Abstract
Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is the best studied baculovirus and most commonly used virus vector for baculovirus expression vector systems. The effect of AcMNPV infection on host cells is incompletely understood. A microarray based on Spodoptera frugiperda ESTs was used to investigate the impact of AcMNPV on host gene expression in cultured S. frugiperda, Sf21 cells. Most host genes were down regulated over the time course of infection, although a small number were up-regulated. The most highly up-regulated genes encoded heat shock protein 70s and several poorly characterized proteins. Regulated genes with the highest score identified by functional annotation clustering included primarily products required for protein expression and trafficking in the ER and golgi. All were significantly down-regulated by approximately 12 hours post infection. Microarray data were validated by qRT-PCR. This study provides the first comprehensive host transcriptome overview of Sf21 cells during AcMNPV infection.
Keywords: Microarray, AcMNPV, Sf21 cells, Baculovirus, host genes, qRT-PCR, Hsp 70, DAVID, transcriptome
Introduction
Baculoviruses infect insects and have enveloped rod-shaped nucleocapsids and double-stranded circular genomes that range from 80 - 180 kb in size. Their replication cycle involves a regulated cascade of gene expression in which early and delayed early virus genes are transcribed by host RNA polymerase II (RNA pol II), whereas late and very late genes are transcribed by a virus encoded RNA polymerase. Baculoviruses that infect Lepidoptera produce two morphological forms budded virus (BV), which spreads the virus from cell to cell in infected insects, and occlusion derived virus (ODV), which spreads the virus between insect hosts (Reviewed in (Rohrmann, 2008)).
Baculoviruses have long been used as microbial insecticides to control insect pests in agriculture and forestry. In the 1980's, they were developed as a powerful eukaryotic protein expression vector (Summers, 2006). The best-studied baculovirus, Autographa californica multicapsid Nucleopolyhedrovirus (AcMNPV) is the type species of the genus alphabaculovirus in the family Baculoviridae (International Committee on Taxonomy of Viruses, 2008; Jehle et al., 2006). It is the prototype and most commonly used virus vector for the baculovirus expression vector system (BEVS). Proteins expressed optimally by BEVS can achieve levels of up to 50% or more of the total cellular protein because of the strong polyhedrin gene (polh) promoter, which is turned on very late in the viral infection cycle (Summers and Smith, 1987). The polh gene encodes for the matrix protein that embeds the ODV in large (1-5μM) protein occlusion bodies, called polyhedra. The polh gene is dispensable for baculovirus infection of cultured insect cells and has been deleted from most baculovirus expression vectors (O'Reilly et al., 1992; Smith et al., 1983). The temporal regulation of the polh promoter has advantages for synthesizing proteins that may be cytotoxic because it acts in a similar manner to a conditional promoter, switching on very late in the infection cycle, after the virus has already replicated, assembled, and released progeny virions in the form of BV.
Since its inception, BEVS has become one of the leading platforms for eukaryotic protein expression in both academic and industrial settings. Proteins expressed in BEVS are glycosylated, phosphorylated, and acetylated at appropriate sites and in most cases are fully functional (Jarvis, 1997). BEVS has many advantages over other expression systems, such as the ability to synthesize multiple proteins for protein complexes and the production of virus-like-particles (VLP). A number of animal vaccines made using BEVS technology are licensed for use around the world (reviewed in (Hu et al., 2008; van Oers, 2006)). BEVS is also a robust platform for the display of expressed eukaryotic protein libraries for screening of ligands and drugs (Makela and Oker-Blom, 2006). BEVS is emerging as one of the best platforms for expressing eukaryotic integral membrane proteins for high-resolution structural studies, which is currently a major challenge in biomedical research.
However, AcMNPV infection impacts host cells in many ways that may influence the expression of heterologous proteins, particularly secretory and integral membrane proteins, which can be more difficult to express than cytoplasmic proteins using BEVS (Higgins et al., 2003; Jarvis and Summers, 1989; Tate et al., 2003; van Oers et al., 2001a). During infection, the nucleus swells, the cell rounds up, and cytoskeletal architecture is modified (Charlton and Volkman, 1991; Roncarati and Knebel-Morsdorf, 1997; Volkman and Zaal, 1990). The cell cycle arrests at G2/M in AcMNPV infected Sf9 cells (Braunagel et al., 1998; Ikeda and Kobayashi, 1999). Host protein synthesis in AcMNPV-infected insect cells is shut down over the course of infection, beginning at approximately 10-12 hpi as the virus takes control of the cell to produce new virions (Carstens et al., 1979; Maruniak and Summers, 1981). By 24 hpi, the majority of newly synthesized proteins are either virus-encoded or virus-induced. The mechanism responsible for the shut off of host protein synthesis is not fully understood but appears to correlate with a reduction in host gene transcripts (Ooi and Miller, 1988; van Oers et al., 2003; van Oers et al., 2001b). A differential display approach was used to identify potentially up-regulated host genes (Nobiron et al., 2003). In this study, among over 3000 cDNA fragments, most were down regulated between 12 and 18 hpi. Only one host gene, heat shock protein 70 cognate (hsc70) mRNA was confirmed to be up-regulated.
To better understand the effects of AcMNPV on host cell gene expression as it relates to BEVS, we performed a more comprehensive study on the effects of AcMNPV infection on host gene expression in Sf21 cells, a widely used host cell line derived from Spodoptera frugiperda pupal ovaries (Vaughn et al., 1977). We used microarrays designed from an extensive S. frugiperda EST database, together with quantitative real time-polymerase chain reaction (qRT-PCR), to investigate the effects of AcMNPV infection on host gene transcription over time up until 48 hpi. Our results showed that transcripts for the majority of host genes declined substantially by 12 hpi. Furthermore, using Database for Annotation, Visualization and Integrated Discovery (DAVID) (Dennis et al., 2003; Huang et al., 2009) to cluster the host genes we identified cellular processes and pathways that were affected by AcMNPV late in infection. The highest scoring cluster was enriched in genes involved in protein trafficking and processing in the ER and Golgi. We expect these data to be useful for optimizing BEVS for expressing foreign proteins, particularly secretory and integral membrane proteins. We also identified several up regulated genes, in addition to hsp70.
Results
Microarray data analysis of Sf21 host genes during AcMNPV infection
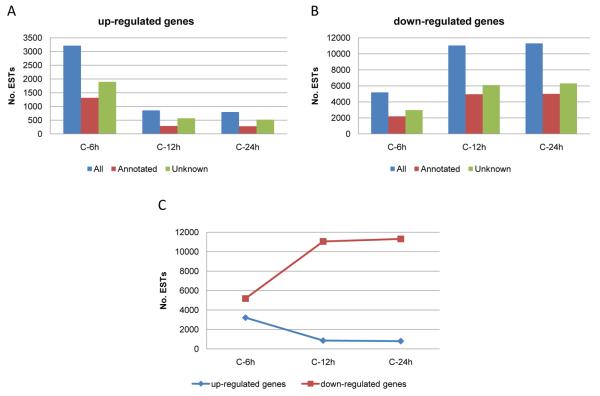
To better understand the effects of AcMNPV infection on host gene expression we compared the transcription profiles of S. frugiperda genes over time in AcMNPV-infected Sf21 cells using microarrays. The microarrays comprised oligonucleotide (60-mer) features and were designed using available S. frugiperda EST data (SPODOBASE, http://bioweb.ensam.inra.fr/spodobase/ (Negre et al., 2006)). The experiment was done in quadruplicates and we employed spike-in mRNAs as internal controls. The expression levels of host genes of Sf21 cells infected with AcMNPV at 6, 12, and 24 hpi were compared to the expression levels of the same genes from the mock infection sample. The comparison was performed with the GeneSpring GX 11 software (Agilent) using ONE-WAY ANOVA method; only genes that showed significant differences in expression levels (p<0.05) were selected. The data showed that the numbers of host genes significantly up-regulated more than 1.2 fold decreased as the infection progressed (Fig. 1A). Although a 1.2 fold change is a low cutoff for microarrays, it was selected in order to identify changes in host gene transcript levels at 6 hpi, which were subtle compared to 12 and 24 hpi. Among host genes whose expression was up regulated during the course of infection, approximately 66% were up regulated at 6 hpi, whereas 18%, and 16% were found to be up-regulated at 12 and 24 hpi, respectively (Fig. 1A). On the contrary, the number of host genes down-regulated more than 1.2 fold increased as the infection progressed. Approximately 19%, 40%, and 41% were down-regulated at 6, 12, and 24 hpi, respectively (Fig. 1B). Thus the trend in up- and down-regulated genes over the time course of infection showed an inverse relationship (Fig. 1C). Because the majority of the genes were represented on the microarray by three different probes, the number of genes represented in Fig. 1 was approximated by dividing the number of probes by 3. Although most of the redundant ESTs (multiple ESTs representing the same gene) present in SPODOBASE were removed before designing the probes to print on the microarray a number were missed resulting in more than three reads for some genes.
