Abstract
The bZip transcription factor MafB is expressed specifically in the myeloid lineage of the hematopoietic system and is up-regulated successively during myeloid differentiation from multipotent progenitors to macrophages. Here we report that this induction reflects an essential role of MafB in early myeloid and monocytic differentiation. We observed that the expression of MafB in transformed chicken hematopoietic precursors dramatically increases the proportion of myeloid colony formation at the expense of multipotent progenitor-type colonies. In addition, the overexpression of MafB in transformed myeloblasts stimulates the rapid formation of macrophages, as judged by morphology, surface marker expression and functional criteria. MafB-induced macrophages exhibit typical levels of phagocytic activity and nitric oxide release after activation by lipopolysaccharide. By contrast, overexpression of the myeloid transcription factor PU.1 in these cells does not induce macrophage differentiation. Furthermore, a dominant-negative allele of MafB inhibits both myeloid colony formation and the differentiation of myeloblasts into macrophages. Taken together, our results indicate that MafB induction is a specific and essential determinant of the monocytic program in hematopoietic cells.
Keywords: hematopoiesis/kreisler/MafB/Maf proteins/macrophages/myeloid differentiation
Introduction
In the adult organism, hematopoietic stem cells of the bone marrow pass through successively more restricted commitment steps to give rise to the different mature cell types of the blood. In the myeloid lineage, myeloblast progenitors differentiate into both neutrophilic granulocytes and monocytes, the latter of which can develop further into tissue macrophages. To decipher the molecular mechanism controlling monocytic differentiation is of significant interest in view of both the multiple immune functions of macrophages (Gordon et al., 1995) and the pathological disturbance of myeloid progenitor cell differentiation in leukemia (Look, 1997).
Lineage-specific gene expression in the hematopoietic system in general and in the myeloid compartment in particular is controlled largely by the combinatorial action of broadly expressed as well as cell type-specific transcription factors (Orkin, 1996; Tenen et al., 1997), involving both overlapping expression profiles and direct protein–protein interactions (Sieweke and Graf, 1998). Knockout and gain-of-function experiments have recently demonstrated a role for the PU.1 and C/EBPα and β transcription factors in myeloid lineage commitment (Scott et al., 1994, 1997; McKercher et al., 1996; Nerlov and Graf, 1998; Nerlov et al., 1998) and in the differentiation of myeloblasts towards granulocytes (Zhang et al., 1997; Radomska et al., 1998). Even though the transcription factors Egr-1, Stat3, Gbx2 and PU.1 have also been implicated in monocytic differentiation (Nguyen et al., 1993; Nakajima et al., 1996; Kowenz-Leutz et al., 1997; Henkel et al., 1999), their function is often redundant, not limited to monocytic differentiation or linked to the mediation of proliferative signals (Scott et al., 1994, 1997; Lee et al., 1996; McKercher et al., 1996; Kowenz-Leutz et al., 1997; Takeda et al., 1998). Thus the critical molecular events that define the monocyte/macrophage differentiation program still remain only poorly understood.
We previously have shown that unlike other myeloid transcription factors, the bZip factor MafB is expressed selectively in monocytes and macrophages but not in other hematopoietic cells (Sieweke et al., 1996; Eichmann et al., 1997; L.M.Kelly, M.H.Sieweke, T.Graf and C.Nerlov et al., submitted). During differentiation, it is not found in multipotent progenitors, but expressed at moderate levels in myeloblasts and strongly up-regulated in monocytes and macrophages (Sieweke et al., 1996). MafB is the avian homologue of the murine kreisler gene product (Eichmann et al., 1997) and belongs to the family of Maf proteins, a subgroup of AP-1-type bZip transcription factors. It can form homodimers via its leucine zipper domain that recognize specific DNA consensus sequences termed Maf response elements or MAREs (Kataoka et al., 1994). Maf proteins have been implicated in several different differentiation processes including the hematopoietic system (Blank and Andrews, 1997; Motohashi et al., 1997). Thus the small Maf protein p18 forms part of the NF–E2 complex, which has been shown to control erythroid and thrombocytic genes and to be required for thrombocytic differentiation (Shivdasani et al., 1995; Kotkow and Orkin, 1996). c-Maf has been shown to induce the differentiation of T helper 2 (Th2) cells (Ho et al., 1996) due to its ability to activate the tissue-specific transcription of the interleukin-4 (IL-4) gene (Kim et al., 1999).
We previously have shown that MafB represses erythroid genes through direct protein interaction with Ets-1, suggesting that the up-regulation of MafB during monocytic differentiation prevents inappropriate erythroid gene expression in this lineage. Indeed, we could show that MafB inhibits erythroid differentiation (Sieweke et al., 1996). The question remained, however, whether MafB could also actively stimulate monocytic differentiation.
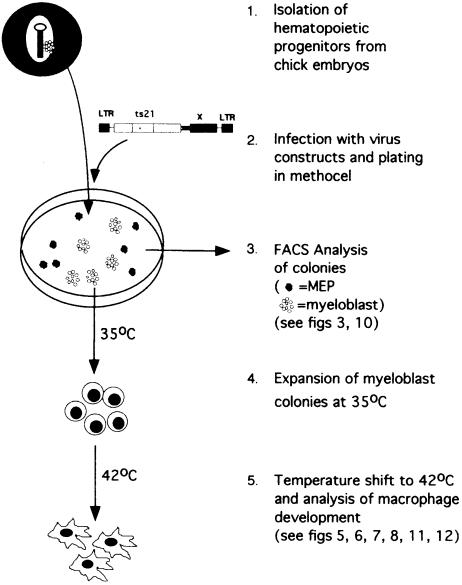
To address this question, we expressed MafB in primary chicken hematopoietic precursor cells transformed by the myb-ets oncogene-containing E26 virus. Infected cells give rise to multipotent progenitor type (MEP) and myeloblast colonies in semisolid medium, which can be expanded in culture due to an increased self-renewal capacity of the cells expressing the oncogene (for a review of the system, see McNagny and Graf, 1996). With the ts21 version of E26, which contains a temperature-sensitive mutation in v-Myb, it is further possible to analyze the macrophage differentiation potential of myeloblast colonies by releasing the oncogene-imposed differentiation block with a temperature shift to 42°C (Beug et al., 1984). Thus the system makes it possible to monitor the effect of the MafB transgene on myeloid differentiation from the multipotent progenitor to the macrophage stage (see Figure 1).
Fig. 1. The ts21E26 differentiation system used in this study.
Here we report that MafB favors myeloid colony formation of ts21E26-transformed hematopoietic progenitors and promotes macrophage differentiation of ts21E26 myeloblasts. Furthermore, a dominant-negative allele of MafB inhibits both processes, indicating that the successive up-regulation of endogenous MafB from the multipotent progenitor to the monocyte stage is required both for the early stages of myeloid differentiation and for the specification of the monocytic program in myeloblasts.
Results
MafB increases the proportion of myeloid colony formation in ts21E26-transformed hematopoietic precursors
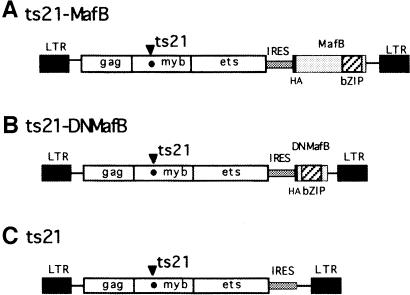
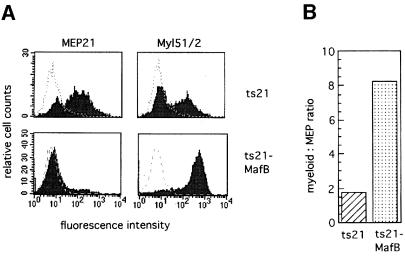
To assess the effect of MafB on myeloid differentiation, we expressed the protein in E26 virus-transformed hematopoietic progenitors. For this purpose, we generated bicistronic retroviral vectors, in which the ts21 version of the E26 myb-ets oncogene is followed by an internal ribosome entry site (IRES) element and the hemagglutinin (HA)-tagged coding sequence of different MafB alleles (see Figure 2). As a source of yolk sac hematopoietic progenitors, we infected suspensions of 2-day-old chick embryos with either a control virus containing the IRES element only or a virus with an IRES-MafB cassette (see Figure 2) and plated the cells in semisolid medium for 2 weeks. In this type of assay, approximately equal numbers of MEP colonies and myeloblast colonies typically are observed with wild-type E26 virus (Graf et al., 1992; McNagny and Graf, 1996). In our experiment, ∼100 colonies of similar size per 35 mm dish developed from both the ts21- and the ts21-MafB-infected cells, but the proportion of myeloid colonies was strongly increased in the ts21-MafB assays compared with the control. To confirm this observation, we pooled the colonies from each plate and analyzed their phenotype by fluorescence activated cell sorting (FACS) with antibodies against the multipotent progenitor marker MEP21 and the myeloid surface antigen Myl51/2. Representative FACS profiles from a ts21 and ts21-MafB colony assay are shown in Figure 3A. We analyzed three assays per condition, and the controls consistently showed the typical mixed population of colonies with both MEP21- and Myl51/2-positive cells. In contrast, of the ts21-MafB virus-infected cells, only very few expressed MEP21 and the vast majority were myeloid (87% on average). Thus in the MafB-expressing cells, the ratio of myeloid- to MEP-type cells had shifted >4-fold in favor of myeloid cells (Figure 3B). The same observations were made in several independent experiments (see also Figure 10). These results thus suggest that MafB favors myeloid differentiation in hematopoietic progenitors.
