Abstract
Bone morphogenetic protein-7 (BMP7) is an endogenous antifibrogenic protein in the kidney which is down regulated in experimental chronic kidney diseases such as obstructive and diabetic nephropathy in parallel with progressively increasing TGFβ. In-vitro studies were performed in Madin-Darby Canine Kidney (MDCK)-cells to identify transcriptional regulators of BMP7. Experiments with various BMP7 promoter fragments (465 to 4267 bp) identify small proximal promoter segments that are transcriptionally activated by high glucose (3.2-fold) but down regulated by TGFβ (0.2-fold) compared to normal glucose. Protein binding to these DNA-segments is increased by high glucose and decreased by TGFβ in a time-dependent, progressive manner. Analysis of BMP7 promoter binding proteins with liquid chromatography/tandem mass spectrometry (LC/MS/MS) identifies seven unique, partially overlapping peptides, spanning 25% of the amino acid sequence of Y-Box protein-1 (YB1). EMSA - Western blot combination experiments confirm that YB1 is a BMP7 promoter binding protein. YB1 knock-down reduces transcriptional responses to high glucose and TGFβ by about one-half, respectively. In addition, high glucose induces but TGFβ reduces nuclear translocation of YB1 from the cytoplasm. These studies identify YB1 as a transcriptional activator of BMP7 and help to explain the progressive decline in renal BMP7 in diabetic nephropathy and other kidney diseases.
Keywords: BMP7, Fibrogenesis, transcriptional activation, diabetic nephropathy, TGFβ
Bone morphogenetic protein-7 (BMP7) is a member of the transforming growth factor-β (TGFβ) superfamily of cysteine-knot cytokines and growth factors. During development BMP7 functions as a morphogen with pivotal roles in eye and kidney development [Dudley et al., 1995]. Post-natally, BMP7-expression decreases and disappears from most tissues where it is developmentally expressed. In adult organisms its expression is limited to few tissues, most notably bone and the kidneys. In normal kidneys BMP7 expression is mainly maintained in glomerular visceral epithelial cells (podocytes) and in distal and connecting tubules and collecting ducts of the cortex, and minimally of the medulla and papilla.
BMP7 binds and activates BMP-receptor heterodimers (or heterotetramers) composed of type II and type I receptors. The latter include activin-like kinases-2, -3 and -6 (Alk2, -3 and -6). In the kidney Alk6 is not expressed, and Alk3 expression is limited to podocytes [Mitu et al., 2007]. In contrast, Alk2 is widely expressed in glomeruli including podocytes and on tubular cells of all nephron segments although there is relative paucity of BMP7 binding in proximal tubules [Bosukonda et al., 2000; Mitu et al., 2007]. Similar to other TGFβ-superfamily proteins BMP7 uses smad signal proteins as the major intracellular receptor-activated substrates. Previous studies have shown that in renal cells smad5 is the preferred BMP7 signaling smad rather than smad1 [Mitu et al., 2007; Wang and Hirschberg, 2004]. As the preferred receptor smad for BMP7, activated smad5 forms a heterdimer with the non-phosphorylated common smad, smad4, and favors its accumulation in the nucleus and association with other transcriptional regulators and support proteins forming larger complexes at target promoter sites.
BMP7 receptors are widely expressed in the kidney [Bosukonda et al., 2000] suggesting autocrine, paracrine and endocrine modes of action of renal BMP7. In the kidney BMP7 functions as an antifibrogenic regulator reducing the profibrogenic activity of TGFβ [Wang and Hirschberg, 2003]. In this context BMP7 acts as an inhibitor of TGFβ smad3 signaling by inducing the inhibitory smad, smad6, downstream of activated smad5 [Wang and Hirschberg, 2004]. Several in-vivo studies in experimental renal diseases in rodents have confirmed the efficacy of BMP7 as an antifibrogenic agent. Streptozotocin-induced diabetic nephropathy is ameliorated in mice expressing transgenic BMP7 in the kidney [Wang et al., 2006]. Moreover, in rats with streptozotocin-induced diabetic nephropathy early interstitial fibrosis is reduced when recombinant BMP7 is administered systemically [Wang et al., 2003]. Most remarkably, when BMP7 administration is delayed and begun several weeks after induction of diabetes and onset of diabetic nephropathy, early interstitial fibrosis is reversed [Wang et al., 2003]. Similarly, the rapid renal fibrogenesis of obstructive nephropathy after unilateral ureteral obstruction is ameliorated by exogenous administration of BMP7 [Hruska et al., 2000].
The BMP7 promoter has been partially characterized especially in the context of expression of BMP7 in osteoblast-like cells. The human BMP7 gene has a single major transcription initiation site [Kawai and Sugiura, 2001]. Little is known about the transcriptional regulation of BMP7. The transcription factor and sonic hedgehog mediator glioma associated oncogene, Gli, transcriptionally activates BMP7 in osteoblasts and COS-7 cells in-vitro in co-transfection experiments [Kawai and Sugiura, 2001]. Whether this regulation is relevant in adult kidney is unknown and perhaps questionable. Retinoic acid (RA) has been found to transcriptionally regulate other TGFB superfamily members BMP2, BMP4 and activin A despite of the absence of known RA-binding motifs [Balmer and Blomhoff, 2002]. RA also activates BMP7 transcription at least in the context of bone metabolism [Paralkar et al., 2002]. Prostaglandin E2 which raises BMP7 levels in bone is not a transcriptional activator but acts on post-transcriptional mechanisms such as mRNA stabilization [Paralkar et al., 2002]. Estradiol has been shown to function as a repressive regulator of BMP7 in estrogen-sensitive tissues; its transcriptional repression, however, may be indirect [Monroe et al., 2000; Ozkaynak et al., 1997].