Fig. 1.
Patterns of host gene expression in AcMNPV-infected Sf21 cells determined by microarray analysis. Probes for host genes designed based on ESTs sequence data from SPODOBASE, http://bioweb.ensam.inra.fr/spodobase/). The approximate number of genes that were A) up-regulated or B) down-regulated during the time course of infection (6, 12, and 24 hpi) compared with the mock-infected controls; C) Chart comparing the expression trends exhibited by up- and down-regulated genes during the time course of infection. Only genes that their level of expression were equal to or more than 1.2 fold change and showed significant difference (p<0.05, ONE WAY ANOVA) were selected.
Functional clustering of differentially regulated host genes at 24 hpi
To determine what cellular processes and pathways might be affected by AcMNPV at late times of infection, DAVID v6.7 was used to analyze microarray data from the 24 hpi time point (http://david.abcc.ncifcrf.gov/ (Dennis et al., 2003; Huang et al., 2009)). However, because of the large number of genes whose expression was influenced by baculovirus infection we focused only on the genes that were well annotated. Thus, unknown genes and genes not identified by the DAVID database were not included in the analysis. A total of 10703 reads representing approximately 3600 genes (individual ESTs and contigs), which consisted of all genes showing down-or up-regulation with more than 15 fold change at 24 hpi compared to mock infection, were uploaded to DAVID. The 15 fold cutoff was selected to limit the number of genes to be analyzed to approximately 3000 entries, the number of genes that DAVID can easily handle. Out of these 3600, 2807 were analyzed by the functional annotation clustering tool, using Bombyx mori as background, and resulted in 52 clusters (Supplementary Table 1). DAVID analysis was unable to cluster genes using other species as background. In Table 1, we present clusters that are most directly related to protein expression and processing, and which could potentially impact gene expression using BEVS. The clusters are listed in descending order of enrichment score (E.S.). The full list of genes with the detailed fold change is presented in Supplementary Table 1. The two highest scoring clusters each comprised genes whose products could affect protein expression. Cluster 1 had an E.S. of 3.7 and included 29 genes encoding products involved in protein localization, transport, or targeting (Table 1, Supplementary Table 1). This cluster is highly enriched in genes encoding proteins required for synthesis and trafficking of secretory and membrane proteins. These include components of the signal recognition particle, members of the Sec61 complex, a translocon-associated protein (TRAP), a SNAP receptor complex member, a member of the transport protein particle (TRAPP) complex, a coatamer protein, a KDEL receptor, and several Rab proteins (Supplementary Table 1). Cluster 2 with E.S. 3.69 included 31 genes whose products were involved in protein translation (Table 1). This cluster was highly enriched in translation initiation factors (Supplementary Table 1). The other selected clusters shown in Table 1 contained genes encoding for proteins with the following functions: a) cluster 5 (E.S. 1.89): endoplasmic reticulum functions, b) cluster 6 (E.S. 1.5): chaperones, c) cluster 15 (E.S. 1.03): protein folding, d) cluster 17 (E.S. 0.91): protein targeting to membranes and localization to organelles, e) cluster 19 (E.S. 0.8): heat shock proteins, f) cluster 24 (E.S. 0.67): regulatory proteins that may affect translation, g) cluster 29 (E.S. 0.5): protein maturation and processing, and h) cluster 43 (E.S. 0.1): mainly ribosomal protein genes. All of the genes in all of these clusters were down regulated. One of these genes, non-clathrin coat protein zeta 1-COP (gi|114051996), a member of clusters 1 and 43, was represented by two different ESTs in SPODOBASE and one of the two ESTs was up-regulated. The expression pattern of non-clathrin coat protein zeta 1-COP is further discussed below.
Table 1.
Selected functional annotation clusters that may influence gene expression in Sf21 cells.
| Cluster No. / Enrichment Score |
Term | No. of genes |
genes % related to the DAVID uploaded genes |
PValue |
|---|---|---|---|---|
| Annotation Cluster 1 | ||||
| 3.70 | protein localization | 27 | 0.96 | 3.50E-07 |
| protein transport | 27 | 0.96 | 3.50E-07 | |
| establishment of protein localization | 27 | 0.96 | 3.50E-07 | |
| cellular protein localization | 17 | 0.61 | 8.70E-06 | |
| intracellular protein transport | 17 | 0.61 | 8.70E-06 | |
| cellular macromolecule localization | 17 | 0.61 | 8.70E-06 | |
| intracellular transport | 19 | 0.68 | 1.55E-05 | |
| protein targeting | 11 | 0.39 | 1.54E-04 | |
| protein targeting to membrane | 9 | 0.32 | 1.12E-03 | |
| protein localization in organelle | 9 | 0.32 | 1.12E-03 | |
| cotranslational protein targeting to membrane | 6 | 0.21 | 1.99E-02 | |
| SRP-dependent cotranslational protein targeting to membrane |
5 | 0.18 | 4.95E-02 | |
| protein targeting to ER | 5 | 0.18 | 4.95E-02 | |
| signal recognition particle | 4 | 0.14 | 8.48E-02 | |
| 7S RNA binding | 4 | 0.14 | 1.06E-01 | |
| Annotation Cluster 2 | ||||
| 3.69 | translation factor activity, nucleic acid binding | 29 | 1.03 | 9.28E-06 |
| protein biosynthesis | 17 | 0.61 | 9.10E-05 | |
| Initiation factor | 20 | 0.71 | 1.80E-04 | |
| translation initiation factor activity | 21 | 0.75 | 2.37E-04 | |
| translational initiation | 10 | 0.36 | 1.03E-02 | |
| Annotation Cluster 5 | ||||
| 1.89 | endomembrane system | 10 | 0.36 | 3.85E-03 |
| endoplasmic reticulum | 12 | 0.43 | 9.97E-03 | |
| endoplasmic reticulum part | 7 | 0.25 | 1.12E-02 | |
| endoplasmic reticulum membrane | 6 | 0.21 | 2.90E-02 | |
| nuclear envelope-endoplasmic reticulum network | 6 | 0.21 | 2.90E-02 | |
| Annotation Cluster 6 | ||||
| 1.52 | Chaperone | 9 | 0.32 | 2.45E-03 |
| unfolded protein binding | 10 | 0.36 | 3.22E-03 | |
| Chaperonin Cpn60/TCP-1 | 4 | 0.14 | 1.47E-01 | |
| Chaperone, tailless complex polypeptide 1 | 4 | 0.14 | 1.47E-01 | |
| Chaperonin TCP-1, conserved site | 4 | 0.14 | 1.47E-01 | |
| Annotation Cluster 15 | ||||
| 1.03 | protein folding | 17 | 0.61 | 1.00E-03 |
| Isomerase | 12 | 0.43 | 2.70E-02 | |
| Rotamase | 6 | 0.21 | 1.90E-01 | |
| peptidyl-prolyl cis-trans isomerase activity | 6 | 0.21 | 2.00E-01 | |
| cis-trans isomerase activity | 6 | 0.21 | 2.00E-01 | |
| PIRSF001467:peptidylprolyl isomerase | 3 | 0.11 | 4.50E-01 | |
| Peptidyl-prolyl cis-trans isomerase, cyclophilin-type | 3 | 0.11 | 6.10E-01 | |
| Annotation Cluster 17 | ||||
| 0.91 | protein targeting to membrane | 9 | 0.32 | 1.12E-03 |
| protein localization in organelle | 9 | 0.32 | 1.12E-03 | |
| membrane-enclosed lumen | 9 | 0.32 | 9.26E-03 | |
| protein transport | 5 | 0.18 | 8.73E-02 | |
| translocation | 4 | 0.14 | 9.95E-02 | |
| intracellular protein transmembrane transport | 4 | 0.14 | 1.19E-01 | |
| protein import | 4 | 0.14 | 1.19E-01 | |
| mitochondrial intermembrane space | 3 | 0.11 | 2.19E-01 | |
