Summary
Cleavage Factor Im (CFIm) is a highly conserved component of the eukaryotic mRNA 3′ processing machinery that functions in sequence-specific poly(A) site recognition through the collaboration of two protein subunits, a 25 kDa subunit containing a nudix domain and a larger subunit of 59, 68, or 72 kDa containing an RNA recognition motif (RRM). Our previous work demonstrated that CFIm25 is both necessary and sufficient for sequence-specific binding of the poly(A) site upstream element UGUA. Here we report the crystal structure of CFIm25 complexed with the RRM domain of CFIm68 and RNA. The CFIm25 dimer is clasped on opposite sides by two CFIm68 RRM domains. Each CFIm25 subunit binds one UGUA element specifically. Biochemical analysis indicates that the CFIm68 RRMs serve to enhance RNA binding and facilitate RNA looping. The intrinsic ability of CFIm to direct RNA looping may provide a mechanism for its function in the regulation of alternative poly(A) site selection.
Introduction
Alternative mRNA processing has come to be recognized as an essential mechanism for the generation of the transcriptome diversity required by complex eukaryotes (Nilsen and Graveley, 2010). Alternative mRNA splicing and 3′ processing both appear to be regulated primarily by an array of RNA:protein interactions that are intimately coupled to each other, as well as to transcription elongation (Licatalosi and Darnell, 2010). Poly(A) site selection within the nascent pre-mRNA requires the participation of three essential multimeric processing factors: CFIm, cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulatory factor (CstF), each of which bind the pre-mRNA in a sequence-specific and cooperative manner (Millevoi and Vagner, 2010). In humans, CPSF and CstF bind the conserved AAUAAA and downstream G/U rich elements that reside within a defined distance relative to the site of endonucleolytic cleavage (Mandel et al., 2008; Shi et al., 2009). In contrast, CFIm binds to UGUA motifs at variable positions upstream of the cleavage site (Hu et al., 2005).
Human CFIm is a heterotetrameric complex composed of a dimer of a highly conserved 25 kDa nudix protein, CFIm25 (encoded by NUDT21), along with two molecules of 59, 68 or 72 kDa (Kim et al., 2010). The 59 and 68 kDa subunits (referred to as CFIm59 and CFIm68, respectively) are encoded by two paralogs, CPSF6 and CPSF7, whereas the 72 kDa form is likely the result of alternative splicing of the CPSF6 transcript (Ruepp et al., 2010). CFIm59 and CFIm68 share a common architecture composed of an N-terminal RNA recognition motif (RRM), a central proline/glycine-rich domain, and a C-terminal domain enriched in arginine/serine, arginine/aspartic acid and arginine/glutamic acid (RS/RD/RE) repeats (Martin et al., 2010)(Figure S1). The combination of an N-terminal RRM and a C-terminal RS/RD/RE domain similar to that of SR splicing regulatory proteins (Dettwiler et al., 2004; Graveley, 2000). Earlier work on the CFIm25/68 complex indicated that both subunits contribute to RNA binding (Dettwiler et al., 2004). The binding of CFIm to UGUA sequences upstream of the poly(A) site contributes to the assembly of a stable mRNA 3′ processing complex, most likely through interactions with the CPSF subunit Fip1 (Venkataraman et al., 2005). CFIm may function in the regulation of alternative mRNA 3′ processing (Kubo et al., 2006; Sartini et al., 2008), as well as in the coupling of splicing and polyadenylation (Millevoi et al., 2006).
We have previously reported the crystal structure of a CFIm25 homodimer/RNA complex (Yang et al., 2010). This structure revealed a mechanism for sequence-specific recognition of the UGUA motif by the nudix domain of CFIm25. These observations raised the question of the contribution of the CFIm68 subunit, specifically the RRM, to RNA binding. The canonical RRM encompasses approximately 90 amino acids and adopts a β1α1β2β3α2β4 topology in which four β strands fold into an anti-parallel β-sheet packed on top of two α-helices (Clery et al., 2008). The solvent-exposed surface of the β-sheet is most commonly utilized for protein:RNA interactions. Conserved aromatic residues, which are responsible for RNA binding, are typically located within two conserved sequence elements, RNP1 (within β3) and RNP2 (within β1) (Clery et al., 2008) (Figure 1D). In addition to the β-sheet surface, the loops connecting the β strands and α helices may also participate in RNA binding (reviewed in (Clery et al., 2008)). In some cases, proteins employ multiple RRM domains to achieve higher affinity and specificity, to bind long RNA sequences or to bind distinct RNA elements (Deo et al., 1999; Handa et al., 1999; Oberstrass et al., 2005; Sickmier et al., 2006; Teplova et al., 2010; Wang and Tanaka Hall, 2001). Although the RRM is the most common RNA-binding domain in humans, some members of this class serve as a platform for protein:protein interactions ((Clery et al., 2008; Fribourg et al., 2003; Kielkopf et al., 2001; Lee et al., 2009) and references within).
Figure 1.
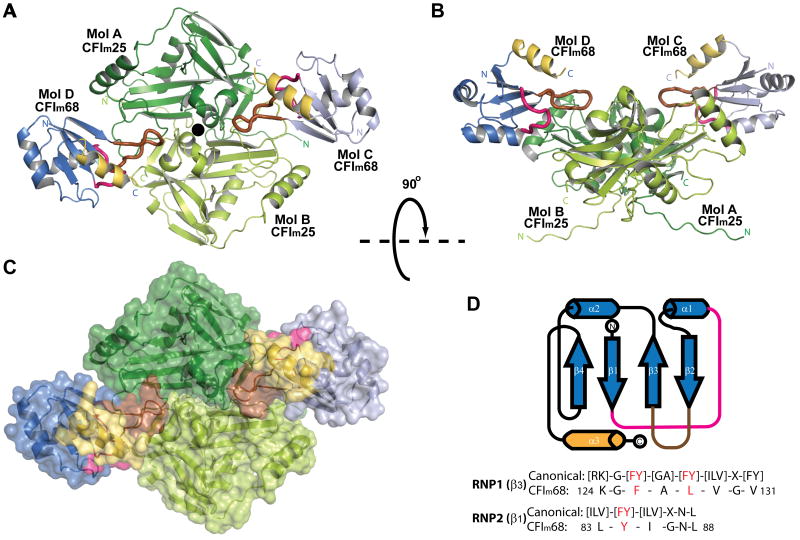
Overall structure of the CFIm complex
(A) Cartoon diagram of the CFIm complex. The CFIm25 homodimer, colored in dark green and lime, is flanked by the RRM domain of two CFIm68 monomers colored in lavender and blue. In CFIm68, loops connecting β1/α1 (residues 87-93) and β2/β3 (residues 115-125) are shown as thicker tubes and colored in magenta and brown, respectively. The RRM C-terminal α-helix (α3, residues 161-172) is highlighted in gold. See also Figure S1 (B) CFIm complex rotated 90° compared to A. (C) Surface representation of the CFIm complex in the same orientation as in A. (D) Topological representation of the secondary structure elements of the CFIm68 RRM. A sequence comparison between CFIm68 and consensus RNP is shown.