In experimental diabetes in rodents renal expression of BMP7 decreases and virtually disappears a few weeks after induction of the disease predating onset and progression of renal interstitial matrix accumulation. This loss of endogenous renal BMP7 is also observed in experimental obstructive nephropathy [Hruska et al., 2000; Morrissey et al., 2002; Wang et al., 2001]. These observations gave rise to the current studies to examine the transcriptional regulation of BMP7 as it relates to high glucose states and increased TGFβ.
Materials & Methods
Cell Culture Model
Studies were performed in Madin-Darby Canine Kidney (MDCK) cells which have originated from and express features of renal distal tubular cells and express high levels of BMP7. Distal tubules and cortical collecting ducts in-vivo are the sites with highest renal BMP7 expression in-vivo in rodents and humans. MDCK-cells were cultured in Eagle’s minimum essential medium with addition of 1.5 g/l NaHCO3, 0.1 mM non-essential amino acids, 1.0 mM Na-pyruvate and penicillin (100 IU/ml), streptomycin (100 μg/ml) and amphotericin B (0.25 μg/ml).
Luciferase Reporter Assays
The 4267 bp upstream (−4264 to +3) of the translation start site of the murine BMP7 gene was generated by polymerase chain reaction (PCR) from mouse genomic DNA and ligated into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA). The identity of the construct was confirmed by DNA sequencing, and the BMP7 promoter region DNA fragment was subcloned into pGL3-basic luciferase reporter vector (Promega Inc, Madison, WI). Then 5′-deletion series of the 4267 bp promoter fragment were generated by restriction enzyme digestion: Mlu I and BstX I for a 3432 bp fragment (−3429 to +3); Mlu I and Nde I for a 2712 bp fragment (−2709 to +3); Mlu I and BstE II for a 1956 bp fragment (−1953 to +3); Sac I for a 1395 bp fragment (−1392 to +3); Kpn I for a 920 bp fragment (−917 to +3), and Mlu I and BssH II for a 465 bp fragment (−462 to +3). The identities of the constructs were confirmed by restriction analysis.
For transient transfection, MDCK cells (ATCC, Manassas, VA) were seeded in 96-well plates at a density of 1.6 × 104 cells per well in EMEM medium (ATCC, Manassas, VA) containing 100 mg/dL D-glucose and 10% FBS. At 75% confluence, 0.3 μg of each of the above luciferase plasmids were transfected using 0.5 μl of Lipofectamine 2000 (Invitrogen, Carlsbad, CA), according to the manufacturer’s protocol. Empty plasmid was used as negative control, and phRL-TK vector (for expression of Renilla, Promega Inc, Madison, WI) was included for normalization of transfection efficiency. Eighteen hours after transfection, the cells were serum-starved with EMEM medium containing 0.1% FBS for 24 hours, and then treated with normal glucose (5.5 mM), TGF-β (1 nM), or high glucose (25 mM) together with glycated albumin (400 μg/ml) for 24 hours. Luciferase activity was measured in cell lysates using the Luciferase Assay System, Renilla Luciferase Assay System (Promega Inc, Madison, WI) and a Femtomaster FB 12 luminometer (Zylux, Maryville, TN).
EMSA
Nuclear proteins were extracted from MDCK cells that were treated with high glucose (25 mM) and glycated albumin (400 μg/ml), or TGF-β (1 nM) using the Nuclear Extract Kit (Active Motif, Carlsbad, CA) and the manufacturer’s protocol. DNA fragments corresponding to 403 bp (−400 to +3), 326 bp (−323 to +3), 195 bp (−192 to +3), 116 bp (−113 to +3), 90 bp (−197 to −108) and 50 bp (−197 to −148; −157 to −108) upstream of the translation start site of the murine BMP7 gene were generated by PCR, gel-purified with the UV-Free Gel Purification kit (Invitrogen, Carlsbad, CA), and labeled with biotin using the Biotin 3′ End DNA Labeling kit (PIERCE, Rockford, IL). The Lightshift Chemiluminescent EMSA kit (PIERCE, Rockford, IL) was used to perform EMSAs, according to the manufacturer’s instruction. Briefly, 1.5 μg nuclear extracts were incubated with 20 fmol biotin-labeled double stranded DNA probes in a 20 μl reaction mixture containing 1 × binding buffer and 50 ng/μl poly(dIx·dC) at room temperature for 20 minutes. For competition analysis, 100-fold molar excess of unlabeled double stranded DNA fragment was added to the nuclear extracts prior to the addition of the labeled probe. Binding reactions were resolved in 5% polyacrylamide gels in 0.5 × TBE, electrophoresed at 100 V for 90 minutes, and transferred to nylon membranes. The biotin-labeled DNA on the membrane was detected by chemiluminescence.
Identification of BMP7 Promoter Binding Proteins by LC/MS/MS
5′ biotinylated 195 bp DNA fragments corresponding to the promoter region (−192 to +3) of the murine BMP7 gene and μMACS Factor Finder kit (Miltenyi Biotec, Auburn, CA, USA) were used to isolate promoter binding proteins from binding reactions, according to the manufacturer’s protocol. Briefly, 25 μg nuclear protein from MDCK cells was incubated with 5 pmol biotinylated DNA fragment of the BMP7 promoter in binding buffer containing binding enhancer and poly(dI·dC) at room temperature for 20 minutes. Then μMACS Streptavidin MicroBeads were added for an additional 10 minutes. The reaction mixture was separated magnetically and resolved by SDS-PAGE. Protein bands were visualized by SYPRO Ruby (Molecular Probes, Eugene, OR, USA) and manually excised.