| mitochondrial intermembrane space protein transporter complex |
3 | 0.11 | 2.19E-01 | |
| organelle envelope lumen | 3 | 0.11 | 2.19E-01 | |
| mitochondrion inner membrane | 5 | 0.18 | 2.32E-01 | |
| membrane organization | 3 | 0.11 | 2.71E-01 | |
| protein targeting to mitochondrion | 3 | 0.11 | 2.71E-01 | |
| protein import into mitochondrial inner membrane | 3 | 0.11 | 2.71E-01 | |
| protein localization in mitochondrion | 3 | 0.11 | 2.71E-01 | |
| mitochondrial transport | 3 | 0.11 | 2.71E-01 | |
| inner mitochondrial membrane organization | 3 | 0.11 | 2.71E-01 | |
| mitochondrial membrane organization | 3 | 0.11 | 2.71E-01 | |
| mitochondrion organization | 3 | 0.11 | 2.71E-01 | |
| IPR004217:Mitochondrial inner membrane translocase complex, Tim8/9/10/13-zinc finger-like |
3 | 0.11 | 3.10E-01 | |
| Zinc finger, Tim10/DDP-type | 3 | 0.11 | 3.10E-01 | |
| mitochondrion | 6 | 0.21 | 7.81E-01 | |
| Annotation Cluster 19 | ||||
| 0.83 | heat shock protein binding | 6 | 0.21 | 4.16E-02 |
| DnaJ | 5 | 0.18 | 9.61E-02 | |
| Heat shock protein DnaJ, N-terminal | 5 | 0.18 | 1.41E-01 | |
| Molecular chaperone, heat shock protein, Hsp40, DnaJ | 4 | 0.14 | 2.68E-01 | |
| Heat shock protein DnaJ | 3 | 0.11 | 4.73E-01 | |
| Annotation Cluster 24 | ||||
| 0.67 | regulation of cellular protein metabolic process | 7 | 0.25 | 2.21E-02 |
| regulation of translation | 5 | 0.18 | 1.09E-01 | |
| posttranscriptional regulation of gene expression | 5 | 0.18 | 1.09E-01 | |
| regulation of translational initiation | 3 | 0.11 | 2.71E-01 | |
| negative regulation of translation | 3 | 0.11 | 2.71E-01 | |
| negative regulation of cellular protein metabolic process | 3 | 0.11 | 2.71E-01 | |
| negative regulation of protein metabolic process | 3 | 0.11 | 2.71E-01 | |
| negative regulation of biosynthetic process | 3 | 0.11 | 4.24E-01 | |
| negative regulation of macromolecule metabolic process | 3 | 0.11 | 4.24E-01 | |
| negative regulation of macromolecule biosynthetic process | 3 | 0.11 | 4.24E-01 | |
| negative regulation of cellular biosynthetic process | 3 | 0.11 | 4.24E-01 | |
| Annotation Cluster 29 | ||||
| 0.57 | protein maturation | 3 | 0.11 | 2.71E-01 |
| signal peptide processing | 3 | 0.11 | 2.71E-01 | |
| protein maturation by peptide bond cleavage | 3 | 0.11 | 2.71E-01 | |
| peptide metabolic process | 3 | 0.11 | 2.71E-01 | |
| protein processing | 3 | 0.11 | 2.71E-01 | |
| Annotation Cluster 43 | ||||
| 0.14 | translation | 46 | 1.64 | 2.87E-01 |
| intracellular non-membrane-bounded organelle | 44 | 1.57 | 6.13E-01 | |
| non-membrane-bounded organelle | 44 | 1.57 | 6.13E-01 | |
| ribonucleoprotein complex | 33 | 1.18 | 6.33E-01 | |
| ribonucleoprotein | 15 | 0.53 | 7.81E-01 | |
| ribosome | 23 | 0.82 | 9.66E-01 | |
| ribosomal protein | 23 | 0.82 | 9.83E-01 | |
| structural constituent of ribosome | 20 | 0.71 | 9.93E-01 | |
| structural molecule activity | 27 | 0.96 | 1.00E+00 |
Bombyx mori was used a s a background in DAVID Bioinformatics Resources 6.7.
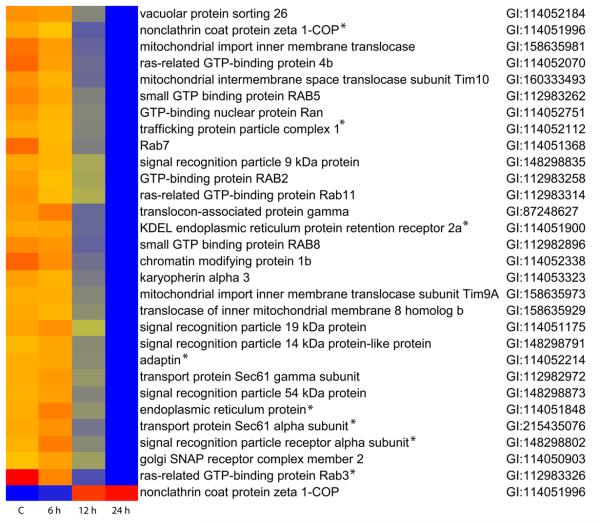
A heat map showing the regulation of the 29 genes forming DAVID cluster 1 throughout the time course of infection is shown in Fig. 2. The levels of expression of most of these genes were dramatically down-regulated at 12 and 24 hpi samples when compared to mock (C) infected sample. At 6 hpi, the level of expression of most of the 29 genes were unchanged, or only slightly changed (≤ 1.5 fold), compared to mock infection. One exception was ras-related GTP-binding protein Rab3 gene, which was down regulated 3.1 fold. Two other genes, ras-related GTP-binding protein 4b and Rab7, showed moderate down regulation, 1.6 and 1.8 fold, respectively. Other than the up-regulated non-clathrin coat protein zeta 1-COP gene (up 6.3 fold), only signal recognition particle receptor alpha subunit showed greater than 1.5 fold (1.6 fold) up regulation at 6 hpi. All the accession numbers for the genes of cluster 1 and the rest of the 52 clusters are included in Supplementary Table 1. The SPODOBASE accession numbers of genes included in Supplementary Table 1 are presented in Supplementary Table 2.
Fig. 2.
Heat map depicts the regulation of the genes representing DAVID-cluster 1. Red color indicates the highest gene expression and the blue color indicates the lowest gene expression. Gene expression was normalized to the spike-in genes on each array. Each gene is represented by one probe showing approximately the median level of expression across all time points. In all cases the median expression values are similar to the mean values. Asterisks denote genes analyzed by qRT-PCR.
Non-clustered and up regulated host genes
Many regulated host genes either were not included in the DAVID functional analysis, or did not form a functional group when analyzed by DAVID. Interestingly, among the strongly down-regulated transcripts were a number encoding for immune functions including, cecropin, hemolin-like protein, β-1,3-glucan recognition protein 2a, peptidoglycan recognition protein, eater, prophenol oxidase activating enzyme 1, and β-1,3-glucan-binding protein (data not shown). The majority of genes found to be up-regulated at 6 hpi were subsequently down regulated by 12 or 24 hpi (Fig. 1 C, Supplemental Tables 1 and 3). Moreover, except for a few genes, the increased expression at 6 hpi was generally low, 2 fold or less. The expression of only a small number of genes was highly up regulated at any time point or up regulated throughout the course of infection. These genes are listed in Table 2. Genes with ambiguous regulation were excluded. The most highly up-regulated genes at 6 hpi were two heat shock 70 proteins (Hsp70) (protein acc. no. gi|229562184 and gi|256862212). However one Hsp70 (protein acc. no. gi|256862212) was associated with two ESTs, which exhibited contrasting expression profiles. Although the best protein annotations (BLASTx) for the EST sequences (SPODOBASE: Sf1P20517-5-1 and Sf2L00008-5-1) were the same, their nucleotide sequences could not be aligned (megablast, discontiguous megablast, or blastn) (Zhang et al., 2000), suggesting they represent different Hsp70 genes or isoforms (data not shown). When a nucleotide BLAST search was done the best match for the up regulated Sf2L00008-5-1 was gi|256862212. Whereas the best match for Sf1P20517-5-1 was gi|167077413, a Hsp70 from the noctuid moth Sesamia nonagrioides (E value 2e-06), and the E value for gi|256862212 was 0.011. For the two up regulated Hsp70's, there was a dramatic increase in transcripts at 6 hpi. Transcripts remained up relative to controls at 12 and 24 hpi, but declined in magnitude (Supplementary Table 3). The down regulated Hsp70 (protein acc. no. gi|256862212, SPODOBASE acc. no. Sf1P20517-5-1) showed gradual down regulation beginning at 6 hpi. Two other ESTs, which could not be annotated (data not shown), as well as a hypothetical protein from Plasmodium knowlesi strain H (protein acc. no. gi|221053556) and a predicted protein from Acyrthosiphon pisum (protein acc. no. gi|193594185) had expression profiles similar to the up regulated Hsp70's (Table 2).
