Abstract
Epithelial tight junctions regulate paracellular diffusion and restrict the intermixing of apical and basolateral plasma membrane components. We now identify a Y-box transcription factor, ZONAB (ZO-1-associated nucleic acid-binding protein), that binds to the SH3 domain of ZO-1, a submembrane protein of tight junctions. ZONAB localizes to the nucleus and at tight junctions, and binds to sequences of specific promoters containing an inverted CCAAT box. In reporter assays, ZONAB and ZO-1 functionally interact in the regulation of the ErbB-2 promoter in a cell density-dependent manner. In stably transfected overexpressing cells, ZO-1 and ZONAB control expression of endogenous ErbB-2 and function in the regulation of paracellular permeability. These data indicate that tight junctions directly participate in the control of gene expression and suggest that they function in the regulation of epithelial cell differentiation.
Keywords: epithelia/MAGUK/tight junctions/Y-box/ZONAB
Introduction
Polarized epithelial cells interact with one another via a specialized intercellular junctional complex that is formed by gap junctions, desmosomes, adherens and tight junctions (TJs) (Farquhar and Palade, 1963). At this junctional complex localize signal transduction molecules and several proteins that are products of oncogenes or tumor suppressor genes, suggesting an involvement of the junctional complex in the control of cell growth and differentiation (Kirkpatrick and Peifer, 1995; Tsukita et al., 1999). Such an involvement has indeed been described for adherens junctions: loss of cadherin-based cell–cell adhesion correlates with tissue disorganization and increase of tumor invasiveness (Birchmeier, 1995; Gumbiner, 1996). β-catenin/armadillo, originally identified as a cytoskeletal linker in adherens junctions, can associate with the product of the APC gene and binds to the transcription factor TCF/LEF-1 (Rubinfeld et al., 1993; Su et al., 1993; Behrens et al., 1996; Huber et al., 1996; Molenaar et al., 1996). β-catenin functions in the transduction of transmembrane signals that regulate cell differentiation and fate (Barth et al., 1997; Bienz, 1998). Gene expression is also modulated by integrin-mediated cell–matrix interactions (Howe et al., 1998).
In vertebrates, TJs regulate diffusion of molecules through the paracellular pathway and restrict apical/basolateral intramembrane diffusion of lipids (Cereijido, 1991; Balda and Matter, 1998; Madara, 1998). Indirect evidence suggests that TJs may also participate in the control of cell growth and differentiation: (i) the N-terminal domain of ZO-1, a peripheral membrane protein of TJs (Stevenson et al., 1986), is homologous to the Drosophila tumor suppressor discs large A (dlg A) (Tsukita et al., 1993; Willott et al., 1993; Woods and Bryant, 1993); (ii) two TJ proteins, ZO-1 and symplekin, have also been reported to localize to the nucleus (Gottardi et al., 1996; Keon et al., 1996); (iii) ZO-1 can interact with the catenin complex under certain conditions, such as MDCK cells lacking intercellular junctions (Rajasekaran et al., 1996; Itoh et al., 1997); and (iv) ZO-1 staining is often reduced in breast cancer cells, and polymorphic markers flanking the ZO-1 gene showed loss of heterozygosity in 23% of breast cancers analyzed (Hoover et al., 1998). Nevertheless, a direct involvement of ZO-1 or any other tight junction-associated protein in the regulation of gene expression has not been demonstrated thus far.
Since the SH3 domain of Drosophila dlg A is critical for its signaling and tumor suppressor function (Hough et al., 1997), we looked for proteins binding to the SH3 domain of ZO-1. We have previously described the interacting protein kinase ZAK (Balda et al., 1996b). We now identify a second interacting protein, ZO-1-associated nucleic acid-binding protein (ZONAB), a Y-box transcription factor that localizes to the nucleus and TJs. ZONAB binds to specific inverted CCAAT box-containing sequences found in the promoters of the genes coding for ErbB-2 and several cell cycle regulators. In vivo, ZO-1 and ZONAB functionally interact in the regulation of ErbB-2 expression. These data indicate that ZO-1 and ZONAB are part of a novel signaling pathway linking TJs to the regulation of gene expression.
Results
Identification of ZONAB
To search for proteins interacting with the SH3 domain of ZO-1, we screened an MDCK expression library with a GST fusion protein containing the SH3 and the third PDZ domain of ZO-1 (GST–PDZ3–SH3). Since the protein kinase ZAK binds to and phosphorylates this fusion protein (Balda et al., 1996b), we used fusion protein radiolabeled by ZAK as a probe.
Screening of 450 000 clones resulted in one positive clone (PS19) that remained positive in further rounds of subcloning. Bacterial extracts were then probed with phosphorylated GST–PDZ3–SH3 in an overlay assay to confirm that the GST fusion protein interacts with the fusion protein of the library clone. Figure 1A shows a signal obtained with the original clone PS19 (lane 1) and with subclones derived from it (lane 2), but not with a clone containing another cDNA producing a fusion protein with a similar apparent molecular weight (lane 3).
Fig. 1. Identification of ZONAB. (A) Induced cultures of clones isolated from expression libraries were extracted, fractionated by SDS–PAGE and transferred to nitrocellulose. The membrane was probed with 32P-labeled GST–PDZ3–SH3 fusion protein and exposed to X-ray film. Shown are lanes corresponding to the original clone positive in the expression screening (lane 1), a subclone derived from it (lane 2) and a negative clone expressing an unrelated fusion protein (lane 3). (B) The cDNA sequence of ZONAB-A and the amino acid sequence of the open reading frame. The in-frame upstream stop codon is printed in italic (DDBJ/EMBL/GenBank accession No. AF171061). (C) The cDNA and corresponding amino acid sequence of an additional domain found in some ZONAB transcripts by RT–PCR (DDBJ/EMBL/GenBank accession No. AF171062). (D) Low confluent MDCK cells were grown for 2 days of culture, and expression of ZONAB was detected by immunoblotting with an affinity-purified polyclonal antibody against the C-terminus (lanes 1–3), ZONAB-A (lane 4) or ZONAB-B (lane 5) (wt, wild-type MDCK cells; T:ZONAB-A and -B, MDCK cells stably overexpressing ZONAB-A or -B). (E) The domain structures of ZONAB-A and -B. The domains are also marked in (A). N-PD, N-terminal proline-rich domain; CSD, cold-shock domain; AD, alternative domain; RP-CD, arginine- and proline-rich conserved domain; C-PD, C-terminal proline-rich domain.
DNA sequencing revealed that the library plasmid isolated from subclone PS19/3 possessed a 1.8 kb insert containing a long open reading frame and additional 3′ sequences (Figure 1B). By 5′-RACE, an additional 74 nucleotides were isolated that contained an in-frame upstream stop codon, indicating that the originally isolated plasmid carried the entire coding sequence. Since the encoded protein binds nucleic acids (see below), we named this protein ZONAB.