With the −192 to +3 bp promoter segment and nuclear extracts from MDCK-cells, three bands were visualized and excised. In-gel proteolytic digestion and liquid chromatography followed by tandem mass spectrometry were performed by NextGen Sciences, Ann Arbor, MI. Briefly, proteolysis was performed on a ProGest digestion robot (Digilab Genomic Solutions, Ann Arbor, MI, USA). Samples were reduced with DTT at 60 °C, alkalinized with iodoacetamide and trypsin digested. Thirty μl of hydrolysate were loaded on a 75 μm C12 vented column at a flow rate of 10 μl/min and eluted at 300 nl/min and a 1 hr gradient and then analyzed on a ThermoFisher Orbitrap XL mass spectrometer. LC/MS/MS-data were analyzed with Mascot (Matrix Science, London, England, UK) searching the Combo Canis Refseq 112307 database containing 67302 entries. Searches were performed with a fragment ion mass tolerance of 0.5 Da and a parent ion tolerance of 10.0 PPM. Iodoacetamine derivative of cysteine was specified in Mascot as fixed modification. Validation of MS/MS-based peptide and protein identifications were performed with Scaffold software (Version 2.01.01, Proteome Software Inc, Portland, OR, USA). Peptide identifications were accepted if established at >80% probability as specified by the Peptide Prophet algorithm [Keller et al., 2002]. Protein identifications were accepted at >90% probability if containing at least 2 identified peptides. The Protein Prophet algorithm was used to assign proteins [Nesvizhskii et al., 2003]. Proteins that contained similar peptides and could not be differentiated based on MS/MS-analysis were grouped to satisfy principles of parsimony. As shown in Results YB1 was identified as a BMP7 promoter binding protein by the presence of seven unique peptides with a probability >95%.
EMSA-Western Blot Analysis of YB1
For confirmation of binding of YB1 to the promoter DNA-segments EMSA-supershift assays were attempted. Two different antibodies were tested in supershift assays, namely a rabbit anti-YB1-C-terminal antibody (Sigma-Aldrich, St. Louis, MO, USA) and a chicken anti-YB1-C-terminal. This latter antibody had been raised against the peptide MSSEAETQQPPAA and was kindly provided as a gift by Isabelle Berquin, Wake Forest University, Winston-Salem, NC. Neither antibody generated supershift bands. Reasons may include masking of the antigen by DNA binding and/or lack of cross-reactivity with canine YB1 (the rabbit antibody reacts with canine YB1 in Western blots but the chicken antibody does not).
In lieu of supershift assays we positively identified the presence of YB1 by a combined EMSA-Western blot approach: Two identical EMSA assays were performed in parallel. Both DNA/nuclear protein mixtures were electrophoresed in the same gel in TBE-buffer together with nuclear proteins without labeled promoter DNA as control. The gel was cut vertically into two equal halves. DNA bands in one half-gel were transferred to nylon membranes and DNA was visualized as in EMSAs described above. The other half-gel was subjected to a buffer exchange by incubation with three exchanges of tris-glycine buffer (30 min each). Proteins were then electro-transferred to nitrocellulose in tris-glycine buffer. Membranes were blocked with 5% dry milk in tris-buffered saline containing Tween-20 and then incubated with rabbit-anti YB1 (1:10,000) in blocking buffer, followed by several washes and incubation with horseradish peroxidase conjugates second antibody. Bands were visualized with enhanced chemiluminescence and captured on X-ray film. This experiment was performed in three independent repeats with similar results.
YB1 Knock-down
For the measurement of BMP7 promoter activity in response to high glucose or TGFβ, YB1 siRNA (S76162, Ambion, Carlsbad, CA, USA) or gapdh siRNA or scrambled siRNA (Ambion), the latter two as controls, were mixed with siPORT NeoFX transfection reagent in Opti-MEM1 medium and dispensed into 96-well plates. Wells were overlayed with cells and incubated for 17 hrs. Cells were then transfected with pGL3 basic or pGL3-BMP7p (−462 to +3 bp) together with the Renilla control vector phRL-TK (Promega, Madison, WI, USA) in lipofectamin 2000 for 6 hrs. Cells were washed and incubated with serum-free MEM/0.1% BSA for 24 hrs, and then treated with this medium (control) or medium containing normal glucose (5.5 mM), high glucose (25 mM) and glycated albumin (400 μg/ml) or TGFβ (1 nM) for 24 hrs. Cells were washed, lysed and luciferase was measured luminometrically.
Results
High Glucose and TGFβ Responsive Promoter
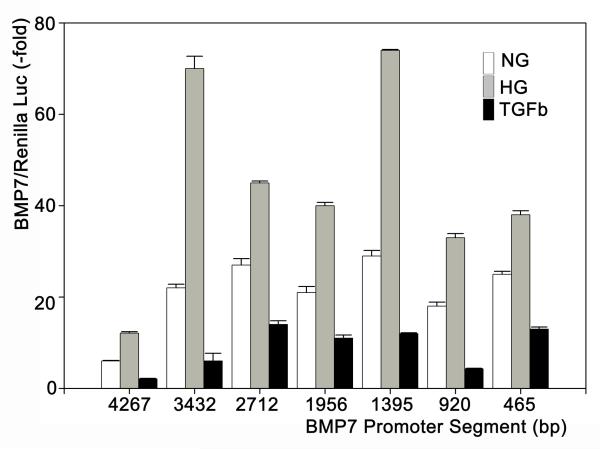
Diabetic nephropathy is associated with exposure of tubular cells to high glucose levels, glycated proteins and TGFβ (which is induced by high glucose and glycated proteins and is a major regulator of many aspects of renal fibrogenesis in diabetes and other chronic renal diseases). We examined the regulation of the murine BMP7 promoter by exposing MDCK-cells that were transfected with the pGL3-Luc containing different portions of the BMP7 promoter between 4267 bp and 465 bp upstream of the translation initiation site. Incubations with serum-free medium containing normal glucose (5.5 mM), high glucose (25 mM) plus glycated albumin (400 μg/ml) or recombinant TGFβ (1 nM) yielded differential activation of the promoter segment as indicated by levels of luciferase activity (Figure 1). All promoter segments between +3 to −462 and +3 to −4264 bp relative to the translation initiation codon resulted in increased activity with high glucose + glycated albumin (HG) compared to normal glucose (NG, Control). In contrast, TGFβ substantially downregulated the promoter activity by 50 – 80% (Figure 1).
