type-specific enhancers in cultured myogenic cells
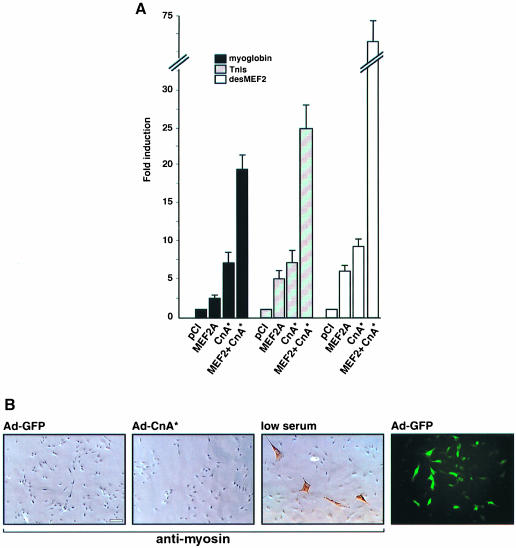
Promoter/reporter constructions were prepared using transcriptional control elements that direct the selective expression of myoglobin in Type I and IIA fibers, or of the slow fiber-specific isoform of troponin I (TnIs), as characterized previously by our laboratory (Chin et al., 1998) and others (Nakayama et al., 1996). In addition, we constructed a synthetic enhancer (desMEF2) designed to read out the transactivating function of MEF2, using three copies of a high affinity MEF2 binding site from the desmin gene (Naya et al., 1999) linked to a minimal promoter. In the absence of exogenous MEF2, forced expression of a constitutively active form of calcineurin (CnA*) up-regulated transcription of these promoter/reporter plasmids by 7- to 10-fold in C2C12 myoblasts (Figure 1A). Forced expression of MEF2A in the absence of calcineurin stimulation produced a detectable but lesser (2- to 6-fold) response. In each case, however, the combined effects of exogenous MEF2A and calcineurin were much greater (20- to 70-fold) than those produced by either stimulus alone. We conclude that MEF2 proteins are capable of transducing calcineurin-generated signals for gene regulation, and that endogenous levels of MEF2 are limiting to this response in this cell background. The effect of calcineurin to up-regulate myoglobin and TnIs enhancer function is not attributable to a generalized stimulation of muscle differentiation. C2C12 myoblasts infected with recombinant adenovirus to express a constitutively active form of calcineurin demonstrate neither an increase in myotube formation nor an induction of endogenous myosin synthesis compared with cells infected with a control virus (Figure 1B).
Fig. 1. The transactivating function of MEF2 is stimulated by calcineurin in C2C12 myogenic cells. (A) Luciferase reporter constructs were prepared using promoter/enhancer elements from the myoglobin or TnI genes, or by three copies of a high affinity MEF2 binding site from the desmin promoter (desMEF2). Expression of luciferase driven by each of these reporter plasmids was determined following co-transfection of expression plasmids containing either no insert (pCI) or cDNAs encoding MEF2A or CnA*. Data are expressed relative to the luciferase activity observed in the control state (pCI co-transfection) and represent mean values (± SEM) from five or more independent experiments. All results are corrected for variations in transfection efficiency by normalization to expression of a co-transfected pCMV-lacZ plasmid. (B) The effect of calcineurin to up-regulate myoglobin and TnIs enhancer function is not attributable to a generalized stimulation of muscle differentiation. The three leftward panels show immunostains using anti-myosin antibody of C2C12 myoblasts infected with an adenoviral vector expressing CnA* (Ad-CnA*), or green fluorescent protein (Ad-GFP), or grown in low serum to promote differentiation. Adenovirus-infected cells are identified by expression of GFP (right panel).
Calcineurin-dependent stimulation of the transactivating function of MEF2 in cultured myocytes is mediated without changes in DNA binding
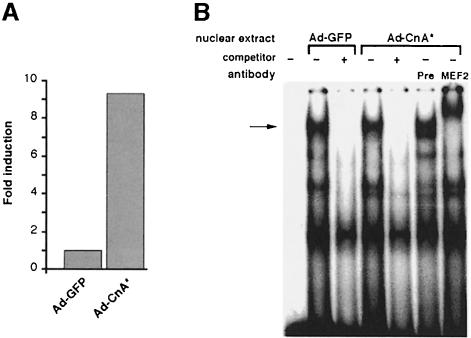
Infection of C2C12 myogenic cells with a recombinant adenovirus engineered to express a constitutively active form of calcineurin augmented the expression of a luciferase reporter gene controlled by the desMEF2 synthetic enhancer (Figure 2A), confirming the results produced by plasmid co-transfection (see Figure 1). Electrophoretic mobility shift assays (EMSA) of nuclear proteins extracted from virally infected cells were used to assess the effects of activated calcineurin on DNA binding by MEF2 (Figure 2B). The most slowly migrating band observed by EMSA was identified as a MEF2–DNA complex by competition with unlabeled probe, and by the change in mobility induced by incubation with anti-MEF2 antibodies. These results support the conclusion that calcineurin-dependent stimulation of the transactivating function of MEF2 can occur in the absence of major changes in DNA binding.
Fig. 2. DNA binding of MEF2 following activation by calcineurin. C2C12 cells were infected with recombinant adenoviral vectors encoding either green fluorescent protein (Ad-GFP) or CnA* (Ad-CnA*). (A) Luciferase reporter gene expression following transfection of the desMEF2 enhancer construct. Data are presented as in Figure 1 and represent mean values of two independent experiments. (B) Electrophoretic mobility shift assays (EMSA) of nuclear proteins from adenovirus-infected cells using a high affinity MEF2 binding site as the labeled oligonucleotide probe. The identity of the MEF2–DNA complex (arrow) is confirmed by the change in mobility in the presence of anti-MEF2 antibodies, and by competition with unlabeled MEF2 binding oligonucleotide (competitor). In contrast to the clear functional effect on transcription, no detectable differences in DNA binding by MEF2 are observed in the presence of activated calcineurin. Pre, preimmune serum.
The phosphorylation status of MEF2 is altered by activated calcineurin
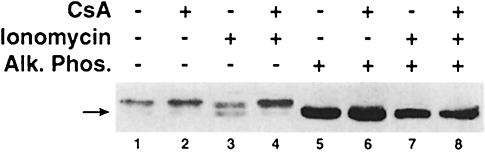
When C2C12 myotubes were exposed to the calcium ionophore ionomycin, a more rapidly migrating form of MEF2A was evident in immunoblot assays (Figure 3, lane 3). The identity of this band as a hypophosphorylated form of MEF2A was confirmed by incubation of protein extracts with alkaline phosphatase (Figure 3, lanes 5–8). Dephosphorylation of MEF2A in the presence of ionomycin requires calcineurin activity, as determined by the inhibitory effect of the specific calcineurin antagonist cyclosporin A (Figure 3, lane 4).