Table 2.
Unclustered genes with significantly up-regulated expression in AcMNPV-infected Sf21 cells.
| Average Fold Changea | |||||
|---|---|---|---|---|---|
| GI acc. No. | protein Descriptionb | Species | 6 hpi | 12 hpi | 24 hpi |
| 110751266 | PREDICTED: similar to interleukin 16 isoform 1 precursor | Apis mellifera | 3.1 | 2.6 | −0.4 |
| 156549977 | PREDICTED: similar to alcohol dehydrogenase | Nasonia vitripennis | 3.3 | 3.6 | −1.6 |
| 183979382 | branched-chain-amino-acid transaminase (a) | Papilio xuthus | 3.8 | 3.6 | −0.4 |
| 183979382 | branched-chain-amino-acid transaminase (b) | Papilio xuthus | 5.1 | 6.2 | 1.9 |
| 157128947 | apolipoprotein D, putative | Aedes aegypti | 5.3 | 3.3 | −0.5 |
| 158302520 | AGAP001116-PA | Anopheles gambiae str. PEST | 5.9 | 3.4 | 0.7 |
| 215820602 | uridine diphosphate glucosyltransferase | Bombyx mori | 6.1 | 4.6 | −1.5 |
| 270010182 | hypothetical protein TcasGA2_TC009549 | Tribolium castaneum | 6.2 | 1.3 | −6.3 |
| 270003309 | hypothetical protein TcasGA2_TC002528 | Tribolium castaneum | 9.2 | 3.2 | −4.4 |
| 193641133 | PREDICTED: similar to elongation factor Tu GTP binding domain containing 1 | Acyrthosiphon pisum | 9.7 | 1.3 | −4.0 |
| 193594185 | PREDICTED: similar to predicted protein (a) | Acyrthosiphon pisum | 13.9 | 9.4 | 2.2 |
| 193594185 | PREDICTED: similar to predicted protein (b) (2 probes) | Acyrthosiphon pisum | 22.9 | 15.0 | 4.4 |
| 221053556 | hypothetical protein | Plasmodium knowlesi strain H | 22.3 | 10.2 | 4.3 |
| 229562184 | heat shock protein 70 | Spodoptera exigua | 37.6 | 4.0 | 1.6 |
| 270010864 | hypothetical protein TcasGA2_TC015905 (a) | Tribolium castaneum | 10.4 | 2.9 | −5.3 |
| 270010864 | hypothetical protein TcasGA2_TC015905 (b) (2 probes) | Tribolium castaneum | 9.3 | 2.9 | −3.1 |
| 195504354 | GE10701 | Drosophila yakuba | −1.1 | 6.0 | 3.4 |
| 167520296 | hypothetical protein | Monosiga brevicollis MX1 | 1.8 | 3.5 | 1.2 |
| 270010547 | hypothetical protein TcasGA2_TC009962 | Tribolium castaneum | 1.3 | 4.7 | 1.8 |
| 157113191 | epoxide hydrolase | Aedes aegypti | 1.3 | 5.4 | 2.2 |
| 242015510 | serine proteinase stubble, putative | Pediculus humanus corporis | 1.6 | 2.3 | −0.5 |
| 221122289 | PREDICTED: similar to arginine and glutamate rich 1 | Hydra magnipapillata | 2.2 | 2.8 | 1.5 |
| 270003926 | hypothetical protein TcasGA2_TC003217 | Tribolium castaneum | 2.0 | 4.0 | 1.7 |
| 7862150 | 3-dehydroecdysone 3alpha-reductase (a) | Spodoptera littoralis | 3.0 | 5.7 | 3.4 |
| 7862150 | 3-dehydroecdysone 3alpha-reductase (b) | Spodoptera littoralis | 2.3 | 2.2 | 3.7 |
| 7862150 | 3-dehydroecdysone 3alpha-reductase © | Spodoptera littoralis | −0.4 | 2.0 | 2.9 |
| 270011314 | hypothetical protein TcasGA2_TC005316 | Tribolium castaneum | 3.2 | 4.5 | 0.4 |
| 170069287 | conserved hypothetical protein | Culex quinquefasciatus | 3.2 | 11.4 | 2.2 |
| 168703125 | hypothetical protein GobsU_26581 | Gemmata obscuriglobus UQM 2246 | 4.1 | 12.8 | 5.3 |
| 57506570 | reverse transcriptase | Bombyx mori | 4.4 | 6.1 | 3.2 |
| 156538499 | PREDICTED: similar to mandelate racemase | Nasonia vitripennis | 4.4 | 6.3 | 1.5 |
| 157783840 | nicotinic acetylcholine receptor alpha 9 subunit (a) | Bombyx mori | 4.6 | 5.8 | 2.1 |
| 157783840 | nicotinic acetylcholine receptor alpha 9 subunit (b) (2 probes) | Bombyx mori | 2.4 | 3.0 | 2.0 |
| 189241946 | PREDICTED: similar to Y26D4A.11 | Tribolium castaneum | 5.2 | 6.8 | 3.6 |
| 237700851 | serine protease 18 (a) | Mamestra configurata | 7.5 | 13.3 | 3.2 |
| 237700851 | serine protease 18 (b) (2 probes) | Mamestra configurata | 0.0 | −0.4 | −3.5 |
| 256862212 * | heat shock protein 70 (a)* | Helicoverpa zea | 93.9 | 11.1 | 1.8 |
| 256862212 | heat shock protein 70 (b) | Helicoverpa zea | −2.2 | −12.3 | −29.2 |
| 116253 | 63 kDa chaperonin, mitochondrial (a) | Heliothis virescens | 1.3 | 11.3 | 4.6 |
| 116253 | 63 kDa chaperonin, mitochondrial (b) | Heliothis virescens | 1.2 | 3.9 | 3.1 |
| 148298724 | cathepsin L-like proteinase | Bombyx mori | 3.3 | 10.8 | 3.4 |
| 158285681 | AGAP007421-PA | Anopheles gambiae str. PEST | 6.6 | 12.5 | 3.0 |
| 224057636 | PREDICTED: protein kinase, cAMP-dependent, catalytic, beta | Taeniopygia guttata | 4.1 | 11.5 | 7.6 |
| 266808630 | diapause bioclock protein-like protein | Helicoverpa armigera | 4.2 | 10.3 | 6.0 |
| 160333383 * | serpin-6 (1 probe)* | Bombyx mori | 1.4 | 137.9 | 64.2 |
| 91076058 | PREDICTED: similar to Bardet-Biedl syndrome 5 | Tribolium castaneum | 0.3 | 1.1 | 2.6 |
| 255977216 | juvenile hormone binding protein | Bombyx mori | 0.5 | 1.6 | 3.2 |
| 108864634 | helicase, putative | Oryza sativa | 0.7 | 2.9 | 3.5 |
| 156540818 | PREDICTED: similar to tyrosine recombinase | Nasonia vitripennis | 1.6 | 4.6 | 7.7 |
Fold change of all probes in the time point for each EST averaged. Unless indicated this comprised values from three probes per time point.