Figure 1. Activation of partial BMP7 promoters by high glucose or TGFβ.
pGL3-Luc reporters were inserted with different portions upstream of the translation start site (+3 bp) and were co-transfected with the Renilla control vector into MDCK-cells which were incubated with normal glucose (5.5 mM), high glucose (25 mM) + glycate albumin (400 μg/ml) or TGFβ (1 nM) (n = 15-20 each). The ratio of the luciferase/renilla luminescence is expressed as multiples of this ratio with the empty pGL3-luc vector.
BMP7-Promoter Binding Proteins
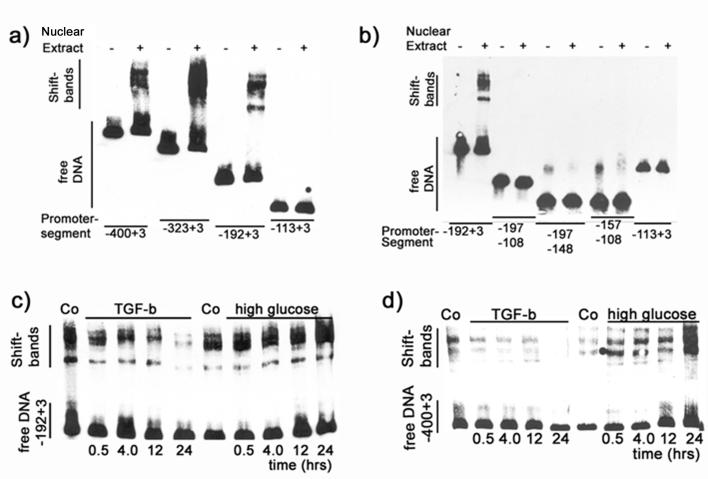
A series of EMSA assays was performed to examine binding of nuclear proteins to different segments of the promoter. MDCK cells were incubated with HG for 24 hours and nuclear proteins were extracted. Promoter DNA segments were biotin-labeled and then incubated with nuclear extracts, electrophoresed and transferred to nylon membranes. DNA was then visualized with streptavidin-luciferase and chemiluminescence. Incubation with nuclear extract yielded two to three shift bands in EMSAs of promoter segments of ≥192 bp (+ 3 bp of the translation start site) (Figure 2a). In contrast, a smaller segment (−113 to +3 bp) did not yield any shift band (Figure 2a). Similarly, portions of the minimal promoter, namely −197 to −108, −197 to −148 and −157 to −108 bp did not show any shift bands under the current conditions (Figure 2b).
Figure 2. High glucose and TGFβ regulate binding of proteins to portions of the BMP7 promoter.
a) Nuclear extracts from cells that were incubated with high glucose + glycated albumin induced shift bands with BMP7 promoter segments ≥195 bp upstream of the translation start site. b) Fractions of the 195 bp promoter segment and a proximal 115 bp promoter segment lack these protein binding sites. c+d) Time course EMSA experiments with the 195 bp promoter (c) and a 403 bp segment (d) show time-dependent increases in the protein binding with high glucose + glycated albumin but decreasing protein binding with extracts from cells that had been incubated with TGFβ.
Time course experiments were performed with nuclear extracts from MDCK-cells that had been incubated with high glucose (25 mM + glycated albumin, 400 μg/ml) or with TGFβ (1 nM) for 0.5, 4, 12 or 24 hours. Nuclear extracts that were obtained from these cells were used in shift assays on two different partial promoters, namely −192 to +3 bp and −400 to +3 bp. Findings between the two promoter segments were similar and show that shift band intensity increased with the period of time of incubation of nuclear extract donor cells with high glucose. In contrast, incubation with TGFβ resulted in a time-dependent decrease in band intensity (Figure 2c). Similar results were observed with a −400 to +3 bp segment of the promoter (Figure 2d). Thus, high glucose increases the levels of BMP7 promoter binding proteins but TGFβ decreases their levels. This finding also concurs with results from the promoter activity assays (Figure 1).
Unbiased identification of BMP7-promoter binding proteins
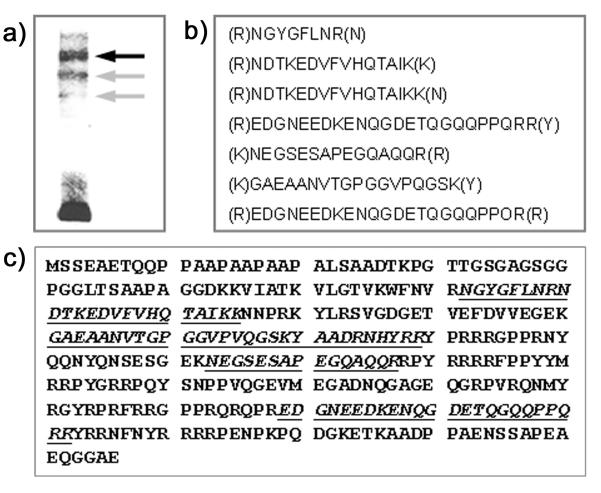
DNA binding proteins that induce shift bands in the EMSA assays likely include transcription regulating proteins. To positively identify proteins causing shifts in the EMSA assays three identifiable bands were excised from the electrophoresis gel. Proteins were in-gel digested with trypsin and analyzed by HPLC followed by tandem mass spectrometry (performed by Nextgensciences Corporation, Ann Arbor, MI, USA). Many peptides were identified in each of the three bands. In the upper band (black arrow in Figure 3a), 71 peptides were identified; 69 peptides were found in the middle band (gray arrow in Figure 3a) and 139 peptides were found in the third band (lower gray arrow in Figure 3a). Most of these peptides were assigned by the Scaffold analysis to structural proteins and in several cases there was ambiguity because peptides were not unique.