Fig. 3. Calcineurin-dependent dephosphorylation of MEF2. Proteins extracted from C2C12 myotubes were separated by SDS–PAGE and immunoblots were probed with anti-MEF2 antibody. A hypophosphorylated form of MEF2A (arrow) is evident in cells exposed to a calcium ionophore (1 µM ionomycin, lane 3). Dephosphorylation of MEF2A in the presence of ionomycin was inhibited by the calcineurin antagonist cyclosporin A (250 nM CsA, lane 4). The identity of the more rapidly migrating band as a hypophosphorylated form of MEF2A was confirmed by incubation of protein extracts with alkaline phosphatase (Alk. Phos., lanes 5–8). Similar findings were observed in each of three independent experiments.
Slow-specific and fast-specific enhancer elements respond differently to MEF2
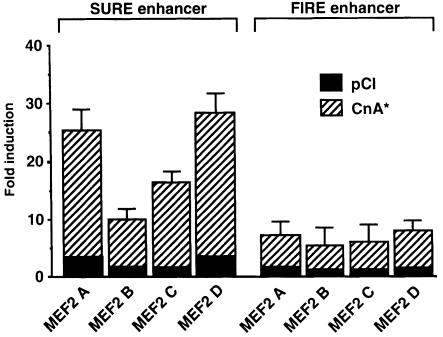
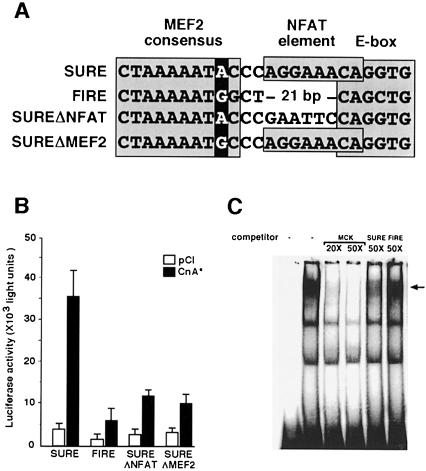
Promoter/reporter constructions were prepared using transcriptional control elements that direct fiber type-specific expression of slow and fast isoforms of troponin I, as characterized previously (Nakayama et al., 1996). In transgenic mice, these SURE (slow upstream regulatory eement) and FIRE (fast intronic regulatory element) enhancers direct expression of reporter genes selectively to slow and fast myofibers, respectively. Forced expression of MEF2 proteins in C2C12 myoblasts stimulated the transcriptional activity of the SURE enhancer to a greater degree than the FIRE enhancer, and this difference was accentuated in the presence of constitutively active calcineurin (Figure 4). All of the major isoforms of MEF2 (A, B, C and D) acted preferentially on the slow fiber-specific transcriptional control region. This selective effect of calcineurin to up-regulate the SURE enhancer is not attributable to a generalized effect on muscle differentiation in this cultured cell system (Figure 1B). On the contrary, the expression of the FIRE enhancer is activated 145-fold in the transition of C2C12 myoblasts to myotubes while the corresponding effect on the SURE enhancer plasmid is 25-fold. If calcineurin were simply accelerating muscle differentiation, then the FIRE enhancer would be expected to exhibit greater calcineurin-dependent activation than SURE, whereas we observed the converse response.
Fig. 4. All MEF2 isoforms enhance calcineurin-dependent transactivation of a SURE enhancer. Expression plasmids containing cDNAs encoding each of the four isoforms of MEF2 were cotransfected with luciferase reporter plasmids controlled by either the SURE or FIRE enhancers (Nakayama et al., 1996). Luciferase activity following forced expression of each MEF2 isoform was measured in the absence (solid bars, pCI) or presence (dashed bars, CnA*) of activated calcineurin. Fold induction is relative to enhancer activity without cotransfected MEF2 or calcineurin. Data are presented as described in Figure 1.
The consensus sequence for MEF2 binding has been defined previously as CT(A/T)4ATAA (Cserjesi and Olson, 1991). Close inspection of the nucleotide sequences of the SURE and FIRE enhancers reveals that the putative MEF2 binding site in FIRE has a G rather than an A in position 9 of the consensus binding motif (Figure 5A). Previous studies of DNA binding by MEF2 proteins (Cserjesi and Olson, 1991) suggest that this nucleotide substitution in FIRE should reduce the binding affinity for MEF2 in comparison with the corresponding MEF2 binding motif found in SURE. As a direct test of this prediction, a variant form of SURE was generated by site-directed mutagenesis to convert the MEF2 binding element of SURE to the sequence found in FIRE (SUREΔMEF2), as shown in Figure 5A. In addition, a mutant was generated to ablate the NFAT binding motif that is unique to SURE (SUREΔNFAT).
Fig. 5. MEF2 binding to slow and fast fiber-specific enhancers. (A) Nucleotide sequence alignment of segments of the rat TnI SURE and quail TnI FIRE enhancers (Nakayama et al., 1996) that include MEF2, NFAT and E box motifs. The putative MEF2 binding site in FIRE has a G nucleotide rather than an A nucleotide in position 9 of the MEF2 consensus binding motif (darkly shaded), and an NFAT consensus binding motif (boxed) found in SURE is not present in FIRE. The sequences of two variant forms of the SURE enhancer generated by site-directed mutagenesis are also illustrated. SUREΔNFAT and SUREΔMEF2 refer to variants in which NFAT and MEF2 binding motifs, respectively, are altered while leaving the remainder of the SURE enhancer sequence intact. The SUREΔMEF2 includes only the indicated single nucleotide substitution within an otherwise intact SURE enhancer. (B) Luciferase reporter constructs were prepared using the SURE or FIRE enhancers, or the indicated mutants thereof. Expression of luciferase following transfection of each of these reporter plasmids was assessed in the presence or absence of CnA*. Data are presented as in Figure 1 and represent mean values of two to four independent experiments. (C) EMSA of nuclear proteins from C2C12 cells using a high affinity MEF2 binding site (MCK-MEF2) as the labeled oligonucleotide probe. Unlabeled oligonucleotides (competitor) representing either the probe sequence (MCK-MEF2) or the MEF2 binding site from the SURE enhancer compete effectively with the labeled probe for binding MEF2, while the MEF2 motif from the FIRE enhancer competes less avidly. The identity of the MEF2–DNA complex (arrow) was confirmed by the change in mobility in the presence of anti-MEF2 antibodies (see Figure 2).
