Abstract
RNA 3′-terminal phosphate cyclases are evolutionarily conserved enzymes catalysing conversion of the 3′-terminal phosphate in RNA to the 2′,3′-cyclic phosphodiester. Their biological role remains unknown. The yeast Saccharomyces cerevisiae contains a gene encoding a protein with strong sequence similarity to the characterized cyclases from humans and Escherichia coli. The gene, named RCL1 (for RNA terminal phosphate cyclase like), is essential for growth, and its product, Rcl1p, is localized in the nucleolus. Depletion or inactivation of Rcl1p impairs pre-rRNA processing at sites A0, A1 and A2, and leads to a strong decrease in 18S rRNA and 40S ribosomal subunit levels. Immunoprecipitations indicate that Rcl1p is specifically associated with the U3 snoRNP, although, based on gradient analyses, it is not its structural component. Most of Rcl1p sediments in association with the 70–80S pre-ribosomal particle and a 10S complex of unknown identity. Proteins similar to Rcl1p are encoded in genomes of all eukaryotes investigated and the mouse orthologue complements yeast strains depleted of Rcl1p. Possible functions of Rcl1p in pre-rRNA processing and its relationship to the RNA 3′-phosphate cyclase are discussed.
Keywords: cyclic phosphate/nucleolus/pre-rRNA processing/snoRNAs
Introduction
RNA cyclases are a family of RNA-modifying enzymes that catalyse the ATP-dependent conversion of the 3′-phosphate to the 2′,3′-cyclic phosphodiester at the end of RNA (Filipowicz et al., 1983; Genschik et al., 1997, 1998). The biological role of cyclases is unknown, although they are thought to be responsible for generating or regenerating cyclic phosphate ends, known to be required by several RNA ligases in both eukaryotes and prokaryotes (Filipowicz and Vicente, 1990; Genschik et al., 1997). These ligases include two tRNA-splicing ligases, and the prokaryotic RNA ligase of unknown function that joins RNA ends via the atypical 2′,5′-phosphodiester (reviewed by Arn and Abelson, 1998; Genschik et al., 1997). Cyclases may also function in the production of cyclic phosphate termini present in U6 spliceosomal RNA (Lund and Dahlberg, 1992; for a discussion of additional possible functions, see Genschik et al., 1997).
The human cDNA and the Escherichia coli gene encoding the cyclase have been cloned and their proteins characterized (Genschik et al., 1997, 1998). The properties and substrate specificities of the two enzymes are nearly identical. They cyclize the 3′-phosphate in RNAs of different sequence and base composition. For both enzymes, cyclization involves formation of the covalent cyclase–AMP intermediate (Filipowicz et al., 1985; Reinberg et al., 1985; Genschik et al., 1998). Experiments performed with the E.coli enzyme have shown that AMP is attached to the conserved His309 via a phosphoamide bond (Billy et al., 1999).
A crystal structure of the E.coli enzyme has recently been determined at 2.1 Å resolution (Palm et al., 2000). The protein consists of two domains. The larger domain, which includes the adenylated His309, contains three repeats of a folding unit previously identified in the bacterial translation initiation factor 3 (IF3). Unexpectedly, the large domain of cyclase appears twice in each of two related enzymes, 5-enol-pyruvylshikimate-3-phosphate synthase (EPSPS) and UDP-N-acetylglucosamine enolpyruvyl transferase (MurA), which are not involved in nucleic acid metabolism (Palm et al., 2000).
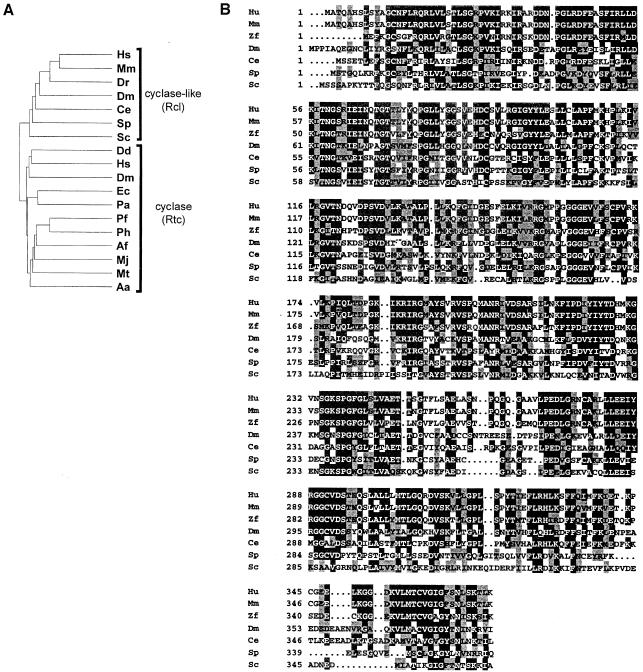
Cloned cyclases have no apparent sequence motifs in common with proteins of known function, including EPSPS and MurA. Database searches showed, however, that genes encoding proteins similar to the human and E.coli enzymes are conserved among Eukarya, Bacteria and Archaea, arguing for an important function of the enzyme in RNA metabolism (Genschik et al., 1997). Similarity analysis of the human and E.coli enzymes, and related proteins from other organisms, indicated that they can be divided into two subfamilies referred to hereafter as cyclases (Rtc, for RNA 3′-terminal phosphate cyclase) and the RNA-3′-terminal-phosphate-cyclase-like (Rcl) proteins (Figure 1A; Billy et al., 1999). The cyclase subfamily includes the enzymes from humans and E.coli, and their likely counterparts in Drosophila melanogaster, Dictyostelium discoideum, and in Archaea and Bacteria. The Rcl subfamily, on the other hand, comprises proteins of only eukaryotic origin. Although clearly falling into distinct groups, proteins of Rtc and Rcl families show a high sequence similarity. For example, the E.coli cyclase and human Rcl are 28% identical and 35% similar; these values are only marginally lower than the 34% identity and 43% similarity between cyclases from these two organisms. The two protein classes share several sequence elements, including a nearly universally conserved sequence RGXXPXGGGXφ (where X stands for any, and φ for hydrophobic amino acids), designated originally as the cyclase signature (Billy et al., 1999; Figure 1).
Fig. 1. Dendritic tree of cyclases (Rtc) and cyclase-like proteins (Rcl) from different organisms (A) and comparison of amino acid sequences of proteins belonging to the Rcl subfamily (B). The compared Rtc proteins originate from the following organisms: Hs, Homo sapiens [SwissProt (SP) O00442]; Dm, Drosophila melanogaster (GenBank SPTREMBL O77264); Dd, Dictyostelium discoideum (SP AAB70847); Ec, E.coli (SP P46849); Pa, Pseudomonas aeruginosa (The Pseudomonas Genome Project); Ph, Pyrococcus horikoshi (Entrez BAA30639); Pf, Pyrococcus furiosus (The Insitute for Genomic Research, Gaithersburg, MD); Af, Archaeoglobus fulgidus (AAB89810); Mj, Methanococcus jannaschii (SP Q60335); Mt, Methanobacterium thermoautotrophicum (AAB86375); Aa, Aquifex aeolicus (AAC06852). Compared Rcl proteins originate from: Hs, H.sapiens (Genschik et al., 1997; GenBank AJ276894); Mm, Mus musculus (GenBank AJ276895); Dr, Danio rerio (zebrafish; GenBank ESTs: AW078116.1 and AW059073.1); Dm, D.melanogaster (SP P56175); Ce, Caenorhabditis elegans (SP Q23400); Sp, Schizosaccharomyces pombe (SP Q09870); Sc, S.cerevisiae (SP Q08096). Multiple sequence alignment was performed with the ClustalW program as described (Genschik et al., 1997). Identical amino acids and amino acids conserved in at least 50% of sequences are indicated by black and grey boxes, respectively.