In this report we present the crystal structure of a complex of human CFIm25 with the RRM domain of CFIm68 and RNA. Combined with the mutational analysis of both protein subunits and their RNA target, the structure reveals a module designed for the sequence-specific recognition of two UGUA sequences separated by an RNA sequence of variable length. The data suggest that the CFIm68 RRM functions to enhance the RNA binding affinity of the CFIm complex and to facilitate the looping of the RNA between the two anti-parallel UGUA binding sites of the CFIm25 dimer.
Results
Structure determination
In order to determine the contribution of CFIm68 to RNA binding, we attempted to crystallize a CFIm25/68 complex using several N-terminal constructs of CFIm68 that encompass the RRM (residues 80-160), a motif known to be required for RNA binding by CFIm (Dettwiler et al., 2004; Yang et al., 2010) (Figure S1). Together with the full-length CFIm25, CFIm25ΔN21 was also used for crystallization trials, since the N-terminal residues 1-21 are disordered in the crystal structure and do not appear to participate in RNA binding (Coseno et al., 2008; Tresaugues et al., 2008; Yang et al., 2010). The best quality crystals were obtained with the CFIm68 N13-235 construct and the Δ21 N-terminal deletion variant of CFIm25, after reductive methylation of both proteins (Schubot and Waugh, 2004). The methylated CFIm25ΔN21/ CFIm68 N13-235 complex diffracted X-rays to 2.90 Å resolution (Table 1). We determined the structure of the complex using molecular replacement phases combined with Single-wavelength Anomalous Diffraction (SAD) phases from an iodide derivative. A complex with a 9-mer RNA containing one UGUA element (UAUUUUGUA) was refined to a resolution of 3.05 Å with good refinement statistics (Table 1).
Table 1. Data Collection and Refinement Statistics.
| CFIm-NaI | CFIm Native | CFIm-RNA | |
|---|---|---|---|
| PDB ID | 3Q2S | 3Q2T | |
| Data collection | |||
| Resolution (Å) | 20-3.2(3.31-3.2) a | 20-2.90(3.00-2.90) | 20-3.05 (3.17-3.05) |
| Space group | P213 | P213 | P213 |
| Cell dimensions | |||
| a= b= c (Å) | 138.59 | 139.32 | 138.41 |
| α= β= γ (°) | 90 | 90 | 90 |
| Total reflections | 66154 (5554) | 454470 (32866) | 622189 (44935) |
| Unique reflections | 14647 (1424) | 20194 (1968) | 16919 (1634) |
| Redundancy | 4.9 (3.9) | 22.5 (16.7) | 36.8 (27.5) |
| I / σI | 9.42 (2.00) | 24.91 (2.01) | 22.04 (2.02) |
| Completeness (%) | 99.2 (97.4) | 100 (100) | 100 (100) |
| Rmerge (%) | 13.7 (66.5) | 9.2 (100) | 14.6 (100) |
| RFriedel on F (%) | 12 (43.2) | 5.9 (38.1) | 6.9 (35.6) |
| Phasing | |||
| No. of iodide sites | 7 | ||
| Figure of merit (Molecular replacement) | 0.46 | ||
| Figure of merit (AutoSol) | 0.74 | ||
| Refinement | |||
| Resolution (Å) | 20-2.9 | 20-3.05 | |
| No. reflections | 31,808 | 27,293 | |
| Rwork / Rfree (%) | 21.17/27.44 b | 21.98/28.51 | |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.005 | 0.006 | |
| Bond angles (°) | 0.934 | 0.959 | |
| B-factors (Å2) | |||
| CFIm25 (Chain A/B) | 61.5/59.3 | 57.1/59.5 | |
| CFIm68 (Chain C/D) | 76.5/78.8 | 69.6/75.7 | |
| RNA (Chain E/F) | N/A | 104.8/121.3 | |
| Water | 46.4 | 54.2 | |
| Ramachandran plot statistics | |||
| Most favored regions | 91.74% | 90.67% | |
| Allowed regions | 7.92% | 9.15% | |
| Disallowed regions | 0.34% | 0.17% | |
Values in parentheses are for the highest-resolution shell.
Rfree was calculated with 5% of the reflections not used in refinement.
The crystal structure reveals a tetrameric CFIm25/68 complex
The CFIm25/68 complex is composed of a dimer of CFIm25 (designated as molecules A and B) and two monomers of CFIm68 (designated as molecules C and D) (Figure 1A, B). The calculated molecular weight of the heterotetramer correlates well with the elution profile of the complex on a gel filtration column (Figure S2). CFIm25 retains the same dimer conformation as observed in the apo and RNA-bound structures (Coseno et al., 2008; Tresaugues et al., 2008; Yang et al., 2010). The overall protein architecture of CFIm25 remains the same as in the previously published unliganded model (3BHO (Coseno et al., 2008)) with an r.m.s. deviation of 0.57 Å calculated on 194 Cα atoms. The most notable movement of CFIm25 was observed in the extended N-terminal segment, which swings ∼20° toward the protein core compared to the unliganded model, possibly due to a different crystal packing environment. Deletion of the N-terminal segment (residues 1-28) does not affect CFIm25/CFIm68 protein complex formation nor RNA binding (data not shown). Therefore the main function of this extended segment may be to interact with other protein binding partners such as U1 snRNP or PABPN1 (Awasthi and Alwine, 2003), which would be consistent with the fact that three of the N-terminal 29 residues are acetylated (Choudhary et al., 2009).
Although diffraction data were collected from a complex containing residues 13-235 of CFIm68 (Figure S2B), the refined electron density map showed interpretable density only for residues 81-172, a segment which comprises the entire RRM domain (residues 81-160). As expected, the RRM domain adopts a canonical β1α1β2β3α2β3 topology in which a four-stranded antiparallel β-sheet packs on top of two helices. An additional C-terminal helix (α3) lies on top of the β-sheet (Figure 1D and 4A). Several RRMs have been identified that possess a similarly positioned C-terminal α3 helix ((Dominguez et al., 2010; Perez-Canadillas, 2006; Schellenberg et al., 2006; Selenko et al., 2003) and references within).
The CFIm25/68 complex revealed an unexpected mode of interaction among the subunits. Instead of interacting with individual CFIm25 monomers, the two CFIm68 monomers wrap around opposite sides of the CFIm25 dimer, simultaneously interacting with both CFIm25 subunits (Figure 1A and B). Such an arrangement is reminiscent of the interactions observed within the CPSF-30 / influenza virus NS1A protein complex (Das et al., 2008). The surface area of the RRM domain is ∼5,140 Å2, nearly a third of which participates in the interaction with CFIm25. A structure of an apo complex between CFIm25 and the RRM domain of CFIm59 was deposited in the Protein Data Bank (Structural Genomics Consortium, Karolinska Institute; PDB ID code 3N9U). The heterotetrameric complex is similar to the CFIm25/CFIm68 complex reported here, with two CFIm59 RRMs flanking the CFIm25 homodimer.