A single nucleotide substitution within the MEF2 binding motif of SURE reduced the response of the SURE enhancer to activated calcineurin in the C2C12 background (Figure 5B). Ablation of the NFAT motif in SURE also reduced the response to activated calcineurin, supporting the notion that both MEF2 and NFAT participate in transducing calcineurin-generated signals to relevant target genes. The prediction that MEF2 would bind preferentially to SURE was confirmed experimentally using EMSA (Figure 5C). Binding of endogenous MEF2 proteins in nuclear extracts from C2C12 myotubes to a radiolabeled oligonucleotide sequence representing a high affinity MEF2 binding site was competed more avidly by unlabeled oligonucleotide representing the MEF2 binding site from SURE than by the corresponding region of FIRE. Thus, MEF2 proteins function more effectively in transcriptional regulation of the SURE enhancer than of the FIRE enhancer, and this differential response is explained, at least in part, by higher affinity binding of MEF2 to SURE than to FIRE.
Calcineurin-dependent stimulation of the transactivating function of MEF2 is amplified by calmodulin-dependent protein kinase (CaMK) activity
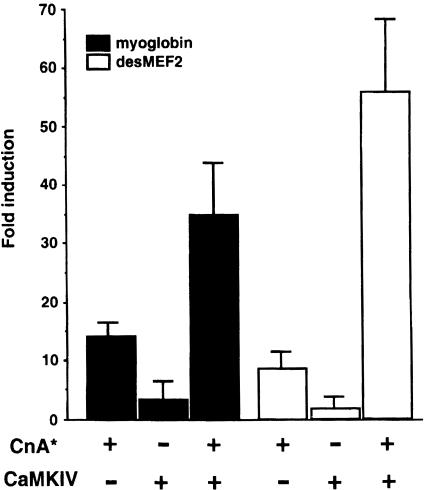
The premise that changes in regulatory pools of intracellular calcium constitute a proximate stimulus by which the firing pattern of motor neurons controls specific programs of gene expression in skeletal myofibers suggests that other calcium-regulated signaling pathways, in addition to calcineurin, may be pertinent to fiber type-specific gene transcription. As an initial test of this concept, we assessed functional interactions between calcineurin and calmodulin-dependent protein kinase IV (CaMKIV) in the control of myoglobin and desMEF2 enhancer function. CaMKIV was shown previously to modify calcineurin-regulated transcriptional responses mediated by the CREB transcription factor in hippocampal neurons (Bito et al., 1996), but studies of combinatorial interactions between calcineurin- and calmodulin-dependent protein kinases in skeletal myocytes have not been reported previously. We observed a pronounced enhancement of calcineurin-induced stimulation of MEF2 transactivator function in the presence of a constitutively active form of CaMKIV. This synergistic effect was evident using either the native myoglobin promoter/enhancer or the synthetic desMEF2 enhancer to detect the response (Figure 6). CaMKIV had only small effects (<5-fold) on transcription of either of these MEF2-dependent enhancers in the absence of concomitant calcineurin activity. In combination, however, CaMKIV and calcineurin evoked a large (35- to 60-fold) response in this assay. These results indicate that MEF2 serves to integrate signaling inputs derived both from calcineurin and from other calcium-regulated pathways. Although these initial studies focused on CaMKIV, it is likely that other isoforms of CaMK that are abundant in skeletal muscle may also prove pertinent to calcium-regulated gene expression in this cell background.
Fig. 6. Calcineurin and CaMKIV act synergistically to enhance the transactivating function of MEF2. Luciferase reporter plasmids controlled by the myoglobin or desMEF2 enhancers were cotransfected into C2C12 cells with expression plasmids encoding either constitutively active calcineurin A subunit or CaMKIV. Data are presented as in Figure 1, and represent results of three or more independent experiments.
MEF2 binding is necessary for slow fiber-specific activity of the SURE enhancer in transgenic mice
Promoter/reporter constructs prepared with native or mutated forms of the SURE enhancer (see Figure 5) were introduced by microinjection into fertilized murine oocytes, and founder transgenic mice (n = 7–12 for each sequence variant) were analyzed for reporter gene expression at 7 weeks of age.
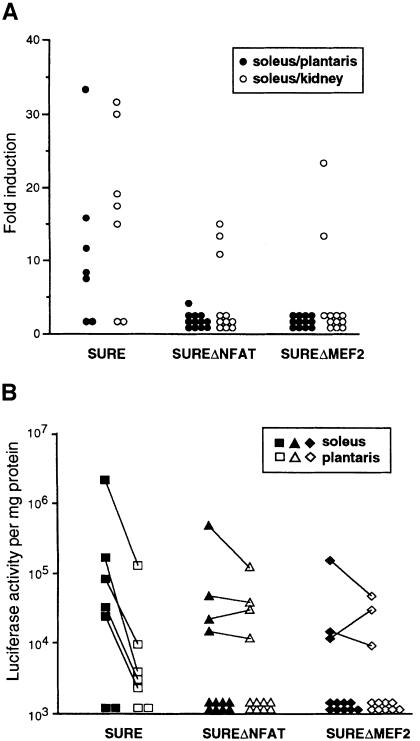
The native SURE element was expressed preferentially in soleus skeletal muscles as compared either with the plantaris skeletal muscle, or with non-muscle tissues such as kidney (Figure 7A). Since the soleus muscle of the mouse contains ∼50% slow, oxidative (Type I) fibers, while myofibers of the plantaris muscle are classified primarily as fast, glycolytic (Type IIb), this result confirms the slow fiber specificity of this transcriptional control element described originally by others (Nakayama et al., 1996). Mutation of the MEF2 binding motif within SURE (SUREΔMEF2) abolished the selective expression of the SURE enhancer in soleus muscles, although this mutant enhancer retained some degree of muscle-specificity (soleus versus kidney in Figure 7A). Disruption of the NFAT binding site (SUREΔNFAT) produced a similar effect. The loss of slow fiber-specific expression of the SUREΔMEF2 and SUREΔNFAT constructs, as compared with the native SURE sequence, was attributable to a decline in expression in the soleus, to an increased expression in the plantaris in some animals, and to a greater probability of almost complete silencing of the transgene (Figure 7B). The high variability in absolute levels of transgene expression among animals with different chromosomal insertion sites (a problem inherent to this experimental design) makes us most confident about conclusions based on ratios (Figure 7A). We interpret these results to indicate that both MEF2 and NFAT participate in regulatory pathways that establish the slow and oxidative myofiber phenotypes.
Fig. 7. Slow fiber-specific activity of the SURE enhancer requires intact NFAT and MEF2 binding motifs. Transgenic mice were generated using luciferase reporter constructs controlled by either the native SURE enhancer, or by two variant forms (SUREΔNFAT or SUREΔMEF2; see Figure 5A). Individual transgenic offspring, each representing a different chromosomal insertion event, were examined at 7 weeks of age for luciferase activity in soleus and plantaris muscles, which are enriched in slow, oxidative or fast, glycolytic myofibers respectively, and in kidney. (A) Data are presented as the ratio of transgene expression in soleus versus plantaris muscles (filled circles) as a measure of slow fiber-specific transcription, and as the ratio of transgene expression in soleus muscle versus kidney (open circles) as a measure of muscle-specific transcription. Each point corresponds to an individual transgenic animal. (B) Data are presented as the log of luciferase activity in the soleus (filled symbols) and plantaris (open symbols) skeletal muscles. Each point corresponds to an individual transgenic animal, and lines connect data from the same animal.