We next raised polyclonal antibodies against peptides containing the predicted N- and C-termini, as well as against a GST fusion protein containing the entire coding sequence. All three antibodies were affinity purified and tested on immunoblots of total MDCK cell extracts. Figure 1D shows that the anti-C-terminus antibody recognized two bands migrating at 47 and 55 kDa (lane 1). The other two antibodies recognized bands migrating at the same molecular weights (not shown). To test whether one of the two bands corresponds to the cloned cDNA, we transferred the coding sequence into a eukaryotic expression plasmid and stably transfected MDCK cells. Immunoblotting of cell extracts demonstrated that the transfected cells exhibited an enhanced expression of the lower molecular weight form (lane 2), indicating that the protein migrating at 47 kDa corresponds to the cloned cDNA.
Because all antibodies recognized the same doublet, we used RT–PCR to test whether MDCK cells express different isoforms of ZONAB. Only primer sets specific for the 3′-half resulted in amplification of two products. Sequencing demonstrated that they only differed in a 204 nucleotide in-frame insertion in the longer product (Figure 1C). Stable expression of the longer cDNA in MDCK cells resulted in an enhanced expression of the higher molecular weight band (Figure 1D, lane 3), indicating that the protein migrating at 55 kDa corresponds to the longer isoform. To confirm this, we generated isoform-specific antibodies. Immunoblotting showed that an antibody against a peptide representing the sequence of the insertion site of the alternative domain recognized only ZONAB-A (Figure 1D, lane 4) and that an antibody against a peptide derived from the alternative domain recognized only ZONAB-B. MDCK cells thus express two ZONAB isoforms: a shorter ZONAB-A and a longer ZONAB-B isoform (Figure 1E).
We searched the DDBJ/EMBL/GenBank and SwissProt databases for homologous sequences and found that ZONAB is homologous to Y-box transcription factors (Matsumoto and Wolffe, 1998). Using ClustalW, we aligned the amino acid sequences of ZONAB-A and -B with the sequences of the most similar Y-box transcription factors obtained from SwissProt. This revealed identities of 75–88% with a group of transcription factors containing human and mouse DNA-binding protein A (DbpA) isoforms, rat YB-2 and rat RYB A, and identities of 50–60% with a group of proteins containing chicken respiratory syncytial virus (RSV) enhancer-binding factor II, human and mouse DNA-binding protein B (DbpB) and YB-1. All of these proteins share a central cold-shock domain as well as a domain rich in arginine and proline residues, and exhibit the greatest differences from each other at the N- and C-termini.
Interaction between endogenous ZONAB and ZO-1 in MDCK cells
We next tested whether the interaction between ZO-1 and ZONAB also occurs in vivo in wild-type MDCK cells. MDCK cell extracts were immunoprecipitated with anti-ZO-1 and anti-ZONAB antibodies, and precipitation of the two proteins was monitored by immunoblotting.
Figure 2A shows the result of such an experiment with wild-type MDCK cells grown to 20 or 80% confluency. ZO-1 was precipitated efficiently by the anti-ZO-1 antibody (lanes 1 and 2) but was absent when no antibody was conjugated to the beads (lanes 3 and 4). ZO-1 was also detected when ZONAB was immunoprecipitated (lanes 5 and 6), indicating that ZO-1 co-immunoprecipitated with ZONAB. Both isoforms of ZONAB could be detected in immunoprecipitates generated with the monoclonal anti-ZO-1 antibody (lanes 9 and 10) and the anti-ZONAB antibody (lanes 13 and 14), but not when no primary antibody was conjugated to the beads (lanes 11 and 12). Only faint signals from the heavy chain were detected when ZO-1-immunoprecipitates were tested without adding primary antibody (lanes 15 and 16). Thus, ZONAB and ZO-1 co-immunoprecipitate with each other, indicating that the two proteins interact in wild-type MDCK cells.
Fig. 2. Interaction between ZO-1 and ZONAB in wild-type MDCK cells. (A) Cleared lysates of MDCK cells that were at a confluency of ∼20 or 80% were loaded on Sepharose beads with covalently conjugated antibody R40.76 against ZO-1, anti-GST–ZONAB antibody or no antibody (----). The immunoprecipitates were analyzed by immunoblotting with antibodies against ZO-1 and ZONAB (anti-C-terminus), or with secondary antibody only (----). (B) Purified GST fusion proteins containing either the third PDZ domain of ZO-1 (GST–PDZ3), the third PDZ and the SH3 domain (GST–PDZ3–SH3) or the SH3 domain only (GST–SH3) were bound to glutathione–agarose and incubated with diluted recombinant histidine-tagged ZONAB-A. Pull-down of ZONAB-A was tested by immunoblotting. The scheme illustrates the domain structure of ZO-1 with the three PDZ domains, the SH3 domain and the guanylate kinase homology domain (GUK).
For the screening of the expression library, we had used a fusion protein containing the SH3 and the third PDZ domain of ZO-1. To test which of the two domains interacts with ZONAB, we performed a pull-down assay in which recombinant purified ZONAB-A was incubated with glutathione beads carrying either GST alone or GST fusion proteins containing different parts of ZO-1. Precipitation of ZONAB was monitored by immunoblotting. Figure 2B shows that amounts of ZONAB above background could be detected in precipitates generated with GST fusion proteins containing the third PDZ and the SH3 domain (lane 3) or the SH3 domain only (lane 4), but not when only the third PDZ domain was linked to GST (lane 2). This indicates that the interaction is mediated by the SH3 domain. Phosphorylation of the fusion protein by ZAK was not required to pull down ZONAB (not shown).
ZONAB is expressed in the nucleus and co-localizes with ZO-1 at intercellular junctions
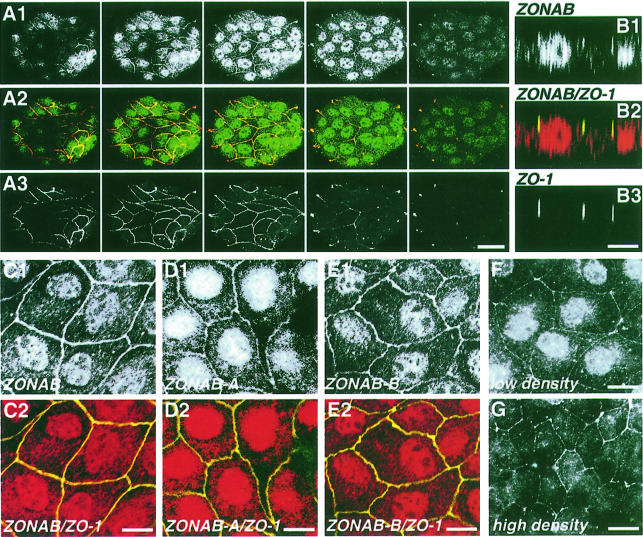
We next determined the subcellular localization of ZONAB by immunofluorescence. In serial confocal xy-sections of wild-type MDCK cells labeled with the anti-ZONAB antibody, ZONAB was detected in the nucleus and at cell–cell contacts (Figure 3A). Co-labeling with an anti-ZO-1 antibody revealed that lateral ZONAB co-localizes with ZO-1. This was supported further by xz-sections showing that the two proteins co-localize at the apical end of the lateral membrane (Figure 3B). Thus, endogenous ZONAB is expressed in the nucleus and is associated with intercellular junctions. Overexpression of ZONAB-A or -B did not change the observed distribution (now shown). Labeling of cells with the isoform-specific antibodies demonstrated that both isoforms co-localize with ZO-1 and are expressed in the nucleus (Figure 3C, D and E).