Where accession numbers are assigned to more than one EST, different ESTs are designated by different letters
expression analyzed by qRT-PCR
Regulation of non-clathrin coat protein zeta 1-COP transcripts
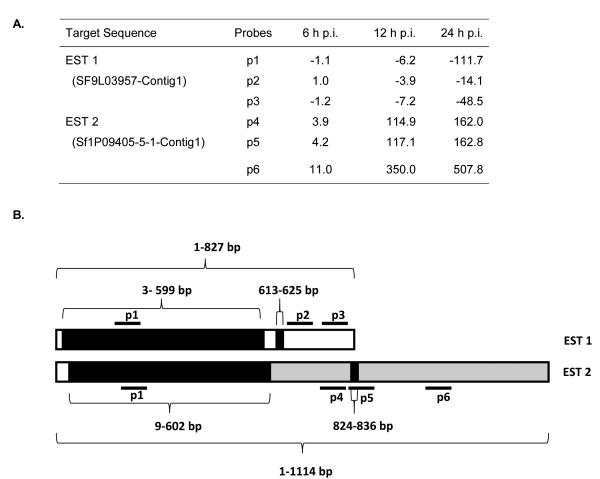
Another up-regulated gene showed a different expression pattern from the Hsp70s. One set of probes corresponding to non-clathrin coat protein zeta 1-COP indicated that the gene was up-regulated slightly at 6 hpi and transcript levels increased dramatically (>100 fold) at 12 and 24 hpi, but another set of probes corresponding to the same gene indicated that the gene was down-regulated (Fig. 2). Two sequences from SPODOBASE (EST1: SF9L03957-Contig1 and EST2: Sf1P09405-5-1-Contig1) represented on the microarray were annotated as non-clathrin coat protein zeta 1-COP and linked to the same protein accession number (gi|114051996). The three probes designed to hybridize to EST1 sequence (p1, 2, and 3) indicated that the expression-level of non-clathrin coat protein zeta 1-COP during viral infection was down regulated comparing to the mock infection. In contrast the three probes designed for EST2 (p4, 5, and 6) showed a substantial increase in transcript levels over time, particularly probe 6 (Fig. 3A). A comparison of the contig nucleotide sequences indicated a high degree of sequence similarity at their 5′-ends, and in another small region further downstream (Fig. 3B, shaded black). EST2 had three fewer nucleotides than EST1 in the 5′ region of similarity, as well as 13 SNPs relative to EST1 (data not shown). We also found that the unique region EST2 shared similarity with AcMNPV DNA (Fig. 3B, shaded gray). Probe 1 (p1) recognized sequence common to both contigs. Whereas p2, and p3 recognized regions that were unique to EST1 (Fig 3B, unshaded), and p4, and p6 recognized sequence unique to EST2 (Fig. 3B, shaded gray). P5 sequence was unique to EST2 except for 13 bp (613-625 bp) of sequence identity shared with EST1 (Fig. 3B). When the unique sequence of EST1 (Fig. 3B, unshaded) was analyzed by nucleotide BLAST, 15% of the fragment length (~33 nucleotide) was similar to tRNA-Leu (trnl) gene of different Rose species. Nucleotide BLAST analysis of all probe sequences also indicated that p1 was the only probe homologous to non-clathrin coat protein zeta 1-COP. Our interpretation of these results is that higher fold reduction in expression indicated by p1, relative to p2 and p3, at 24 hpi is most likely due to hybridization of transcripts corresponding to both EST1 and EST2, resulting in a greater difference in intensity between control and 24 hpi reads. The dramatic increase in expression levels indicated by probes 4, 5, and especially 6, is most likely due to cross hybridization of AcMNPV transcripts with sequence similarity.
Fig. 3.
Analysis of non-clathrin coat protein zeta 1-COP gene expression. Non-clathrin coat protein zeta 1-COP gene is represented by two different ESTs in SPODOBASE. A) SF9L03957-Contig1 (EST1) and Sf1P09405-5-1-Contig1 (EST2) that exhibit divergent expression patterns on microarrays. The fold change in expression for each probe, designated p1 - p6, at each time point is the average from four independent biological replicates. B) Diagram shows the relationship between EST1 and 2 and the locations of the microarray probes. Sequence comparisons reveal that both contigs share two common regions of nearly identical nucleotide sequence, indicated by the shaded black bars. The remaining sequences are unique to each EST, and depicted by either unfilled white or shaded grey bars. Probe 1 (p1) matches both contig sequences, p2-p3 are specific for the unique region of EST1, and p4 and p6 are specific for the unique region of EST2. Most of p5 is specific for the unique region of EST2, however it overlaps the small region of sequence identity shared with EST 1 (13 identical nucleotides).
Validation of microarray data using qRT-PCR
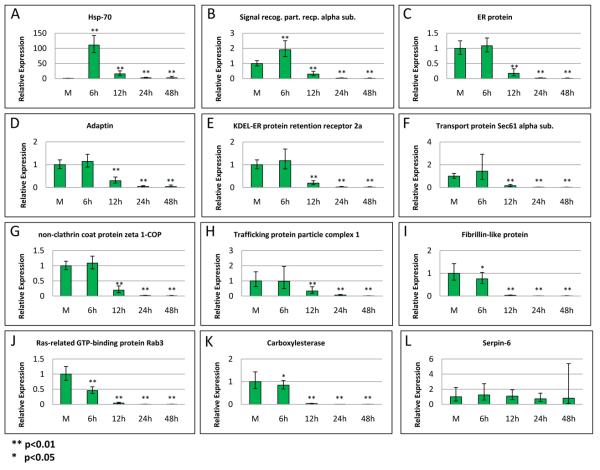
We used quantitative real-time PCR (qRT-PCR) to validate microarray results. For this analysis we selected eight genes from cluster 1 (Fig. 2, asterisks), a highly down regulated, >1000 fold at 24 hpi, carboxylesterase (gi|114050871) gene from cluster 46 (Supplemental Table 1), an unclustered gene encoding a fibrillin-like protein (gi|112983550) that was highly down regulated at 24 hpi (375 fold), and two unclustered genes, Hsp70 and serpin 6 that were up-regulated (Table 2, asterisks). Expression levels of these genes were analyzed in two technical replicates for four biological replicates in mock-infected cells and at 6, 12, 24, and 48 hpi in AcMNPV-infected cells (Fig. 4). All the data from qRT-PCR showed that the expression level of the selected genes was regulated in the same manner as shown by the microarray analysis, except for the serpin-6 gene (Table 3). Microarray data showed that signal recognition particles receptor alpha subunit, ER protein, Adaptin, KDEL-ER protein retention receptor 2a, transport protein Sec61 alpha subunit, and non-clathrin coat protein zeta 1-COP were first up-regulated at 6 hpi and subsequently down-regulated beginning at 12 hpi (Table 3). Although the qRTPCR showed the same pattern, the up-regulation observed at 6 hpi was not significantly different from mock-infected controls except for the signal recognition particle receptor alpha subunit (Fig. 4, B, C, D, E, F, and G). In all of these cases, microarray data indicated less than 2 fold up-regulation of transcript levels relative to mock-infected cells, with the highest fold difference recorded for signal recognition particle receptor alpha subunit. The primers used for qRT-PCR analysis of the non-clathrin coat protein zeta 1-COP were designed from the common region of both contigs (Fig. 3B, shaded black). The results showed down-regulation of expression from 12-48 hpi (Fig. 4, G). This indicates that the up-regulation of non-clathrin coat protein zeta 1-COP transcripts observed on microarrays is due either to specific up-regulation of transcripts corresponding to EST2 or cross hybridization of AcMNPV transcripts. The strong up-regulation of Hsp70 expression at 6 hpi shown by microarray results was confirmed, as was its continued, but declining, up-regulated expression at 12 and 24 hpi, relative to mock infection. The qRT-PCR analysis extended these findings, indicating significant up-regulated expression relative to mock-infected controls even at 48 hpi. Although microarray data suggested that the gene encoded for serpin-6 was significantly up regulated (Table 3), qRT-PCR data showed no significant change in its level of expression up to 48 hpi (Fig. 4, L). Upon reviewing the microarray data, we found that serpin 6 was represented by three probes, but only one showed up regulation by more than 1.2 fold (the cutoff fold change), the signals recorded for the other two probes were not significantly different from the controls. The apparent up regulation of serpin 6 detected by this probe is likely due to the result of aberrant hybridization. Moreover, the other serpin family members (serpin 2, 4A, 5, 7, 8, 10, 11, 13, 14, 27, 31) included in the microarray were down regulated. Down regulated expression from the genes encoding for Fibrillin-like protein, Ras-related GTP-binding protein, and carboxylesterase (B. mori) starting from 6 hpi was also confirmed (Fig. 4, I, J, and K). A comparison between the qRT-PCR data and the microarray data was established to demonstrate the average fold change corresponding to the level of expression of the selected genes (Table 3). The data from both expression analyses were in agreement and the fold change calculated in Table 3 was remarkably close between the two sets, considering the difference in approaches.
Fig. 4.