The transactivating function of MEF2 within skeletal muscles of adult animals is detectable selectively in the soleus muscle
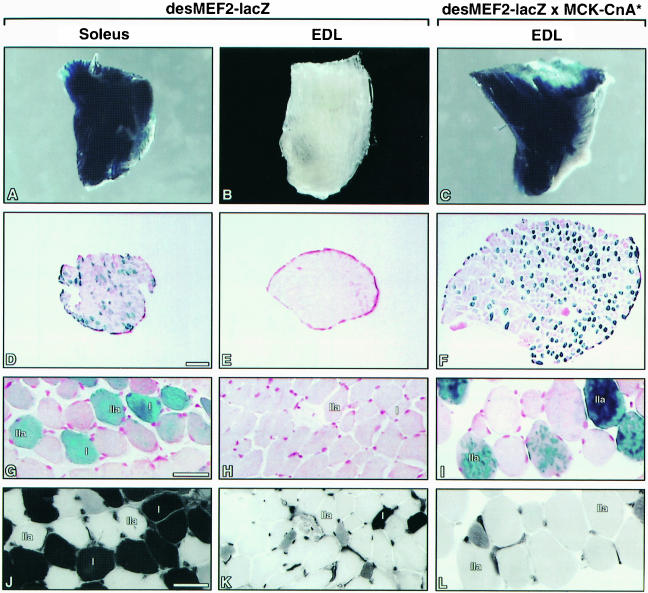
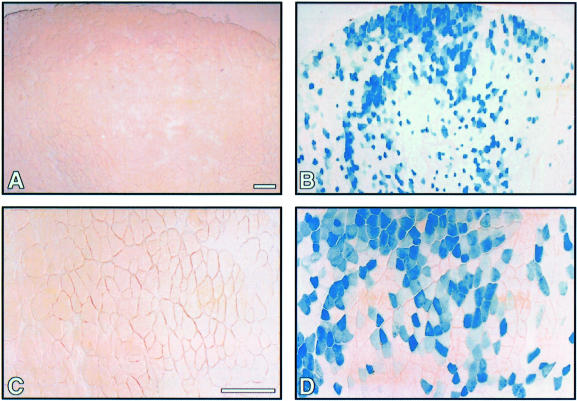
A line of transgenic mice was generated to harbor a lacZ reporter gene controlled by a minimal promoter and three repeats of a high affinity MEF2 binding site from the desmin gene (desMEF2-lacZ; Naya et al., 1999). During embryonic development, expression of lacZ was observed throughout skeletal, cardiac and smooth muscle lineages, mirroring expression of MEF2 proteins. After birth, however, consistently high levels of lacZ expression in this animal are restricted predominantly to the brain (Naya et al., 1999). Notably, β-galactosidase activity is not detectable in cardiac muscle and most skeletal muscles of these animals, despite the clear presence of MEF2 proteins within these tissues. Careful analysis of skeletal muscles of a large number of desMEF2-lacZ transgenic mice revealed that β-galactosidase activity is expressed selectively in soleus muscles (Figure 8A and D), while muscles such as the extensor digitorum longus (EDL) that consist predominantly of fast fibers are negative (Figure 8B and E). This phenotype was incompletely penetrant, in that grossly positive β-galactosidase staining in the soleus was observed in eight of 55 animals bearing this transgene, as compared with none of 55 animals in which β-galactosidase activity was detectable in the EDL. In cross-sections of these soleus muscles, we observed that the MEF2-dependent reporter gene was activated both in Type I and in Type IIa fibers (Figure 8G and J).
Fig. 8. The transactivating function of MEF2 is detected selectively in soleus muscles of transgenic mice, and is up-regulated by activated calcineurin. Soleus and EDL muscles were dissected from the hindlimbs of desMEF2-lacZ transgenic mice, or from doubly transgenic mice carrying both the desMEF-lacZ transgene and an MCK-CnA* transgene (see Material and methods), and stained for β-galactosidase activity. The gross appearance of the muscles (A–C), and photomicrographs of serial cross sections stained for β-galactosidase activity (D–I) or myosin ATPase activity (J–L) are shown. The distribution of β-galactosidase appeared to be uniform along the length of positively staining fibers (not shown). (D–F) Bar, 200 µm; (G–L) bar, 40 µm.
The transactivating function of MEF2 in skeletal muscle is increased by activated calcineurin and by motor nerve stimulation
Another transgenic experiment showed that calcineurin can stimulate the transcriptional activation function of MEF2 within intact muscles, as well as in cultured myocytes. The desMEF2-lacZ transgenic line was crossed to a line of transgenic mice that express a constitutively active form of calcineurin under the control of the muscle isoenzyme of creatine kinase (MCK) enhancer. In doubly transgenic animals, desMEF lacZ expression is observed in fibers of the EDL (four of 10 animals examined) (Figure 8C and F), whereas this muscle was uniformly negative (none of 55 animals examined) in desMEF2-lacZ mice in the absence of the MCK–calcineurin transgene. Animals with the desMEF2-lacZ transgene were also subjected to 5 days of motor nerve stimulation. This stimulus uniformly increased the transactivating function of MEF2 in a large proportion of fibers (Figure 9).
Fig. 9. The transactivating function of MEF2 is up-regulated by electrical stimulation of the motor nerve. Gastrocnemius muscles were dissected from the hindlimbs of desMEF2-lacZ transgenic mice that were subjected to 5 days of motor nerve stimulation (see Materials and methods). Photomicrographs of muscle cross sections stained for β-galactosidase activity representing the unstimulated contralateral limb (A and C) and the stimulated limb (B and D) of a single animal are shown at two different magnifications (bar, 100 µm). Similar results were observed in each of three animals subject to this stimulation protocol.
Discussion
The principal conclusion of this study is that MEF2 proteins function in skeletal muscles as downstream effectors of calcium-regulated signaling pathways that are triggered initially by motor nerve activity, thereby contributing to fiber type-specific gene regulation. New findings identify MEF2 not only as a downstream transducer of calcineurin signaling in skeletal myocytes, but also as a point of integration of other calcium-regulated signaling pathways. Functional activation of MEF2 correlates with its dephosphorylation by a calcium-regulated, calcineurin-dependent mechanism. Based on the results of studies performed both in cultured myocytes and in transgenic mice, we propose that MEF2 serves as a nodal point in the molecular signaling pathways by which motor nerve activity controls distinctive programs of gene expression in myofibers.