Figure 3. Identification of Y-Box Protein-1 (YB1) as a BMP7-promoter Binding Protein.
a) DNA-protein complexes after incubation of nuclear proteins from MDCK-cells that had been exposed to high glucose + glycated albumin with the −192 to +3 bp BMP7-promoter fragment were electrophoresed. 3 bands were individually excised, trypsin-digested and analyzed with HPLC/MS/MS. b) In one band (dark arrow in a), seven unique peptides were identified. c) These peptides (underlined) are unique for YB1, are partially overlapping and cover 25% of the YB1 amino acid sequence.
One protein was positively identified with highest probability (>95%) by the presence of 7 unique, partially overlapping peptides that were present only in the prominent band (black arrow in Figure 3a). These peptides are listed in Figure 3b. This protein was identified with near certainty (probability >95%) as Y-box protein-1 (YB1). The 7 peptides (Figure 3b) cover about 25% of the amino acid sequence of canine YB1 and are partially overlapping (Figure 3c).
Confirmation of YB1 as a BMP7 Promoter Binding Protein
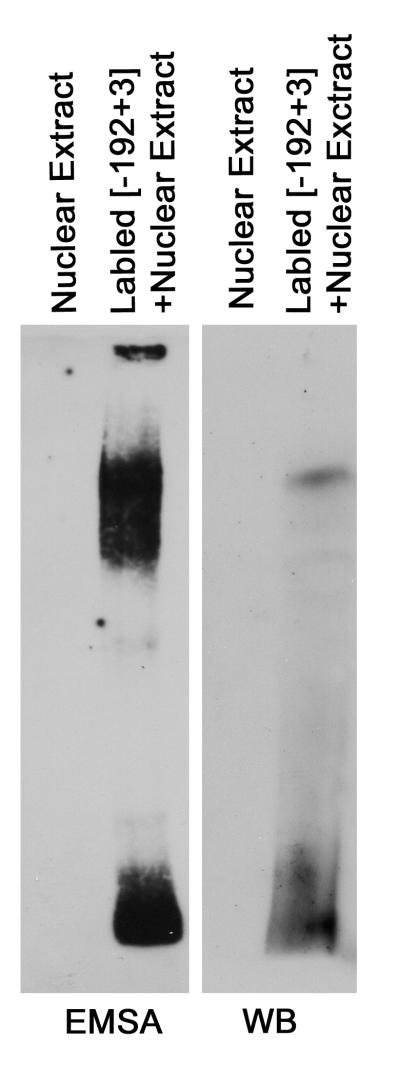
To positively demonstrate binding of YB1 to the BMP7 promoter segments supershift assays were attempted but were unsuccessful, probably because of limitations in the available YB1 antibodies. In lieu of EMSA supershifts, assays were performed using nuclear extracts from HG-exposed MDCK cells and the biotin-conjugated PLoS One192 to +3 bp promoter segment in duplicate in the same gel. After electrophoresis the gel was cut in two halves. One half-gel was processed for visualization of the labeled DNA as in the above EMSAs. The second half-gel underwent buffer exchange, and Western blotting with anti-YB1 was performed after protein transfer to nitrocellulose membranes. The findings demonstrate a single band in the Western blot that corresponds to the band in the EMSA assay (Figure 4). These results confirm that YB1 is indeed a BMP7-promoter binding protein.
Figure 4. Confirmation of YB1 as a BMP7 Promoter Binding Protein.
Nuclear extracts from high glucose/glycated albumin - treated cells were incubated with the biotinylated −192 to +3 bp promoter segment and separated by SDS-page in duplicate. The gel was cut into identical halves and each half-gel was either processed for EMSA (left) or Western Blot with anti-YB1 antibody (right). Findings demonstrate that the top band in the EMSA contains YB1.
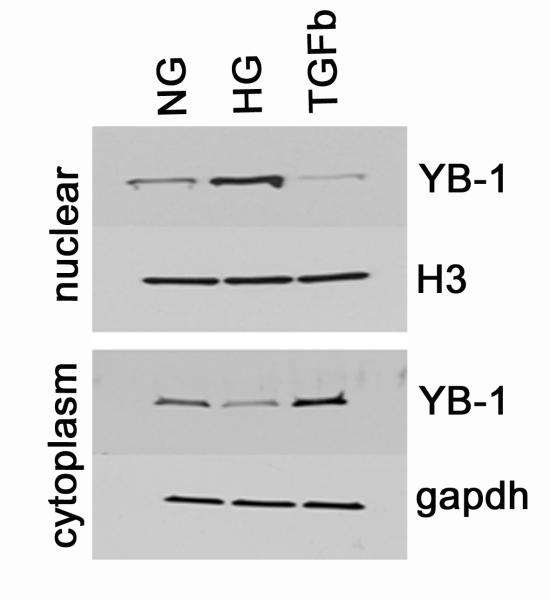
Nuclear Translocation of YB1
Nuclear and cytoplasmic levels of YB1 were examined by Western blot analysis after separation of nuclei and nuclear protein extraction from MDCK cells that had been incubated with normal glucose (5.5 mM), high glucose/glycated albumin (25 mM/400 μg/ml) or TGFβ (1 nM). As shown in Figure 5, nuclear YB1 levels increase with HG but decrease with TGFβ compared to incubation with normal glucose levels. YB1 levels in the cytoplasm of these same cells show the opposite pattern: During incubation with high glucose cytoplasmic YB1 levels decrease but they increase compared to NG during exposure to TGFβ. These results indicate nuclear translocation of YB1 with HG and translocation in the reverse direction with TGFβ (Figure 5).