Validation of regulated gene expression by qRT-PCR. Genes were selected from DAVID-cluster 1, indicated by asterisks in Fig. 2, along with several additional genes, which displayed substantial up or down regulation on microarrays. RNA was extracted from mock-infected Sf21 cells harvested at 12 hpi and from infected cells harvested at 6, 12, 24, or 48 hpi. Primers are listed in Supplemental Table 4. Each value represents two technical replicates of each of four biological replicates. Standard deviation is indicated by bars. Relative expression was calculated based on 2−ΔΔCt method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). Double asterisks (**) indicate that p<0.01, while one asterisk (*) indicates that p<0.05.
Table 3.
comparison between the average fold change of microarray and qRT-PCR
| h p.i. |
qRT-PCR (Fold Change) |
Microarray (Fold Change) |
|||
|---|---|---|---|---|---|
| KDEL-ER protein retention receptor 2a | 6h | 1.18 | (+) | 1.03 | (+) |
| 12h | 5.16 | (−) | 6.61 | (−) | |
| 24h | 45.35 | (−) | 92.09 | (−) | |
| 48h | 75.06 | (−) | NA | ||
| Adaptin | 6h | 1.14 | (+) | 1.05 | (+) |
| 12h | 3.39 | (−) | 5.01 | (−) | |
| 24h | 22.01 | (−) | 55.44 | (−) | |
| 48h | 25.55 | (−) | NA | ||
| ER protein | 6h | 1.09 | (+) | 1.41 | (+) |
| 12h | 5.70 | (−) | 4.42 | (−) | |
| 24h | 55.82 | (−) | 25.81 | (−) | |
| 48h | 128.41 | (−) | NA | ||
| Signal recog. part. recp. alpha sub. | 6h | 1.91 | (+) | 1.58 | (+) |
| 12h | 3.28 | (−) | 4.80 | (−) | |
| 24h | 32.56 | (−) | 48.52 | (−) | |
| 48h | 59.16 | (−) | NA | ||
| Ras-related GTP-binding protein Rab3 | 6h | 2.19 | (−) | 3.11 | (- ) |
| 12h | 23.63 | (−) | 36.01 | (−) | |
| 24h | 203.93 | (−) | 415.06 | (−) | |
| 48h | 316.66 | (−) | NA | ||
| Transport protein Sec61 alpha sub. | 6h | 1.44 | (+) | 1.27 | (+) |
| 12h | 6.52 | (−) | 5.90 | (−) | |
| 24h | 52.14 | (−) | 41.43 | (−) | |
| 48h | 112.75 | (−) | NA | ||
| Trafficking protein particle complex 1 | 6h | 1.02 | (−) | 1.13 | (−) |
| 12h | 2.98 | (−) | 5.09 | (−) | |
| 24h | 15.65 | (−) | 56.12 | (−) | |
| 48h | 145.86 | (−) | NA | ||
| Fibrillin-like protein | 6h | 1.32 | (−) | 1.64 | (−) |
| 12h | 31.25 | (−) | 33.79 | (−) | |
| 24h | 148.08 | (−) | 377.07 | (−) | |
| 48h | 183.56 | (−) | NA | ||
| Hsp-70 | 6h | 110.75 | (+) | 93.93 | (+) |
| 12h | 15.95 | (+) | 11.08 | (+) | |
| 24h | 3.10 | (+) | 1.78 | (+) | |
| 48h | 2.79 | (+) | NA | ||
| Serpin-6 | 6h | 1.24 | (+) | 1.45 | (+) |
| 12h | 1.09 | (+) | 137.94 | (+) | |
| 24h | 1.38 | (−) | 64.18 | (+) | |
| 48h | 1.22 | (−) | NA | ||
| Carboxylesterase | 6h | 1.18 | (−) | 1.54 | (−) |
| 12h | 27.68 | (−) | 41.53 | (−) | |
| 24h | 294.64 | (−) | 1160.95 | (−) | |
| 48h | 396.31 | (−) | NA | ||
| non-clathrin coat protein zeta 1-COP | 6h | 1.09 | (+) | 1.11 | (+) |
| 12h | 5.15 | (−) | 5.77 | (−) | |
| 24h | 64.28 | (−) | 58.11 | (−) | |
| 48h | 138.00 | (−) | NA |
(+) up regulation
(−) down regulation
Discussion
Microarray analysis of the changes in the levels of host gene transcripts in AcMNPV-infected Sf21 cells over time extends previous studies on changes in host gene expression during infection. Studies on individual proteins suggested that AcMNPV down regulates the expression of host protein synthesis at the transcriptional level (Ooi and Miller, 1988; van Oers et al., 2003; van Oers et al., 2001b). A subsequent study using differential display in Sf9 cells found that transcripts of most host genes declined by 12-18 hpi, ribosomal RNA levels remained constant, and the expression of only one host gene, hsc70 increased (Nobiron et al., 2003). In the present study, we found that among approximately 42,000 probes for host genes included on our arrays approximately 70% showed differential regulation, with nearly all of these down regulated. The remaining probes represent genes that were either not expressed in Sf21 cells or were not regulated. Numerous genes were up regulated moderately at 6 hpi (less than 2 fold), although most subsequently declined in abundance, and were significantly lower than mock-infected controls by 12 or 24 hpi. Similar to the differential display study, we found that two Hsp70s transcripts were up regulated. They displayed strong up regulation at 6 hpi, which gradually declined but remained significantly higher than mock-infected controls up until at least 48 hpi. AcMNPV infection of Sf9 cells induced the expression or up regulated at least three Hsp70 proteins, as determined by two-dimensional PAGE followed by western blots (Lyupina et al., 2010). Two of these were also induced by heat shock. Up regulated expression of two Hsp70s was also observed in AcMNPV-infected Hv-Am1 cells, a Heliothis virescens cell line that is permissive for AcMNPV (Popham et al., 2010). In the present study we found additional genes that were significantly up regulated over the course of infection, although many of these could not be annotated. Four of these genes displayed expression patterns that were similar to the Hsp70 genes. Whereas other up regulated genes were most strongly up regulated at 12 hpi, for example, 63 kDa mitochondrial chaperonin, cathepsin L-like protease, and diapause bioclock protein-like protein (Table 2). Four were found to be most up-regulated at 24 hpi, juvenile hormone binding protein, a putative helicase, a predicted protein similar to tyrosine recombinase (Table 2) and non-clathrin coat protein zeta 1-COP (Fig. 3A). The most highly up regulated gene at 24 hpi was non-clathrin coat protein zeta 1-COP. Upon further analysis, probes for one EST were found to share similarity with AcMNPV, suggesting that the apparent up regulation was most likely due to cross hybridization with viral transcripts (Fig. 3). The other genes that were up regulated at 24 hpi and those strongly up regulated earlier in the infection cycle (6 or 12 hpi) may play roles in the virus replication cycle. In particular, viruses frequently utilize HSPs to support their own replication (reviewed by (Mayer, 2005)). In the case of AcMNPV, inhibition of inducible HSP70s significantly reduced viral DNA synthesis (Lyupina et al., 2010).
When we compare the results of studies of AcMNPV regulation of host gene expression with those of Bombyx mori nucleopolyhedrovirus (BmNPV) there are some differences. Seven up-regulated host genes were identified in BmNPV-infected BmN-4 cells by subtractive hybridization at the early stage of infection (2 and 6 hpi) (Iwanaga et al., 2007). These did not include Hsc70 or any other Hsp70 members. Only four genes were down regulated, including RNA pol II. In addition, a microarray analysis of BmNPV-infected NIAS-Bm-oyangi2 cells identified 35 significantly up-regulated host genes, and 17 down-regulated genes, many of them unknown (Sagisaka et al., 2010). The fold changes were also not high as the fold change that detected in AcMNPV-infected Sf21 cells. These studies indicate that there may be both virus- and host cell-specific differences in the regulation of host gene expression.
The results of microarray analysis were confirmed by qRT-PCR, with only one exception, and the relative expression levels were remarkable similar, validating the microarray data (Table 3). The only gene whose regulation as shown by microarray analysis was not confirmed by qRT-PCR, serpin-6, was represented by three probes on the microarray. However, only one probe indicated differential regulation. This suggested that the microarray hybridization with this probe was probably an anomaly and the expression of this gene is not differentially regulated in AcMNPV-infected Sf21 cells. One possible explanation is that this probe may cross hybridize with viral transcripts. A BLASTn search against AcMNPV revealed several regions of nucleotide similarity with this probe located in close proximity on the viral genome. The qRT-PCR results indicated that serpin-6 was not differentially regulated in AcMNPV-infected Sf21 cells. This was in agreement with the other probes for this EST, which did not show differential regulation.