The transcriptional response evoked by activated calcineurin is transduced by MEF2 to complex native promoter/enhancer elements from the myoglobin or TnIs genes, and to a synthetic enhancer (desMEF2) constructed solely from MEF2 binding elements. Although MEF2 acts synergistically with NFAT to transactivate enhancers that bind both proteins, our data using the desMEF2 reporter plasmid demonstrate that DNA binding by NFAT is not absolutely required for the ability of MEF2 to transduce calcineurin-dependent signals to target genes.
MEF2 has been implicated previously as a component of calcium-regulated signaling pathways in non-muscle cell types (Liu et al., 1997; Mao and Wiedmann, 1999; Mao et al., 1999; Youn et al., 1999). In particular, our current data should be compared with a recent report that the function of MEF2 as a transcriptional activator is regulated by calcineurin in cultured neurons (Mao and Wiedmann, 1999). Both studies demonstrate functional up-regulation of the transactivating function of MEF2 in the presence of activated calcineurin, corresponding to dephosphorylation of MEF2 through a calcineurin-dependent mechanism. However, calcineurin was observed to stimulate DNA binding of MEF2 in cerebellar granule neurons (Mao and Wiedmann, 1999), while our studies of skeletal myocytes point to a response in which the transactivating function of MEF2 is augmented in the absence of detectable changes in DNA binding. Such differences among cell types in the terminal events of otherwise similar calcineurin-dependent signaling pathways may contribute to the distinctive programs of gene expression characteristic of calcium-regulated gene expression in each cell background. Additional studies are required to determine whether MEF2 is a direct substrate for the protein phosphatase activity of calcineurin, and to define more completely the functional roles of phosphorylation at specific amino acid residues of MEF2 proteins in different cell types. Our new data are the first to demonstrate an important role for MEF2 in transducing calcium-regulated signals within skeletal muscle, and provide a rationale for more detailed studies of MEF2 phosphorylation in the context of different signaling inputs that alter muscle phenotype.
The ability of MEF2 to integrate signals from multiple calcium-regulated signaling proteins, in this case calcineurin and CaMK, is another important feature of our current findings. Previous studies demonstrate that this integrative function of MEF2 also extends to mitogen-activated protein kinase (MAPK) pathways (Mao et al., 1999; Yang et al., 1999; Zhao et al., 1999). For example, five consensus MAPK phosphorylation sites are conserved within C-terminal domains of MEF2 proteins, and p38/Erk5 has been shown to act directly on specific residues of MEF2C and MEF2A, thereby augmenting their transactivating function (reviewed in Black and Olson, 1998; Zhao et al., 1999). Recently, we and others (Miska et al., 1999) have found that MEF2 associates with specific isoforms of histone deacetylase (HDAC), and that HDAC–MEF2 complexes bind DNA and function as transcriptional repressors. Both CaMKIV and CaMKI serve to up-regulate MEF2-dependent transcription by relieving this repression (our unpublished data). Although our current studies did not examine MAPK signaling or HDACs in the context of skeletal muscle, it is likely that future investigations will reveal an increasingly complex repertoire of signaling inputs acting upon MEF2, the relative balance of which determines the ultimate physiological response.
Our findings illustrate other previously unrecognized features of MEF2-dependent gene regulation in skeletal muscle. Direct comparison of well defined enhancer elements from genes encoding slow and fast muscle isoforms of troponin I demonstrated higher affinity binding of MEF2 to the SURE enhancer. More avid binding of MEF2 to the SURE sequence, by comparison with binding to the FIRE sequence, correlated with a greater responsiveness of SURE to transactivation by MEF2. This result appears to be based on a single nucleotide substitution in the MEF2 binding region of SURE. This distinction—lower affinity MEF2 binding in FIRE versus SURE enhancer elements—while pertinent to expression of troponin I isoforms, is unlikely to represent a universal feature that distinguishes fiber type-selective enhancers. Regulatory regions of certain other fast fiber-selective genes (e.g. MCK) that respond only minimally to calcineurin activation in myotubes (Chin et al., 1998) include high affinity binding sites for MEF2 (Gosset et al., 1989). We propose, therefore, two independent mechanisms by which MEF2 contributes to fiber type-specific gene transcription. Weak binding of MEF2 to certain fast fiber-specific enhancers (e.g. FIRE) represents a mechanism that is relevant only to a subset of fast-specific genes. A second, and more generalized, mechanism is based on modulation of the transcriptional activation function of MEF2 by signals that are triggered selectively in slow and oxidative fast fibers.
This notion is supported by examination of transgenic mice developed for the present study. A high affinity binding site for MEF2 appears to be necessary to generate a slow or oxidative fiber-specific pattern of transgene expression. Even a single nucleotide substitution within the MEF2 binding motif of the SURE enhancer, converting it to identity with the lower affinity MEF2 recognition sequence from FIRE, abolished slow fiber specificity of this transcriptional control region. Ablation of the NFAT binding site within the SURE enhancer produced similar functional consequences. This latter result speaks to the complementary role of NFAT in promoting expression of slow or oxidative fiber-specific genes (Chin et al., 1998). The results of our mutational analysis of the SURE enhancer in transgenic mice differ somewhat from findings reported recently by others (Calvo et al., 1999) who generated a variant of the SURE enhancer that lacked the NFAT binding motif but retained soleus-specific expression. The basis for these discordant results with respect to NFAT site mutations within the SURE enhancer is unclear, although we speculate that the specific transgene constructs examined by this group may contain additional MEF2 sites within core promoter or flanking regions linked to the SURE enhancer. Since the desMEF-lacZ transgene is expressed selectively in the soleus muscle, and is activated by either calcineurin or motor nerve stimulation in the absence of NFAT binding sites, it follows that slow or oxidative fiber-specific transcription of artificially constructed regulatory elements can occur in the absence of DNA binding by NFAT. Our viewpoint, however, continues to be that both MEF2 and NFAT are important transducers of signals that control fiber type-selective programs of gene expression in skeletal muscles.
This position is supported by new data presented in this manuscript, and by additional findings to be reported elsewhere (Naya et al., 2000; Rothermel et al., 2000). Here we show that the function of MEF2 as a transcriptional activator in skeletal muscles of intact mice is increased by forced expression of a constitutively active form of calcineurin, or by motor nerve stimulation. Since these stimuli are sufficient to provoke fiber type transformation, these results lend further support to the conclusion that MEF2 functions as a downstream effector of calcineurin-dependent signaling pathways that participate in the control of fiber type-specific gene expression. We do not know why in resting animals MEF2 transcriptional activity is detectable only in a subset of Type I and IIa fibers, and primarily in the soleus muscle. It appears that a sufficient stimulus to activate MEF2 to detectable levels in resting animals occurs primarily in the soleus, except when the intensity of the stimulus is increased (as provided here by the activated calcineurin transgene or by motor nerve stimulation). Future studies may determine that MEF2 participates in activity-dependent mechanisms that control fiber type-specific gene expression, while redundant control mechanisms (acting independently of MEF2) take precedence in maintaining specialized characteristics of different fiber types in resting muscles. Alternatively, perhaps only transient periods of MEF2 activation provoked by locomotor activity, and detected only infrequently in resting animals, may be sufficient to maintain oxidative fiber phenotypes once they have been established.