The CFIm25/68 interface
The interactions of CFIm68 with CFIm25 are mediated primarily by the protruding loops connecting β1 to α1 and β2 to β3 of the CFIm68 RRM (Figure 2A and B). A variety of interactions were observed at the CFIm25/68 interface, including aromatic stacking and hydrogen-bonding via both side chain and main chain atoms (Figure 2C). The contacts between the CFIm25 dimer and each of the CFIm68 monomers are identical, unless otherwise noted.
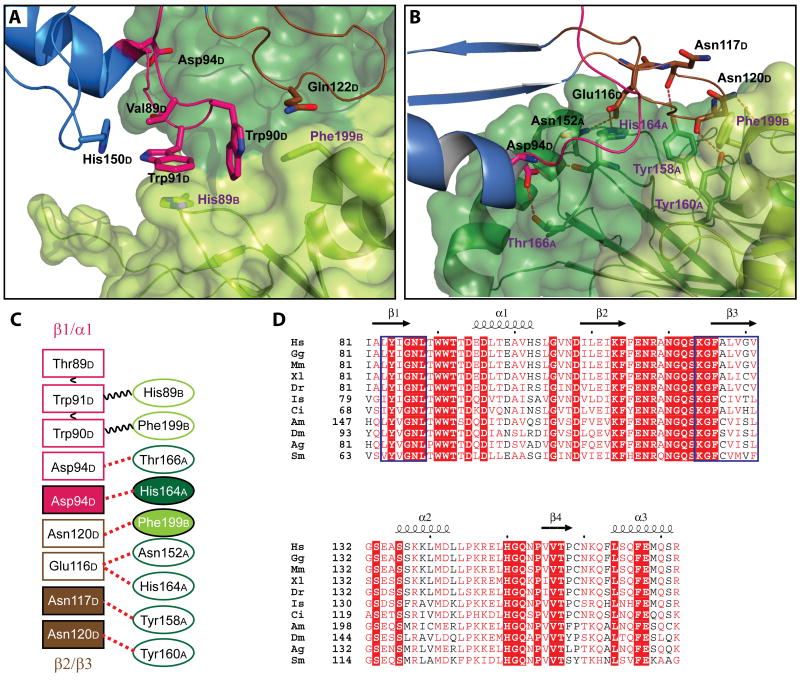
Figure 2.
Interactions between the CFIm25 homodimer and one CFIm68 subunit.
Close-up view of interactions between the CFIm25 dimer and a CFIm68 monomer (Mol D) mediated by (A) hydrophobic stacking and (B) hydrogen bonding. The proteins are colored as in Figure 1. Residues involved in protein-protein interactions are shown as stick model and colored according to the molecule and domain they belong to. See also Figure S2. (C) Schematic representation of the CFIm25-CFIm68 interactions. CFIm25 residues are represented with ellipses, CFIm68 residues with rectangles. Stacking interactions are represented by black wavy lines and hydrogen bonds by red dashed lines. Filled boxes represent main chain interactions, whereas open boxes indicate side chain interactions. Residues are colored as in A and B. (D) A sequence alignment of the CFIm68 RRM homologs from various species listed below. Consensus RNPs are delineated by the blue boxes. Residues with more than 70% conservation are colored in red. Strictly conserved residues are in the filled red boxes. Species abbreviations are as follows: Hs, Homo sapiens; Gg, Gallus gallus; Mm, Mus musculus; Xl, Xenopus laevis; Dr, Danio rerio; Is, Ixodes scapularis; Ci, Ciona intestinalis; Am, Apis mellifera; Dm, Drosophila melanogaster; Ag, Anopheles gambiae; Sm, Schistosoma mansoni. See also Figure S6 for a more complete alignment.
Hydrophobic stacking interactions are the dominant forces at the interface of the CFIm68 β1/α1 loop and the CFIm25 dimer. Trp91 of molecule D (Trp91D) is enclosed in a tight cavity composed of aromatic and aliphatic side chains (Figure 2A). Trp91D is sandwiched between His89 from molecule B of CFIm25 (His89B) and Thr89D. His150D and Trp90D are oriented perpendicularly to the pyrrole and benzene rings of Trp91D, respectively. Trp90D is also in the middle of a perpendicular stacking arrangement, with Trp91D stacking on the benzene ring on one side and Gln122D and Phe199B stacking on the other side. All residues that contribute to this multi-layer stacking interaction (Trp90D, Trp91D, His89B and Phe199B) are highly conserved (Figure 2D,(Yang et al., 2010)). Besides these hydrophobic interactions, the side chain of Asp94D interacts with the hydroxyl group of Thr166A. Further hydrogen bonding between the main chain amide of Asp94D and carbonyl of His164A couples the two CFIm subunits (Figure 2B).
In order to assess the contribution of the CFIm68 β1/α1 loop to complex assembly, a double mutant, CFIm68 W90A/W91A and a triple mutant, W90A/W91A/D94A, were constructed. Unexpectedly, both protein variants were still able to form a stable complex with CFIm25 based on gel filtration analysis (Figure S2A). This finding was confirmed by the co-elution of both subunits from a Ni-NTA column, in which the W90A/W91A and W90/W91A/D94A variants were His-tagged, whereas CFIm25 was untagged (data not shown). The CFIm25 residues (His89 and Phe199) that interact with CFIm68 Trp90/Trp91 were subsequently mutated to alanines. We found that W90A/W91A was still capable of binding to the CFIm25 H89A/F199A double variant based on gel-filtration (Figure S2A) and Ni-NTA co-elution (data not shown). This finding suggests that the interactions observed between loop β1/α1 and CFIm25 are not essential for CFIm complex formation.
The CFIm68 RRM harbors another loop (β2/β3) that interacts with CFIm25. Unlike the hydrophobic core formed by the CFIm68 β1/α1 loop and molecule B of CFIm25, the β2/β3 loop participates primarily in hydrogen bonding interactions with molecule A (Figure 2B). The side chain of Glu116D forms hydrogen bonds with His164A and Asn152A. All three of these residues are highly conserved. Interestingly, in addition to side chain interactions, hydrogen bonding mediated by main chain atoms is prevalent at the β2/β3-CFIm25 interface. Main chain carbonyl groups of Asn117D and Asn120D interact with the hydroxyl group of Tyr158A and Tyr160A, respectively. The side chain of Asn120A connects to the carbonyl group of Phe199B.