To gain further insight into the biological function of proteins belonging to the Rtc/Rcl family, we have analysed properties of the Saccharomyces cerevisiae cyclase-like protein, Rcl1p, and the effect of its genetic depletion on RNA metabolism in yeast. We demonstrate that Rcl1p is an essential nucleolar protein required for pre-rRNA processing at sites A0, A1 and A2. A fraction of Rcl1p is associated with U3 small nucleolar (sno)RNP, a central component of the 18S rRNA processing machinery in yeast and vertebrates (reviewed by Venema and Tollervey, 1995; Kressler et al., 1999). However, Rcl1p is not a structural component of U3 snoRNP and its association with U3 snoRNP occurs, most probably, in large macromolecular complexes representing nascent ribosomes.
Results
Alignment of the Rcl proteins
The alignment of proteins belonging to the Rcl family is shown in Figure 1B. In addition to sequences present in different databases, human and mouse proteins, deduced from cDNAs characterized in this work are included. The human and mouse proteins are 97% identical, while the mouse and yeast proteins are 38% identical and 54% similar. Proteins of the Rcl subfamily have no motifs in common with proteins of known function other than cyclases. Importantly, Rcl proteins lack a conserved histidine, which acts as an adenylate acceptor and is required for phosphate cyclization (Billy et al., 1999), suggesting that these proteins either use another amino acid for adenylation or have no cyclase activity (see Discussion).
RCL1 is an essential gene
The gene encoding the cyclase-like protein in S.cerevisiae, named RCL1 (YOL010w), is predicted to encode a 40 kDa protein. The rcl1 null allele was constructed by replacing the entire RCL1 open reading frame (ORF) with the TRP1 marker using a PCR-based procedure. All dissected tetrads from sporulated heterozygous diploids gave two or fewer viable spores that were all auxotrophic for tryptophan. Microscopic analysis showed that the spores deleted for RCL1 germinate but cell division stops after three to five generations. One of the heterozygous RCL1/rcl1::TRP1 strains was transformed by the centromeric plasmid pFL38-RCL1, harbouring the RCL1 gene and URA3 marker. After selection for uracil prototrophy, cells were sporulated and dissected. Six complete tetrads analysed showed a 2:2 segregation of TRP1:Δtrp1 and cosegregation of TRP1 and URA3 markers (data not shown). Hence, RCL1 is essential for cell viability, a conclusion also reached recently in the yeast ORF disruption project (Winzeler et al., 1999).
Conditional RCL1 alleles
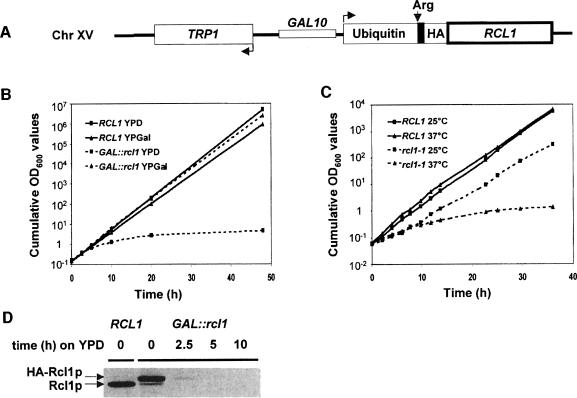
To analyse Rcl1p function, two conditional systems were established. In the first, the RCL1 ORF, modified at the N-terminus by addition of the destabilizing cassette containing ubiquitin and the influenza haemagglutinin (HA) tag (Jenny et al., 1996), was put under the control of the GAL10 promoter (Figure 2A). To characterize the effect of Rcl1p depletion, the wild-type and conditional mutant strains were pre-grown on a galactose-containing medium (YPGal) and then transferred to a glucose medium (yeast extract/peptone/dextrose; YPD). From ∼5 h after transfer, the doubling time of the GAL::rcl1 strain progressively increased, while the parent wild-type strain maintained a constant doubling time of 1.9 h (Figure 2B). Western analysis performed with α-Rcl1p antibodies (Abs) showed that, following 5 h growth on YPD, the GAL::rcl1 cells were depleted of Rcl1p (Figure 2D).
Fig. 2. Conditional alleles. (A) Scheme of the GAL::rcl1 allele. The RCL1 ORF is fused to the ubiquitin, HA-tag and LacI (shown in black) regions. Following removal of ubiquitin, the fusion protein starts with the destabilizing amino acid, arginine (Arg). (B) Growth curves of GAL::rcl1 and the wild-type strain in YPD and YPGal. Cells were shifted to non-permissive conditions at time 0. (C) Growth curves of rcl1-1 ts and wild-type strains at 25 and 37°C. (D) Depletion of HA-Rcl1p analysed by Western blotting with α-Rcl1p Abs. Positions of the HA-Rcl1p and wild-type Rcl1p proteins are indicated.
A thermosensitive (ts) rcl1-1 allele was also generated and introduced into the RCL1 locus. Sequence analysis indicated two amino acid changes: L279S and D319G. No attempt was made to find out whether both are required for the ts phenotype. Comparison of wild-type and rcl1-1 strains indicated that the mutant grows more slowly even at the permissive temperature of 25°C. At 37°C, the rcl1-1 strain displayed a very severe growth defect (Figure 2C).
Rcl1p and its mouse orthologue are functionally conserved nucleolar proteins
Possible functions of proteins belonging to Rtc and Rcl families include generation and/or maintenance of cyclic phosphate ends in tRNA half molecules undergoing splicing, and formation of cyclic ends in the spliceosomal RNA U6 (see Introduction). Using the GAL::rcl1 strain, we tested the effect of Rcl1p depletion on tRNA and mRNA processing. No appreciable inhibition of splicing of the two intron-containing tRNAs investigated, tRNATrpCCA and tRNAProUGG, was observed. Likewise, no effect on splicing of two intron-containing pre-mRNAs, actin pre-mRNA and CYH2 pre-mRNA, was detected when GAL::rcl1 cells were grown on YPD for up to 48 h (data not shown).
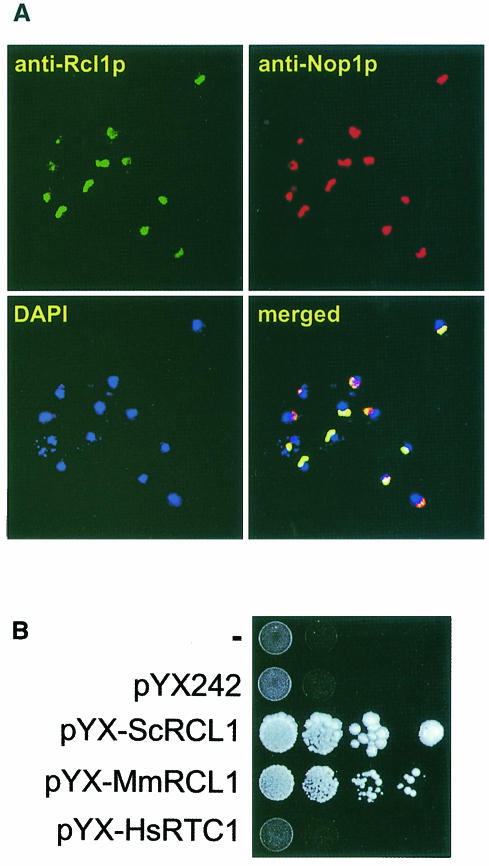
The cellular localization of Rcl1p was investigated by indirect immunofluorescence, using rabbit α-Rcl1p and FITC-coupled α-rabbit Abs. As a control, Nop1p (fibrillarin), a nucleolar protein (Venema and Tollervey, 1995), was visualized with the mouse monoclonal Ab (mAb) A66 and α-mouse Abs conjugated to Texas Red (TR). DNA was stained with 4′,6-diamidine-2-phenylindole (DAPI) to localize the nucleus. As shown in Figure 3A, treatment with both Rcl1p and Nop1p Abs showed a typical crescent-shaped staining, characteristic of nucleolar proteins. Merging of all three images revealed co-localization of Rcl1p and Nop1p, while DAPI staining was primarily confined to the nucleoplasm. Similar results were obtained when the HA-tagged Rcl1p was localized in the GAL::rcl1 strain grown in YPGal, using α-HA mAbs (data not shown).