The majority of regulated gene transcripts were down regulated. When we analyzed the genes that were most highly down regulated by functional clustering, the cluster with highest enrichment score comprised mainly genes involved in protein trafficking in the ER and golgi. It is possible that these enrichment scores are a reflection of the B. mori genes that are best annotated, the background used in the DAVID analysis. However down regulation of transcripts for proteins involved in trafficking of secretory and membrane proteins in the ER is consistent with the relative ease in BEVS expression of cytoplasmic proteins compared with integral membrane proteins and secretory proteins, especially those that are highly glycosylated (Higgins et al., 2003; Jarvis and Summers, 1989; Stults et al., 1994; Tate et al., 2003; van Oers et al., 2001a). In most cases, proteins are expressed in BEVS under control of the polh promoter. Although this results in high levels of expression, our results suggest that it may not be optimal for expressing secretory or integral membrane proteins. For example, the amounts of specific chaperones available late in infection appear to be insufficient for proper co-translational processing of integral membrane proteins during synthesis. The serotonin transporter, with twelve membrane-spanning domains, was expressed to high levels, but as a heterogeneous mix containing a large proportion of non-glycosylated and inactive protein (Tate et al., 2003). Co-expressing molecular chaperones, in particular calnexin increased its functional expression (Tate et al., 2003; Tate et al., 1999a; Tate et al., 1999b). Simply expressing a potassium channel from an earlier promoter increased the amount of functional protein produced (Higgins et al., 2003). The yield of functional channels was further improved by co-expression of calnexin. In the present study, microarrays included three probes for a putative calnexin precursor and its transcripts were down regulated approximately 70 fold by 24 hpi (data not shown). The levels of two chaperones were examined directly in BEVS (Yun et al., 2005). Yun and colleagues found that transcripts of the protein disulfide isomerase gene (PDI) could only be detected on Northern blots during the first day of infection in Sf9 cells expressing secreted GFP from the polh promoter. Moreover its protein levels declined over three days. In the present study, transcripts of PDI, as well as PDI-like protein ERp57 gene were both strongly down regulated by 24 hpi (Supplementary Table 1). Yun et al. also found that the steady state levels of calnexin gradually declined over four days. Interestingly, expression of another chaperone, calreticulin, was highly induced at 24 hpi following AcMNPV infection of Hv-AM1 cells (Popham et al., 2010). Up regulated calreticulin expression was represented by five spots on a two-dimensional gel, two of them highly up regulated. In the present study, two calreticulin genes (protein acc. no. gi|17826933 and gi|28804517) were represented on our microarrays, and transcripts for both were down-regulated, 75 fold (data not shown) and greater than 100 fold by 24 hpi (Supplementary Table 1), respectively.
Despite the compromised secretory system suggested by this study, as of October 2010, BEVS was used successfully to express several types of membrane proteins to sufficient levels for high-resolution structural studies (http://blanco.biomol.uci.edu/Membrane_proteins_xtal.html) (White, 2004). These include a monotopic membrane protein (Sidhu et al., 2010), five G-protein coupled receptors (GPCRs) (Cherezov et al., 2007; Chien et al., 2010; Hanson et al., 2008; Jaakola et al., 2008; Rasmussen et al., 2007; Warne et al., 2008; Wu et al., 2010a), four ion channels (Gonzales et al., 2009; Jasti et al., 2007; Kawate et al., 2009; Sobolevsky et al., 2009; Wu et al., 2010b; Yuan et al., 2010), an aquaporin (Hiroaki et al., 2006; Tani et al., 2009), and a gap junction (Maeda et al., 2009). However, many membrane proteins are not expressed well using BEVS. What is more surprising is that often, very similar types of membrane protein are expressed to very different levels but the reasons for this are not clear (Grisshammer and Tate, 1995; Massotte, 2003; Sarramegna et al., 2003). Expressing integral membrane proteins from the polh promoter can be an advantage when they adversely affect host cell physiology. However, our results suggest that by late times in the baculovirus infection cycle the necessary proteins and machinery for efficient and high-level expression may be insufficient for many proteins that are synthesized in the ER. Whether the reduction in transcripts for secretory system components results in a sufficient loss of available proteins to affect function remains to be determined. In at least one case, the levels of a host protein, TATA-binding protein, increased up to 72 hpi even as its transcripts decreased in AcMNPV-infected Sf21 cells (Quadt et al., 2002). However our study suggests that to improve expression levels of some secretory and especially integral membrane proteins in BEVS, it will be necessary to balance the timing of expression to maximize the availability of cellular proteins and machinery, while minimizing potential physiological effects on the cell. Another approach is to determine which critical cellular proteins may be insufficient and supplement them by expressing them from an unaffected promoter either from the virus or in stably transformed cells.
De novo synthesis of host proteins is gradually shut down in AcMNPV-infected cells over the course of infection and correlates with the reduction in transcripts (van Oers et al., 2003; van Oers et al., 2001b). The reduction in host protein synthesis also correlates with expression of viral genes required for DNA replication (Schultz and Friesen, 2009). RNAi knock down of these genes prevents global protein synthesis shut down, suggesting that it could result from a host DNA damage response. The mechanism responsible for the reduction in host cell transcripts is unknown, but possibilities include degradation of transcripts, inactivation of host RNA pol II, or both (Ooi and Miller, 1988). The timing of the shut down of host transcription corresponds to the beginning of the late phase of the baculovirus lifecycle when late virus genes are transcribed by a virus RNA polymerase instead of host RNA pol II. To date, a host shut-off factor has not been identified among baculovirus genes or proteins. However the rapid reduction of most host transcripts we observed during the course of AcMNPV-infection seems unlikely to result simply from a switch from host to virus RNA polymerase and strongly suggests that a virus host shut-off factor exists.
We were surprised to find that genes for immune proteins were expressed in Sf21 cells. These cells were derived from pupal ovaries and have not been previously characterized as having an immune function. Regulated transcripts represented genes involved in both humoral (cecropin and prophenol oxidase activating protein) and cellular responses (eater). Although antimicrobial peptides such as cecropin are produced primarily in the fat body, they are also produced in hemocytes (Lavine et al., 2005). Pattern recognition receptors (β-1,3-glucan-binding protein) are expressed in all immune tissues, hemocytes, fat body, and epidermis (Ochiai and Ashida, 2000). However, Eater, a scavenger receptor involved in phagocytosis is expressed exclusively in hemocytes (Kocks et al., 2005). This suggests the possibility that Sf21 cells arose from hemocytes. All of these genes were down regulated in AcMNPV-infected Sf21 cells beginning at 6 hpi, suggesting that by reducing immune protein transcripts, AcMNPV may inhibit host immune response.
In this study we extended the findings of Nobiron et al. (Nobiron et al., 2003) and report additional Hsp70 family members, as well as several other genes that were up regulated in AcMNPV-infected Sf21 cells. Further characterization of these genes may provide new insights on the roles of cellular gene products in baculovirus life-cycles. The dramatic down regulation of most cellular gene transcripts, especially secretory pathway genes, suggests that the difficulty in expressing many secretory and membrane proteins is the result of severely compromised cellular structure late in infection. This work serves as a comprehensive transcriptome platform that not only confirms host transcription shut off, but also provides a close estimate of the magnitude of these shut off for many genes (fold change). The effects of the reduced transcripts on protein levels and cellular function remain to be investigated.
Materials and Methods
Cell line and virus
Spodoptera frugiperda IPL-Sf21 cells (Vaughn et al., 1977) were maintained at 27 °C in TC-100 media (Sigma) supplemented with 10% fetal bovine serum (Atlanta Biologicals). The virus used in this study was the wild-type AcMNPV strain L1 (Lee and Miller, 1978). Infection was performed at MOI of 10 and polyhedra were observed microscopically starting at 24 hpi and at 48 hpi all Sf21 cells showed polyhedra.
Microarray Design
A 44K Agilent oligonucleotide microarray chip was designed to include most of the known ESTs of Spodoptera frugiperda. The ESTs sequences were obtained from SPODOBASE (http://bioweb.ensam.inra.fr/spodobase/ (Negre et al., 2006). Each array included 14,619 ESTs of Spodoptera frugiperda and most of the AcMNPV ORFs. Each EST was represented by at least one 60-mer oligo but most of the time by three different 60-mer oligos (total 42,334 oligos) targeting different sites of each EST.