Fig. 2. Retroviral vectors used. The constructs are based on the ts21 version of E26 virus containing a temperature-sensitive point mutation in v-Myb. At the 3′ end of the coding sequence, an IRES-driven expression cassette was added to create the viral vectors (A) ts21-MafB, expressing full-length MafB, (B) ts21-DNMafB, expressing an N-terminally truncated dominant-negative allele of MafB, and (C) the ts21 vector control construct containing the IRES element only.
Fig. 3. Phenotype of transformed colonies obtained from ts21- or ts21-MafB-infected chicken hematopoietic progenitor cells. (A) FACS profiles of cells from pooled colonies of methylcellulose cultures using monoclonal antibodies against the multipotent progenitor antigen MEP21 or the myeloid antigen Myl51/2. The dotted line overlay shows the staining with an isotype-matched control monoclonal antibody against an intracellular antigen. (B) Graph showing the ratio of Myl51/2-positive to MEP21-positive cells calculated from the average of three colony assays each from ts21- and ts21-MafB-infected cells.
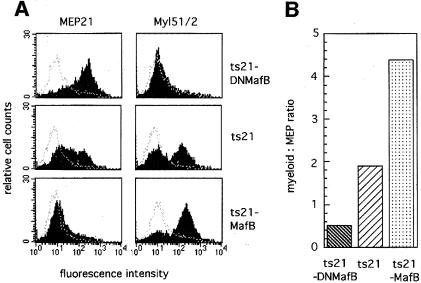
Fig. 10. Phenotype of transformed colonies obtained from chicken hematopoietic progenitor cells infected with ts21-DNMafB, ts21 or ts21-MafB viruses. (A) FACS profiles of cells from pooled colonies of methylcellulose cultures using monoclonal antibodies against the multipotent progenitor antigen MEP21 or the myeloid antigen Myl51/2. The dotted line overlay shows the staining with an isotype-matched control monoclonal antibody against an intracellular antigen. (B) Graph showing the ratio of Myl51/2-positive to MEP21-positive cells calculated from the average of three colony assays each from ts21-DNMafB-, ts21- and ts21-MafB-infected cells.
MafB stimulates macrophage differentiation in ts21 myeloblasts
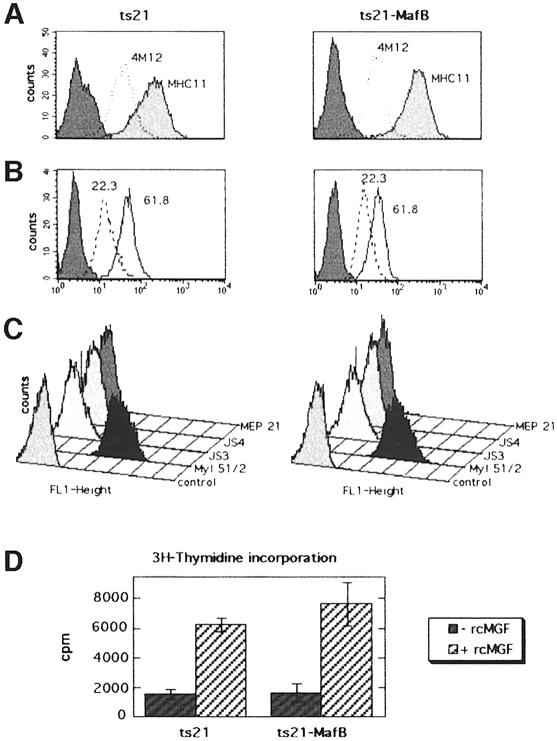
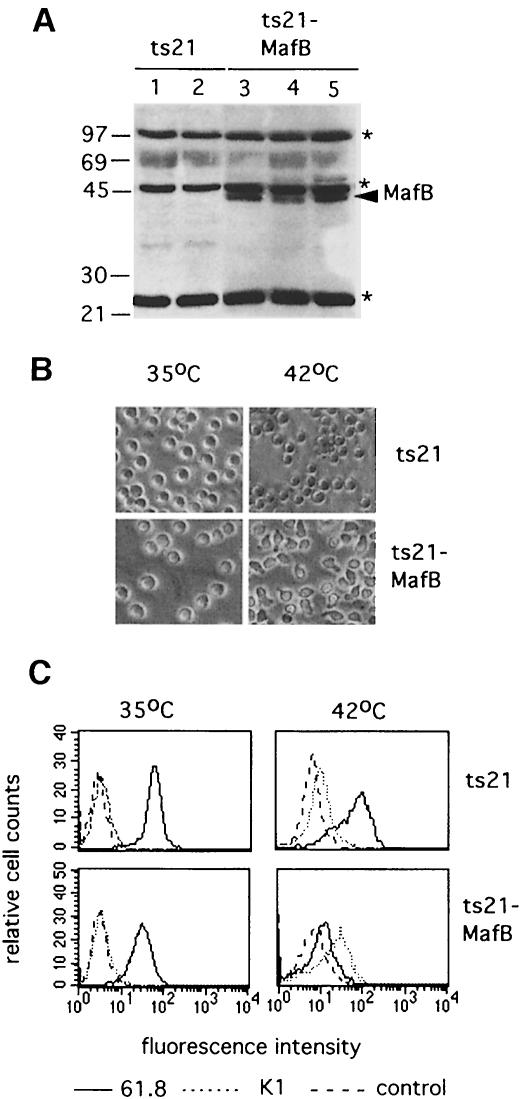
A dramatic increase in MafB expression is observed in the late stages of monocytic differentiation from myeloblasts to macrophages (Sieweke et al., 1996). To investigate the functional significance of this induction, we analyzed whether overexpression of MafB in myeloblasts influenced macrophage differentiation. For this purpose, we expanded individual myeloblast colonies at 35°C from ts21 control- or ts21-MafB-infected methylcellulose cultures. As determined by FACS analysis, both ts21 control and ts21-MafB clones expressed the myeloid surface antigens 4M12, cIa (MHC II; Figure 4A), 22.3, 61.8 (Figure 4B) and 51/2 (Kornfeld et al., 1983; Ewert et al., 1984; McNagny et al., 1992) but not the erythroid antigens JS3 and JS4 (Schmidt et al., 1986) or the multipotent progenitor antigen MEP21 (McNagny et al., 1992) (Figure 4C). In addition, both ts21 control and ts21-MafB clones were dependent on the avian myeloid-specific growth factor cMGF (Figure 4D). Thus both control and ts21-MafB clones had an indistinguishable myeloblast phenotype at 35°C. The expression of MafB in the individual ts21-MafB clones was verified by Western blotting with an antibody against an HA tag fused to the N-terminus of MafB (Figure 5A).
Fig. 4. Phenotype of expanded ts21 and ts21-MafB myeloblast clones. FACS profiles of representative ts21 or ts21-MafB myeloblast clones stained with antibodies against the myeloid markers 4M12 and cIa (MHC II) (A), 61.8 and 22.3 (B) and 51/2, the erythroid markers JS3 and JS4 and the multipotent progenitor/thrombocytic marker MEP21 (C). (D) Dependence of ts21 or ts21-MafB myeloblast clones on the avian myeloid growth factor cMGF. The rate of DNA synthesis in six ts21 and six ts21-MafB myeloblast clones was determined, culturing an equal number of cells in the absence or presence of recombinant cMGF for 48 h, pulse labeling with [3H]thymidine and measuring the radioactivity incorporated into DNA. Error bars indicate the SEM.