Additional protein variants were constructed to determine the contribution of the CFIm68 β2/β3 loop to CFIm complex assembly. Since most of the interactions mediated by the β2/β3 loop are main-chain interactions (Figure 2B), we engineered a loop deletion variant of CFIm68 that removed 7 residues from the tip of the loop (residues 116-122) (Figure S2D). The resulting variant (CFIm68 Δloop) no longer bound CFIm25, based on gel filtration analysis (Figure S2A). Moreover, when the RNA binding properties of CFIm68 Δloop/CFIm25 complex were evaluated by EMSA, the observed shifted band was consistent with the CFIm25/RNA complex (data not shown). We note that the loss of the interaction between the CFIm 25 and 68 kDa subunits is unlikely to be due to large scale unfolding of the CFIm68 Δloop variant since the limited proteolysis profile of this protein was essentially identical to that of wild type CFIm68 (Figure S2C). In addition to the β2/β3 loop deletion in CFIm68 we also mutated two residues in CFIm25 that interact with β2/β3 to alanine (Y158A, Y160A). The resulting variant no longer forms a complex with CFIm68. Taken together these results indicate that β2/β3 is essential for CFIm25/68 complex formation.
Specific recognition of two UGUA sequences by the CFIm25 dimer within the CFIm25/68 complex
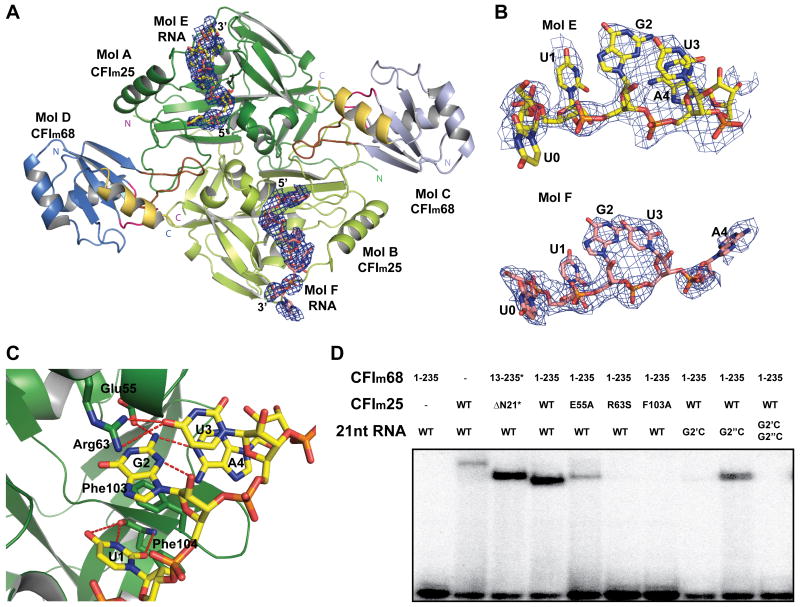
We have previously shown that the CFIm25 homodimer alone is sufficient for the sequence-specific binding of UGUA (Yang et al., 2010). Within the CFIm25/68 complex, however, the CFIm68 subunit contributes to the affinity of RNA binding as illustrated by a decrease in the Kd for binding of a poly(A) polymerase alpha (PAPOLA) poly(A) site upstream element (GGGUGUAAAACAGAUGAUGUAU) from 645 nM for the CFIm25 homodimer alone to 222 nM for the CFIm25/68 complex (Figure S3). To understand the nature of the contribution of the CFIm68 subunit to RNA binding, we soaked a 9-mer RNA (UAUUUUGUA) into a preformed CFIm25/68 crystal. Unlike the CFIm25/RNA dimer, in which only one UUGUAU was observed per CFIm25 dimer due to crystal packing, we observed clear density for an RNA molecule bound in each of the RNA binding pockets of the CFIm25 dimer within the CFIm25/68 complex (Figure 3A and B). The interactions between CFIm25 and UGUA within the CFIm25/68 complex are similar to those previously reported for the CFIm25 dimer (Yang et al., 2010). Briefly, U1, G2 and U3 are recognized via hydrogen bonding interactions with Phe104, Glu55 and Arg63, respectively. Additional three-layer stacking forces provided by Phe103, G2 and U3 further stabilize the protein-RNA complex (Figure 3C). In Mol A, A4 is specifically recognized via an intramolecular Watson–Crick/sugar-edge base pairing between G2 and A4, as previously observed (Auweter et al., 2006; Yang et al., 2010). In Mol B, however, A4 swings away from G2 and forms hydrogen bond with the hydroxyl group of Tyr54 through its N1 atom (Figure 3B). The difference in the position of A4 in the two CFIm25 monomers may be due to the fact that the RNA was soaked into preformed crystals rather than co-crystallized.
Figure 3.
The CFIm complex binds two UGUA elements simultaneously.
(A) Overview of the CFIm-UAUUUUGUA complex. RNA molecules are shown as stick model and colored in yellow and salmon. Simulated annealing omit map for the RNA molecules is shown as a blue mesh and contoured a 2.5σ. (B) A close up view of the bound RNA molecules. Density was observed for the UGUA elements and the ribose of the base preceding the first U (U0). (C) A close up view of CFIm25 interacting with UGUA elements in Mol A. Residues participating in RNA binding are shown in stick model representation. Hydrogen bonds are represented by red dashed lines. (D) Electrophoretic mobility shift assays (EMSA) of CFIm variants in complex with various PAPOLA RNA sequence variants. An asterisk (*) indicates that the protein underwent reductive methylation. A single prime (′) represents the first UGUA element, and double prime (″) represents the second UGUA element. See also Figure S3.
To validate our structural observations, the RNA binding properties of various protein variants were analyzed by EMSA. The three key CFIm25 residues, namely Glu55 and Arg63, which participate in the specific recognition of UGUA, and Phe103 which stabilizes the RNA, are highly conserved (Yang et al., 2010). Mutating each residue abrogated RNA binding (Figure 3D) and thus confirmed that the CFIm25 dimer is responsible for sequence-specific RNA recognition in the context of the CFIm25/68 complex. In addition, the 21 nt RNA (Figure 4B) encompassing the PAPOLA poly(A) site upstream element (Yang et al., 2010) was used to confirm that two UGUA elements within a single RNA can be bound by the CFIm25/68 complex. A single G2C mutation at the first or second UGUA element significantly decreased the RNA binding affinity, and the combination of both mutations essentially eliminated RNA binding (Figure 3D).
Figure 4.