Fig. 3. Nucleolar localization of Rcl1p and complementation of the yeast GAL::rcl1 mutant by the mouse Rcl1p-like protein. (A) Rcl1p in the BMA41 strain was detected by a rabbit α-Rcl1p Ab, followed by a goat FITC-coupled α-rabbit Ab. Nop1p was detected by mouse mAb A66 followed by a goat TR-conjugated α-mouse Ab. Chromatin was stained with DAPI. Merging of all three images is shown in the bottom right panel. (B) The GAL::rcl1 strain was transformed with either pYX-ScRCL1, pYX-MmRCL1, pYX-HsRTC1 or the empty vector pYX242; upper row, untransformed cells. Complementation was assayed by monitoring growth on YPD plates. Western blotting with Abs against the human cyclase verified that it is expressed in cells transformed with pYX-HsRTC1 (data not shown).
Rcl1p-like proteins are conserved among eukaryotes (Figure 1). We have found that mouse orthologue of Rcl1p, named mRcl1, also localizes to the nucleolus when expressed as the C-terminal fusion with green fluorescent protein in HeLa and NIH 3T3 cells (data not shown). As shown in Figure 3B, mRcl1 expressed from the 2µ expression vector pYX242 (row pYX-MmRCL1) complemented growth of the Gal::rcl1 strain on YPD. Expression of the human RNA 3′-phosphate cyclase, a nucleoplasmic protein (Genschik et al., 1997), did not complement yeast cells depleted of Rcl1p (row pYX-HsRTC1).
We conclude that Rcl1p and the related mouse protein, mRcl1, are both localized to the nucleolus and probably perform a similar function.
Rcl1p is required for 18S rRNA processing
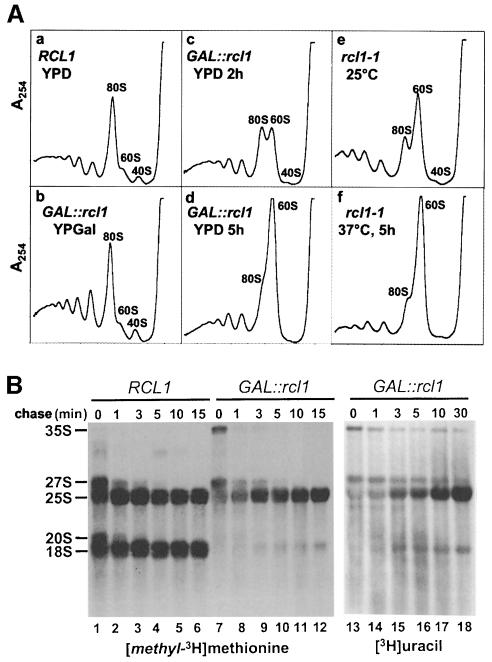
The nucleolus is a site of ribosome biogenesis. To test whether depletion or inactivation of Rcl1p has an effect on the ribosome status of the cell, we analysed profiles of polysomes isolated from conditional rcl1 strains. The profile of the GAL::rcl1 strain grown in YPGal was identical to that of the wild-type strain grown in YPD (Figure 4A, panels a and b). Analysis of polysomes from GAL::rcl1 grown in YPD for 2–5 h revealed rapid disappearance of free 40S subunits, accompanied by a gradual increase in the amount of free 60S subunits and a decrease in polysome levels (panels c and d). Consistent with the observed drop in polysome levels, a shift of GAL::rcl1 cells to the glucose-containing minimal medium (SD) resulted in a gradual decrease in [35S]methionine incorporation into total protein (40% decrease after 5 h in SD compared with the wild-type strain; data not shown).
Fig. 4. Polysome profile (A) and pulse–chase (B) analyses. (A) Polysome profiles of GAL::rcl1 (panels b–d), rcl1-1 ts (panels e–f) mutant strains and a wild-type (panel a) strain grown under permissive and/or restrictive conditions. Time of growth on YPD or at the restrictive temperature is indicated. Polysome profiles of wild-type cells grown at 30°C (panel a), and at 25 and 37°C (not shown) are similar. (B) Pulse–chase labelling of rRNA with [methyl-3H]methionine (lanes 1–12) and [3H]uracil (lanes 13–18) in the wild-type and GAL::rcl1 strains. Labelling (3 min) was performed following 5 h growth on YPD.
Polysome profiles of the rcl1-1 ts mutant were also analysed. Consistent with the partial growth defect of this strain at permissive temperature (see Figure 2C), a deficit of free 40S and an excess of free 60S subunits was already evident with cells grown at 25°C (Figure 4A, panel e). A shift to 37°C for 5 h resulted in a further increase in free 60S subunits and diminished polysome levels (panel f).
Analysis of rRNA levels in the GAL::rcl1 strain indicated that a shift from YPGal to YPD resulted in a gradual decrease in the 18S:25S rRNA ratio from 1 (at the time of transfer) to 0.2 (20 h after transfer; see Figure 6G). To distinguish whether the observed drop in 18S rRNA and 40S subunit levels is due to decreased 18S rRNA synthesis or to its destabilization, metabolic labelling was performed. The GAL::rcl1 cells, maintained for 5 h on a glucose medium to deplete Rcl1p, were pulse-labelled with either [methyl-3H]methionine or [3H]uracil. An excess of unlabelled methionine or uracil was then added and the cells incubated for the time indicated (Figure 4B). In the wild-type strain, the 20S and 27S RNAs, which represent precursors of 18S and 25S rRNAs, respectively (for the diagram of pre-rRNA processing see Figure 5), were rapidly chased into mature rRNAs (Figure 4B, lanes 1–6; for the [3H]uracil labelling, see Supplementary material available at The EMBO Journal Online). In the GAL::rcl1 strain, 25S rRNA accumulation was only marginally delayed, but the labelling of 20S pre-rRNA and 18S rRNA was dramatically reduced. Comparison of labelling with [methyl-3H]methionine (Figure 4B, lanes 7–12) and [3H]uracil (lanes 13–18) indicated that cells depleted of Rcl1p are not impaired in rRNA methylation. Metabolic labelling of 5.8S and 5S rRNAs was also analysed; depletion of Rcl1p had no significant effect on synthesis of these RNAs (data not shown).
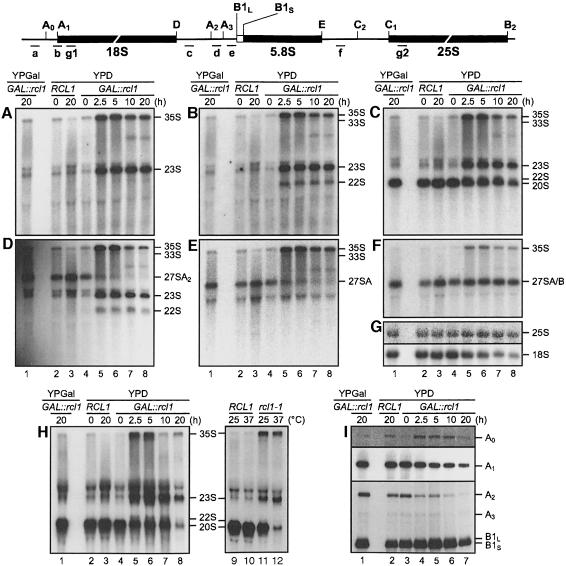
Fig. 6. Depletion or inactivation of Rcl1p affects pre-rRNA processing at sites A0, A1 and A2 (A–G and H, lanes 1–8). Northern analysis of pre-rRNAs accumulating in the wild-type strain grown in YPD (lanes 2 and 3) and GAL::rcl1 strain grown either in YPGal (lanes 1) or YPD (lanes 4–8). The following oligonucleotide probes (indicated schematically above the gels) were used: (A) probe a, the 5′ETS upstream of A0; (B) probe b, junction of 5′ETS and 18S rRNA; (C and H) probe c, the ITS1 upstream of A2; (D) probe d, between A2 and A3; (E) probe e, the ITS1 downstream of A3; (F) probe f, the ITS2 upstream of C2; (G) probes g1 and g2, complementary to 18S and 25S rRNA, respectively; (H) lanes 9–12, processing intermediates accumulating in the rcl1-1 ts mutant grown at permissive or restrictive temperature (37°C, 5 h). (I) Primer extension analysis of RNAs accumulating in the GAL::rcl1 strain grown either in YPGal (lane 1) or YPD (lanes 3–7), and in the wild-type strain grown in YPD (lane 2). Oligo g1 was used for primer extension to sites A0 and A1, and oligo f for extension to sites A2, A3 and B1L/S. Positions of primer extension stops corresponding to different pre-rRNA cleavage sites are indicated. The A0 and A1 panels represent different exposures of the same gel.