Fig. 3. Subcellular distribution of ZONAB in subconfluent wild-type and transfected MDCK cells. Wild-type MDCK cells (A–F, low density; G, high density) were fixed and permeabilized with Triton X-100/ethanol. (A) Serial confocal sections taken at 1 µm intervals along the apical–basal axis of an island of cells stained with anti-GST–ZONAB (A1) and anti-ZO-1 (A3) antibodies (bar, 40 µm). (B) Confocal xz-sections of cells stained with the anti-N-terminus ZONAB (B1) and anti-ZO-1 (B3) antibodies. Confocal sections of cells stained with the anti-N-terminus ZONAB (C), anti-ZONAB-A (D) or anti-ZONAB-B (E) antibody; also shown are overlays with ZO-1 staining (C2, D2 and E2) (bars, 10 µm). (F and G) Epifluorescence images of samples labeled with the anti-GST–ZONAB antibody. Note that cells grown to a high density are taller; hence, the focus has been set to the basal end of the junctions to have the nucleus in the same focal plane. This causes the irregular appearance of the junctional staining (bar, 10 µm). Similar ZONAB and ZO-1 staining patterns were observed in cells fixed with methanol and in stable cell lines overexpressing ZONAB.
Previously, nuclear localization of ZO-1 had been observed in growing cells (Gottardi et al., 1996). We could not detect specific nuclear staining of ZO-1 in our cells using poly- and monoclonal antibodies, different fixation and permeabilization conditions (including those described by Gottardi et al., 1996), and MDCK cells stably overexpressing human ZO-1. Even though we cannot exclude that small amounts of ZO-1 are in the nucleus in our cells, our results suggest that most of the ZONAB molecules bound to ZO-1 are associated with TJs.
Overexpressed ZONAB and ZO-1 regulate paracellular permeability
To analyze the functions of ZONAB and ZO-1 in vivo, we produced stable MDCK cells lines overexpressing ZONAB-A and ZO-1 either alone or together. This resulted in cell lines overexpressing ZONAB-A at least 10-fold, and ZO-1 by 2- to 5-fold (based on immunoblots, see below).
First, we analyzed transepithelial electrical resistance (TER) of different clones overexpressing ZO-1 (four clones), ZONAB (two clones) or ZO-1 and ZONAB (six clones). We did not observe significant changes in TER (two experiments in triplicate; determinations after 4, 6 and 8 days of plating, and with or without overnight induction with sodium butyrate to induce higher expression levels). Thus, the transfected cells were still able to form functional TJs.
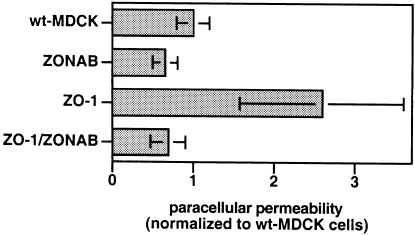
Since TJs are not simple diffusion barriers but are semipermeable, we used the same filter cultures to determine paracellular flux of [3H]mannitol. Figure 4 shows that only clones overexpressing ZO-1 alone and not clones overexpressing ZONAB, or ZO-1 and ZONAB exhibited a 2- to 3-fold increase in paracellular flux. This indicates that the relative expression levels of ZONAB and ZO-1 regulate paracellular permeability.
Fig. 4. Regulation of paracellular permeability by ZO-1 and ZONAB. Wild-type and stably transfected MDCK cells overexpressingZONAB-A, ZO-1 or both proteins were plated on filters. TER was measured every day and was not significantly affected by transfection of the two proteins (not shown). After 8 days, the cells were incubated overnight with sodium butyrate to induce higher expression levels of the transfected proteins; this also did not result in differences in TER (not shown). Paracellular flux of [3H]mannitol was then measured during 3 h and normalized to the values obtained from wild-type MDCK cells. Shown are means ± 1 SD of two experiments with four clones overexpressing ZO-1, two clones overexpressing ZONAB, and six clones stably overexpressing both proteins. The absolute numbers were 582 ± 116 c.p.m. for wild-type cells, 378 ± 87 c.p.m. for ZONAB-overexpressing cells, 1513 ± 582 c.p.m. for ZO-1-overexpressing cells and 401 ± 122 c.p.m. for the double transfectants.
ZONAB interacts with promoter sequences of genes coding for ErbB-2 and cell cycle regulatory proteins
ZONAB is homologous to Y-box transcription factors. Since Y-box transcription factors can bind to inverted CCAAT box sequences (Sommerville and Ladomery, 1996; Matsumoto and Wolffe, 1998), we tested with gel shift assays whether recombinant ZONAB-A interacts with double-stranded phosphorylated oligonucleotides containing such a sequence. We started with oligonucleotides derived from the promoters of the ErbB-2 gene and the multidrug resistance gene-1 (MDR1); both are known to interact with Y-box transcription factors in vitro (Sakura et al., 1988; Ohga et al., 1998).
Figure 5 shows that GST–ZONAB, but not GST alone, caused a shift of the radiolabeled ErbB-2 promoter oligonucleotides and that the amount of shifted oligonucleotides depended on the amount of protein added. If GST–ZONAB was added to single-stranded oligonucleotides, little binding could be detected (not shown). Binding of radiolabeled ErbB-2 promoter oligonucleotides could be quenched specifically with unlabeled ErbB-2 promoter oligonucleotides.
Fig. 5. ZONAB interacts with specific promoter sequences in vitro. (A) Phosphorylated double-stranded oligonucleotides derived from the ErbB-2 promoter were incubated with different concentrations of GST or GST–ZONAB (2.0, 1.0 and 0.5 µg) in the absence or presence of increasing amounts of unlabeled double-stranded oligonucleotides derived from the ErbB-2 or MDR-1 promoter. Complex formation was analyzed by non-denaturing gel electrophoresis and autoradiography. (B) Binding of ZONAB to inverted CCAAT boxes with different flanking sequences was assayed by direct binding and by the ability to compete for binding of phosphorylated ErbB-2 double-stranded oligonucleotides. The sequences are derived from DDBJ/EMBL/GenBank entries of the genes for ErbB-2 (J05264), p21WAF1/CIP1 (U50603), p27KIP1 (U77914), proliferating cell nuclear antigen (J05614; PCNA), thymidine kinase (M13643; TK-A and TK-B), MDR-1 (L07624), DNA polymerase α (M64481; DPOL), 70 kDa heat-shock protein (M19865; hsp70) and topoisomerase IIa (X66794; TopoII).