Fig. 5. (A) Western blot of cellular extracts from myeloblast clones expanded from ts21- and ts21-MafB-transformed colonies using a monoclonal antibody directed against the HA epitope tag in the ts21-MafB construct. Two ts21 control clones and three ts21-MafB clones are shown. The arrowhead indicates MafB protein, the asterisks indicate background bands detected in all samples. The size of molecular weight standards is shown on the left. (B) Phase contrast photomicrograph of a representative ts21 and ts21-MafB clone each maintained at 35°C or shifted to 42°C for 48 h. (C) FACS profiles of representative ts21 and ts21-MafB clones maintained at 35°C or shifted to 42°C for 48 h using the myeloblast-specific antibody 61.8 (–––) the macrophage antigen-detecting antibody K1 (- - - -) and an isotype-matched control antibody against an intracellular antigen (— — —).
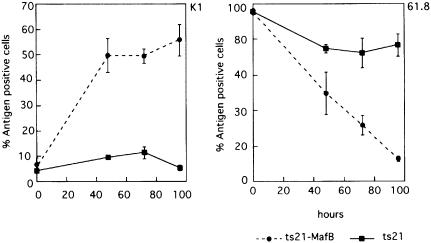
Equal numbers of cells from ts21 control and ts21-MafB clones were either maintained at 35°C or their differentiation block was released by a temperature shift to 42°C. After 24 h at 42°C, we observed adherent cells in the ts21-MafB clones, and after 48 h at 42°C the majority of the MafB-expressing cells had assumed a macrophage phenotype. By contrast, at the same time points, no such morphological changes were observed in the control cells (Figure 5B). To verify that these phenotypic changes were indeed due to macrophage differentiation, we analyzed the cells by FACS with the monoclonal antibodies 61.8, which labels myeloblast but not macrophages (Beug et al., 1987), and K1, which in the myeloid lineage is specific for macrophages but not present on myeloblasts (Kaspers et al., 1993). As shown in Figure 5C, both MafB-expressing and control myeloblasts grown at 35°C were positive for 61.8 and negative for K1. After 48 h at 42°C, however, the MafB-expressing cells had down-regulated the 61.8 antigen and the majority were positive for K1. By contrast, in the control cells, only small changes were observed. When these changes in surface marker expression were monitored over a period of 4 days after release of the differentiation block, MafB-expressing cells already showed strong K1 expression after 48 h and rapidly down-regulated the myeloblast antigen 61.8, resulting in an almost complete shut down after 96 h. By contrast, in the control cells, even after 96 h, only a small proportion of cells had lost myeloblast marker expression or had induced K1 expression (see Figure 6). The analysis of a total of >20 individual MafB-expressing clones in several separate experiments consistently revealed rapid changes in phenotype and surface marker expression after temperature shift. Together, these results indicate that MafB stimulates macrophage differentiation in ts21 myeloblasts after release of their differentiation block.
Fig. 6. Time course for the expression of the macrophage antigen K1 and the myeloblast antigen 61.8 after temperature shift to 42°C of ts21 control clones (–––) and ts21-MafB clones (------). The average of antigen-positive cells from four ts21-MafB clones and three ts21 control clones is shown. Error bars indicate the SEM.
MafB-induced macrophages are functional phagocytes
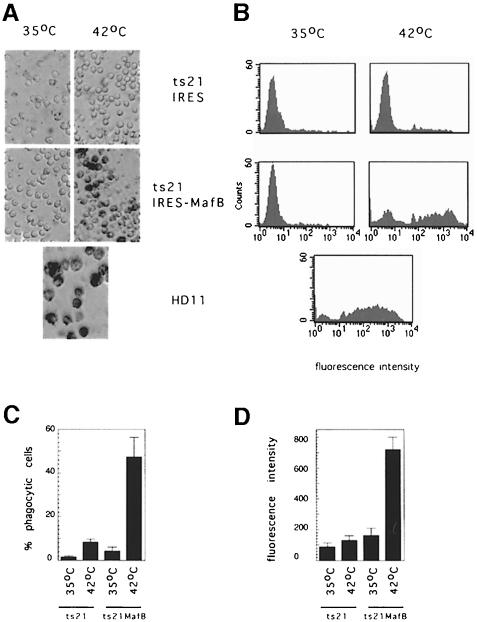
To verify that MafB induced the full gene expression program of macrophages rather than selectively affecting the regulation of a set of marker genes, we tested whether the cells differentiated from the MafB-expressing myeloblasts exhibited functional characteristics of macrophages. One hallmark of macrophage function is the ability to phagocytose foreign objects. To monitor this function, we incubated MafB-expressing and control cells with colored latex beads and observed the amount of beads internalized after 2 h. As shown in Figure 7A, the MafB-expressing cells at 42°C but not the control cells show a high degree of phagocytosis, comparable to that observed in the macrophage cell line HD11 under the same conditions. To quantify phagocytosis and use this function as a measure of the degree of differentiation, we incubated control, MafB-expressing and HD11 cells with fluorescent phycoerythrin-conjugated beads for 2 h. FACS analysis of the cells confirmed that the MafB-induced macrophage clones were highly phagocytic, at a level comparable to the macrophage cell line HD11 (Figure 7B). As shown in Figure 7C and D, both the percentage of fluorescence-positive cells, as a measure of the total number of phagocytic cells, and the mean fluorescence intensity of positive cells, as a measure of the average number of beads internalized per cell, were highly elevated in the ts21-MafB cells at 42°C. By contrast, in the control condition, only a small percentage of cells were phagocytic and to a much lesser degree. In summary, these results indicate that MafB expression induced the differentiation of ts21 myeloblasts to functional phagocytes.
Fig. 7. Phagocytic capacity of ts21-MafB and ts21 control clones. (A) Photomicrograph of representative ts21-MafB and ts21 control clones incubated with blue latex beads for 2 h after culture at 35°C or 48 h at 42°C. HD11 macrophages are shown as a positive control. (B) FACScan analysis of representative ts21-MafB and ts21 control clones at 35°C or 48 h after temperature shift to 42°C incubated with phycoerythrin (PE)-conjugated latex beads for 2 h. HD11 macrophages are shown as a positive control. (C) Percentage of fluorescence-positive cells, indicating the proportion of phagocytic cells, using the assay shown in (B). (D) Mean fluorescent intensity of PE-positive cells as a measure of the average phagocytic activity per cell, using the assay shown in (B). For each condition, the average of four clones each is shown in (C) and (D). Error bars indicate the SEM.
MafB-induced macrophages mount a response to LPS activation
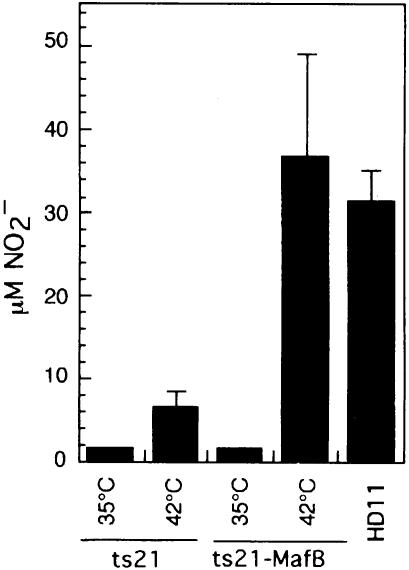
As a further criterion for the full repertoire of macrophage function, we analyzed the ability of MafB-induced macrophages to respond to a challenge with lipopolysaccharide (LPS). When macrophages encounter bacteria, they mount a rapid bacteriotoxic response that includes the release of nitric oxide (NO). In cell culture, the bacterial challenge can be mimicked by stimulation with the bacterial cell wall component LPS, and the degree of the macrophage response can be quantified by measuring the concentration of nitrite in the medium as a direct indicator of NO released by the cells. To analyze the response to LPS, we incubated an equal number of cells from control and MafB-expressing clones at 35 or 42°C, stimulated them with LPS after 48 h and measured the NO2– concentration in the medium 18 h later. As a positive control, HD11 macrophages were analyzed for NO production under the same conditions. As shown in Figure 8, the ts21-MafB cells at 42°C mounted a strong response resulting in an NO2– concentration of 35 µM in the medium, even slightly higher than the concentration of 32 µM NO2– observed for HD11 cells. By comparison, the ts21 control clones at 42°C showed a dramatically lower, yet detectable, response to LPS (6.5 µM NO2–). Thus also by an additional functional criterion, MafB expression in myeloblasts stimulates the macrophage-specific differentiation program.