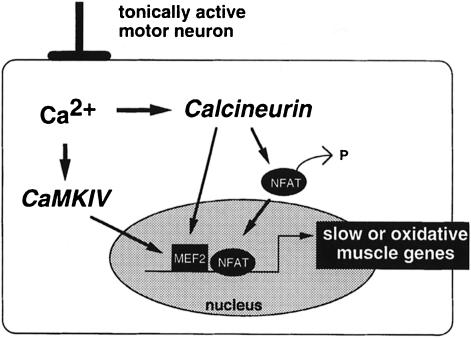
We previously proposed a mechanistic model to account for the effects of specific patterns of motor nerve activity in the control of specialized properties of skeletal myofiber subtypes (Chin et al., 1998). Based on the new information reported here, we have refined our model for the control of fiber type-specific gene expression by calcium signaling (Figure 10). A major new feature is that MEF2, in addition to NFAT, is capable of transducing signals initiated by calcineurin in skeletal myocytes. In addition, we propose that MEF2 serves to integrate multiple signaling inputs evoked by motor nerve stimulation that amplify calcineurin-generated responses. Our studies in cultured myocytes document the existence of the signaling mechanisms proposed in this model, and our studies in transgenic mice described here and elsewhere (Naya et al., 2000) support its relevance to fiber type determination in intact animals. The experiments reported in this paper have focused on skeletal muscle, but it is likely that MEF2 proteins also function in cardiac muscle to transduce signals generated by calcineurin-dependent signaling cascades that modulate gene expression and hypertrophic growth (Molkentin et al., 1998).
Fig. 10. A revised mechanistic model for control of slow or oxidative fiber-specific gene expression by motor nerve activity. Findings presented here extend the model we proposed previously (Chin et al., 1998) to emphasize the importance of MEF2 as a downstream effector of calcium-dependent changes in gene expression provoked by a tonically active pattern of motor neuron firing. Signals generated by calcineurin augment the transactivating function of MEF2 in a manner that also is increased by concomitant activation of CaMKIV, and by synergistic interactions with NFAT.
Molecular details of the signaling mechanisms proposed here remain to be elucidated, and additional relationships between these pathways and other proteins that govern muscle phenotype undoubtedly will be revealed. Our model should not be interpreted to suggest that other transcription factors and other signaling pathways are unimportant with respect to fiber type-specific gene expression. On the contrary, a number of interesting observations suggest a role for transcriptional regulatory proteins of the bHLH (Hughes et al., 1999), GATA (Musaro et al., 1999), SIX (Spitz et al., 1998) and nuclear receptor (Spitz et al., 1999) protein families. In addition, this current model is not yet sufficiently complete to explain the manner in which calcineurin may, under certain conditions, contribute to hypertrophic growth rather than fiber type transformation of skeletal myofibers, as described in recent reports in which muscle growth was stimulated by IGF-1 (Musaro et al., 1999; Semsarian et al., 1999). Irrespective of these limitations, the new conclusions presented here provide a fresh perspective on fiber type-specific gene regulation, and support a conceptual framework on which a more complete understanding can be constructed as additional experimental results become available.
Materials and methods
Plasmid constructs
Luciferase reporter plasmids were constructed in pGL3 (Promega) by inserting promoter/enhancer regions from genes encoding human myoglobin (Devlin et al., 1989) or the SURE and FIRE isoforms of TnIs (Nakayama et al., 1996). In addition, a synthetic enhancer constructed from three copies of a high affinity MEF2 binding sequence from the desmin promoter [termed desMEF2 (Naya et al., 1999)] was linked to a minimal promoter (hsp70) and inserted into pGL3. β-galactosidase reporter plasmids pCMV-lacZ (Grayson et al., 1995) and desMEF2-lacZ (Naya et al., 1999), and expression vectors encoding MEF2A (Black et al., 1995), MEF2B (Yu et al., 1992), MEF2C (Black et al., 1995), MEF2D (Black et al., 1995), or constitutively active forms of calcineurin or CaMKIV (Ho et al., 1996), were previously described. PCR-based mutagenesis of the SURE enhancer to generate the SUREΔMEF2 and SUREΔNFAT luciferase reporter plasmids was performed as described (Yang et al., 1997) and mutations were confirmed by DNA sequencing.
Tissue culture, cell transfection, virus infection and reporter gene assays
C2C12 myoblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum and antibiotics (100 U of penicillin and 100 µg of streptomycin per ml). Myotube formation was induced by switching confluent cells to differentiation media [DMEM supplemented with 2% heat-inactivated horse serum, 10 µg/ml insulin (Gibco-BRL) and 10 µg/ml transferrin (Gibco-BRL)] for 48–72 h. Differentiation was assessed by the formation of multinucleated myotubes and by immunohistochemistry using anti-myosin antibodies (MF-20; Developmental Studies Hybridoma Bank, University of Iowa). For transient transfection assays, C2C12 cells were plated 12 h before transfection in six-well tissue culture dishes at ∼1.0 ×105 cells per well, and transfected with 1.6 µg of plasmid DNA using Lipofectamine Plus (Gibco-BRL). Cells were harvested 40 h after transfection. Luciferase and β-galactosidase assays of whole cell extracts were conducted as previously described (Grayson et al., 1995; Yang et al., 1997).
Recombinant, replication-defective adenoviruses encoding either green fluorescent protein (Ad-GFP) or CnA* (Ad-CnA*) were generated using standard techniques (Gerard and Meidell, 1995). For gene transfer using adenoviral vectors, C2C12 cells were first transfected with reporter plasmid as already described, and then exposed to the adenoviral suspension at a multiplicity of infection (m.o.i.) of 10–100. Cells were harvested 24 h after infection. For immunostaining, C2C12 myoblasts were fixed in 4% paraformaldehyde, rinsed with phosphate-buffered saline (PBS), incubated with the monoclonal anti-myosin antibody (MF-20 supernatant) for 1 h, rinsed in PBS, incubated with anti-mouse IgG conjugated to horseradish peroxidase and detected with DAB chromogen tablets (Dako).
Phosphorylation status of MEF2
Ionomycin was added to the media of C2C12 myotubes 48 h after differentiation at a final concentration of 1 µM, and cells were harvested 4.5 h later. Cyclosporin A was included at a final concentration of 250 nM. Nuclear protein extracts were prepared as described previously (Yang et al., 2000) and subjected to immunoblot analysis using a polyclonal antibody recognizing MEF2A (Santa Cruz Biotechnology) and the ECL detection system (Amersham). Aliquots of each nuclear protein extract were incubated with alkaline phosphatase (Roche Molecular Biochemicals) at a concentration of 100 U/ml for 60 min at 37°C.