Figure 5. YB1 undergoes Nuclear Translocation.
During incubation with high glucose + glycated albumin (HG), there is translocation of YB1 from the cytoplasm to the nucleus compared to incubation with normal glucose (NG). In contrast, TGFβ favors cytoplasmic accumulation of YB1 and reduces nuclear translocation.
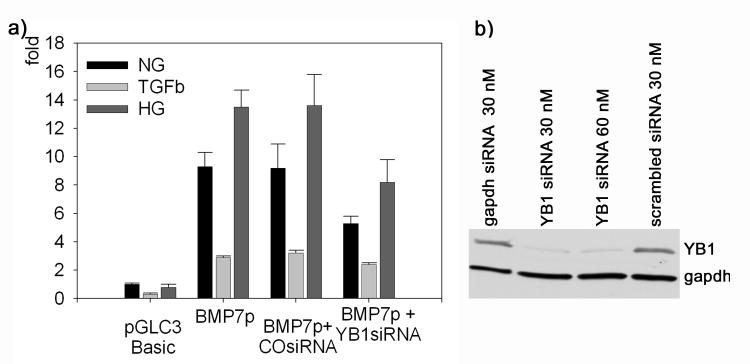
YB1-Knock-down Reduces BMP7 Promoter Activity
As shown above, high glucose up-regulates and TGFβ down-regulates the transcriptional activity of proximal portions of the BMP7 promoter in parallel with nuclear or cytoplasmic accumulation of YB1, respectively, which binds to these partial promoters. To directly examine the contribution of YB1 to the high glucose and TGFβ - dependent transcriptional regulation, the 465 bp partial BMP7 promoter/luc construct was transfected into cells with or without YB1 knock-down using specific siRNA. Findings indicate that YB1 knock-down partially blocks the transcriptional activity response to normal glucose, high glucose and TGFβ, respectively (Figure 6). These findings confirm that YB1 is a transcriptional regulator of BMP7 with relevance to diabetes and diabetic nephropathy.
Figure 6. Knock-down of YB1 blocks transcriptional Regulation of BMP7.
a) Transcriptional efficiency of the partial BMP7 promoter (BMP7p, −462 to +3 bp) with or without YB1 knock-down (YB1 siRNA) or scrambled siRNA control (CO siRNA) during incubation with normal glucose (NG), TGFβ or high glucose + glycated albumin (HG); n = 10-15 each. b) Efficiency of the YB1 siRNA (Western blot).
Discussion
BMP7 is an antifibrogenic regulator in the kidney and functions by opposing profibrogenic signals of TGFβ [Mitu et al., 2007; Wang et al., 2003; Wang et al., 2006; Wang and Hirschberg, 2003; Zeisberg et al., 2003]. Its levels are reduced early in chronic renal diseases and virtually disappear not only in diabetic nephropathy but also in nondiabetic chronic renal diseases with fibrosis such as unilateral obstructive nephropathy [Hruska et al., 2000; Wang et al., 2001]. The present experimental studies were performed to identify transcriptional regulators. YB1 was found to be a BMP7-promoter regulating protein as described here.
Thus far, only one BMP7-regulating transcription factor has been identified, namely retinoic acid [Balmer and Blomhoff, 2002; Paralkar et al., 2002]. Estrogen is an important repressor of BMP7 in the endometrium and other estrogen-sensitive tissues, perhaps even bone but its action may be indirect and mediated by estrogen-dependent transcriptional repressors [Monroe et al., 2000; Ozkaynak et al., 1997]. Sonic hedgehog (SHH)-regulated Gli’s have been found in-vitro in co-transfection experiments to transcriptionally upregulate BMP7 [Kawai and Sugiura, 2001]. This may be important during development where SHH is widely expressed in bone and kidney but its relevance in adult kidney is quite questionable given the absence of SHH. There is surprisingly little known about the transcriptional regulation of BMP7 in extra-osseous tissues such as kidney and in disease states such as diabetes mellitus and chronic kidney disease.
It is somewhat surprising that high glucose actually increased the transcriptional activation of the BMP7-promoter given that in-vivo in diabetic nephropathy, BMP7 expression progressively declines. One explanation is that emerging TGFβ in diabetic nephropathy becomes the dominant regulator. This view is supported by the previous findings that the decline in BMP7 expression in experimental diabetic nephropathy occurs slowly over several weeks and parallels the emergence and rise in renal TGFβ levels in experimental nephropathy [Wang et al., 2006; Wang et al., 2001]. Indeed, Yang and associates examined the levels of BMP7 in streptozotocin-induced diabetic nephropathy in rats and found an early (<2 wks) increase in renal expression of BMP7 but subsequently a chronically progressing decrease below the expression in control animals [Yang et al., 2007]. Thus, findings from in-vivo experimental studies in diabetes and diabetic nephropathy in rodents are highly compatible with the present results.