We next tested Y-box-containing double-stranded oligonucleotides derived from different promoters, identified by searching DDBJ/EMBL/GenBank, for binding to ZONAB-A using the gel shift assay (direct binding and competition with radiolabeled ErbB-2 oligonucleotides). We classified the sequences according to the observed results (Figure 5B). Of all the sequences tested, ZONAB bound with the highest affinity to the double-stranded oligonucleotides derived from the ErbB-2 promoter. The next best oligonucleotides were derived from the promoters of the cell cycle regulators p21WAF1/CIP1, p27KIP1 and proliferating cell nuclear antigen. Similar binding was observed with double-stranded oligonucleotides containing either one of the two inverted CCAAT boxes of the thymidine kinase promoter. Because ZONAB does not bind to all sequences containing inverted CCAAT boxes, it is clear that the flanking sequences are also important for ZONAB binding. A consensus sequence for ZONAB binding is not yet evident from the analyzed oligonucleotides.
Regulation of the ErbB-2 promoter by ZONAB and ZO-1
The in vitro interaction between ZONAB and specific promoter sequences suggests that ZONAB is involved in the regulation of specific genes. To test this, we performed reporter gene assays using a full-length ErbB-2 promoter driving the expression of firefly luciferase (Ertl and Gill, 1993). We mixed this plasmid with a control reporter plasmid containing a cytomegalovirus promoter driving the expression of renilla luciferase, and expression vectors containing the ZO-1 and/or the ZONAB-A cDNAs. These plasmids were then co-transfected into MDCK cells plated at low or high density, and the expression of the luciferases was assayed with a dual luciferase assay. Expression of renilla luciferase was used to normalize the values obtained from firefly luciferase.
Figure 6A shows that co-transfection of expression vector containing the ZONAB cDNA did not affect the activity of the ErbB-2 promoter in subconfluent MDCK cells. In contrast, co-transfection of the ZO-1 plasmid resulted in a concentration-dependent increase in firefly luciferase expression. This effect could be counteracted by co-transfecting ZONAB, indicating a functional interaction between ZO-1 and ZONAB in the regulation of ErbB-2 promoter activity.
Fig. 6. Regulation of the ErbB-2 promoter by ZO-1 and ZONAB. Low confluent (A, C, E and G) and confluent (B and F) MDCK cells were co-transfected with a plasmid carrying a fragment of the human ErbB-2 promoter driving the expression of firefly luciferase and a control plasmid carrying a CMV promoter driving the expression of renilla luciferase. (A–C) the luciferase reporter plasmid contained the entire 3.6 kb ErbB-2 promoter fragment. (D–G) The reporter plasmids contained a shorter fragment of the ErbB-2 promoter with a functional (D: ErbB-2: S) or mutated ZONAB-binding site (D: ErbB-2: S-D, deleted; ErbB-2: S-A, substitution of the inverted CCAAT box). The reporter plasmids were transfected together with empty pCB6 and/or the indicated amounts of vectors resulting in the expression of ZO-1, ZONAB-A, antisense ZONAB RNA (ZONABas), an HA-tagged fragment of ZO-1 containing the third PDZ and the SH3 domain (HA-PDZ3–SH3), or the SH3 domain only (HA-SH3). Empty pCB6 was used to adjust the total DNA concentrations to the same value in all samples. Expression of the luciferases was assayed with a dual-luciferase assay system. (A–C) The ratios obtained (firefly luciferase divided by renilla luciferase) are expressed as a percentage of those obtained by co-transfecting empty pCB6. (D–F) Stimulation or inhibition, respectively, was calculated with respect to control transfections performed with each reporter plasmid in the presence of empty pCB6. The values represent means ± 1 SD of at least two independent experiments performed in duplicate.
Since ZO-1 and cytoplasmic ZONAB are associated with intercellular junctions, we tested whether the effects of ZO-1 and ZONAB on the activity of the ErbB-2 promoter depend on the cell density. Figure 6B shows that in confluent MDCK cells, co-transfection of ZONAB inhibited firefly luciferase expression, while overexpression of ZO-1 did not significantly affect ErbB-2 promoter activity. Co-transfection of ZO-1 counteracted the ZONAB effect. Hence, the response to ZO-1 and ZONAB transfection depends on the cell density.
To test whether ZONAB is required for ErbB-2 promoter activity, we co-transfected the reporter plasmids together with a plasmid expressing antisense ZONAB RNA. In stable cell lines expressing ZONAB antisense RNA, the expression levels of ZONAB-A and -B are reduced to a similar extent (by ∼70%, not shown). In subconfluent cells, expression of antisense ZONAB RNA resulted in enhanced luciferase expression (Figure 6C), suggesting that ZONAB may function as an inhibitor.
To test whether the SH3 domain of ZO-1, which is sufficient for ZONAB binding in vitro, is also sufficient for stimulation of the ErbB-2 promoter in subconfluent cells, we co-transfected plasmids coding for hemagglutinin (HA)-tagged truncated versions of ZO-1 that contain the SH3 domain. Figure 6C shows that expression of proteins containing the third PDZ and the SH3 domain of ZO-1 (HA-PDZ3–SH3) or the SH3 domain only (HA-SH3) was sufficient to stimulate the ErbB-2 promoter. Although inclusion of the PDZ3 domain resulted in an enhanced effect, this result indicates that expression of the SH3 domain of ZO-1 is sufficient for ErbB-2 promoter stimulation, as it is for ZONAB binding in vitro.
To analyze whether the ZONAB-binding site in the ErbB-2 promoter is required for the observed responses, we constructed new reporter plasmids (Figure 6D). Plasmid ErbB-2: S contains nucleotides –396 to –150 of the ErbB-2 promoter, which include the ZONAB-binding site as well as the main regulatory elements of the ErbB-2 promoter. In plasmid ErbB-2: S-D, the ZONAB binding was deleted, and in ErbB-2: S-A the inverted CCAAT box was substituted.
We then repeated the reporter assays under the same conditions as above. Luciferase expression from ErbB-2: S was stimulated by the expression of ZONAB antisense RNA in low confluent cells (Figure 6E) and inhibited by the co-transfection of ZONAB-A in confluent cells (Figure 6F). In contrast, the plasmids with a mutated ZONAB-binding site did not respond to co-expression of ZONAB antisense RNA in low confluent cells (Figure 6E) and ZONAB-A in confluent cells (Figure 6F). These data indicate that ZONAB regulates ErbB-2 promoter activity by interacting with the promoter region containing the inverted CCAAT box. Co-transfection of pCB6 HA-SH3 only caused a significant increase in luciferase expression in low confluent cells when a reporter plasmid containing a functional ZONAB-binding site was analyzed (Figure 6F), indicating that ZO-1 indeed regulates ErbB-2 promoter activity via the ZONAB-binding site.
ZO-1 and ZONAB regulate endogenous ErbB-2 expression
We next used the cell lines overexpressing ZO-1 and ZONAB to analyze whether they also regulate the expression of endogenous ErbB-2. Because ZONAB binds to promoters of genes coding for a growth factor co-receptor and cell cycle regulators, we plated cells at low confluency and synchronized them by culturing in 0.1% fetal calf serum (FCS). Half of the samples were then stimulated with 10% FCS, while the other half were harvested directly. Equal amounts of protein of total cell extracts were loaded on SDS–gels, and expression of ZO-1, ZONAB and ErbB-2 was analyzed by immunoblotting.