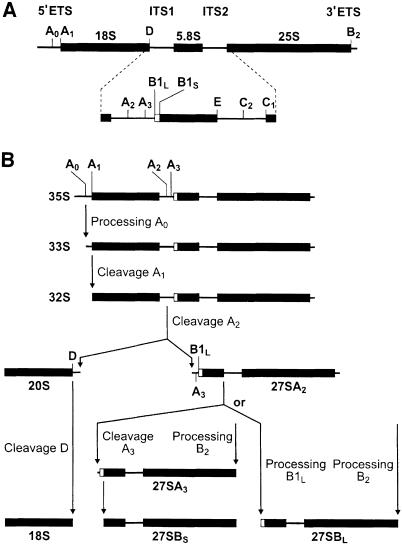
Fig. 5. Pre-rRNA processing pathway in yeast. (A) Structure of 35S pre-rRNA. (B) Physiological pathway of 35S pre-rRNA processing. Final steps of 5.8 and 25S maturation are not shown. Processing sites and intermediates are indicated. For more details, see Venema and Tollervey (1995); Kressler et al. (1999).
Taken together, these results indicate that depletion or inactivation of Rcl1p leads to decreased levels of 40S subunits, resulting in an accumulation of free 60S ribosomes and a fall in the amount of polysomes, and that these effects are due to a defect in 18S rRNA synthesis.
Depletion of Rcl1p results in defective processing at sites A0, A1 and A2
In yeast, 18S, 5.8S and 25S rRNAs are derived from a long 35S precursor that additionally contains 5′ and 3′ external transcribed sequences (5′ETS and 3′ETS) and internal transcribed spacers, ITS1 and ITS2 (Figure 5; Venema and Tollervey, 1995). Physiologically, the 35S pre-rRNA is first cleaved at the A0 site in 5′ETS, yielding 33S pre-rRNA. 33S RNA is then processed rapidly at sites A1 and A2 to generate 20S pre-rRNA, which is further processed into the mature 18S rRNA in the cytoplasm. Cleavage at A2 also generates 27SA2 pre-rRNA, which, upon further processing, yields 5.8S and 25S rRNAs, components of the 60S subunit. Hence, cleavage at A2 effectively separates processing and assembly of 40S and 60S components (Figure 5).
To define steps in pre-rRNA maturation affected by Rcl1p depletion, we analysed steady-state levels of processing intermediates in the GAL::rcl1 strain by Northern blot analysis using oligonucleotide probes complementary to different regions in the 35S pre-rRNA (Figure 6). Probing with oligonucleotide a (Figure 6A), specific for the region upstream of A0, indicated that the shift to YPD results in accumulation of 23S pre-rRNA. Since this RNA was also visualized with probes b, c and d (Figure 6B–D), complementary to regions across site A1, and upstream and downstream of site A2, respectively, it most likely extends from the transcription start site to A3. A similar aberrant processing intermediate accumulates upon depletion of other factors affecting cleavages at sites A0–A2 (Venema and Tollervey, 1995; Kressler et al., 1999). The 22S product, visualized with probes b–d, but not probes a and e (Figure 6A–E), most probably represents aberrant pre-rRNA extending from sites A0 to A3. This product, diagnostic of the lower severity of the processing defect at A0 than at sites A1 and A2, has been identified before (see Discussion). Hybridization with probe c (Figure 6C) demonstrated a gradual decrease, but not elimination, of 20S pre-rRNA, a direct precursor of the mature 18S rRNA (for better resolution of 20S and 22S products, see Figure 6H). Persistence of the 20S pre-rRNA suggests that some processing at sites A1 and A2, likely caused by a low level of HA–Rcl1p expression under restrictive conditions, is taking place. This is consistent with the results of pulse–chase experiments showing that traces of 18S rRNA are formed in the GAL::rcl1 strain grown on YPD (see Figure 4B). Hybridization with probes d and e revealed a decrease in the pre-rRNA 27SA2, consistent with the inhibition of cleavage at A2 (Figure 6D and E), while hybridization with probe f, specific for ITS2, indicated that there is no defect in accumulation of the 27S pre-rRNA processed downstream of A2, acting as a precursor to 5.8S and 25S rRNAs (Figure 6F). All probes revealed increased accumulation of the unprocessed 35S pre-rRNA and probes b–f also revealed a slight increase in the amount of 33S pre-rRNA in cells grown on YPD.
Processing intermediates accumulating in the rcl1-1 ts mutant were also analysed, using the oligonucleotide c specific for the D/A2 region. Consistent with the data for the GAL::rcl1 strain, growth of rcl1-1 at 37°C resulted in accumulation of 35S and 23S pre-rRNAs and a strong decrease in the 20S pre-rRNA. Similar, although less pronounced, changes were seen in rcl1-1 cells grown at 25°C (Figure 6H, lanes 9–12). Longer exposure of the film (not shown) indicated that low levels of the 22S pre-rRNA were also present in rcl1-1 cells grown at either 25 or 37°C.
Defects in pre-rRNA processing were further analysed by primer extension (Figure 6I). Consistent with the accumulation of 23S and 22S pre-rRNAs, depletion of Rcl1p resulted in a pronounced reduction of products extending to the A2 site. Levels of products cleaved at A0 were slightly increased, confirming that processing at this site is less affected than at sites A1 and A2. A gradual drop in the level of the cDNA extending to site A1 confirmed the observed net decrease in mature 18S rRNA levels. There were no significant changes in the level of RNA processed at sites A3, and B1L and B1S; additional analyses (not shown) demonstrated that the ratio of RNAs processed at sites B1L and B1S remained unchanged and that all cleavages investigated were correct at the nucleotide level.
We conclude that depletion or inactivation of Rcl1p leads to inhibition of processing at sites A0, A1 and A2, required for synthesis of 18S rRNA, processing at A0 being less affected than that at sites A1 and A2.
Rcl1p associates with but is not a structural component of U3 snoRNP
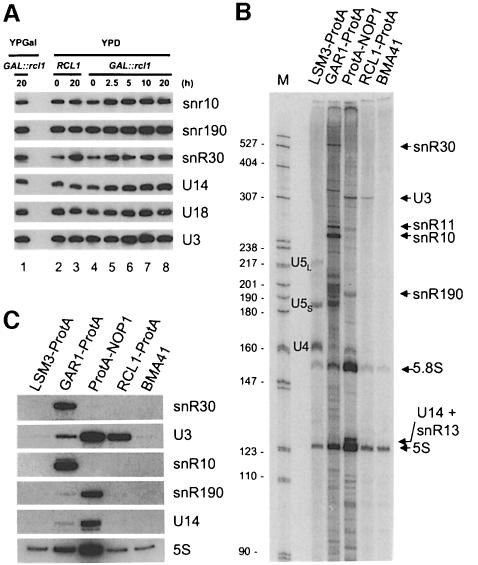
Genetic depletion of four snoRNAs (U3, U14, snR30 and snR10) inhibits or delays processing of 18S rRNA in yeast (Tollervey, 1987; Li et al., 1990; Hughes and Ares, 1991; Morrissey and Tollervey, 1993; Venema and Tollervey, 1995). We therefore investigated whether depletion of Rcl1p affects the accumulation of these or other snoRNAs; the snoRNAs investigated included some transcribed from independent genes as monomeric (U3, snR10 and snR30) or dimeric (U14 and snR190) RNAs, or processed from introns (U24) (reviewed by Maxwell and Fournier, 1995; Tollervey and Kiss, 1997). Levels of none of the snoRNAs assayed decreased when the GAL::rcl1 strain was grown in YPD (Figure 7A). Likewise, RNase MRP, RNase P and U6 RNAs showed no decrease (data not shown).