The CFIm68 RRM facilitates RNA looping
(A) A close up view of the CFIm68 RRM (Mol D). The β-sheet is colored in blue, α1 and α2 in green, C-terminal helix α3 in gold, loop β1/α1 in pink, loop β2/β3 in brown and the other loops in white. Mutated residues are shown as stick models. (B) EMSA data of CFIm in complex with PAPOLA RNA variants with spacers of various lengths. UGUA elements are highlighted in red. Inserted nucleotides and nucleotides moved from their original position are shown in green. EMSA data were plotted based on bound fractions relative to the wild type PAPOLA 21mer. Error bars represent the standard deviation from triplicate experiments. (C) Bar graph representation of the EMSA of CFIm68 RRM variants in complex with wild type CFIm25 and 21 mer PAPOLA RNA. Error bars represent the standard deviation from triplicate experiments. (D) EMSA of CFIm68 RRM variants in complex with wild type CFIm25 and PAPOLA RNA variants with spacers of various lengths, or PAPOLG. Error bars represent the standard deviation from duplicate experiments. All bound fractions in figure (B), (C) and (D) were plotted relative to the wild type complex and wild type 21mer PAPOLA RNA. The error bars represent the standard deviation. See also Figure S5. (E) and (F) Protein residues that were subjected to alanine substitution are shown in surface representation. The red color represents a decrease greater than 80% in RNA binding efficiency by CFIm68 E111A, W90A/W91A/D94A and W90A/W91A/N117A/R118A; magenta represents a 50% decrease by CFIm25 E154A, and pink represents a less than 40% decrease by CFIm68 Y84A/L128A. Green indicates an increase in RNA binding efficiency by CFIm68 F126A.
Taken together, the RNA binding data obtained with protein and RNA sequence variants support the crystal structure and confirm the sequence-specific binding of UGUA by each of the CFIm25 subunits within the CFIm25/68 complex.
CFIm68 RNP1 and RNP2 are not critical for RNA binding
The anti-parallel orientation of the two CFIm25 subunits dictates that the RNA connecting the two bound UGUA sequences must form a loop in order to accommodate the 180° turn of the RNA. The looping of the RNA between two UGUA sequences is analogous to the RNA loops created by the polypyrimidine tract binding protein 1 (PTB1), which possesses a set of anti-parallel RRMs (Oberstrass et al., 2005) and the zipcode-binding protein 1 (ZBP1), which possesses a set of anti-parallel KH-domains (Chao et al., 2010). The ability of CFIm to facilitate the formation of an RNA loop is consistent with the variable spacing of UGUA elements within mammalian poly(A) sites (Venkataraman et al., 2005). The stoichiometry of the CFIm heterotetramer-21 nt PAPOLA RNA interaction was determined by EMSA to be 1:1, by fitting the data to a quadratic model of saturable ligand binding (Chao et al., 2010; Rambo and Doudna, 2004). The 1:1 protein:RNA ratio supports the hypothesis that an RNA loop is formed between two UGUA elements (Figure S4).
Our initial determination of the distance between two UGUA elements that would be compatible with binding by the CFIm25 dimer alone revealed a maximal loop size of 9 nucleotides (Yang et al., 2010). A single nucleotide insertion in the loop separating the two UGUA sequences of the PAPOLA poly(A) site upstream element reduced its binding to the CFIm25 dimer to nearly undetectable levels (data not shown). In contrast, insertion of 2 to 6 nucleotides within the loop of the same RNA enhanced its binding by the CFIm25/68 complex (see below) (Figure 4B). This finding suggested that the larger CFIm subunit participates in RNA looping and we set out to elucidate which parts of the CFIm68 RRM contribute to this process.
The top side of the RRM motif presents a typical RNA binding surface (Clery et al., 2008). A quick inspection of the CFIm68 RRM structure revealed that a C-terminal α3 helix is tethered to the β-sheet of the RRM (Figure 4A). A third C-terminal helix appended to the canonical RRM fold is commonly observed among reported RRM structures ((Clery et al., 2008; Dominguez et al., 2010; Handa et al., 1999; Wang and Tanaka Hall, 2001) and references therein). In some cases, the α3 helix completely occludes the surface of the β-sheet, including the key aromatic residues of RNP1 and RNP2 that are critical for RNA binding (Dominguez et al., 2010). In CFIm68, the residues predicted to specifically interact with RNA, Tyr84 and Phe126 match the RRM consensus, whereas Leu128 differs from the consensus aromatic residue within RNP1 (Figure 1D and 4A). A Phe to Leu mutation at the equivalent structural position in the U1A spliceosomal protein leads to more than 100-fold decrease in RNA-binding affinity (Shiels et al., 2002). Alanine substitution mutations were introduced for Tyr84, Phe126, and Leu128, and a double alanine mutant for Tyr84 and Leu128. As illustrated in Figure 4C, the impact of each of these changes on RNA binding was rather modest. RNA binding by the CFIm25/68 complex containing the Y84A/L128A double mutant was reduced by ∼40%, while the F126A mutant exhibited a ∼40% enhancement of RNA binding. These results suggested that the RRM β-sheet is not essential for RNA binding by the CFIm25/68 complex.
The β1/α1 and β2/β3 loops of CFIm68 direct RNA looping
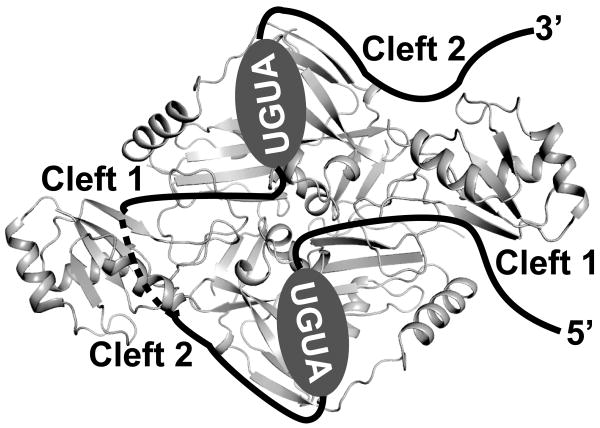
The loops that connect the β-strands and α-helices of a variety of RRMs have been shown to contribute to RNA binding (Clery et al., 2008; Dominguez et al., 2010). Within the CFIm complex, the CFIm68 β1/α1 and β2/β3 loops reside within clefts formed at the interface of the CFIm68 and CFIm25 subunits. Intersubdomain (Song et al., 2003) and intersubunit (Kurimoto et al., 2001) clefts have previously been found to participate in RNA binding. The two clefts that flank the CFIm68 RRM are denoted in Figure 4E. The CFIm68 β2/β3 loop resides within cleft 1, which we will refer to as the ‘entry cleft’, based on its position relative the 5′ end of the bound RNA. The CFIm68 β1/α1 loop resides within cleft 2, which will be referred to as the ‘exit cleft’, due to its proximity to the 3′ end of the bound RNA. As a symmetrical complex, CFIm thus possesses two entry clefts and two exit clefts.