YB1 belongs to the superfamily of cold shock domain proteins which are phylogenetically highly conserved. YB1 has pleiotropic functions which include transcriptional regulation and translational silencing [Evdokimova et al., 2006]. In the latter context YB1 is present in cytoplasmic mRNA processing bodies (P-bodies) as well as in the cytoplasm [Yang and Bloch, 2007]. Several transcriptional targets of YB1 have been identified although its name is derived from its binding to the Y-box sequence (CTGATTGGCCAA) in the promoter of MHC class II genes [Didier et al., 1988]. Several other genes containing a Y-box are upregulated by YB1 [Wolffe, 1994]. These also include, among other, the cell cycle regulators cyclin A and B1 as well as DNA-polymerase-α [En-Nia et al., 2005; Jurchott et al., 2003]. The BMP7-promoter does not contain a Y-box (or its reverse, a CCAAT-box). However, YB1 has previously been shown to also bind and regulate non-Y-box promoters such as the epidermal growth factor receptor and this transcription factor recognizes target sites other than the classic Y-box [Kolluri et al., 1992; To et al., 2007]. The chemokine CCL5 (RANTES) in monocytes/macrophages as well as the HGF-receptor MET in breast cancer and normal mammary epithelial cells are also transcriptionally regulated by YB1 despite of the absence of a classic Y-box [Finkbeiner et al., 2009; Raffetseder et al., 2009].
Given the presence of YB1 in the cytoplasm, in P-bodies where it represses translation of some mRNA-species, and in the nucleus where it engages in transcriptional regulation as well as in modulation of secondary nucleic acid structure [Skabkin et al., 2001], the question arises how its distribution between these compartments is regulated. Several different mechanisms have been proposed each of which may be context dependent including a role for p53, the splicing factor SRp30c during stress and/or by casein kinase II-dependent phosphorylation of CKII consensus sequences in the C-terminal portion of YB1 [Raffetseder et al., 2003; Skabkin et al., 2001; Valiahdi, 2006; Zhang et al., 2003]. Findings from the present experiments indicate that YB1 is indeed shuttled between the two compartments as indicated by data in Figure 5 showing a reverse mirror image between the nuclear and cytoplasmic levels of YB1: During incubation with HG cytoplasmic YB1 decreases and it accumulates in the nucleus. The opposite is found during incubation with TGFβ.
In summary, the present experiments demonstrate that in MDCK cells, a model of distal nephron - derived epithelial cells which efficiently express BMP7 in-vivo, high glucoses up-regulates but TGFβ down-regulates the transcriptional activity of BMP7. This is consistent with in-vivo findings in models of diabetic nephropathy. YB1 is identified as a high glucose and TGFβ - regulated transcriptional activator of BMP7 where high glucose increases but TGFβ reduces nuclear translocation of YB1. The current data also provide evidence that there are other, currently unidentified BMP7 transcriptional regulators. Since BMP7 has emergent as an important anti-fibrogenic cytokine in chronic kidney disease, knowledge of its transcriptional regulation may have relevance for the development of targeted therapies.
Acknowledgements
These studies were supported by grants from the Juvenile Diabetes Research Foundation (1-2004-78) and the National Institutes of Health (DK63360). The authors express their appreciation to Isabelle Berquin, PhD, Winston-Salem, NC, for the kind gift of the chicken anti-YB1 antibody that was used in some of the experiments.
References
- Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- Bosukonda D, Shih MS, Sampath KT, Vukicevic S. Characterization of receptors for osteogenic protein-1/bone morphogenetic protein-7 (OP-1/BMP-7) in rat kidneys. Kidney Int. 2000;58:1902–1911. doi: 10.1111/j.1523-1755.2000.00362.x. [DOI] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988;85:7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AT, Lyons KM, Robertson EJ. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 1995;9:2795–2807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- En-Nia A, Yilmaz E, Klinge U, Lovett DH, Stefanidis I, Mertens PR. Transcription factor YB-1 mediates DNA polymerase alpha gene expression. J Biol Chem. 2005;280:7702–7711. doi: 10.1074/jbc.M413353200. [DOI] [PubMed] [Google Scholar]
- Evdokimova V, Ovchinnikov LP, Sorensen PH. Y-box binding protein 1: providing a new angle on translational regulation. Cell Cycle. 2006;5:1143–1147. doi: 10.4161/cc.5.11.2784. [DOI] [PubMed] [Google Scholar]
- Finkbeiner MR, Astanehe A, To K, Fotovati A, Davies AH, Zhao Y, Jiang H, Stratford AL, Shadeo A, Boccaccio C, Comoglio P, Mertens PR, Eirew P, Raouf A, Eaves CJ, Dunn SE. Profiling YB-1 target genes uncovers a new mechanism for MET receptor regulation in normal and malignant human mammary cells. Oncogene. 2009;28:1421–1431. doi: 10.1038/onc.2008.485. [DOI] [PubMed] [Google Scholar]
- Hruska KA, Guo G, Wozniak M, Martin D, Miller S, Liapis H, Loveday K, Klahr S, Sampath TK, Morrissey J. Osteogenic protein-1 prevents renal fibrogenesis associated with ureteral obstruction. Am J Physiol Renal Physiol. 2000;279:F130–143. doi: 10.1152/ajprenal.2000.279.1.F130. [DOI] [PubMed] [Google Scholar]