Fig. 7. Rcl1p is specifically associated with but is not a structural component of U3 snoRNP. (A) Depletion of Rcl1p has no effect on snoRNA levels. For lane description, see Figure 6. (B and C) Rcl1p immunoprecipitates U3 snoRNA. In (B) RNA isolated from IP pellets was labelled with [5′-32P]pCp and fractionated by 6% PAGE. U6 snRNA immunoprecipitated by Lsm3p is not labelled with [5′-32P]pCp (Lund and Dahlberg, 1992). In (C) the same material was separated by 8% PAGE and analysed by Northern blotting. The amount of U3 precipitated by Rcl1p-ProtA is three times less than that precipitated by ProtA-Nop1p. This is consistent with the observations that only ∼50% of U3 RNA is associated with large complexes co-sedimenting with Rcl1p (see E) and that the efficiency of Rcl1p-ProtA binding to IgG–Sepharose is lower than that of other ProtA fusions, including ProtA-Nop1p (data not shown). (D) Co-immunoprecipitation of U3 snoRNP proteins and Rcl1p. Eluates from IPs carried out with the strains indicated were analysed by Western blotting using α-Rcl1p Ab (left panel) and non-immune Ab detecting only ProtA-tagged proteins (right panel). (E) Sedimentation of Rcl1p and U3snoRNP on 10–30% glycerol gradients run for long (upper panel) and short (lower panel) times. Positions of 80S, 60S and 40S ribosomes and protein markers (from Pharmacia Biotech) run in parallel gradients (19S thyroglobulin, 669 kDa; 11.3S catalase, 232 kDa; 8S aldolase, 158 kDa; 4.3S bovine serum albumin, 67 kDa) are indicated. Apparently faster sedimentation of the 10S Rcl1p peak upon shorter centrifugation (lower panel) may be due to either poor resolution of different Rcl1p-containing complexes or retention of loosely associated protein(s) that is lost during prolonged centrifugation. We have confirmed that the 43S peak corresponds to the precursor of the 40S subunit by demonstrating that it co-fractionates with 20S precursor rRNA; its association with U3 RNA suggests that it represents a nuclear form of the 43S particle (Trapman et al., 1975).
We next investigated whether Rcl1p is associated with any of the snoRNAs. A strain was constructed that expresses, from the chromosome, Rcl1p containing two IgG-binding protein A domains at the C-terminus (Rcl1p-ProtA). The strain was used for immunoprecipitation (IP) reactions, together with strains expressing other ProtA-tagged proteins: nucleolar proteins Nop1p and Gar1p, known to be associated with C/D and H/ACA class snoRNAs, respectively (Balakin et al., 1996; Ganot et al., 1997 and references therein), and a nucleoplasmic protein Lsm3p, associated with U6 snRNA, and also indirectly with U4 and U5 snRNAs (Salgado-Garrido et al., 1999). Extracts from these and wild-type strains were incubated with the IgG–Sepharose beads, and RNA isolated from the IP pellets was labelled with [5′-32P]pCp and analysed by polyacrylamide gel electrophoresis (PAGE). As shown in Figure 7B, the IP pellet from the Rcl1p-ProtA strain is enriched in U3 snoRNA. For a better estimate of the enrichment, RNAs recovered from the same IP pellets were analysed on Northern blots, using probes specific for U3, U14 and snR190 RNAs (C/D class members), snR10 and snR30 RNAs (H/ACA class members), and 5S rRNA. This analysis confirmed that Rcl1p is associated with U3 and not other snoRNAs tested (Figure 7C). Quantification of the hybridization data indicated that U3 RNA is enriched in the Rcl1p-ProtA pellet 22- and 44-fold relative to U14 and 5S RNAs, respectively, when compared with pellets from Lsm3p-ProtA or wild-type strains.
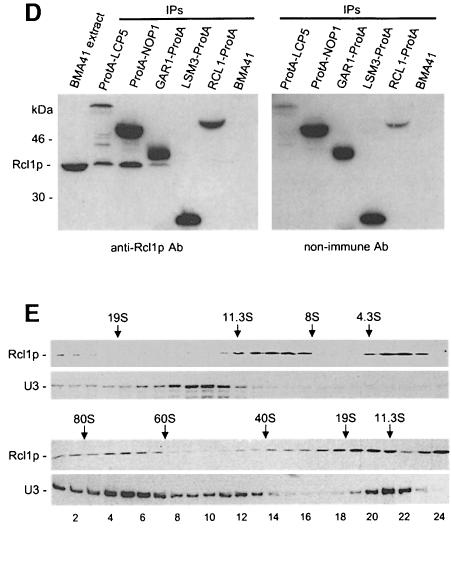
We tested whether proteins known to be associated with U3 snoRNP could co-precipitate Rcl1p. In addition to the strains listed above, a strain expressing ProtA-Lcp5p was also analysed; Lcp5p is required for 18S rRNA processing and specifically associates with U3 snoRNA (Wiederkehr et al., 1998). Eluates from the IPs were separated by SDS–PAGE and analysed by Western blotting, using either α-Rcl1p (Figure 7D, left panel) or control non-immune (right panel) Abs. Rcl1p co-immunoprecipitated with Lcp5 and Nop1p, two proteins known to associate with U3 snoRNP. A weak Rcl1p signal present in the Gar1p-ProtA lane is unlikely to be significant since Gar1p is known to co-precipitate non-specifically with small amounts of C/D class snoRNAs (Balakin et al., 1996; Ganot et al., 1997; see also Figure 7C). No Rcl1p was detected in eluates originating from Lsm3p-ProtA or wild-type strains.
Rcl1p association with U3 snoRNP was further studied by gradient sedimentation. Extracts from the BMA41 strain were separated on a glycerol gradient and fractions analysed by Western and Northern blotting, using α-Rcl1p Abs and the U3 RNA-specific oligonucleotide probe, respectively. As seen in Figure 7E, Rcl1p did not co-fractionate with the U3 snoRNP monoparticle, which sediments at ∼12S (Fabrizio et al., 1994), in gradients centrifuged for either long (upper panel) or short (lower panel) periods. Inspection of the Rcl1p distribution indicated that, in addition to the free protein present near the top of the gradient, Rcl1p is associated with complexes sedimenting at ∼10S (Figure 7E, upper panel; see also legend), and 70–80S; a fraction of Rcl1p also reproducibly sedimented in the 20–40S region (Figure 7E, lower panel). Consistent with its role in 18S rRNA processing, the U3 snoRNA was found to co-sediment with complexes of 70–80S and 43S, likely representing ribosome maturation intermediates, before and after cleavage at site A2, respectively (Trapman et al., 1975; Venema and Tollervey, 1995). It should be noted that in order to minimize non-specific interactions, gradient analyses were carried out at 150 mM NaCl. Trapman et al. (1975) have reported that the large pre-ribosomal particle sediments at 90S at 10 mM KCl, but its sedimentation is significantly slower at higher salt concentrations.
We conclude that Rcl1p associates specifically with U3 snoRNP but is not its structural component. Co-immunoprecipitation of Rcl1p and U3 snoRNP is most readily explained by their interaction within the large complex representing a nascent ribosome.