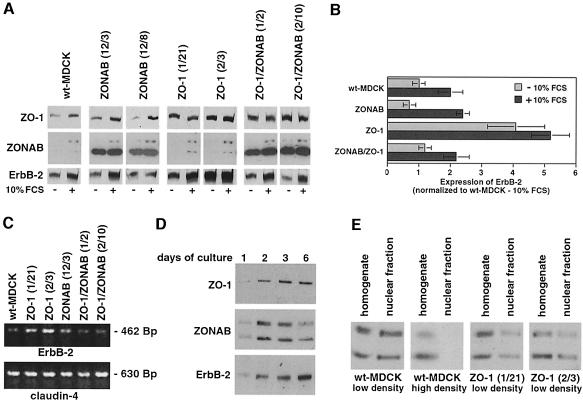
Figure 7A shows that all cells expressed ZO-1 with and without serum stimulation. ZO-1 expression increased upon serum stimulation in wild-type and ZONAB-transfected MDCK cells but not in cells overexpressing ZO-1, where expression of ZO-1 was already high in serum-depleted cells (on average, ZO-1 was overexpressed 4-fold in serum-depleted cells; co-transfection of ZONAB did not affect the overexpression of ZO-1). Wild-type and ZO-1-overexpressing cells expressed little ZONAB in 0.1% serum, but both isoforms were detected in longer exposures than that shown in Figure 7A. Expression of both ZONAB isoforms was induced upon serum stimulation. Overexpression of ZONAB-A alone or together with ZO-1 resulted in large increases in the expression of ZONAB-A but did not affect the amounts of expressed ZONAB-B. ErbB-2 was overexpressed ∼4-fold in serum-depleted clones overexpressing ZO-1 alone but was not affected in clones overexpressing ZONAB-A alone or together with ZO-1 (Figure 7A and B). Comparable results were obtained when the levels of ErbB-2 mRNA were analyzed by RT–PCR (Figure 7C). These data indicate that ZO-1 and ZONAB indeed regulate expression of endogenous ErbB-2.
Fig. 7. Regulation of endogenous ErbB-2 expression by ZO-1 and ZONAB. (A) Wild-type and stably transfected MDCK cells overexpressing ZO-1 and/or ZONAB were synchronized by serum starvation and then collected directly or incubated for 24 h with 10% FCS. Equal amounts of protein were loaded on SDS–gels, and expression of ZO-1, ZONAB and ErbB-2 was quantified by immunoblotting and densitometric scanning. The shown film of the immunoblot with anti-ZONAB antibody is derived from a short exposure to avoid overexposure of lanes derived from cell lines overexpressing ZONAB-A. After longer exposure times, both isoforms of ZONAB were detected in serum-depleted cells without overexpression. (B) The levels of ErbB-2 expression were normalized to the expression in wild-type MDCK cells after serum starvation (three clones of each type of transfection were analyzed in at least two independent experiments). (C) The mRNA levels of ErbB-2 and claudin-4 in cells synchronized by serum starvation were determined by RT–PCR. (D) Total extracts of MDCK cells cultured for 1, 2, 3 and 6 days were separated by SDS–PAGE (equal amounts of protein were loaded in each lane), transferred to nitrocellulose and the expression levels of ZO-1, ZONAB and ErbB-2 were determined by immunoblotting. Cells cultured for 6 days reached full confluency. (E) Nuclear fractions were isolated from wild-type and ZO-1-overexpressing MDCK cells grown to low or high densities. The levels of ZONAB in homogenates and nuclear fractions were determined by immunoblotting.
Our results suggest a model in which ZONAB functions as a repressor of ErbB-2 expression that is sequestered to TJs by ZO-1 binding. Therefore, we compared the expression levels of ZO-1, ZONAB and ErbB-2 in MDCK cells at different densities. Figure 7D shows that ZO-1 levels are low in growing cells and are maximal in confluent cells, while ZONAB expression is high during growth and low in confluent cells. Interestingly, ErbB-2 levels parallel those of ZO-1, supporting the model. This is also supported by fractionation experiments (Figure 7E) and immunofluorescence (Figure 3F and G) showing that the amount of ZONAB detected in the nucleus is high in growing cells (low density) but low in confluent (high density) cultures. Moreover, overexpression of ZO-1 also resulted in reduced amounts of ZONAB-A and -B recovered in the nuclear fraction.
Discussion
We have identified a transcription factor that binds to the SH3 domain of the tight junction protein ZO-1. In MDCK cells, the two proteins functionally interact to modulate the expression of the erbB-2 proto-oncogene, indicating a direct linkage between TJs and the regulation of gene expression.
ZONAB is homologous to Y-box proteins, a group of transcription factors containing a central cold-shock domain followed by a domain rich in arginine and proline residues (Sommerville and Ladomery, 1996; Matsumoto and Wolffe, 1998). ZONAB shows most identity with human DbpA, and with rat YB-2 and RYB A. Although one might expect a higher degree of identity between mammalian homologs, it could be that ZONAB is the dog homolog of one of these proteins. This is supported by the finding that human DbpA also binds to the ErbB-2 promoter in vitro (Sakura et al., 1988). Nevertheless, DbpB also interacts with the ErbB-2 promoter but is only 52% identical to ZONAB. None of the previously known Y-box proteins is known to associate with intercellular junctions or other plasma membrane domains.
MDCK cells express two isoforms of ZONAB that differ from one another in a 68 amino acid insertion just after the cold-shock domain in the ZONAB-B isoform. Although both isoforms are expressed at higher levels in growing cells and co-immunoprecipitate with ZO-1 (although with different efficiencies), the insertion may affect the functional properties of ZONAB. Since all of the expression experiments described here testing a function of ZONAB were done with the ZONAB-A isoform, it is not clear whether functional differences indeed exist between the two isoforms. In reporter assays with the ErbB-2 promoter, however, co-transfection of the two ZONAB isoforms gave comparable results (not shown).
ZO-1 and ZONAB interact in the regulation of the paracellular permeability
TJs regulate the diffusion of molecules through the paracellular pathway by acting as semipermeable diffusion barriers (Cereijido, 1991). Measurements of TER and paracellular permeability of MDCK cell monolayers stably overexpressing ZO-1 and/or ZONAB showed that the two proteins functionally interact in the regulation of paracellular permeability. One can think of two possibilities of how overexpression of ZO-1 increases selective paracellular permeability. Since ZO-1 binds directly to occludin (Furuse et al., 1994; Fanning and Anderson, 1998), a transmembrane protein critical for the regulation of paracellular permeability (Matter and Balda, 1999), one possibility is that ZO-1 and ZONAB directly regulate paracellular permeability via occludin. Alternatively, ZO-1 and ZONAB may modulate the expression of proteins involved in paracellular permeability.
Regulation of ErbB-2 expression by ZO-1 and ZONAB
Of all the inverted CCAAT box sequences tested, ZONAB showed strongest binding to that found in the ErbB-2 promoter. In vivo, ZO-1 and ZONAB functionally interact in the regulation of ErbB-2 promoter activity.