EMSA
A synthetic oligonucleotide representing a high affinity MEF2 binding motif (5′-GATCCTCTAAAAATAACCCT-3′) from the MCK gene (Gosset et al., 1989) was annealed to its complementary strand, labeled with polynucleotide kinase and [γ-32P]ATP (3000 Ci/mmol; Amersham) and incubated with nuclear protein extracts (15 µg) from C2C12 myotubes in binding buffer [20 mM HEPES pH 7.6, 50 mM KCl, 10% glycerol, 0.2 mM EDTA, 1 mM DTT and 1 µg of poly(dI–dC) per lane] for 20 min at room temperature. Competitive binding assays were conducted under the same conditions, with the addition of 0.4–1 pmol (20- to 50-fold molar excess) of unlabeled competitor oligonucleotides that included the following sequences:
MCK-MEF2 (5′-GATCCTCTAAAAATAACCCT-3′);
SURE-MEF2 (5′-GATCTCCTAAAAATACGATC-3′);
FIRE-MEF2 (5′-GATCTTCTAAAAATGGGATC-3′).
The identity of MEF2–DNA complexes was verified by pre-incubation of nuclear protein extracts with anti-MEF2 antibody (Santa Cruz) or pre-immune serum for 20 min. Complexes were resolved on 4% polyacrylamide gels at 4°C in 45 mM Tris–45 mM borate–1 mM EDTA buffer, dried and visualized using PhosphorImager analysis.
Transgenic mice
Transgenic mice containing a lacZ gene under the control of three copies of desmin MEF2 binding elements were described previously (Naya et al., 1999). Muscles from hind limbs of animals at 7 weeks of age were dissected and stained for β-galactosidase as described elsewhere (Naya et al., 1999). Other features of transgenic mice that express a constitutively active form of calcineurin under the control of a muscle-specific enhancer from the MCK gene are also described elsewhere (Naya et al., 2000). After removal of prokaryotic sequences, luciferase transgenes controlled by the SURE enhancer, or by the variant forms SUREΔNFAT or SUREΔMEF2, were introduced by microinjection into fertilized oocytes of C57B6/C3H mice. The resulting transgenic offspring were identified by Southern blot or PCR analysis of genomic DNA. Tissues were harvested from transgenic animals at 7–10 weeks of age and assayed for expression of luciferase (Yang et al., 1997), or processed for β-galactosidase assay (Naya et al., 1999) or ATPase myosin activity (Chin et al. 1998). All experiments involving animals were reviewed and approved by the Institutional Animal Care and Research Advisory Committee.
Electrical stimulation of the hindlimb musculature
Electrical stimulation was conducted on anesthetized mice for 30 min three times daily, separated by 4 h intervals for 5 consecutive days. Contractions were elicited by stimulation of the sciatic nerve at supramaximal voltage (Grass stimulator, model S48) every 30 s with a 1 s, 100 Hz train of pulses that were 0.1 ms in duration. Immediately at the conclusion of the final pacing period, muscles were excised, weighed and frozen in melting isopentane pre-cooled with liquid nitrogen.
Acknowledgments
Acknowledgements
We thank Caroline Humphries for expert technical assistance and Dr A.Buonanno for providing a plasmid containing the SURE enhancer. H.W. and F.J.N. were supported by fellowship awards from the NIH, and T.A.M. is a Pfizer Fellow of the Life Sciences Research Foundation. This work was supported by the grants from the National Institutes of Health (NIH) (R.S.W. and E.N.O.), the Robert A.Welch Foundation (E.N.O.), the D.W.Reynolds Foundation (R.S.W. and E.N.O.) and the NSERC of Canada (R.N.M.).
References
- Bito H., Deisseroth,K. and Tsien,R.W. (1996) CREB phosphorylation and dephosphorylation: a Ca2+- and stimulus duration-dependent switch for hippocampal gene expression. Cell, 87, 1203–1214. [DOI] [PubMed] [Google Scholar]
- Black B.L. and Olson,E.N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol., 14, 167–196. [DOI] [PubMed] [Google Scholar]
- Black B.L., Martin,J.F. and Olson,E.N. (1995) The mouse MRF4 promoter is trans-activated directly and indirectly by muscle-specific transcription factors. J. Biol. Chem., 270, 2889–2892. [DOI] [PubMed] [Google Scholar]
- Blagden C.S., Currie,P.D., Ingham,P.W. and Hughes,S.M. (1997) Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes Dev., 11, 2163–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S., Venepally,P., Cheng,J. and Buonanno,A. (1999) Fiber-type-specific transcription of the troponin I slow gene is regulated by multiple elements. Mol. Cell. Biol., 19, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin E.R. et al. (1998) A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev., 12, 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserjesi P. and Olson,E.N. (1991) Myogenin induces the myocyte-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol. Cell. Biol., 11, 4854–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B.H., Wefald,F.C., Kraus,W.E., Bernard,T.S. and Williams,R.S. (1989) Identification of a muscle-specific enhancer within the 5′ flanking region of the human myoglobin gene. J. Biol. Chem., 264, 13896–13900. [PubMed] [Google Scholar]
- DiMario J.X., Fernyak,S.E. and Stockdale,F.E. (1993) Myoblasts transferred to the limbs of embryos are committed to specific fibre fates. Nature, 362, 165–167. [DOI] [PubMed] [Google Scholar]
- Dunn S.E., Burns,J.L. and Michel,R.N. (1999) Calcineurin is required for skeletal muscle hypertrophy. J. Biol. Chem., 274, 21908–21912. [DOI] [PubMed] [Google Scholar]
- Garry D.J., Bassel-Duby,R.S., Richardson,J.A., Grayson,J., Neufer,P.D. and Williams,R.S. (1996) Postnatal development and plasticity of specialized muscle fiber characteristics in the hindlimb. Dev. Genet., 19, 146–156. [DOI] [PubMed] [Google Scholar]
- Gerard R.D. and Meidell,R.S. (1995) Adenovirus vectors. In Rickwood,D. and Hames,B.D. (eds), DNA Cloning—A Practical Approach: Mammalian Systems. IRL Press, Oxford, UK, pp. 285–306. [Google Scholar]