Discussion
The studies described here represent the first demonstration of the biological function for proteins belonging to the Rtc/Rcl family. It is shown that yeast Rcl1p, a protein structurally similar to the human and bacterial RNA 3′-terminal phosphate cyclases, is an evolutionarily conserved and essential nucleolar protein required for pre-rRNA processing at sites A0, A1 and A2. A fraction of Rcl1p is specifically associated with U3 snoRNP, established as a central component of the 18S rRNA processing machinery in yeast and vertebrates. However, Rcl1p is not a structural protein of the ∼12S U3 monoparticle, but probably interacts with it at the level of the nascent ribosome. In addition to the association with large structures, Rcl1p co-sediments with a 10S complex of unknown identity. Association of Rcl1p with multiple structures suggests that it plays an important and dynamic role in pre-rRNA processing in the nucleolus.
The defect in pre-rRNA processing caused by inactivation of Rcl1p strongly resembles that brought about by the genetic depletion of U3 snoRNA and its associated components, and also other essential snoRNPs (U14 and snR30; depletion of snR10 also delays cleavages at sites A0–A2; Maxwell and Fournier, 1995; Venema and Tollervey, 1995), and non-snoRNP transacting factors. The latter include putative ATP-dependent RNA helicases (de la Cruz et al., 1999), Dim1p methylase (Lafontaine et al., 1995) and several proteins with no obvious enzymic function (Venema and Tollervey, 1995; Kressler et al., 1999). The best explanation for the similar outcome of mutations in so many different components is their participation in the assembly of the multi-RNP processing complex, sometimes referred to as a processome, responsible for co-ordinate cleavage of pre-rRNA at sites A0, A1 and A2. That all the above-mentioned factors assist the cleavage reactions directly is unlikely; many are probably needed to assemble the nascent ribosomal particle that acts as a processing substrate (Maxwell and Fournier, 1995; Venema and Tollervey, 1995; Kressler et al., 1999).
The conclusion that depletion of Rcl1p impairs pre-rRNA processing at sites A0–A2 is supported by the accumulation of unprocesssed 35S pre-rRNA, the disappearance of the 27SA2 intermediate, and generation of aberrant 23S and 22S products. 23S and 22S represent RNAs extending from the 5′ end to A3, and from A0 to A3, respectively. Accumulation of the 22S RNA indicates that A0 cleavage is less strongly affected than cleavages at sites A1 and A2. Uncoupling of site A0 cleavage from those at sites A1 and A2 has been demonstrated for mutations in several other processing factors (Venema and Tollervey, 1995; Kressler et al., 1999), including the C-terminal truncation of the U3-associated protein Mpp10p (Lee and Baserga, 1997). Mutations preventing the interaction of U3 snoRNA with the 18S region of pre-rRNA also affect the cleavage at sites A1 and A2 but not at A0 (Hughes, 1996; Sharma and Tollervey, 1999).
Much evidence, from both yeast and vertebrates, points to U3 as a key snoRNP contributing to the assembly of the pre-rRNA processing complex. Its early interaction with 5′ETS appears to orchestrate assembly of other processing components (Kass and Sollner-Webb, 1990; Beltrame and Tollervey, 1992; Mougey et al., 1993). In addition, specific base-pair interactions between U3 RNA and pre-rRNA have been shown to be required for processing at sites A0, A1 and A2 (Beltrame and Tollervey, 1995; Sharma and Tollervey, 1999 and references therein). Our results, indicating that Rcl1p associates with U3 RNP but that this interaction seems to occur in the processome rather than the U3 monoparticle, are consistent with the assembly of the 18S rRNA processing complex on a nascent ribosome being a very dynamic and step-wise process, likely to involve many as yet uncharacterized factors. Notably, sedimentation analysis has revealed that much Rcl1p sediments at 10S, probably complexed with other proteins. We have identified a novel essential protein specifically interacting with Rcl1p, the significance of which is being investigated (F.Nasr and W.Filipowicz, unpublished results). Our results emphasize the importance of gradient sedimentation analysis for assigning specific association of proteins with snoRNP particles. Five proteins, Sof1p, Mpp10p, Lcp5p, Imp3p and Imp4p (Jansen et al., 1993; Dunbar et al., 1997; Wiederkehr et al., 1998; Lee and Baserga, 1999) were identified previously as specific U3 snoRNP-associated proteins by immunoprecipitation but none has yet been demonstrated to co-sediment with U3 RNP on gradients. Importantly, as with Rcl1p, none of these proteins is required to maintain the U3 RNA integrity.
We initiated the study of Rcl1p because of its structural similarity to the RNA 3′-terminal phosphate cyclase, an enzyme we had already characterized in humans and E.coli (see Introduction). Since cyclases appear to be conserved among Eukarya, Bacteria and Archaea (Genschik et al., 1997; Figure 1), and Rcl1p is the only protein encoded in the yeast genome having similarity with them, we anticipated Rcl1p to be the yeast counterpart of the human and E.coli enzymes. However, the following results (unpublished data) argue that Rcl1p may not have cyclase activity. (i) Recombinant Rcl1p cannot cyclize the 3′-terminal phosphate in the model substrates, CCCCACCCCGp and AAAACAAAAGp, when tested in the presence of different divalent cations and NTPs as potential co-factors. (ii) No nucleotidylation of the protein was observed in the presence of α-32P-labelled ATP, GTP or UTP. (iii) Rcl1p and its likely counterparts in other eukaryotes lack a histidine residue equivalent to the E.coli His309, which is conserved in all members of the Rtc family, and which acts as an adenylate acceptor during the cyclization reaction (Billy et al., 1999). Although these results do not exclude a cyclase activity of Rcl1p (for example, both nucleotidylation and cyclization could require a specific RNA substrate), they make it very unlikely. To approach the substrate specificity question, we investigated whether U3 or other essential snoRNAs in yeast might contain the 2′,3′-cyclic phosphate termini: no evidence was found. Likewise, we have confirmed that U6 snRNA in yeast, in contrast to other eukaryotes (Lund and Dahlberg, 1992), contains a monoester and not a 2′,3′-cyclic phosphate at the 3′-end (unpublished data).
Is Rcl1p merely a structural component of the rRNA processing complex, the integrity of which is disrupted by Rcl1p depletion or inactivation, or does Rcl1p play a more active role in 18S rRNA maturation? Crystallography of the E.coli cyclase (Palm et al., 2000), followed by modelling of the human enzyme (Billy et al., 1999) and yeast Rcl1p (K.Mizuguchi, personal communication), have revealed that all these proteins consist of two domains. The smaller is related to domains found in proteins such as thioredoxin or glutathione S-transferase (GST), while the larger consists of three repeats of the folding unit identified previously in the bacterial translation factor IF3, ribosomal protein S8 (rpS8) and some other proteins acting on nucleic acids. Interestingly, the large domain of the Rtc/Rcl proteins is structurally highly similar to the domains of two related proteins, EPSPS and MurA, involved in peptidoglycan and chorismate synthesis, respectively (Palm et al., 2000). Both these enzymes are composed of two related domains, each consisting of three IF3 repeats, arranged similarly to those in the large domain of the Rtc/Rcl proteins. In common with the cyclase, MurA and EPSPS use phosphate-containing molecules as substrates: phosphoenolpyruvate (PEP) and UDP-GlcNAc (MurA), and PEP and 3-phosphoshikimate (EPSPS). The catalytic sites of these enzymes and that of the cyclase are likely to be located in structurally equivalent areas (Palm et al., 2000).
These structural similarities suggest that, during evolution, the IF3/rpS8 domain, probably representing a very ancient RNA-binding fold, has evolved into the cyclase by undergoing triplication and acquiring an additional domain. Duplication of the large domain present in the cyclase would then lead to proteins with functions unrelated to the cyclase, and RNA metabolism in general, such as MurA and EPSPS. Proteins of the Rcl1p family are much more closely related to cyclases (see Figure 1 and Introduction) than MurA and EPSPS are. While the latter proteins have no discernible sequence similarity with cyclases and Rcl1p, even the most evolutionarily distant members of the Rtc and Rcl families show as much as 28% sequence identity. In the light of the aforementioned propensity of the triplicated IF3 unit to serve as a catalytic domain in different enzymes, it is tempting to propose that proteins of the Rcl1p family, while retaining a potential to act on RNA, have gained a new catalytic function during eukaryote evolution. Nothing is known about factors catalysing cleavages at sites A0, A1 and A2, and it will be interesting to study whether Rcl1p may have endonucleolytic or another enzymatic activity, contributing to pre-rRNA processing. Structural information available for Rtc/Rcl proteins should help to explore these possibilities.