To examine the contribution of the CFIm68 β1/α1 and β2/β3 loops to RNA binding, the two highly conserved tryptophan residues within the β1/α1 loop, Trp90 and Trp91, were substituted with alanines. The W90A/W91A double mutation decreased the binding affinity of the CFIm complex for the wild type 21 mer PAPOLA RNA by 70% (Figure 4D and Figure S5). Alanine substitution mutations were also introduced at two positions within the β2/β3 loop, Asn117 and Arg118, and position Glu111 of the β2 strand, which also resides within the entry cleft along with the β2/β3 loop (Figure 4A). Although the N117A/R118A double mutant conferred a modest reduction in binding compared to the wild type RNA, the E111A mutation caused an 85% decrease. Moreover, an alanine substitution of CFIm25 Glu154, which also resides within the entry cleft, exhibited a 50% decrease in RNA binding. Taken together, these data indicate that both clefts contribute to RNA binding (Figure 4E and 4F). Furthermore, the positions of the altered residues suggest a potential path for the RNA loop that connects the two UGUA elements. Rather than traversing the β-sheet surface, the mutation data suggest that the RNA may be directed beneath the RRM.
To investigate the potential role of the CFIm68 β1/α1 and β2/β3 loops in directing RNA looping, we asked if the impact of the cleft residue mutations might be influenced by the length of the RNA loop. One might expect that the impact of the cleft mutations might be most pronounced for an RNA a with minimal loop sequence, as the path of the RNA between the two UGUA binding sites would be highly constrained. The PAPOLA 21nt RNA represents such a minimal substrate, as a deletion of 2nt (-2 loop RNA) dramatically reduced binding of the CFIm25/68 complex, whereas increasing the size of the RNA loop by 2, 4 or 6 nt significantly enhanced binding (Figure 4B). We therefore examined the impact of the cleft mutations on the binding of the PAPOLA +6 RNA. We also examined the binding of an RNA containing a PAPOLG poly(A) site upstream element (Venkataraman et al., 2005) in which the two UGUA elements are separated by 12 nt (rather that 9 nt in the wild type PAPOLA RNA). As illustrated in Figure 4D, the RNAs containing the larger loops were efficiently bound by the wild type CFIm complex, and were less sensitive to mutations within the clefts than the wild type PAPOLA RNA. A notable exception was the W90A/W91A/N117A/R118A complex, in which alterations have been made within both clefts. This variant complex exhibited a dramatic reduction in the binding of each of the RNAs tested (Figure 4D).
Lastly, we examined the impact of W90A/W91A mutations in the context of a D94A mutation. Asp94 resides on the bottom surface of the CFIm68 RRM, facing away from the β sheet surface. As illustrated in Figure 4C, the combined D94A/W90A/W91A mutations had a severe impact on the binding of the wild type PAPOLA RNA, but caused only a modest reduction in the PAPOLA +6 and PAPOLG RNAs. Taken together, the RNA binding data strongly suggest that the path of the RNA between the two bound UGUA elements extends beneath the CFIm68 RRM domain (Figure 5).
Figure 5.
Model illustrating how CFIm68 may facilitate RNA looping. RNA is shown as a red line. UGUA elements are highlighted by ovals. Dashed lines indicate that the RNA is below the RRM surface. See also Figure S4
Discussion
The structural analysis of a CFIm25/68 tetramer bound to two UAUUUUGUA molecules has revealed a complex in which two distinct domains, an RRM and a nudix domain, together form a module capable of both sequence-specific ssRNA binding and RNA looping. In this unique complex, the CFIm25 nudix domain provides sequence-specificity for UGUA, while the CFIm68 RRM domain enhances RNA binding and facilitates RNA looping. This structure provides a key insight into the ability of CFIm to contribute to the assembly of an mRNA 3′ processing complex through the recognition of multiple poly(A) site upstream elements.
The structure of the CFIm25/68 complex revealed an unusual mode through which the CFIm68 RRM participates in protein:protein interactions. Within the heterotetramer, the RRM of each CFIm68 subunit interacts with both CFIm25 subunits, using the β1/α1 and β2/β3 loops of the RRM (Figure 1A and 4A). Of the two loops only β2/β3 appears to be essential for the formation of the CFIm25/68 complex. The β1/α1 and β2/β3 loops also contribute a set of highly conserved residues to two clefts formed at the interface of the RRM and the CFIm25 homodimer (Fig. 4E and 4F). Our data suggest that the clefts function to guide the RNA to each of the UGUA binding pockets of the CFIm25 homodimer and to direct the looping of the intervening RNA sequence.
A set of charged residues within cleft 1 (the RNA entry cleft), provided by both the CFIm 25 and CFIm68 subunits, appears to present a non-specific RNA binding surface that facilitates the interaction of the 5′ UGUA sequence with the CFIm25 RNA binding pocket. Single substitution mutations of CFIm68 E111A or CFIm25 E154A led to a 85% and a 50% decrease, respectively, in RNA binding affinity by the CFIm complex. In addition, binding of the PAPOLA RNA not only required two UGUA elements, but also the presence at least three nucleotides preceding the 5′ UGUA (data not shown). These residues appear to be docked within the entry cleft. Residues within the entry cleft preceding the second UGUA binding pocket would likewise be expected to facilitate RNA binding in a similar manner.
Cleft 2 (the RNA exit cleft) encompasses a set of highly conserved aromatic residues contributed by both the CFIm25 and CFIm68 subunits. The CFIm68 β1/α1 loop residues, Trp90 and Trp91, appear to be a unique feature of this RRM, conserved even within the CFIm68 subunit of the early diverging eukaryote, Trichomonas vaginalis. Substitution of both of these tryptophans by alanine led to a significant decrease in RNA binding. Aromatic residues located within the β1/α1 loops of several RRMs have been found to be crucial for RNA binding (reviewed in (Clery et al., 2008)). Furthermore, the combination of the W90A/W91A and N117A/R118A mutations was found to nearly abolish RNA binding (Figure 4D). Finally, the elimination of PAPOLA RNA binding by the combination of W90A/W91A with D94A, a residue that resides on the bottom surface of the RRM, provides strong evidence for RNA:protein interactions within the exit cleft. Taken together the mutagenesis data illuminate a likely path of the RNA that, unexpectedly, takes it beneath the CFIm68 RRM as illustrated in Figure 5.
Whereas the β-sheet surface of the CFIm68 RRM would be expected to participate directly in RNA binding, this does not appear to be the case for the poly(A) site upstream element RNAs that we have examined. A notable feature of the CFIm68 RRM is the presence of a C-terminal α3 helix that is tethered to the β-sheet and appears to partially occlude RNA access to the β-sheet. A variety of RRM structures have been reported that contain a C-terminal α3 helix ((Allain et al., 1997; Clery et al., 2008; Dominguez et al., 2010; Nagai et al., 1990; Oubridge et al., 1994; Perez Canadillas and Varani, 2003; Schellenberg et al., 2006; Selenko et al., 2003) and references within). In the case of hnRNP F, the α3 helix makes numerous hydrophobic contacts with the β-sheet and occludes the RNA binding surface (Dominguez et al., 2010). In addition, alanine substitutions the RNP1 and RNP2 motifs of CFIm68 (at residues that would be predicted to be essential for RRM function in RNA binding) had only a modest impact (Figure 4B). Thus the primary contribution of the CFIm68 RRM to RNA binding appears to be mediated by β2/β3 and β1/α1 loop residues that reside within the entry and exit clefts, respectively.