The effect of ZO-1 and ZONAB on ErbB-2 promoter activity was found to depend on the confluency of the cells. Overexpression of ZONAB did not affect promoter activity in subconfluent cells that have high levels of endogenous ZONAB, but it inhibited promoter activity in confluent cells that have low levels of endogenous ZONAB. Overexpression of ZO-1 stimulated promoter activity in subconfluent cells but had no effect in confluent cells. Hence, ZONAB could be a repressor that is sequestered to TJs by overexpression of ZO-1, resulting in stimulation of ErbB-2 expression. Such a model is compatible with the activation of the promoter by the expression of antisense ZONAB RNA and by the reduced nuclear expression of ZONAB in confluent and ZO-1-overexpressing cells.
ErbB-2 is a tyrosine kinase co-receptor important for epithelial differentiation and morphogenesis (Alroy and Yarden, 1997; Montesano et al., 1997; Niemann et al., 1998). The fact that ZO-1 and ZONAB functionally interact in the regulation of ErbB-2 expression suggests that the two proteins might be part of a signaling system important for epithelial differentiation and morphogenesis. Such a function of ZO-1 in differentiation is supported by the finding that a functional homolog of ZO-1 in Drosophila, the tamou gene product, is involved in the specification of sensory organ precursor cells (Takahisa et al., 1996).
Taken together, our experiments demonstrate that ZO-1 and the associated Y-box transcription factor ZONAB function in the regulation of expression of the erbB-2 proto-oncogene, indicating the existence of a direct pathway by which tight junctions can modulate the expression of specific genes. Signaling along the ZO-1–ZONAB pathway is itself regulated by the cell density. ZONAB also binds to promoter elements of several cell cycle regulators in gel shift assays. Hence, it is conceivable that tight junctions and the ZO-1– ZONAB pathway also function in the regulation of cell cycle progression.
Materials and methods
Isolation of ZONAB and RT–PCR
Phosphorylated GST–PDZ3–SH3 fusion protein (containing the third PDZ and the SH3 domain of ZO-1) was generated by incubating 200–300 µg of fusion protein bound to glutathione beads with MDCK cell extract (from 143 cm2 confluent MDCK cells) to bind ZAK (Balda et al., 1996b). After washing, 250 µCi of [γ-32P]ATP and kinase buffer were added to allow phosphorylation. The beads were washed five times, and the phosphorylated fusion protein was eluted with 0.3 ml of phosphate-buffered saline (PBS) containing 0.25% SDS.
MDCK expression libraries constructed in pTEX were generously provided by Dr K.K.Stanley (The Heart Research Institute, Sydney, Australia; Herz et al., 1990). The libraries were plated and prepared as described for immunological screenings (Sambrook et al., 1989). The filters were blocked by incubating with PBS containing 5% low fat dry milk and 0.1% Tween-20 (PMT). After 1 h, fresh PMT was added containing 10 µg/ml purified GST, and blocking was continued for another hour. The phosphorylated GST–PDZ3–SH3 fusion protein (1–3 µg/ml) was then added to the same solution. After incubating overnight, the filters were washed with 0.1% Tween-20 in PBS, and bound phosphorylated fusion protein was visualized by autoradiography. Isolated clones were retested with overlays in which total extracts of induced bacterial cultures were probed with phosphorylated fusion protein as described above. The insert of the pTEX vector was transferred to BSSK+ for sequencing. The first 74 nucleotides of the 5′ end of the ZONAB mRNA were obtained by 5′-RACE (Gibco-BRL). The additional domain of ZONAB-B was cloned by RT–PCR using Pfu polymerase (Stratagene) and sets of primers resulting in the amplification of either the N- or the C-terminal half of ZONAB.
RT–PCR for ErbB-2 and claudin-4 expression was done with MMLV reverse transcriptase (Roche Diagnostics) and Taq polymerase (Gibco-BRL) using total RNA as template. For ErbB-2, a 462 bp fragment close to the 3′ end of the coding sequence was amplified. For claudin-4, the entire coding sequence of 630 bp was amplified.
Expression plasmids and transfection
To express ZONAB, cDNAs corresponding to ZONAB-A or -B were cloned into pCB6. For the antisense ZONAB plasmid, the PS19/3 insert was inserted in the antisense orientation into pCB6. To produce GST– and histidine–ZONAB-A in Escherichia coli, the fragment corresponding to the coding sequence was cloned into pGEX-2T and pRSET-A. The pCB6-ZO-1 construct was kindly provided by Drs A.S.Fanning and J.M.Anderson (Yale University, New Haven, CT). The plasmids coding for GST fusion proteins containing different portions of ZO-1 were as described (Balda et al., 1996b). To generate pCB6-HA-PDZ3-SH3 and pCB6-HA-SH3, the corresponding pieces were amplified by PCR and used to replace the occludin sequence in pCB6-HAoccludin (Balda et al., 1996a). MDCK (strain 2) cells were transfected, selected and cloned as previously described (Balda et al., 1996a). Cloned MDCK cells were tested for expression by immunofluorescence and immunoblotting.
Antibodies and immunofluorescence
Rabbit anti-ZONAB antibodies were generated against recombinant GST–ZONAB-A and peptides corresponding to the N- or the C-terminus of ZONAB. Isoform-specific antibodies were generated against a peptide spanning the insertion site of the alternative domain and a peptide representing an internal sequence of the alternative domain. All sera were affinity purified using either the histidine-tagged ZONAB-A fusion protein or the peptide used as an antigen. Monoclonal antibody R40.76 and polyclonal antibody 8040 against ZO-1 were described previously (Anderson et al., 1988; Balda and Anderson, 1993). The antibody against ErbB-2 (C-18) was purchased from Santa Cruz Biotechnology.
For immunofluorescence, cells grown on glass coverslips were fixed and permeabilized with Triton X-100 and 95% ethanol, or with methanol at –20°C without pre-extraction (Balda et al., 1996a). In additional control experiments, the cells were fixed either with ethanol/acetone (Balda et al., 1996a) or with paraformaldehyde and then extracted with Triton X-100 (Gottardi et al., 1996). The samples were incubated with antibodies and mounted as previously described (Balda et al., 1996a).
Analysis of TJ functions
Cells were plated on tissue culture-treated polycarbonate filters (Costar Transwells) with a pore size of 0.4 µm and a diameter of 12 mm. TER and paracellular flux of [3H]mannitol were measured as described (Balda et al., 1996a).
Immunoprecipitations, pull-down assay, immunoblots and subcellular fractionation
Immunoprecipitations were performed at 4°C. Wild-type MDCK cells were harvested and lysed with extraction buffer [PBS containing 1% Triton X-100, 40 µg/ml phenylmethylsulfonyl fluoride (PMSF), 10 mM sodium fluoride, 10 mM pyrophosphate and 1 mM sodium vanadate]. After centrifugation, the supernatants were transferred to tubes containing Sepharose beads with covalently conjugated antibodies. After incubating for 2 h, the beads were washed twice with extraction buffer and once with PBS. Bound proteins were eluted by boiling in SDS–PAGE sample buffer lacking dithiothreitol (DTT) to avoid partial elution of the antibody. The eluates were transferred to fresh tubes, 0.1 M DTT was added and the samples were analyzed by SDS–PAGE and immunoblotting.