- Gosset L.A., Kelvin,D.J., Sternberg,E.A. and Olson,E.N. (1989) A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol., 9, 5022–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson J., Williams,R.S., Yu,Y.T. and Bassel-Duby,R. (1995) Synergistic interactions between heterologous upstream activation elements and specific TATA sequences in a muscle-specific promoter. Mol. Cell. Biol., 15, 1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N., Gullberg,M. and Chatila,T. (1996) Activation protein 1-dependent transcriptional activation of interleukin 2 gene by Ca2+/calmodulin kinase type IV/Gr. J. Exp. Med., 184, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.M., Chi,M.M., Lowry,O.H. and Gundersen,K. (1999) Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. J. Cell Biol., 145, 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Liu,P., Borras,A., Chatila,T. and Speck,S.H. (1997) Cyclosporin A sensitive induction of the Epstein–Barr virus lytic switch is mediated via a novel pathway involving a MEF-2 family member. EMBO J., 16, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z. and Wiedmann,M. (1999) Calcineurin enhances MEF2 DNA binding activity in calcium-dependent survival of cerebellar granule neurons. J. Biol. Chem., 274, 31102–31107. [DOI] [PubMed] [Google Scholar]
- Mao Z., Bonni,A., Xia,F., Nadal-Vicens,M. and Greenberg,M.E. (1999) Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science, 286, 785–790. [DOI] [PubMed] [Google Scholar]
- Miska E.A., Karlsson,C., Langley,E., Nielsen,S.J., Pines,J. and Kouzarides,T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D., Lu,J., Antos,C.L., Markham,B., Richardson,J., Robbins,J., Grant,S.R. and Olson,E.N. (1998) A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell, 93, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musaro A., McCullagh,K.J., Naya,F.J., Olson,E.N. and Rosenthal,N. (1999) IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature, 400, 581–585. [DOI] [PubMed] [Google Scholar]
- Nakayama M., Stauffer,J., Cheng,J., Banerjee-Basu,S., Wawrousek,E. and Buonanno,A. (1996) Common core sequences are found in skeletal muscle slow- and fast-fiber-type-specific regulatory elements. Mol. Cell. Biol., 16, 2408–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya F.J., Wu,C., Richardson,J.A., Overbeek,P. and Olson,E.N. (1999) Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development, 126, 2045–2052. [DOI] [PubMed] [Google Scholar]
- Naya F.J., Mercer,B., Shelton,J., Richardson,J.A., Williams,R.S. and Olson,E.N. (2000) Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem., 275, 4545–4548. [DOI] [PubMed] [Google Scholar]
- Ontell M.P., Sopper,M.M., Lyons,G., Buckingham,M. and Ontell,M. (1993) Modulation of contractile protein gene expression in fetal murine crural muscles: emergence of muscle diversity. Dev. Dynam., 198, 203–213. [DOI] [PubMed] [Google Scholar]
- Pette D. and Staron,R.S. (1997) Mammalian skeletal muscle fiber type transitions. Int. Rev. Cytol., 170, 143–223. [DOI] [PubMed] [Google Scholar]
- Pette D. and Vrbova,G. (1992) Adaptation of mammalian skeletal muscle fibers to chronic electrical stimulation. Rev. Physiol. Biochem. Pharmacol., 120, 115–202. [DOI] [PubMed] [Google Scholar]
- Romanul F.C. and Van der Meulen,J.P. (1966) Reversal of the enzyme profiles of muscle fibres in fast and slow muscles by cross-innervation. Nature, 212, 1369–1370. [DOI] [PubMed] [Google Scholar]
- Rothermel B., Vega,R.B., Yang,J., Wu,H., Bassel-Duby,R. and Williams,R.S. (2000) A protein encoded within the Down Syndrome Critical Region is enriched in striated muscles and inhibits calcineurin signaling. J. Biol. Chem., 275, 8719–8725. [DOI] [PubMed] [Google Scholar]
- Saltin B. and Gollnick,P.D. (1983) Skeletal muscle adaptability: significance for metabolism and performance. In Peachey,L.D.D. (ed.), Handbook of Physiology: Skeletal Muscle. American Physiological Society, Bethesda, MD, pp. 555–632. [Google Scholar]
- Schiaffino S. and Reggiani,C. (1996) Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev., 76, 371–423. [DOI] [PubMed] [Google Scholar]
- Semsarian C., Wu,M.J., Ju,Y.K., Marciniec,T., Yeoh,T., Allen,D.G., Harvey,R.P. and Graham,R.M. (1999) Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature, 400, 576–581. [DOI] [PubMed] [Google Scholar]
- Spitz F., Demignon,J., Porteu,A., Kahn,A., Concordet,J.P., Daegelen,D. and Maire,P. (1998) Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc. Natl Acad. Sci. USA, 95, 14220–14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Demignon,J., Kahn,A., Daegelen,D. and Maire,P. (1999) Developmental regulation of the aldolase A muscle-specific promoter during in vivo muscle maturation is controlled by a nuclear receptor binding element. J. Mol. Biol., 289, 893–903. [DOI] [PubMed] [Google Scholar]
- Stockdale F.E. (1997) Mechanisms of formation of muscle fiber types. Cell Struct. Funct., 22, 37–43. [DOI] [PubMed] [Google Scholar]
- Vrbova G. (1963) The effect of motoneurone activity on the speed of contraction of striated muscle. J. Physiol., 169, 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.S. and Neufer,P.D. (1996) Regulation of gene expression in skeletal muscle by contractile activity. In Rowell L.B. and Shepard,J.T. (eds), The Handbook of Physiology: Integration of Motor, Circulatory, Respiratory and Metabolic Control During Exercise. American Physiology Society, Bethesda, MD, pp. 1124–1150. [Google Scholar]
- Williams R.S., Salmons,S., Newsholme,E.A., Kaufman,R.E. and Mellor,J. (1986) Regulation of nuclear and mitochondrial gene expression by contractile activity in skeletal muscle. J. Biol. Chem., 261, 376–380. [PubMed] [Google Scholar]
- Yang Q., Bassel-Duby,R. and Williams,R.S. (1997) Transient expression of a winged-helix protein, MNF-β, during myogenesis. Mol. Cell. Biol., 17, 5236–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Kong,Y., Rothermel,B., Garry,D.J., Bassel-Duby,R. and Williams,R.S. (2000) The winged helix/forkhead protein myocyte nuclear factor β (MNF-β) forms a co-repressor complex with mammalian Sin3B. Biochem. J., 345, 335–343. [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Galanis,A. and Sharrocks,A.D. (1999) Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell. Biol., 19, 4028–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn H.D., Sun,L., Prywes,R. and Liu,J.O. (1999) Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science, 286, 790–793. [DOI] [PubMed] [Google Scholar]
- Yu Y.-T., Breitbart,R.E., Smoot,L.B., Lee,Y., Mahdavi,V. and Nadal-Ginard,B. (1992) Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcriptional factors. Genes Dev., 6, 1783–1798. [DOI] [PubMed] [Google Scholar]
- Zhao M., New,L., Kravchenko,V.V., Kato,Y., Gram,H., di Padova,F., Olson,E.N., Ulevitch,R.J. and Han,J. (1999) Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol., 19, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]