Fig. 8. NO production by LPS-stimulated cells from ts21 and ts21-MafB clones. Equal number of cells were incubated at 35 or 42°C and stimulated with LPS after 48 h. NO release was measured 18 h later as NO2– concentration in the medium. Bars show the average data from four clones. NO production of LPS-stimulated HD11 macrophages is shown as a positive control (average of two experiments). Error bars indicate the SEM.
Expression of a dominant-negative MafB mutant in hematopoietic progenitors inhibits myeloid colony formation
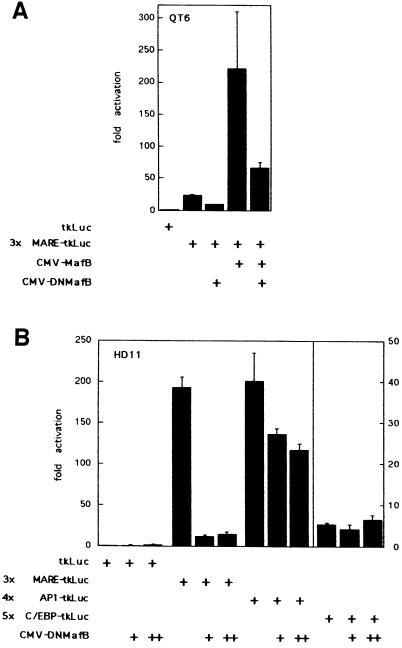
To determine whether MafB is not only capable of stimulating myeloid differentiation but whether its function is also required in this process, it was necessary to inactivate endogenous MafB activity. Our experimental approach was to generate a dominant-negative MafB allele by constructing an N-terminally truncated molecule (DNMafB) containing the DNA-binding and leucine zipper (bZip) domain but devoid of effector functions (see Figure 2). This type of mutant is able to bind to itself and to full-length MafB via its leucine zipper (Kataoka et al., 1994) and should thus act as a competitive inhibitor by preventing access of the endogenous MafB protein to its natural target genes. As shown in Figure 9A, transient co-transfections in QT6 cells confirmed that DNMafB could indeed inhibit the transactivation potential of full-length MafB by ∼3.5-fold on a synthetic reporter containing three MAREs. To confirm that DNMafB could also inhibit endogenous MafB activity, we transfected HD11 macrophages, which express high levels of MafB (Sieweke et al., 1996). As shown in Figure 9B, in these cells DNMafB repressed the 3×MARE reporter up to 15-fold. To analyze the specificity of the dominant-negative construct, we also transfected reporter constructs containing multimerized binding sites for other bZip factors expressed in the myeloid lineage. As shown in Figure 9B, DNMafB repressed an AP-1 reporter construct only minimally (1.7-fold), and a C/EBP reporter not at all. This indicated that DNMafB indeed inhibits endogenous MafB activity but does not significantly interfere with AP-1 or C/EBP proteins.
Fig. 9. Repression of MafB transactivation potential by DNMafB. (A) QT6 cells were co-transfected with 0.2 µg of a luciferase reporter construct containing three multimerized Maf-responsive elements (3×MARE-tkLuc) together with 0.5 µg of CMV promoter expression constructs for MafB and DNMafB, as indicated. (B) HD11 macrophages were co-transfected with 0.5 µg of luciferase reporter constructs containing multimerized response elements for either Maf (3×MARE-tkLuc), AP-1 (4×AP1-tkLuc) or C/EBP proteins (5×C/EBP-tkLuc) and with no, 0.5 µg (+) or 1 µg (++) of DNMafB constructs as indicated. Luciferase activities are expressed as fold activation over activity of a reporter containing no response elements (tkLuc). Assays were performed in duplicate and normalized to β-galactosidase activity derived from a co-transfected LTR-lacZ vector. Bars indicate the SEM. Results are representative of at least two independent experiments.
To assess its biological effects, we inserted DNMafB into the ts21E26-based retroviral vector (Figure 2) and infected embryonic hematopoietic progenitor cells with this virus construct as well as with ts21-MafB and the ts21 control vector. The cells were again plated in semisolid medium for 2 weeks and the phenotype of the developing colonies was then determined by FACS with antibodies against the MEP marker MEP21 and the myeloid marker Myl51/2. Figure 10A shows representative FACS profiles from a pooled colony assay of each condition. In the ts21-DNMafB assays, almost all cells were positive for MEP21 and only a few myeloid cells could be detected. As observed before, control cells expressed both myeloid and MEP markers, whereas the vast majority of ts21-MafB-infected cells were myeloid. Compared with the control, expression of DNMafB thus resulted in the development of a significantly reduced number of myeloid colonies, whereas MafB expression led to strongly increased myeloid colony formation, as reflected in the corresponding myeloid to MEP cell ratio (Figure 10B). Together, these results indicate that the repression of endogenous MafB activity inhibits myeloid colony formation in ts21E26-transformed hematopoietic progenitors.
A dominant-negative MafB mutant inhibits macrophage differentiation
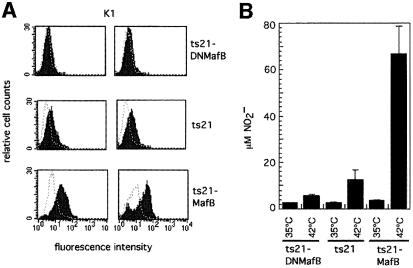
We observed in the previous assays that a small proportion of the ts21 myeloblast control cells could differentiate into macrophages, albeit at a dramatically lower rate and with significantly slower kinetics than the MafB clones (see Figures 6 and 8). Together with the fact that endogenous MafB is up-regulated in ts21 control cells shifted to 42°C (Sieweke et al., 1996), this suggested that MafB expression is not only sufficient but also is required for monocytic differentiation. To analyze whether the inhibition of endogenous MafB by DNMafB in these clones would compromise their ability to differentiate further into macrophages after release of the differentiation block, we isolated a number of myeloid colonies from ts21-DNMafB-infected cells. The expanded clones were positive for the myeloid markers Myl51/2, 61.8 and 22.3 but negative for the multipotent progenitor marker MEP21 and expressed DNMafB (data not shown). We then shifted two individual clones each from ts21-DNMafB-, ts21 control- and ts21-MafB-infected cells to 42°C for an extended period of 6 days to allow for a higher proportion of the control cells to develop into macrophages. Monitoring the expression of the macrophage-specific K1 antigen, we observed a high proportion of K1-positive macrophages in the MafB-expressing cells, but also weakly K1-positive cells in the ts21 control clones. By contrast, ts21-DNMafB clones were completely K1 negative (Figure 11A). No effect on the expression of the 61.8 myeloblast antigen was detectable. The repression of macrophage development by the dominant-negative allele was confirmed further when assaying the presence of macrophages by their ability to respond to LPS. Here we observed a strong release of NO in the medium of activated MafB-expressing cells and a 6-fold weaker response in the control clones. By contrast, in the DNMafB-expressing clones, the NO response was reduced further to almost background levels (Figure 11B). Together, these results indicate that the inhibition of the endogenous MafB also prevents the differentiation of ts21 myeloblasts into macrophages.
Fig. 11. Effect of dominant-negative MafB (DNMafB) on macrophage differentiation. (A) Myeloblast colonies from ts21-DNMafB-, ts21- and ts21-MafB-infected cells were expanded at 35°C and then shifted for 6 days to 42°C. FACS profiles for the macrophage antigen K1 from two individual clones each are shown. The dotted line overlay indicates the staining with an isotype-matched control monoclonal antibody against an intracellular antigen. Note that the ts21 profiles show weakly K1-positive cells but ts21-DNMafB clones are completely negative for K1 expression. (B) NO production by LPS-stimulated cells from ts21-DNMafB, ts21 and ts21-MafB clones. Equal numbers of cells were incubated at 35 or 42°C and stimulated with LPS after 48 h. NO release was measured 18 h later as NO2– concentration in the medium. The average data from three to five clones are shown, with error bars indicating the SEM.