For pull-down assays, GST and GST fusion proteins were coupled to glutathione–agarose (Balda et al., 1996b) and incubated with purified histidine-tagged ZONAB-A that had been diluted in 0.5× PBS containing 0.5% Triton X-100 and pre-adsorbed with empty glutathione–agarose. After 2 h, the samples were washed with the same buffer and analyzed by SDS–PAGE and immunoblotting.
For immunoblots of total cell extracts, cells were harvested in cold PBS containing 40 µg/ml PMSF, 5 mM sodium fluoride, 5 mM pyrophosphate and 1 mM sodium vanadate, pelleted and solubilized by boiling in 60 mM Tris (pH 6.8) containing 1% SDS, 10 mM EDTA and 30% glycerol. The protein concentrations were determined by performing a DC protein assay (Bio-Rad). β-mercaptoethanol (5% final concentration) and Bromophenol blue were added, and the samples were separated on 6–20% gradient gels and transferred to nitrocellulose. The above-described antibodies were used to detect endogenous and transfected proteins using horseradish peroxidase (HRP)-conjugated secondary antibodies and the enhanced chemiluminescence detection system (Amersham).
Nuclear fractions were prepared essentially as described (Spector et al., 1998). Briefly, cells were homogenized with a tight fitting glass–Teflon Potter homogenizer in 0.25 M sucrose, 10 mM NaCl, 2 mM MgCl2, 40 µg/ml PMSF, 10 mM Tris, pH 7.5 (the efficiency was checked by phase contrast microscopy and was ∼90% of clean nuclei for all cell lines). The homogenates were centrifuged at 1000 g for 10 min. The nuclear pellets were resuspended in the homogenization buffer, loaded on a 1.8 M sucrose (in 10 mM NaCl, 1.5 mM MgCl2, 10 mM Tris pH 7.5) cushion, and centrifuged for 45 min at 32 000 r.p.m. in a TST55.5 rotor.
Gel mobility assay
The oligonucleotides shown in Figure 5B were used for DNA binding studies. Double-stranded oligonucleotides were labeled with [γ-32P]ATP. Radiolabeled oligonucleotides (1 pmol) were incubated with fusion protein or GST (0.5–2 µg) at 25°C for 60 min in 20 mM Tris–HCl, pH 8, containing 100 mM KCl, 2 mM MgCl2 and 10% glycerol. The mixtures were fractionated on 7% non-denaturing acrylamide gels.
Reporter gene assays
MDCK cells were plated into 12-well dishes at a low (32 000 cells/well) or high (240 000 cells/well) concentration the day before transfection by calcium phosphate precipitation (Matter et al., 1992). The plasmid pXp1 (Ertl and Gill, 1993) containing a 3.6 kb fragment of the human ErbB-2 gene promoter driving the transcription of firefly luciferase was kindly provided by Dr G.N.Gill (University of California, San Diego, CA). An pXb-1-based reporter plasmid containing only the ErbB-2 promoter sequences from –396 to –150 (pXb-1 ErbB-2: S) was generated by PCR. The same strategy was used to generate pXb-1 ErbB-2: S-D, which contains nucleotides –396 to –168 and lacks the inverted CCAAT box, and pXb-1 ErbB-2: S-A, which contains the same region of the promoter as pXb-1 ErbB-2: S but the inverted CCAAT box was substituted (ATTGG to AAAAA). Promoter activities were determined with the dual-luciferase reporter assay system (Promega). The pXp1 plasmids (1.5 µg/well) were co-transfected with the indicated expression vectors and the control plasmid pRL-CMVΔ (0.6 µg/well). pRL-CMVΔ contains the renilla luciferase gene and was produced from pRL-CMV (Promega) by removing the fragment between the NheI and PstI sites, which contains an inverted CCAAT box sequence. The total DNA concentrations of all samples were equalized with empty pCB6. The incubations of the cells with the precipitates were done overnight, and expression of the luciferases was assayed 26–30 h after removing the precipitates.
Acknowledgments
Acknowledgements
We dedicate this work to the memory of Thomas Kreis, our colleague and friend, who tragically died in an airplane crash on September 2, 1998. We thank Drs K.K.Stanley, G.N.Gill, A.S.Fanning, J.M.Anderson and S.Ishii for providing valuable materials, and Dr J.A.Whitney for critical reading of the manuscript. K.M. is a fellow of the START (Swiss Talents in Academic Research and Teaching) program of the Swiss National Science Foundation. This research was supported by the Swiss National Science Foundation and the Canton de Genève.
References
- Alroy I. and Yarden,Y. (1997) The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand–receptor interactions. FEBS Lett., 410, 83–86. [DOI] [PubMed] [Google Scholar]
- Anderson J.M., Stevenson,B.R., Jesaitis,L.A., Goodenough,D.A. and Mooseker,M.S. (1988) Characterization of ZO-1, a protein component of the tight junction from mouse liver and Madin–Darby canine kidney cells. J. Cell Biol., 106, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda M.S. and Anderson,J.M. (1993) Two classes of tight junctions are revealed by ZO-1 isoforms. Am. J. Physiol., 264, C918–C924. [DOI] [PubMed] [Google Scholar]
- Balda M.S. and Matter,K. (1998) Tight junctions. J. Cell Sci., 111, 541–547. [DOI] [PubMed] [Google Scholar]
- Balda M.S., Whitney,J.A., Flores,C., González,S., Cereijido,M. and Matter,K. (1996a) Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical–basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol., 134, 1031–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda M.S., Anderson,J.M. and Matter,K. (1996b) The SH3 domain of the tight junction protein ZO-1 binds to a serine protein kinase that phosphorylates a region C-terminal to this domain. FEBS Lett., 399, 326–332. [DOI] [PubMed] [Google Scholar]
- Barth A.I.M., Näthke,I.S. and Nelson,W.J. (1997) Cadherins, catenins and APC protein: interplay between cytoskeletal complex and signaling pathways. Curr. Opin. Cell Biol., 9, 683–690. [DOI] [PubMed] [Google Scholar]
- Behrens J., von Kries,J.P., Kuhl,M., Bruhn,L., Wedlich,D., Grosschedl,R. and Birchmeier,W. (1996) Functional interaction of β-catenin with the transcription factor LEF-1. Nature, 382, 638–642. [DOI] [PubMed] [Google Scholar]
- Bienz M. (1998) TCF: transcriptional activator or repressor. Curr. Opin. Cell Biol., 10, 366–372. [DOI] [PubMed] [Google Scholar]
- Birchmeier W. (1995) E-cadherin as a tumor (invasion) suppressor gene. BioEssays, 17, 97–99. [DOI] [PubMed] [Google Scholar]
- Cereijido M. (1991) Evolution of ideas on the tight junction. In Cereijido,M. (ed.), Tight Junctions. CRC Press, Inc., Boca Raton, FL, pp. 1–13. [Google Scholar]
- Ertl A.P. and Gill,G.N. (1993) Structural features of the 5′ region of the human erbB-2 gene. Gene, 136, 361–364. [DOI] [PubMed] [Google Scholar]