Materials and methods
Strains, media and genetic methods
The following yeast strains were used: wild type, BMA41 (MATa/α, leu2-3 112/leu2-3 112, his3-11,15/his3-11,15, ade2-1/ade2-1, ura3-1/ura3-1, trp1Δ/trp1Δ, can1-100/can1-100); BMA41-1a, as BMA41 but MATa; rcl1-1, as BMA41-1a but rcl1-1; GAL::rcl1, as BMA41-1a but GAL10::RCL1, TRP1; and RCL1-ProtA, as BMA41-1a but Kl-URA3:: RCL1-ProtA. The RCL1-ProtA strain was constructed by transforming BMA41-1a with a DNA fragment amplified by PCR using appropriate 63mer oligonucleotides as primers (see Supplementary material) and plasmid pBS1365 (Puig et al., 1998) as a template. Yeast strains expressing proteins tagged with ProtA were kindly provided by T.Kiss and M.Caizergues-Ferrer (GAR1-ProtA and ProtA–NOP1; Ganot et al., 1997), B.Seraphin (LSM3-ProtA; Salgado-Garrido et al., 1999) and T.Wiederkehr (ProtA-LCP5; Wiederkehr et al., 1998). Genetic manipulations and preparation of standard yeast media followed established procedures (Brown and Tuite, 1998).
Sequences of human and mouse Rcl cDNAs
The EST (DDBJ/EMBL/GenBank accession No. AA087160) encoding the mouse Rcl1p-like protein, mRcl1p, was obtained from the IMAGE consortium and fully sequenced on both strands. An EST (DDBJ/EMBL/GenBank accession No. AA219360) encoding uncomplete human Rcl1p was obtained from the same source; the missing N-terminal sequence was identified by sequencing the PCR fragment amplified from a HeLa cDNA library. Sequences of mouse and human cDNAs are deposited in the DDBJ/EMBL/GenBank with accession Nos AJ276895 and AJ276894, respectively.
Construction of plasmids
The RCL1 (YOL010w) gene with its flanking regions was PCR-amplified from the cosmid pEOA215 (kindly provided by B.Dujon, Pasteur Institute, Paris) and cloned in XhoI and XbaI sites in pBluescript SKII (Stratagene). The insert was sequenced on both strands. The RCL1 gene with its promoter was recloned, as a KpnI fragment, in pFL38 vector (centromeric, with URA3 marker; Bonneaud et al., 1991), yielding pFL38-RCL1, and as a BamHI PCR-amplified fragment in pFL36 vector (centromeric, with LEU2 marker; Bonneaud et al., 1991), yielding pFL36-RCL1. Both plasmids complemented the rcl1Δ::TRP1 strain.
The region encoding mRcl1 was PCR amplified using the IMAGE cDNA clone as a template and oligonucleotides ATAGAATTCATGGC GACCCAGGCGCACTCACTC and ATAGAATTCGGATCACTTGAGG GTCTTGCTCAG (EcoRI sites are underlined) as primers, and cloned into the EcoRI site of pYX242 (Novagen), resulting in pYX-MmRcl1. Plasmids pYX-ScRcl1 and pYX-HsRtc1 were obtained similarly by cloning the PCR-amplified yeast RCL1 and human RTC1 (Genschik et al., 1997) coding regions into the EcoRI site of pYX242. Proteins expressed from the pYX242-based plasmids harbour a His6-tag at the N-terminus.
Gene disruption
Deletion disruption of RCL1 was obtained by a PCR-based method (Baudin et al., 1993) with TRP1 as a selection marker. Briefly, two synthetic 59mer oligonucleotides, each containing 42 nt complementary to the upstream or downstream non-coding region of RCL1, were used to amplify the TRP1 marker cassette from the plasmid pFL35 (Bonneaud et al., 1991). The PCR product was additionally re-amplified with 21mer oligonucleotides corresponding to the 5′-termini of the long primers used in the first amplification and used to transform the yeast diploid strain BMA41. Trp+ transformants were tested for correct integration by PCR and Southern blotting.
Construction of conditional mutants
To generate the GAL::rcl1 strain, the promoter of RCL1 on a chromosome was replaced by the inducible GAL10 promoter fused to the destabilizing and HA-epitope-bearing cassette. Two 60mer oligonucleotides were used to amplify the TRP1-GAL10-Ubiquitin-HA-tag cassette carried on the plasmid YipGUR (Jenny et al., 1996). The amplified fragment was used to transform BMA41-1a, grown on YPGal. Trp+ transformants were analysed by PCR and Southern blotting. Correct in-frame integration was confirmed by sequencing the PCR-amplified DNA and Western blots.
The ts mutant was generated by PCR as previously described (Brown and Tuite, 1998) using pFL36-RCL1 plasmid as a template. After yeast transformation and selection, a plasmid complementing the rcl1Δ::TRP1 strain at 28°C but not at 37°C was isolated. The insert-bearing rcl1-1 allele was subsequently used for transformation to isolate the strain in which the rcl1-1 allele replaces the wild-type copy.
Heterocomplementation
The GAL::rcl1 strain was transformed with either pYX-ScRcl1, pYX-MmRcl1, pYX-HsRtc1 or the empty vector pYX242. Transformants were selected on SGal-TL plates. Drop-tests were performed from independent colonies at either permissive conditions (on SGal-TL) or restrictive conditions (on SD-TL and YPD plates). Complementation was monitored by analysing growth of the transformants under restrictive conditions.
RNA extraction, Northern blotting and primer extension
RNA from yeast cells and IP pellets was isolated using the standard hot-phenol procedure. RNA from gradient fractions was isolated by proteinase K treatment (50 µg/ml) in the presence of 0.5% SDS at 55°C, and subsequent phenol–chloroform extraction. For analysis of small species, RNA was separated on 8 or 10% PAGE–8 M urea gels, and electroblotted to Hybond-N+ membrane, using 0.5× TBE as a transfer buffer. For analysis of high-molecular-weight RNAs, they were separated on 1.2% agarose gels containing formaldehyde, and electroblotted as indicated above. Northern hybridizations were performed in 6× SSPE containing 0.5% SDS, 5× Denhardt’s solution at 37°C; membranes were washed in 6× SSPE at 8°C below the calculated melting temperature. The following oligonucleotide probes complementary to pre-rRNAs (see Figure 6) were used: a, CGCTGCTCACCAATGG; b, CCAGATAACT ATCTTAAAAG; c, GCTCTCATGCTCTTGCC; d, ATGAAAACTCC ACAGTG; e, CCAGTTACGAAAATTCTTG; f, GGCCAGCAATTT CAAGTTA; g1, CATGGCTTAATCTTTGAGAC; g2, GCTCTTTG CTCTTGCC. The oligonucleotide probes specific for snoRNAs were: U3, TAGATTCAATTTCGGTTT; U14, TCACTCAGACATCCTAGG; snR190, GTCGAATCGGACGAGG; snR10, AATTTGTTCTCCAG TCCAAGC; snR30, GCCGTTGTCCGAAGCGCC; U18, GTCAGAT ACTGTGATAGTC.
For primer extension, oligonucleotides g1 and f were used to analyse cleavage sites A0 and A1, and A2, A3 and B1L/S, respectively. Reactions were performed using total RNA as template and the SuperScript II reverse transcriptase (Life Technologies) according to the manufacturer’s recommendations. Transcriptions with oligo g1 contained 100-fold more primer than those with oligo f, to ascertain the molar excess over the 18S rRNA template. To identify positions of the extension stops, sequencing reactions with primers g1 and f were performed on rDNA carried by plasmids kindly provided by P.Linder.