- Jurchott K, Bergmann S, Stein U, Walther W, Janz M, Manni I, Piaggio G, Fietze E, Dietel M, Royer HD. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem. 2003;278:27988–27996. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- Kawai S, Sugiura T. Characterization of human bone morphogenetic protein (BMP)-4 and -7 gene promoters: activation of BMP promoters by Gli, a sonic hedgehog mediator. Bone. 2001;29:54–61. doi: 10.1016/s8756-3282(01)00470-7. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kolluri R, Torrey TA, Kinniburgh AJ. A CT promoter element binding protein: definition of a double-strand and a novel single-strand DNA binding motif. Nucleic Acids Res. 1992;20:111–116. doi: 10.1093/nar/20.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitu GM, Wang S, Hirschberg R. BMP7 is a podocyte survival factor and rescues podocytes from diabetic injury. Am J Physiol Renal Physiol. 2007;293:F1641–F1648. doi: 10.1152/ajprenal.00179.2007. [DOI] [PubMed] [Google Scholar]
- Monroe DG, Jin DF, Sanders MM. Estrogen opposes the apoptotic effects of bone morphogenetic protein 7 on tissue remodeling. Mol Cell Biol. 2000;20:4626–4634. doi: 10.1128/mcb.20.13.4626-4634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J, Hruska K, Guo G, Wang S, Chen Q, Klahr S. Bone morphogenetic protein-7 improves renal fibrosis and accelerates the return of renal function. J Am Soc Nephrol. 2002;13(Suppl 1):S14–21. [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Ozkaynak E, Jin DF, Jelic M, Vukicevic S, Oppermann H. Osteogenic protein-1 mRNA in the uterine endometrium. Biochem Biophys Res Commun. 1997;234:242–246. doi: 10.1006/bbrc.1997.6624. [DOI] [PubMed] [Google Scholar]
- Paralkar VM, Grasser WA, Mansolf AL, Baumann AP, Owen TA, Smock SL, Martinovic S, Borovecki F, Vukicevic S, Ke HZ, Thompson DD. Regulation of BMP-7 expression by retinoic acid and prostaglandin E(2) J Cell Physiol. 2002;190:207–217. doi: 10.1002/jcp.10048. [DOI] [PubMed] [Google Scholar]
- Raffetseder U, Frye B, Rauen T, Jurchott K, Royer HD, Jansen PL, Mertens PR. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J Biol Chem. 2003;278:18241–18248. doi: 10.1074/jbc.M212518200. [DOI] [PubMed] [Google Scholar]
- Raffetseder U, Rauen T, Djudjaj S, Kretzler M, En-Nia A, Tacke F, Zimmermann HW, Nelson PJ, Frye BC, Floege J, Stefanidis I, Weber C, Mertens PR. Differential regulation of chemokine CCL5 expression in monocytes/macrophages and renal cells by Y-box protein-1. Kidney Int. 2009;75:185–196. doi: 10.1038/ki.2008.457. [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Evdokimova V, Thomas AA, Ovchinnikov LP. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J Biol Chem. 2001;276:44841–44847. doi: 10.1074/jbc.M107581200. [DOI] [PubMed] [Google Scholar]
- To K, Zhao Y, Jiang H, Hu K, Wang M, Wu J, Lee C, Yokom DW, Stratford AL, Klinge U, Mertens PR, Chen CS, Bally M, Yapp D, Dunn SE. The phosphoinositide-dependent kinase-1 inhibitor 2-amino-N-[4-[5-(2-phenanthrenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]phen yl]-acetamide (OSU-03012) prevents Y-box binding protein-1 from inducing epidermal growth factor receptor. Mol Pharmacol. 2007;72:641–652. doi: 10.1124/mol.107.036111. [DOI] [PubMed] [Google Scholar]
- Valiahdi M. [Accessed January 4, 2010];The intracellular localisation of chk-YB1 protein depends on a casein kinase II consensus sequence in the C-terminal part of the protein. Fifth AACR International Conference on Frontiers in Cancer Prevention Research. 2006 http://aacrmeetingabstracts.org/cgi/content/abstract/2006/3/A70.
- Wang S, Chen Q, Simon TC, Strebeck F, Chaudhary L, Morrissey J, Liapis H, Klahr S, Hruska KA. Bone morphogenic protein-7 (BMP-7), a novel therapy for diabetic nephropathy. Kidney Int. 2003;63:2037–2049. doi: 10.1046/j.1523-1755.2003.00035.x. [DOI] [PubMed] [Google Scholar]
- Wang S, de Caestecker M, Kopp JB, Mitu G, LaPage J, Hirschberg R. Renal bone morphogenetic protein-7 protects against diabetic nephropathy. J Am Soc Nephrol. 2006;17:2504–2512. doi: 10.1681/ASN.2006030278. [DOI] [PubMed] [Google Scholar]
- Wang S, Hirschberg R. BMP7 antagonizes TGF-beta -dependent fibrogenesis in mesangial cells. Am J Physiol Renal Physiol. 2003;284:F1006–1013. doi: 10.1152/ajprenal.00382.2002. [DOI] [PubMed] [Google Scholar]
- Wang S, Hirschberg R. Bone morphogenetic protein-7 signals opposing transforming growth factor beta in mesangial cells. J Biol Chem. 2004;279:23200–23206. doi: 10.1074/jbc.M311998200. [DOI] [PubMed] [Google Scholar]
- Wang S, LaPage J, Hirschberg R. Loss of tubular bone morphogenetic protein-7 (BMP7) in diabetic nephropathy. J Am Soc Nephrol. 2001;12:2392–2399. doi: 10.1681/ASN.V12112392. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- Yang Q, Han B, Xie RJ, Cheng ML. Changes of bone morphogenetic protein-7 and inhibitory Smad expression in streptozotocin-induced diabetic nephropathy rat kidney. Sheng Li Xue Bao. 2007;59:190–196. [PubMed] [Google Scholar]
- Yang WH, Bloch DB. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA. 2007;13:704–712. doi: 10.1261/rna.411907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Homer C, Edwards SJ, Hananeia L, Lasham A, Royds J, Sheard P, Braithwaite AW. Nuclear localization of Y-box factor YB1 requires wild-type p53. Oncogene. 2003;22:2782–2794. doi: 10.1038/sj.onc.1206357. [DOI] [PubMed] [Google Scholar]