MafB function in macrophage differentiation does not involve PU.1
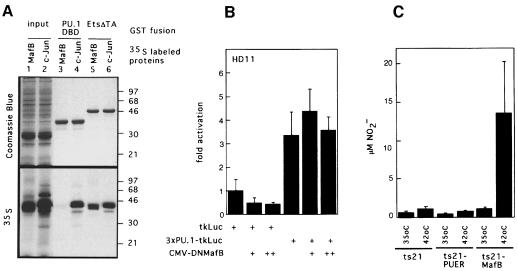
We previously reported a direct interaction of MafB with Ets-1 (Sieweke et al., 1996). Since experiments in knockout mice have shown that in the absence of the Ets factor PU.1 no mature macrophages develop, we investigated whether the effect of MafB in macrophage differentiation involved a physical or functional interaction with PU.1. Using biochemical interaction experiments with GST fusion proteins of the DNA-binding domains of Ets-1 or PU.1, we observed that in vitro translated, [35S]methionine-labeled MafB interacts with the DNA-binding domain of Ets-1 but not with that of PU.1 (Figure 12A). In the same assay, the bZip factor c-Jun bound strongly to both Ets-1 and PU.1, as reported before (Bassuk and Leiden, 1995). This indicated that MafB interacts specifically with Ets-1 but not PU.1.
Fig. 12. Analysis of physical and functional interaction of PU.1 with MafB in macrophage differentiation. (A) Analysis of MafB interaction with Ets-1 and PU.1 in GST pull-down assays. In vitro translated [35S]methionine-labeled MafB (lane 1) or c-Jun (lane 2) were incubated with affinity matrix-bound GST fusion proteins of the PU.1 DNA-binding domain (PU.1-DBD, lanes 3 and 4) or the C-terminal portion of Ets-1 including its DNA-binding domain (Ets-ΔTA, lanes 5 and 6), washed, resuspended in SDS sample buffer and analyzed by SDS–PAGE. Coomassie Blue staining and autoradiography of the same gel are shown. Molecular weight markers (in kDa) are indicated on the right. (B) Effect of DNMafB on PU.1 activity in HD11 macrophages. HD11 macrophages were co-transfected with 0.5 µg of a luciferase reporter construct containing three multimerized PU.1 response elements (3×PU.1-tkLuc), with no, 0.5 µg (+) or 1 µg (++) of DNMafB constructs as indicated. Luciferase activities are expressed as fold activation over activity of a reporter containing no response elements (tkLuc). The assay was performed in duplicate and normalized to β-galactosidase activity derived from a co-transfected LTR-lacZ vector. Bars indicate the SEM. Results are representative of three independent experiments. (C) NO production by temperature-shifted ts21 myeloblasts expressing PU.1 or MafB. Equal numbers of cells from ts21, ts21-PUER and ts21-MafB clones were incubated at 35 or 42°C in the presence of 1 µM β-estradiol and stimulated with LPS after 48 h. NO release was measured 18 h later as NO2– concentration in the medium. The average data from four clones each are shown, with error bars indicating the SEM.
Despite the lack of direct physical interaction, we further investigated whether the inhibition of macrophage differentiation by the dominant-negative MafB allele could be due to an indirect repressive effect on PU.1 activity. For this purpose, we transfected a PU.1 reporter construct into HD11 macrophages in the absence or presence of increasing amounts of DNMafB. As shown in Figure 12B, PU.1 activity in macrophages was not affected by DNMafB.
Since PU.1, like MafB, favors myeloid commitment in multipotent progenitors (Nerlov and Graf, 1998), we wanted to determine whether PU.1 could also drive the later steps of monocyte/macrophage differentiation in myeloblasts. Therefore, we expanded ts21-myeloblast clones expressing an estrogen receptor hormone-binding domain fusion of PU.1 (PUER). This PU.1 construct has been shown to be inducible upon β-estradiol addition to the medium (Nerlov and Graf, 1998). Neither at 35°C nor at 42°C did we observe macrophage differentiation in the ts21-PUER clones after β-estradiol addition to the medium. By contrast, also under these conditions, ts21-MafB clones differentiated into macrophages at 42°C. The presence of macrophages was quantified by measuring the amount of NO released into the medium after LPS stimulation. As shown in Figure 12C, only in ts21-MafB clones shifted to 42°C, but not in ts21 control or ts21-PUER clones, was a significant amount of NO detected.
Together, our results indicate that the role of MafB at later stages of myelomonocytic differentiation is clearly distinct from and independent of that of PU.1.
Discussion
In the present study, we have shown that the up-regulation of MafB expression during myelomonocytic differentiation reflects an essential role in monocyte/macrophage specification. We demonstrate that overexpression of MafB favors myeloid colony formation in hematopoietic progenitors and macrophage differentiation in myeloblasts transformed by ts21E26 virus, whereas a dominant-negative MafB allele inhibits these processes. By contrast, overexpression of PU.1 in this system does not induce macrophage differentiation of myeloblasts. Together, our results provide evidence that MafB is both necessary and, within the myeloid context, sufficient for the induction of monocytic differentiation.
MafB selectively stimulates monocyte/macrophage differentiation
We have used the ts21E26 system to study the role of MafB in myeloid differentiation. Because the myb-ets oncogene of the virus increases the self-renewal capacity of hematopoietic progenitor cells without abolishing their differentiation potential (Graf et al., 1992; McNagny and Graf, 1996), it was possible to use transformed primary cells, a distinct advantage over cell line systems immortalized by unknown genetic changes. Since we observed that MafB binds to cellular Ets-1 (Sieweke et al., 1996) but not to the viral Ets protein encoded by E26 (our unpublished results), a direct cross-talk between MafB and the Myb-Ets oncoprotein is unlikely, making the system a useful tool for our purposes.
In the hematopoietic system, proliferation and differentiation typically are linked phenomena in that cells become growth arrested during terminal differentiation. Several lines of evidence indicate, however, that MafB does not influence differentiation indirectly through an effect on cellular growth but is a bona fide selector gene of monocyte/macrophage differentiation. Thus the inhibition of proliferation is not sufficient to induce macrophage differentiation in ts21E26 myeloblasts (Beug et al., 1987), and additional signals appears to be required for this process. Our observations indicate that this is provided by the induction of MafB. Consistent with this, bone marrow-derived ts21E26 myeloblasts, used in previous studies (Beug et al., 1984, 1987; Frykberg et al., 1988), express higher endogenous levels of MafB (our unpublished observation) and differentiate more efficiently than the yolk sac-derived control clones used here, yet still with dramatically slower kinetics and to a lower degree than the ts21-MafB clones. Together with the observed inhibitory effect of the dominant-negative MafB allele, this indicates that the levels of MafB activity determine the degree of macrophage differentiation.
Most importantly, however, the differentiation-promoting stimulus of MafB is cell type specific. Even though ts21E26-transformed myeloblast have not lost the intrinsic potential to differentiate to granulocytes (Frampton et al., 1996), we did not observe any granulocytic differentiation in the ts21-MafB clones (our unpublished observation). Furthermore, we have reported previously that MafB inhibits the differentiation of erythroid cells without affecting their proliferation (Sieweke et al., 1996). Taken together, these observations indicate that MafB is a specific and direct selector gene for monocytic differentiation.
Dual function of MafB in myeloid differentiation
In this study, we have shown that the expression of MafB in ts21E26-transformed multipotent progenitors leads to their preferential differentiation towards myeloid cells as indicated by the increased formation of myeloblast colonies at the expense of multipotent progenitor-type colonies. This suggests that MafB is not only required for late stages of monocytic differentiation but also plays a role in myeloid lineage commitment of multipotent progenitor cells. Our observations could be explained either by the repression of the progenitor program or by the activation of myeloid differentiation in the precursor cells. The two mechanisms may not be mutually exclusive and could occur simultaneously under physiological conditions. In fact, our previous results suggest that one important function of MafB during monocytic differentiation is the repression of genes that are incompatible with the monocytic program. Thus, MafB can repress genes required for erythroid differentiation, such as the transferrin receptor (TfR), through the direct interaction and repression of Ets-1 (Sieweke et al., 1996). Consistent with this observation, the increase of MafB expression during myeloid differentiation inversely correlates with the successive down-regulation of the TfR gene (Omary et al., 1980; Sieff et al., 1982). It has also been reported that Ets-1 in conjunction with c-Myb activates the early myeloid gene CD13 (Shapiro, 1995) and that the transactivation activity of this complex can be inhibited by c-Maf (Hedge et al., 1998), a Maf family protein related to MafB. It is therefore possible that MafB-repressed Ets-1 targets also include genes specific for multipotent progenitors and early myeloid cells that have to be shut down as differentiation proceeds successively towards monocytes.