Anti-Rcl1p Abs and immunoprecipitations
The synthetic peptide corresponding to residues 355–367 of Rcl1p was coupled to KLH via an N-terminal cysteine included in the peptide, and used to immunize rabbits (Eurogentec). Polyclonal Abs were purified from serum using a Sulfolink peptide-affinity column (Pierce). IPs were performed as previously described (Ganot et al., 1997). Buffers contained either 500 mM (for IP of RNA) or 250 mM (for IP of proteins) potassium acetate during binding and washing steps.
Gradient analyses
Polysome profiles were analysed as previously described by Zanchin et al. (1997) with minor changes. Twenty-five OD254 units of the yeast extract were layered on 15–50% (w/v) sucrose gradients, which were centrifuged in a Beckman SW41Ti rotor at 38 000 r.p.m. for 3 h at 4°C.
To analyse the distribution of Rcl1p and the U3 RNA, 100 OD600 units of the BMA41 cells were lysed with glass beads by vigorous vortexing in a buffer containing 20 mM Tris–HCl pH 8, 5 mM MgCl2, 150 mM NaCl, 0.2% Triton X-100, 1 mM dithiothreitol (DTT) and the Complete protease inhibitor cocktail (Roche Molecular Biochemicals). The extract was clarified by centrifugation at 10 000 g for 10 min and layered on a 10–30% glycerol gradient prepared in the lysis buffer (with the concentration of Triton X-100 reduced to 0.1% and protease inhibitors omitted). Gradients were centrifuged at 25 000 r.p.m. for 14 h (short run) or 36 000 r.p.m. for 24 h (long run).
Pulse–chase labelling and immunolocalization
Labelling was done according to Zanchin et al. (1997). For its details and immunolocalization, see Supplementary material.
Supplementary material
Supplementary material to this paper is available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank J.Aris, M.Caizergues-Ferrer, T.Kiss, P.Linder, L.Minvielle-Sebastia, T.Wiederkher, W.Keller and B.Seraphin for strains and reagents, D.Tollervey, L.Minvielle-Sebastia, P.Linder and A.Wlodawer for valuable advice and discussions, K.Mizuguchi for Rcl1p modelling, and P.Genschik and G.Thomas for comments on the manuscript.
References
- Arn E.A. and Abelson,J.N. (1998) RNA ligases: function, mechanism and sequence conservation. In Simons,R.W. and Grunberg-Manago,M. (eds), RNA Structure and Function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 695–726. [Google Scholar]
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in S.cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M. and Tollervey,D. (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M. and Tollervey,D. (1995) Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E., Hess,D., Hofsteenge,J. and Filipowicz,W. (1999) Characterization of the adenylation site in the RNA 3′-terminal phosphate cyclase from E.coli. J. Biol. Chem., 274, 34955–34960. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos,O., Li,G.Y., Labouesse,M., Minvielle-Sebastia,L. and Lacroute,F. (1991) A family of low and high copy replicative, integrative and single-stranded S.cerevisiae/E.coli shuttle vectors. Yeast, 7, 609–615. [DOI] [PubMed] [Google Scholar]
- Brown A.J.P. and Tuite,M. (1998) Yeast Gene Analysis. Academic Press, London, UK. [Google Scholar]
- de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in S.cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. [DOI] [PubMed] [Google Scholar]
- Dunbar D.A., Wormsley,S., Agentis,T.M. and Baserga,S.J. (1997) Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol., 17, 5803–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Esser,S., Kastner,B. and Luhrmann,R. (1994) Isolation of S.cerevisiae snRNPs: comparison of U1 and U4/U6.U5 to their human counterparts. Science, 264, 261–265. [DOI] [PubMed] [Google Scholar]
- Filipowicz W. and Vicente,O. (1990) RNA 3′-terminal phosphate cyclase from HeLa cells. Methods Enzymol., 181, 499–510. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Konarska,M., Gross,H.J. and Shatkin,A.J. (1983) RNA 3′-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res., 11, 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Strugala,K., Konarska,M. and Shatkin,A.J. (1985) Cyclization of RNA 3′-terminal phosphate by cyclase from HeLa cells proceeds via formation of N (3′)pp (5′)A activated intermediate. Proc. Natl Acad. Sci. USA, 82, 1316–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- Genschik P., Billy,E., Swianiewicz,M. and Filipowicz,W. (1997) The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria and Archaea. EMBO J., 16, 2955–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P., Drabikowski,K. and Filipowicz,W. (1998) Characterization of the E.coli RNA 3′-terminal phosphate cyclase and its σ54-regulated operon. J. Biol. Chem., 273, 25516–25526. [DOI] [PubMed] [Google Scholar]
- Hughes J.M. (1996) Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol., 259, 645–654. [DOI] [PubMed] [Google Scholar]
- Hughes J.M. and Ares,M.J. (1991) Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J., 10, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Tollervey,D. and Hurt,E.C. (1993) A U3 snoRNP protein with homology to splicing factor PRP4 and G β domains is required for ribosomal RNA processing. EMBO J., 12, 2549–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Minvielle-Sebastia,L., Preker,P.J. and Keller,W. (1996) Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science, 274, 1514–1517. [DOI] [PubMed] [Google Scholar]
- Kass S. and Sollner-Webb,B. (1990) The first pre-rRNA-processing event occurs in a large complex: analysis by gel retardation, sedimentation and UV cross-linking. Mol. Cell. Biol., 10, 4920–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Linder,P. and de la Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in S.cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D., Vandenhaute,J. and Tollervey,D. (1995) The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev., 9, 2470–2481. [DOI] [PubMed] [Google Scholar]
- Lee S.J. and Baserga,S.J. (1997) Functional separation of pre-rRNA processing steps revealed by truncation of the U3 small nucleolar ribonucleoprotein component, Mpp10. Proc. Natl Acad. Sci. USA, 94, 13536–13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J. and Baserga,S.J. (1999) Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol., 19, 5441–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.D., Zagorski,J. and Fournier,M.J. (1990) Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in S.cerevisiae. Mol. Cell. Biol., 10, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E. and Dahlberg,J.E. (1992) Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNAs. Science, 255, 327–330. [DOI] [PubMed] [Google Scholar]
- Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- Morrissey J.P. and Tollervey,D. (1993) Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol., 13, 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougey E.B., O’Reilly,M., Osheim,Y., Miller,O.L., Beyer,A. and Sollner-Webb,B. (1993) The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev., 7, 1609–1619. [DOI] [PubMed] [Google Scholar]
- Palm J.G., Billy,E., Filipowicz,W. and Wlodawer,A. (2000) Crystal structure of RNA 3′-terminal phosphate cyclase, an ubiquitous enzyme with unusual topology. Structure, 8, 13–23. [DOI] [PubMed] [Google Scholar]
- Puig O., Rutz,B., Luukkonen,B.G., Kandels-Lewis,S., Bragado-Nilsson,E. and Seraphin,B. (1998) New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast, 14, 1139–1146. [DOI] [PubMed] [Google Scholar]
- Reinberg D., Arenas,J. and Hurwitz,J. (1985) The enzymatic conversion of 3′-phosphate terminated RNA chains to 2′,3′-cyclic phosphate derivatives. J. Biol. Chem., 260, 6088–6097. [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson,E., Kandels-Lewis,S. and Seraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K. and Tollervey,D. (1999) Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol., 19, 6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. (1987) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J., 6, 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- Trapman J., Retel,J. and Planta,R.J. (1975) Ribosomal precursor particles from yeast. Exp. Cell Res., 90, 95–104. [DOI] [PubMed] [Google Scholar]
- Venema J. and Tollervey,D. (1995) Processing of pre-ribosomal RNA in S.cerevisiae. Yeast, 11, 1629–1650. [DOI] [PubMed] [Google Scholar]
- Wiederkehr T., Pretot,R.F. and Minvielle-Sebastia,L. (1998) Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA, 4, 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Zanchin N.I., Roberts,P., DeSilva,A., Sherman,F. and Goldfarb,D.S. (1997) S.cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell. Biol., 17, 5001–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]