- Fanning A.S. and Anderson,J.M. (1998) The tight junction protein ZO-1 establishes a link between the membrane protein occludin and the actin cytoskeleton. J. Biol. Chem., 273, 29745–29753. [DOI] [PubMed] [Google Scholar]
- Farquhar M.G. and Palade,G.E. (1963) Junctional complexes in various epithelia. J. Cell Biol., 17, 375–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M., Itoh,M., Hirase,T., Nagafuchi,A., Yonemura,S., Tsukita,S. and Tsukita,S. (1994) Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J. Cell Biol., 127, 1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi C.J., Arpin,M., Fanning,A.S. and Louvard,D. (1996) The junction-associated protein, zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell–cell contacts. Proc. Natl Acad. Sci. USA, 93, 10779–10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B.M. (1996) Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell, 84, 345–357. [DOI] [PubMed] [Google Scholar]
- Herz J., Flint,N., Stanley,K., Frank,R. and Dobberstein,B. (1990) The 68 kDa protein of signal recognition particle contains a glycine-rich region also found in certain RNA-binding proteins. FEBS Lett., 276, 103–107. [DOI] [PubMed] [Google Scholar]
- Hoover K.B., Liao,S.Y. and Bryant,P.J. (1998) Loss of the tight junction MAGUK ZO-1 in breast cancer: relationship to glandular differentiation and loss of heterozygosity. Am. J. Pathol., 153, 1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough C.D., Woods,D.F., Park,S. and Bryant,P.J. (1997) Organizing a functional junctional complex requires specific domains of the Drosophila MAGUK Discs large. Genes Dev., 11, 3242–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe A., Aplin,A.E., Alahari,S.K. and Juliano,R.L. (1998) Integrin signaling and cell growth control. Curr. Opin. Cell Biol., 10, 220–231. [DOI] [PubMed] [Google Scholar]
- Huber O., Korn,R., McLaughlin,J., Ohsugi,M., Herrmann,B.G. and Kemler,R. (1996) Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech. Dev., 59, 3–10. [DOI] [PubMed] [Google Scholar]
- Itoh M., Nagafuchi,A., Moroi,S. and Tsukita,S. (1997) Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α-catenin and actin filaments. J. Cell Biol., 138, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keon B.H., Schäfer,S., Kuhn,C., Grund,C. and Franke,W.W. (1996) Symplekin, a novel type of tight junction plaque protein. J. Cell Biol., 134, 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick C. and Peifer,M. (1995) Not just glue: cell–cell junctions as cellular signaling centers. Curr. Opin. Genet. Dev., 5, 56–65. [DOI] [PubMed] [Google Scholar]
- Madara J.L. (1998) Paracellular permeability. Annu. Rev. Physiol., 60, 143–159. [DOI] [PubMed] [Google Scholar]
- Matsumoto K. and Wolffe,A.P. (1998) Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol., 8, 318–323. [DOI] [PubMed] [Google Scholar]
- Matter K. and Balda,M.S. (1999) Occludin and the functions of tight junctions. Int. Rev. Cytol., 186, 117–146. [DOI] [PubMed] [Google Scholar]
- Matter K., Hunziker,W. and Mellman,I. (1992) Basolateral sorting of LDL receptor in MDCK cells: the cytoplasmic domain contains two tyrosine-dependent targeting determinants. Cell, 71, 741–753. [DOI] [PubMed] [Google Scholar]
- Molenaar M., van de Wetering,M., Oosterwegel,M., Peterson-Maduro,J., Godsave,S., Korinek,V., Roose,J., Destree,O. and Clevers,H. (1996) XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell, 86, 391–399. [DOI] [PubMed] [Google Scholar]
- Montesano R., Soriano,J.V., Pepper,M.S. and Orci,L. (1997) Induction of epithelial branching tubulogenesis in vitro.J. Cell Physiol., 173, 152–161. [DOI] [PubMed] [Google Scholar]
- Niemann C., Brinkmann,V., Spitzer,E., Hartmann,G., Sachs,M., Naundorf,H. and Birchmeier,W. (1998) Reconstitution of mammary gland development in vitro: requirement of c-met and c-erbB2 signaling for branching and alveolar morphogenesis. J. Cell Biol., 143, 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohga T., Uchiumi,T., Makino,Y., Koike,K., Wada,M., Kuwano,M. and Kohno,K. (1998) Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J. Biol. Chem., 273, 5997–6000. [DOI] [PubMed] [Google Scholar]
- Rajasekaran A.K., Hojo,M., Huima,T. and Rodriguez-Boulan,E. (1996) Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J. Cell Biol., 132, 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B., Souza,B., Albert,I., Muller,O., Chamberlain,S.H., Masiarz,F.R., Munemitsu,S. and Polakis,P. (1993) Association of the APC gene product with β-catenin. Science, 262, 1731–1734. [DOI] [PubMed] [Google Scholar]
- Sakura H., Maekawa,T., Imamoto,F., Yasuda,K. and Ishii,S. (1988) Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene, 73, 499–507. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Sommerville J. and Ladomery,M. (1996) Masking of mRNA by Y-box proteins. FASEB J., 10, 435–443. [DOI] [PubMed] [Google Scholar]
- Spector D.L., Goldman,R.D. and Leinwand,L.A. (1998) Cells: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Stevenson B.R., Siliciano,J.D., Mooseker,M.S. and Goodenough,D.A. (1986) Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol., 103, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L.K., Vogelstein,B. and Kinzler,K.W. (1993) Association of the APC tumor suppressor protein with catenins. Science, 262, 1734–1737. [DOI] [PubMed] [Google Scholar]
- Takahisa M., Togashi,S., Suzuki,T., Kobayashi,M., Murayama,A., Kondo,K., Miyake,T. and Ueda,R. (1996) The Drosophila tamou gene, a component of the activating pathway of extramacrochaetae expression, encodes a protein homologous to mammalian cell–cell junction-associated protein ZO-1. Genes Dev., 10, 1783–1795. [DOI] [PubMed] [Google Scholar]
- Tsukita S., Itoh,M., Nagafuchi,A., Yonemura,S. and Tsukita,S. (1993) Submembranous junctional plaque proteins include potential tumor suppressor molecules. J. Cell Biol., 123, 1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S., Furuse,M. and Itoh,M. (1999) Structural and signalling molecules come together at tight junctions. Curr. Opin. Cell Biol., 11, 628–633. [DOI] [PubMed] [Google Scholar]
- Willott E., Balda,M.S., Fanning,A.S., Jameson,B., van Itallie,C. and Anderson,J.M. (1993) The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppressor protein of septate junctions. Proc. Natl Acad. Sci. USA, 90, 7834–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.F. and Bryant,P.J. (1993) ZO-1, DlgA and PSD-95/SAP90: homologous proteins in tight, septate and synaptic junctions. Mech. Dev., 44, 85–89. [DOI] [PubMed] [Google Scholar]