MafB cannot only act as an inhibitor but can also transactivate promoters containing Maf consensus-binding sites (MAREs; Kataoka et al., 1994; this study). We have shown here that dominant-negative MafB (DNMafB) acts as a competitive inhibitor of transactivation for exogenously transfected MafB as well as for the endogenous factor in macrophages, whereas the activity of other endogenous bZip proteins expressed in myeloid cells, such as AP-1 and C/EBP proteins, or PU.1 is not affected significantly. In addition, from a broad panel of AP-1 and Maf proteins tested, the MafB bZip domain has been shown previously to only dimerize with itself, c-Maf and Fos proteins, but not with MafF, MafG, MafF or Jun proteins (Kataoka et al., 1994, 1995; Matsushima-Hibiya et al., 1998). Since we could not detect c-Maf expression in our system (unpublished results), DNMafB appears to target specifically MafB homodimers or MafB/Fos heterodimers. Thus the inhibition of both myeloid colony formation and macrophage differentiation of myeloblasts by DNMafB indicates that the transactivation of direct MafB target genes is important for myeloid differentiation. Although the relevant target genes that are activated by MafB during the myeloid differentiation process remain to be identified, our observations further support the concept that transcription factors can be both activators and repressors, depending on the promoter context and the presence of particular interaction partners (Sieweke and Graf, 1998). Regardless of the precise molecular mechanism involved, our results indicate that MafB cannot only repress erythroid differentiation but can also actively promote monocytic differentiation. This suggests that MafB acts as a dual function selector gene that can both activate the correct gene expression program for monocyte/macrophage differentiation and inhibit that of other lineages.
Integration of MafB with other myeloid transcription factors
Several other transcription factors besides MafB have been implicated in early and late stages of myeloid differentiation (Tenen et al., 1997). Thus both PU.1 and C/EBPβ can induce myeloid lineage commitment in multipotent progenitors (Nerlov and Graf, 1998; Nerlov et al., 1998). Together with our finding that MafB also plays a role in early myeloid differentiation, these observations suggest a partial functional overlap between the three proteins in myeloid lineage commitment. Clear differences and distinct functions, however, become apparent at later stages of myeloid differentiation. PU.1 knockout mice lack both mature granulocytes and macrophages (Scott et al., 1994, 1997; McKercher et al., 1996). Consistent with this, in semisolid culture, fetal or neonatal liver cells from these mice give rise only to myeloid colonies at reduced frequency, containing predominantly cells of immature phenotype. Upon stimulation with myeloid cytokines, cells resembling direct granulocytic and monocytic precursors could be obtained, but the late stages of differentiation were still blocked in both of these lineages (DeKoter et al., 1998; Henkel et al., 1999). Despite the requirement of PU.1 for macrophage differentiation, it is not sufficient for this process. Although PU.1 can induce myeloid lineage commitment efficiently in multipotent progenitors (Nerlov and Graf, 1998), we have shown in this study that, in contrast to MafB, it cannot drive monocyte/macrophage differentiation in myeloblasts. PU.1–/– myeloid precursors may thus already express an altered complement of other myeloid transcription factors, that precludes further differentiation to mature macrophages.
C/EBPβ knockout mice show only late functional defects in macrophages (Screpanti et al., 1995; Tanaka et al., 1995), suggesting that this factor is required later than PU.1 during myelomonocytic differentiation. Furthermore, we have observed that the macrophage-specific expression of MafB depends, at least in part, on the combinatorial action of C/EBPβ and SP1 or SP3 proteins (L.M.Kelly, M.H.Sieweke, T.Graf and C.Nerlov, submitted), suggesting that C/EBPβ acts upstream of MafB. However, both the partially overlapping functions of the three proteins during early myeloid differentiation, and the expression and function of PU.1 and C/EBPβ in hematopoietic cells other than the myelomonocytic lineage argue against a simple linear hierarchy of the three proteins. Thus PU.1 is also required during the differentiation of lymphoid cells (Scott et al., 1994, 1997; McKercher et al., 1996), and gain-of-function experiments suggest a principal role for C/EBPβ in eosinophilic differentiation (Nerlov et al., 1998). By contrast, MafB appears to specify monocyte/macrophage differentiation selectively.
Other transcription factors have also been shown to play a role in later stages of myeloid differentiation, yet MafB appears to be unique in specifically inducing the monocyte/macrophage differentiation program. Thus C/EBPα has been implicated in directing neutrophil and eosinophil granulocyte differentiation, based on both gene inactivation studies in mice (Zhang et al., 1997) and gain-of-function experiments (Nerlov et al., 1998; Radomska et al., 1998). The zinc finger protein Egr-1 has been shown in different cell lines to either block granulocytic differentiation (Nguyen et al., 1993) or stimulate monocytic differentiation (Krishnaraju et al., 1998), but Egr-1–/– mice did not show any defect in the macrophage lineage (Lee et al., 1996). Stat3 appears to mediate the anti-proliferative effect of IL-6 in M1 cell differentiation (Nakajima et al., 1996). However, it is also essential for IL-6-mediated anti-apoptotic signals in T cells (Takeda et al., 1998), indicating that it is involved in the mediation of cytokine signals rather than in the autonomous specification of macrophage differentiation. The homeobox gene GBX2 induces a monoblastic phenotype in myeloblasts transformed by the avian E26 retrovirus through activation of the cMGF gene, an avian homolog of granulocyte colony-stimulating factor (G-CSF)/IL-6. However, it is also expressed in non-myeloid hematopoietic cells and absent from macrophages. Consistent with the latter observation, GBX2-induced monoblasts do not proceed to develop a macrophage phenotype unless treated with TPA (Kowenz-Leutz et al., 1997).
In summary, these observations are consistent with a model where lineage choice is not determined by individual master regulators but by the combinatorial action of several transcription factors, involving both functional overlaps and the formation of protein complexes (Sieweke and Graf, 1998). MafB appears to assume an essential and unique function for the specification of monocytic differentiation within the network of myeloid transcription factors. It will be interesting to decipher which synergistic and antagonistic interactions with other factors are required for this role at different stages of differentiation.
Materials and methods
Virus constructs
A fragment coding for the HA epitope sequence MGYPYDVPDYASH was inserted in front of the start codon of the full-length quail MafB cDNA (Sieweke et al., 1996) by PCR to generate BSSK-HAMafB. A fragment containing the HA-MafB fusion was then inserted into the cytomegalovirus (CMV) promoter-driven expression construct RC/CMV (InVitrogen) or behind an encephalomyocarditis (EMC) virus IRES element in the E26 proviral vector pMI3-IRES (Rossi et al., 1996), which had been modified to contain the ts21 mutation of E26 (Frykberg et al., 1988). The dominant-negative construct was generated by religating a blunted HincII–BstXI digest of BSSK-HAMafB, resulting in the fusion of amino acids 195–311 of MafB to its N-terminus and the HA tag. This construct was then inserted into RC/CMV or the ts21 version of pMI3-IRES as above. The presence of the ts21 point mutation and the coding region junctions were verified by sequencing. The ts21-PUER construct is the ts21 version of the described pMI3-IRES-PUER (Nerlov and Graf, 1998). Standard methods of molecular cloning were used to generate the constructs (Sambrook et al., 1989).
Cell culture and colony assays
Pools from 2-day chick embryo blastoderms (stages 11–13) were prepared from an outbred flock of white Leghorn (Lohmann and Co., Cuxhaven, Germany). The target cells for E26 transformation are contained in the yolk sac surrounding the embryo (T.Graf, unpublished results). These cells were infected by co-cultivation with the avian retroviral packaging cell line Q2bn (Stoker and Bissell, 1988), transfected with the proviral DNA constructs as described (Rossi et al., 1996) and plated in methylcellulose at 37°C. Two weeks after infection, the colonies from each assay were pooled and phenotyped by indirect immunofluorescence (IIF) on a Becton-Dickinson FACScan (Graf et al., 1992). The MEP21, MYL51/2, cIa (chicken MHC II), 4M12, 22.3, 61.8, JS3, JS4 and K1 monoclonal antibodies have been described (Kornfeld et al., 1983; Ewert et al., 1984; Schmidt et al., 1986; Beug et al., 1987; McNagny et al., 1992; Kaspers et al., 1993). For analysis of individual myeloblast clones infected with the different ts21 virus constructs, colonies were picked and expanded at 35°C in the presence of recombinant cMGF, an avian IL-6/G-CSF homolog (Leutz et al., 1989). Equal numbers of cells were seeded at 35 or 42°C and analyzed by IIF or functional macrophage assays as indicated. Within one assay, only clones of the same age were compared. The origin of the HD11 macrophage cell line has been described (Beug et al., 1979). [3H]Thymidine incorporation assays for growth factor dependence were performed as described (Rossi et al., 1996), except that recombinant, rather than crude, cMGF was used (Leutz et al., 1989) and cells were pulse labeled with 10 rather than 5 µCi of [3H]thymidine.