Based on our characterization of the structure and function of CFIm68, we have been able to define a set of search parameters to identify likely orthologs of CFIm68. The identification of potential CFIm68 orthologs by amino acid sequence alignment alone is hampered by the ubiquity of the RRM domain coupled with the two flexible protein domains: a central proline/glycine rich domain and a C-terminal RS/RD/RE domain. Our functional data allowed us to constrain our BLAST search to those RRM-containing proteins that possessed two adjacent tryptophans within the β1/α1 loop, accompanied by a central proline/glycine domain and/or a C-terminal RS/RD/RE domain. As illustrated in Figure S6, likely orthologs were identified in both metazoans and protists. Most strikingly, all of these organisms also possess a clear CFIm25 ortholog. In contrast, both S. cerevisiae and S. pombe, which lack a CFIm25 ortholog (Yang et al., 2010), also lack a CFIm68 ortholog that meets our search criteria. These observations support the hypothesis that the CFIm complex is an ancient mRNA processing factor that has been lost in some protists.
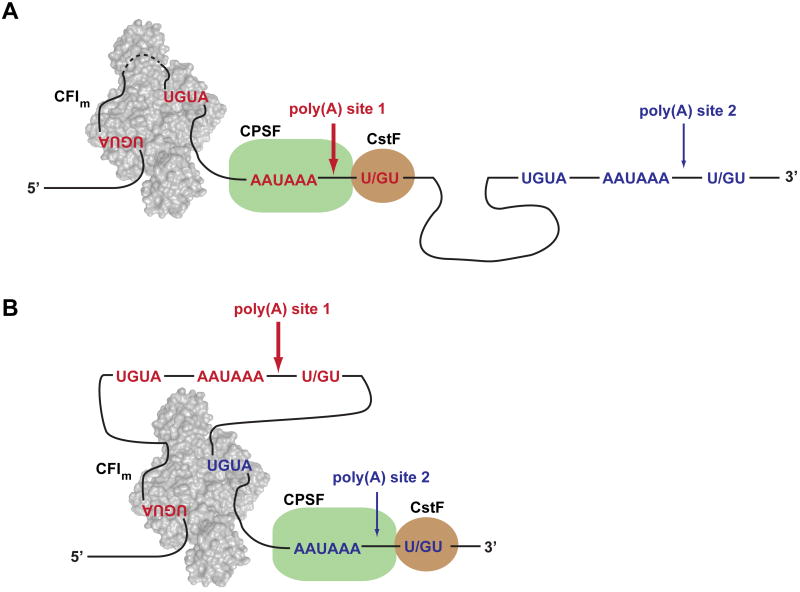
CFIm has been implicated in the regulation of alternative polyadenylation in Hela cells (Kubo et al., 2006) and male germ cells (Sartini et al., 2008). Based on our structural and biochemical data, we propose a model for the function of CFIm in the regulation of alternative mRNA 3′ processing (Figure 6). We postulate that the inherent ability of CFIm to bind two UGUA elements (separated by a sequence of variable length) and thereby loop out the intervening RNA may allow CFIm to pair alternative sets of UGUA poly(A) site upstream elements. In this fashion, the binding of different combinations of UGUA elements may influence the choice of alternative poly(A) sites. Moreover, the potential long range looping of RNA sequences may permit CFIm to loop out an entire poly(A) site and thereby facilitate the selection of a downstream poly(A) site. Such a mechanism may account for the observation that depletion of CFIm in vivo led to a shift to the use of promoter-proximal poly(A) sites (Kubo et al., 2006). A similar regulatory mechanism has been proposed for the splicing regulator pyrimidine track binding protein (PTB) (Oberstrass et al., 2005). The anti-parallel organization of RRM3 and RRM4 of PTB may allow the protein to loop out an entire exon through the binding of polypyrimidine tracts that flank the exon, leading to the exclusion of the exon from the mature mRNA (Oberstrass et al., 2005). As pointed out by Chao et al. (Chao et al., 2010), an additional consequence of RNA looping is the juxtaposition of RNA sequences that may constitute a signal that is recognized by additional regulatory factors.
Figure 6.
Model illustrating how CFIm may mediate alternative polyadenylation. (A) Cleavage reaction occurs at poly(A) site 1. (B) Cleavage reaction occurs at poly(A) site 2.
In summary, our structural and biochemical data provide a new insight into the mechanism by which CFIm functions in the recognition of mammalian poly(A) sites. An intriguing question that remains, however, is the role and potential interaction partner(s) of the CFIm68 β-sheet surface. Conservation of the residues within the CFIm68 RNP1 and RNP2 motifs suggests that this surface has a conserved function in mRNA processing.
Experimental Procedures
Protein purification
CFIm25, CFIm68 and their variants were purified as described in the supplemental Experimental Procedures. CFIm complexes were assembled by mixing CFIm25 and CFIm68 at 1:1 molar ratio and purified by a cation exchange column (SP column, GE Healthcare). Pure fractions were concentrated and eluted on a gel filtration column (Superdex 200 10/30, GE Healthcare). CFIm complexes were concentrated to about 5 mg/ml, flash frozen, and stored at -80°C.
Reductive methylation and crystallization
Reductive methylation treatment was performed as described in the supplemental Experimental Procedures (Rayment, 1997; Schubot and Waugh, 2004). The methylated complexes were eluted from the Superdex 200 10/30 column, concentrated to about 10 mg/ml and stored at 4°C prior to crystallization. Hanging drops were set up by mixing 1 μl of the methylated CFIm complex with 1 μl of crystallization solution consisting of 50 mM HEPES pH 7.0, 200 mM magnesium formate and 20% (w/v) PEG 3350Complete X-ray diffraction data sets were recorded at 100 K at λ= 1.5418 Å on our laboratory MAR345 detector (MAR Research). Data were processed using the HKL2000 suite of programs (Otwinowski and Minor, 1997). CFIm complex crystals belong to the space group P213, with the cell parameters (a = b = c = 139.3 Å). One CFIm25 dimer and two CFIm68 monomers are present in the asymmetric unit, which corresponds to a calculated solvent content of 46%.