Western blots
A total of 1 × 106 cells were lysed in 20 µl sample buffer (50 mM Tris–HCl pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.02% bromophenol blue). Lysates were separated by 12.5% SDS–PAGE (Harlow and Lane, 1988) and transferred to an Immobilon P membrane (Millipore) with a Bio-Rad dry blotter at 2.5 mA/cm2 using 25 mM Tris-base, 192 mM glycine, 20% (v/v) methanol as transfer buffer. Membranes were blocked in Tris-buffered saline (TBS) with 4% (w/v) dry milk overnight and stained with the mouse monoclonal antibody 12CA5-I against the HA epitope (Berkeley Antibody Co.) and a secondary goat anti-mouse peroxidase-coupled antibody (Amersham). Antibody incubations were for 1 h in TBS with 4% (w/v) dry milk followed by three washes of 15 min in TBS with 0.2% Triton X-100. For detection, we used the ECL chemiluminescent peroxidase substrate kit from Amersham.
Assays for macrophage function
To analyze LPS activation of macrophages, 5 × 105 cells per 24-well dish were cultured for 48 h at 35 or 42°C and then stimulated with 10 µg/ml LPS from Escherichia coli O26:B6 (Sigma) for 18 h. As a direct indication of the NO released in response to the stimulation, we determined the NO2– concentration in the medium using Griess-Ilosvay reagent (Merck). A 250 µl aliquot of cell culture supernatant was cleared by centrifugation, incubated with 750 µl of reagent for 5 min in the dark and the optical absorption was measured at 546 nm. For the phagocytosis assay, the cells were incubated with a suspension of 0.6 µm diameter blue carboxylated latex beads (Seradyn) or phycoerythrin-conjugated 1 µm diameter latex sulfate beads (Molecular probes) for 2 h at a 0.05% dilution followed by three extensive medium washes. Cells incubated with fluorescent beads were analyzed on a Becton-Dickinson FACScan.
Transient transfection assays
QT6 cells (Moscovici et al., 1977) were seeded at 2.5 × 105 cells/35 mm plate, cultured overnight and transfected by the calcium phosphate co-precipitation procedure (Graham and van der Eb, 1973), using 0.2 µg of reporter and 0.5 µg of expression plasmids per sample. HD11 cells were seeded at 2.5 × 105 cells/35 mm plate, cultured overnight and transfected with Lipofectamin™ or Lipofectin™ (both Gibco-BRL), following the instructions of the supplier and using 8 µl of reagent, 0.5 µg of reporter and 0.5–1 µg of DNMafB plasmid as indicated. Cell lysates were prepared 36 (HD11) or 48 h (QT6) after transfection and assayed for luciferase activity as described (deWet et al., 1987). Transfection efficiency was normalized by assaying for β-galactosidase activity expressed from 0.5 µg of co-transfected RSV-β-gal plasmid (Bonnerot et al., 1987) as described (Herbomel et al., 1984). Each data point was obtained by averaging of duplicate samples and is representative of a set of at least two independent experiments. MafB and DNMafB were expressed from the CMV promoter in RC/CMV (Invitrogen). The Maf-responsive luciferase reporter contained three multimerized MAREs as defined in Kataoka et al. (1994). The PU.1 reporter construct has been described (Nerlov and Graf, 1998); the AP-1 and C/EBP reporters correspond to the described 4×A and 5×C reporters, respectively, that contain multimerized elements from the cMGF promoter (Sterneck et al., 1992). All reporter constructs were generated in the same TK81luc background (Lim et al., 1992).
Protein interaction assays
The protocols both for the interaction assays with GST fusion proteins and the plasmids GST–EtsΔTA, GST–PU.1-DBD and for MafB in vitro translation have been described (Sieweke et al., 1996, 1998). Full-length chicken c-Jun was in vitro translated from a T7 promoter in a bluescriptSK– ™ vector.
Acknowledgments
Acknowledgements
We thank Dr Claus Nerlov for the E26ts21-PUER plasmid, Dr Achim Leutz for the AP-1 and C/EBP reporter constructs, Dr Dirk Bohmann for the c-Jun plasmid, Dr Ludwig Englmeier for critical reading of the manuscript, and Corrine Bezier for expert help with the graphic design of the figures.
References
- Bassuk A.G. and Leiden,J.M. (1995) A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity, 3, 223–237. [DOI] [PubMed] [Google Scholar]
- Beug H., von Kirchbach,A., Döderlein,G., Conscience,J.-F. and Graf,T. (1979) Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell, 18, 375–390. [DOI] [PubMed] [Google Scholar]
- Beug H., Leutz,A., Kahn,P. and Graf,T. (1984) Ts mutants of E26 leukemia virus allow transformed myeloblasts, but not erythroblasts or fibroblasts to differentiate at the nonpermissive temperature. Cell, 39, 579–588. [DOI] [PubMed] [Google Scholar]
- Beug H., Blundell,P. and Graf,T. (1987) Reversibility of differentiation and proliferative capacity in avian myelomonocytic cells transformed by tsE26 leukemia virus. Genes Dev., 1, 277–286. [DOI] [PubMed] [Google Scholar]
- Blank V. and Andrews,N.C. (1997) The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci., 22, 437–441. [DOI] [PubMed] [Google Scholar]
- Bonnerot C., Rocancourt,D., Briand,P., Grimber,G. and Nicolas,J.F. (1987) A β-galactosidase hybrid protein targeted to nuclei as a marker for developmental studies. Proc. Natl Acad. Sci. USA, 84, 6795–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKoter R.P., Walsh,J.C. and Singh,H. (1998) PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J., 17, 4456–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deWet J.R., Wood,K.V., DeLuca,M., Helinski,D.R. and Subramani,S. (1987) Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann A., Grapin-Botton,A., Kelly,L., Graf,T., Le Douarin,N.M. and Sieweke,M. (1997) The expression pattern of the mafB/kr gene in birds and mice reveals that the kreisler phenotype does not represent a null mutant. Mech Dev., 65, 111–122. [DOI] [PubMed] [Google Scholar]
- Ewert D.L., Munchus,M.S., Chen,C.L. and Cooper,M.D. (1984) Analysis of structural properties and cellular distribution of avian Ia antigen by using monoclonal antibody to monomorphic determinants. J. Immunol., 132, 2524–2530. [PubMed] [Google Scholar]
- Frampton J., Ramqvist,T. and Graf,T. (1996) v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev., 10, 2720–2731. [DOI] [PubMed] [Google Scholar]
- Frykberg L., Metz,T., Brady,G., Introna,M., Beug,H., Vennstrom,B. and Graf,T. (1988) A point mutation in the DNA binding domain of the v-myb oncogene of E26 virus confers temperature sensitivity for transformation of myelomonocytic cells. Oncogene Res., 3, 313–322. [PubMed] [Google Scholar]
- Gordon S., Clarke,S., Greaves,D. and Doyle,A. (1995) Molecular immunobiology of macrophages: recent progress. Curr. Opin. Immunol., 7, 24–33. [DOI] [PubMed] [Google Scholar]
- Graf T., McNagny,K., Brady,G. and Frampton,J. (1992) Chicken ‘erythroid’ cells transformed by the Gag-Myb-Ets-encoding E26 leukemia virus are multipotent. Cell, 70, 201–213. [DOI] [PubMed] [Google Scholar]
- Graham F.L. and van der Eb,A.J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology, 52, 456. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) In Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hedge S.P., Kumar,A., Kurschner,C. and Shapiro,L.H. (1998) c-Maf interacts with c-Myb to regulate transcription of an early myeloid gene during differentiation. Mol. Cell. Biol., 18, 2729–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel G.W., McKercher,S.R., Leenen,P.J. and Maki,R.A. (1999) Commitment to the monocytic lineage occurs in the absence of the transcription factor PU.1. Blood, 93, 2849–2858. [PubMed] [Google Scholar]
- Herbomel P., Bourachot,B. and Yaniv,M. (1984) Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell, 39, 653–662. [DOI] [PubMed] [Google Scholar]
- Ho I.C., Hodge,M.R., Rooney,J.W. and Glimcher,L.H. (1996) The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell, 85, 973–983. [DOI] [PubMed] [Google Scholar]
- Kaspers B., Lillehoj,H.S. and Lillehoj,E.P. (1993) Chicken macrophages and thrombocytes share a common cell surface antigen defined by a monoclonal antibody. Vet. Immunol. Immunopathol., 36, 333–346. [DOI] [PubMed] [Google Scholar]
- Kataoka K., Fujiwara,K.T., Noda,M. and Nishizawa,M. (1994) MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol. Cell. Biol., 14, 7581–7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K., Igarashi,K., Itoh,K., Fujiwara,K.T., Noda,M., Yamamoto,M. and Nishizawa,M. (1995) Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell. Biol., 15, 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.I., Ho,I.C., Grusby,M.J. and Glimcher,L.H. (1999) The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity, 10, 745–751. [DOI] [PubMed] [Google Scholar]
- Kornfeld S., Beug,H., Doederlein,G. and Graf,T. (1983) Detection of avian hematopoietic cell surface antigens with monoclonal antibodies to myeloid cells. Their distribution on normal and leukemic cells of various lineages. Exp. Cell Res., 143, 383–394. [DOI] [PubMed] [Google Scholar]
- Kotkow K.J. and Orkin,S.H. (1996) Complexity of the erythroid transcription factor NF-E2 as revealed by gene targeting of the mouse p18 NF-E2 locus. Proc. Natl Acad. Sci. USA, 93, 3514–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowenz-Leutz E., Herr,P., Niss,K. and Leutz,A. (1997) The homeobox gene GBX2, a target of the myb oncogene, mediates autocrine growth and monocyte differentiation. Cell, 91, 185–195. [DOI] [PubMed] [Google Scholar]
- Krishnaraju K., Hoffman,B. and Liebermann,D.A. (1998) The zinc finger transcription factor Egr-1 activates macrophage differentiation in M1 myeloblastic leukemia cells. Blood, 92, 1957–1966. [PubMed] [Google Scholar]
- Lee S.L., Wang,Y. and Milbrandt,J. (1996) Unimpaired macrophage differentiation and activation in mice lacking the zinc finger transplantation factor NGFI-A (EGR1). Mol. Cell. Biol., 16, 4566–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutz A. et al. (1989) Molecular cloning of the chicken myelomonocytic growth factor. EMBO J., 8, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim F., Kraut,N., Framptom,J. and Graf,T. (1992) DNA binding by c-Ets-1, but not v-Ets, is repressed by an intramolecular mechanism. EMBO J., 11, 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look A.T. (1997) Oncogenic transcription factors in the human acute leukemias. Science, 278, 1059–1064. [DOI] [PubMed] [Google Scholar]
- Matsushima-Hibiya Y., Nishi,S. and Sakai,M. (1998) Rat maf-related factors: the specificities of DNA binding and heterodimer formation. Biochem. Biophys. Res. Commun., 245, 412–418. [DOI] [PubMed] [Google Scholar]
- McKercher S.R. et al. (1996) Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J., 15, 5647–5658. [PMC free article] [PubMed] [Google Scholar]
- McNagny K.M. and Graf,T. (1996) Acute avian leukemia viruses as tools to study hematopoietic cell differentiation. Curr. Top. Microbiol. Immunol., 212, 143–162. [DOI] [PubMed] [Google Scholar]
- McNagny K.M., Lim,F., Grieser,S. and Graf,T. (1992) Cell surface proteins of chicken hematopoietic progenitors, thrombocytes and eosinophils detected by novel monoclonal antibodies. Leukemia, 6, 975–984. [PubMed] [Google Scholar]
- Moscovici C., Moscovici,M.G., Jiminez,H., Lai,M.M.C., Hayman,M.J. and Vogt,P.T. (1977) Contineous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell, 11, 95–103. [DOI] [PubMed] [Google Scholar]
- Motohashi H., Shavit,J.A., Igarashi,K., Yamamoto,M. and Engel,J.D. (1997) The world according to Maf. Nucleic Acids Res., 25, 2953–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K. et al. (1996) A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J., 15, 3651–3658. [PMC free article] [PubMed] [Google Scholar]
- Nerlov C. and Graf,T. (1998) PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev., 12, 2403–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C., McNagny,K.M., Döderlein,G., Kowenz-Leutz,E. and Graf,T. (1998) Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes Dev., 12, 2413–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.Q., Hoffman Liebermann,B. and Liebermann,D.A. (1993) The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell, 72, 197–209. [DOI] [PubMed] [Google Scholar]
- Omary M.B., Trowbridge,I.S. and Minowada,J. (1980) Human cell-surface glycoprotein with unusual properties. Nature, 286, 888–891. [DOI] [PubMed] [Google Scholar]
- Orkin S.H. (1996) Development of the hematopoietic system. Curr. Opin. Genet. Dev., 6, 597–602. [DOI] [PubMed] [Google Scholar]
- Radomska H.S., Huettner,C.S., Zhang,P., Cheng,T., Scadden,D.T. and Tenen,D.G. (1998) CCAAT/enhancer binding protein α is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol., 18, 4301–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F., McNagny,M., Smith,G., Frampton,J. and Graf,T. (1996) Lineage commitment of transformed haematopoietic progenitors is determined by the level of PKC activity. EMBO J., 15, 1894–1901. [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) In Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmidt J.A., Marshall,J., Hayman,M.J., Doderlein,G. and Beug,H. (1986) Monoclonal antibodies to novel erythroid differentiation antigens reveal specific effects of oncogenes on the leukaemic cell phenotype. Leuk. Res., 10, 257–272. [DOI] [PubMed] [Google Scholar]
- Scott E.W., Simon,M.C., Anastasi,J. and Singh,H. (1994) Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science, 265, 1573–1577. [DOI] [PubMed] [Google Scholar]
- Scott E.W., Fisher,R.C., Olson,M.C., Kehrli,E.W., Simon,M.C. and Singh,H. (1997) PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity, 6, 437–447. [DOI] [PubMed] [Google Scholar]
- Screpanti I. et al. (1995) Lymphoproliferative disorder and imbalanced T-helper response in C/EBP β-deficient mice. EMBO J., 14, 1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L.H. (1995) Myb and Ets proteins cooperate to transactivate an early myeloid gene. J. Biol. Chem., 270, 8763–8771. [DOI] [PubMed] [Google Scholar]
- Shivdasani R.A., Rosenblatt,M.F., Zucker-Franklin,D., Jackson,C.W., Hunt,P., Saris,C.J. and Orkin,S.H. (1995) Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell, 81, 695–704. [DOI] [PubMed] [Google Scholar]
- Sieff C., Bicknell,D., Caine,G., Robinson,J., Lam,G. and Greaves,M.F. (1982) Changes in cell surface antigen expression during hemopoietic differentiation. Blood, 60, 703–713. [PubMed] [Google Scholar]
- Sieweke M.H. and Graf,T. (1998) A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev., 8, 545–551. [DOI] [PubMed] [Google Scholar]
- Sieweke M.H., Tekotte,H., Frampton,J. and Graf,T. (1996) MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell, 85, 49–60. [DOI] [PubMed] [Google Scholar]
- Sieweke M.H., Tekotte,H., Jarosch,U. and Graf,T. (1998) Cooperative interaction of Ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. EMBO J., 17, 1728–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterneck E., Muller,C., Katz,S. and Leutz,A. (1992) Autocrine growth induced by kinase type oncogenes in myeloid cells requires AP-1 and NF-M, a myeloid specific, C/EBP-like factor. EMBO J., 11, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker A.W. and Bissell,M.J. (1988) Development of avian sarcoma and leukosis virus based vector packaging cell lines. J. Virol., 62, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Kaisho,T., Yoshida,N., Takeda,J., Kishimoto,T. and Akira,S. (1998) Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol., 161, 4652–4660. [PubMed] [Google Scholar]
- Tanaka T. et al. (1995) Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell, 80, 353–361. [DOI] [PubMed] [Google Scholar]
- Tenen D.G., Hromas,R., Licht,J.D. and Zhang,D.E. (1997) Transcription factors, normal myeloid development and leukemia. Blood, 90, 489–519. [PubMed] [Google Scholar]
- Zhang D.E., Zhang,P., Wang,N.D., Hetherington,C.J., Darlington,G.J. and Tenen,D.G. (1997) Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl Acad. Sci. USA, 94, 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]