Structure determination and refinement
The CFIm complex structure was solved by a combination of molecular replacement and Single-wavelength Anomalous Diffraction (SAD). The initial molecular replacement (MR) search was carried out using MOLREP (Vagin and Teplyakov, 1997) and the apo structure of CFIm25 (PDB ID code 3BHO (Coseno et al., 2008)) excluding the extended N-terminal segment (residues 20-33) in the search model. The CFIm25 dimer was readily identified by molecular replacement, but the density from the CFIm68 subunit was more ambiguous. We therefore collected X-ray data on an iodide derivative in an attempt to improve phases. Seven iodide sites were located using an isomorphous difference Fourier (Fo-Fo) map calculated using data from the unliganded CFIm complex and iodide derivative, and phases from the molecular replacement model (Rould, 2006). Phenix.AutoSol was used to refine the iodide sites. The combined SAD and MR phases yielded an overall FOM of 0.74 (Adams et al., 2002). The model of CFIm68 was built into the improved electron density map using Phenix.AutoBuild. Further improvement of the model was made using the program COOT (Emsley et al., 2010), followed by iterative cycles of positional and B-factor refinement performed with Phenix.Refine (Adams et al., 2002). The overall quality of the final model was evaluated using both PROCHECK (Laskowski et al., 1993) and MOLPROBITY (Davis et al., 2007). The models comprise residues 21-227 for both CFIm25 monomers and residues 81-172 for both CFIm68 monomers. Gly21 is a residue left after the TEV protease digestion, and coincidently corresponds to the 21st amino acid of CFIm25, which happens to be a glycine.
The co-crystallization attempts to obtain CFIm in complex with RNA of various lengths failed to yield any complex crystals. The complex with RNA was therefore obtained by soaking a 9-mer RNA (UAUUUUGUA) (0.25mM) into pre-formed CFIm25/68 complex crystals 30 minutes prior to the cryocooling step. The crystals were cryocooled as described above and a 3.05 Å data set was collected at 100 K on our laboratory X-ray source. The structure was solved using difference Fourier methods (Rould, 2006) and refined as described above for the CFIm complex. We obtained clear density for the UGUA element, but not the rest of the nucleotides. A similar outcome was observed with data collected on a crystal soaked with a 6-mer RNA (UUGUAU), which is not included in this report due to its lower diffraction quality (3.3 Å). All molecular structure figures were generated with PyMOL (DeLano, 2008)
Gel electrophoretic mobility shift assays
Gel electrophoretic mobility shift assays (EMSA) were performed as described before (Yang et al., 2010) (See details in the supplemental Experimental Procedures). Stoichiometry experiments were performed using EMSA protocols, with the following modifications: Cold RNA (4 μM) was added to the binding reaction and CFIm (initial concentration 20 μM) was serially diluted 3:1 with buffer in order to increase the number of data points (Chao et al., 2010; Rambo and Doudna, 2004). Data were fit to a quadratic model of saturable binding, which yielded the stoichiometric equivalence point (Chao et al., 2010; Rambo and Doudna, 2004).
Supplementary Material
Acknowledgments
We are grateful to Dr. Walter Keller and Georges Martin (Biozentrum, University of Basel, Switzerland) for providing the pET-CFIm68 plasmid and for advice and discussions, Samantha Ogilvie for help with gel shift assays, Dr. Pierre Aller and Karl Zahn for assistance with data collection and Drs. Mark Rould, Kayo Imamura, Frédérick Faucher and Brian Eckenroth for help with structure determination and refinement. Part of this work was supported by a National Institutes of Health grant [GM62239 to S.D.].
Footnotes
Accession numbers: The coordinates and structure factors of CFIm and CFIm-RNA complex have been deposited in the RCSB Protein Data Bank under the accession code 3Q2S and 3Q2T, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Allain FH, Howe PW, Neuhaus D, Varani G. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16:5764–5772. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S, Alwine JC. Association of polyadenylation cleavage factor I with U1 snRNP. RNA. 2003;9:1400–1409. doi: 10.1261/rna.5104603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Coseno M, Martin G, Berger C, Gilmartin G, Keller W, Doublié S. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 2008;36:3474–3483. doi: 10.1093/nar/gkn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, et al. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A. 2008;105:13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: 2008. http://www.pymol.org. [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Fisette JF, Chabot B, Allain FH. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol. 2010;17:853–861. doi: 10.1038/nsmb.1814. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- Kim S, Yamamoto J, Chen Y, Aida M, Wada T, Handa H, Yamaguchi Y. Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes Cells. 2010;15:1003–1013. doi: 10.1111/j.1365-2443.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34:6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Fukai S, Nureki O, Muto Y, Yokoyama S. Crystal structure of human AUH protein, a single-stranded RNA binding homolog of enoyl-CoA hydratase. Structure. 2001;9:1253–1263. doi: 10.1016/s0969-2126(01)00686-4. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Lee JH, Rangarajan ES, Yogesha SD, Izard T. Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure. 2009;17:833–842. doi: 10.1016/j.str.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Ostareck-Lederer A, Chari A, Neuenkirchen N, Dettwiler S, Blank D, Ruegsegger U, Fischer U, Keller W. Arginine methylation in subunits of mammalian pre-mRNA cleavage factor I. RNA. 2010;16:1646–1659. doi: 10.1261/rna.2164210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J. 2006;25:3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Canadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Doudna JA. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry. 2004;43:6486–6497. doi: 10.1021/bi049912u. [DOI] [PubMed] [Google Scholar]
- Rayment I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 1997;276:171–179. [PubMed] [Google Scholar]
- Rould MA. The same but different: isomorphous methods for phasing and high-throughput ligand screening. Methods Mol Biol. 2006;364:159–182. doi: 10.1385/1-59745-266-1:159. [DOI] [PubMed] [Google Scholar]
- Ruepp MD, Schümperli D, Barabino SML. mRNA 3′ end processing and more—multiple functions of mammalian cleavage factor I-68. Wiley Interdisciplinary Reviews - RNA. 2010 doi: 10.1002/wrna.35. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- Sartini BL, Wang H, Wang W, Millette CF, Kilpatrick DL. Pre-messenger RNA cleavage factor I (CFIm): potential role in alternative polyadenylation during spermatogenesis. Biol Reprod. 2008;78:472–482. doi: 10.1095/biolreprod.107.064774. [DOI] [PubMed] [Google Scholar]
- Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Luhrmann R, Glover JN, MacMillan AM. Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci U S A. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubot FD, Waugh DS. A pivotal role for reductive methylation in the de novo crystallization of a ternary complex composed of Yersinia pestis virulence factors YopN, SycN and YscB. Acta Crystallogr D Biol Crystallogr. 2004;60:1981–1986. doi: 10.1107/S0907444904023005. [DOI] [PubMed] [Google Scholar]
- Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Kramer A, Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels JC, Tuite JB, Nolan SJ, Baranger AM. Investigation of a conserved stacking interaction in target site recognition by the U1A protein. Nucleic Acids Res. 2002;30:550–558. doi: 10.1093/nar/30.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresaugues L, Stenmark P, Schuler H, Flodin S, Welin M, Nyman T, Hammarstrom M, Moche M, Graslund S, Nordlund P. The crystal structure of human cleavage and polyadenylation specific factor-5 reveals a dimeric Nudix protein with a conserved catalytic site. Proteins. 2008;73:1047–1052. doi: 10.1002/prot.22198. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- Yang Q, Gilmartin GM, Doublié S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci U S A. 2010;107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.