Total RNA extraction
A total of 2 × 106 Sf21 cells were seeded into 60mm dish and infected with either TC-100 medium (Mock-infection) or wild-type AcMNPV strain L1 at MOI of 10. Cells were collected at 6, 12, 24 and 48 hpi. Mock-infection cells were collected at 12 h. Four biological repeats were set up for each time point and mock-infection. Total RNA were purified by RNAeasy Mini Kits (Qiagen) according to the manual. The quality of total RNA were assessed by Agilent 2100 Bioanalyzer using Agilent RNA 6000 Nano LabChip kit.
Sample preparation and hybridization
One-Color Spike-in Mix was added to 500 ng of total RNA and the complementary RNAs were synthesized for mock infection and for three AcMNPV-infected samples (6, 12, 24 hpi) following the recommended methods described in One-Color Microarray-Based Gene Expression Analysis Manual (Agilent). Complementary RNAs were purified by RNAeasy Mini Kits (Qiagen) and quantified by NanoDrop ND1000 (Thermo Scientific). A total of 1.65 μg of each of the Cy3-labeled linearly amplified cRNAs were fragmented by the fragmentation mix and applied to the microarrays for hybridization at 65 °C for 19 hours. Then the slides were washed and handled according to the protocol (One-Color Microarray-based Gene Expression Analysis, Agilent).
Data analysis
The microarray slides were scanned by Agilent scanner (G2505B) and data were extracted by Agilent Feature Extraction Software (v9.5.1). Quality Control ( QC ) reports were verified for all the arrays before the data analysis. Then GeneSpring GX11software (Agilent) was used to analyze the data. The data was normalized to the Spike-in genes in each of the arrays. The level of gene expression was calculated as fold change and only genes that their level of expression changed with more than 1.2 fold and with a p<0.05 were considered for the analysis. This was performed by One-Way ANOVA (GeneSpring GX11software, Agilent).
Quantitative real-time Polymerase chain reaction (qRT-PCR)
To confirm microarray data, 12 genes were selected to perform qRT-PCR (asterisks, Fig 2 and Table 2). A total of 26 forward and reverse primers (Supplementary Table 4) were designed for these 12 genes in addition to S. frugiperda 28S ribosomal RNA gene using Primer Express Software (Applied Biosystems). S. frugiperda 28S gene was used as an internal control for normalization. The same total RNA samples that were used for microarray experiment were used in the mock-, and 6, 12, and 24 hpi-infected samples for qRT-PCR. The 48 hpi samples were collected and processed at the same time as the microarray samples. A total of 1 μg of the DNase-treated total RNA was used for cDNA synthesis following the manual of the iScript cDNA synthesis kit (Bio-Rad). The qPCR was conducted using Power SYBR Green PCR Master Mix (Applied Biosystems) for the mock infected as well as the 6, 12, 24, and 48 h.p.i. samples. Four biological samples for each time point of infection and mock infection were used in the qRT-PCR. In addition, each of the biological samples was conducted in duplicate. For each sample, 5 ng RNA-equivalent cDNA were used as a template using 300 nM of each primer (forward & reverse) in addition to the 5X Power SYBR Green PCR Master Mix in a 15 μl reaction. The relative expression of each gene was calculated according to the 2−ΔΔCt method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008). The relative expression of mock infection sample was considered to be 1 and the relative expression of the rest of the samples was calculated accordingly. A Student's t-test was performed on the data to determine whether the difference of the relative gene expression between the infected samples and the mock-infected samples were significant.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant 5R01GM086719 to SMT. We thank Dr. Philippe Fournier, Unité Biologie Intégrative et Virologie des Insectes, Université Montpellier II, Montpellier, France, for graciously providing SPODOBASE sequence files and Dr. Jeff Landgraf at the Research Technology Support Facility (RTSF) at Michigan State University for assistance with the GeneSpring data analysis software package.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Braunagel SC, Parr R, Belyavskyi M, Summers MD. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G(2)/M phase. Virology. 1998;244(1):195–211. doi: 10.1006/viro.1998.9097. [DOI] [PubMed] [Google Scholar]
- Carstens EB, Tjia ST, Doerfler W. Infection of Spodoptera-Frugiperda Cells with Autographa- Californica Nuclear Polyhedrosis-Virus .1. Synthesis of Intracellular Proteins after Virus-Infection. Virology. 1979;99(2):386–398. doi: 10.1016/0042-6822(79)90017-5. [DOI] [PubMed] [Google Scholar]
- Charlton CA, Volkman LE. Sequential Rearrangement and Nuclear Polymerization of Actin in Baculovirus-Infected Spodoptera-Frugiperda Cells. Journal of Virology. 1991;65(3):1219–1227. doi: 10.1128/jvi.65.3.1219-1227.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta(2)-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien EYT, Liu W, Zhao QA, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, Stevens RC. Structure of the Human Dopamine D3 Receptor in Complex with a D2/D3 Selective Antagonist. Science. 2010;330(6007):1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(9) [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460(7255):599–U62. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisshammer R, Tate CG. Overexpression of Integral Membrane-Proteins for Structural Studies. Quarterly Reviews of Biophysics. 1995;28(3):315–422. doi: 10.1017/s0033583500003504. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 angstrom structure of the human beta(2)-adrenergic receptor. Structure. 2008;16(6):897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins MK, Demir M, Tate CG. Calnexin co-expression and the use of weaker promoters increase the expression of correctly assembled Shaker potassium channel in insect cells. Biochimica Et Biophysica Acta-Biomembranes. 2003;1610(1):124–132. doi: 10.1016/s0005-2736(02)00715-0. [DOI] [PubMed] [Google Scholar]
- Hiroaki Y, Tani K, Kamegawa A, Gyobu N, Nishikawa K, Suzuki H, Walz T, Sasaki S, Mitsuoka K, Kimura K, Mizoguchi A, Fujiyoshi Y. Implications of the aquaporin-4 structure on array formation and cell adhesion. Journal of Molecular Biology. 2006;355(4):628–639. doi: 10.1016/j.jmb.2005.10.081. [DOI] [PubMed] [Google Scholar]
- Hu YC, Yao K, Wu TY. Baculovirus as an expression and/or delivery vehicle for vaccine antigens. Expert Review of Vaccines. 2008;7(3):363–371. doi: 10.1586/14760584.7.3.363. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Kobayashi M. Cell-cycle perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology. 1999;258(1):176–188. doi: 10.1006/viro.1999.9706. [DOI] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses, I . Virus Taxonomy 2008. Vol. 2009. International Committee on Taxonomy of Viruses (ICTV); 2008. [Google Scholar]
- Iwanaga M, Shimada T, Kobayashi M, Kang W. Identification of differentially expressed host genes in Bombyx mori nucleopolyhedrovirus infected cells by using subtractive hybridization. Applied Entomology and Zoology. 2007;42(1):151–159. [Google Scholar]
- Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. The 2.6 Angstrom Crystal Structure of a Human A(2A) Adenosine Receptor Bound to an Antagonist. Science. 2008;322(5905):1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis DL. Baculovirus expression vectors. In: Miller LK, editor. The Viruses. Plenum Press; New York: 1997. pp. 389–431. [Google Scholar]
- Jarvis DL, Summers MD. Glycosylation and Secretion of Human-Tissue Plasminogen- Activator in Recombinant Baculovirus-Infected Insect Cells. Molecular and Cellular Biology. 1989;9(1):214–223. doi: 10.1128/mcb.9.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9A resolution and low pH. Nature. 2007;(7160):316. doi: 10.1038/nature06163. + [DOI] [PubMed] [Google Scholar]
- Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, Rohrmann GF, Theilmann DA, Thiem SM, Vlak JM. On the classification and nomenclature of baculoviruses: A proposal for revision. Archives Of Virology. 2006;151(7):1257–1266. doi: 10.1007/s00705-006-0763-6. [DOI] [PubMed] [Google Scholar]
- Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009;460(7255):592–U55. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, Strom C, Conto SL, Hetru C, Stuart LM, Stehle T, Hoffmann JA, Reichhart JM, Ferrandon D, Ramet M, Ezekowitz RAB. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123(2):335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Lavine MD, Chen G, Strand MR. Immune challenge differentially affects transcript abundance of three antimicrobial peptides in hemocytes from the moth Pseudoplusia includens. Insect Biochemistry and Molecular Biology. 2005;35(12):1335–1346. doi: 10.1016/j.ibmb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Lee HH, Miller LK. Isolation of Genotypic Variants of AutographaCalifornica Nuclear Polyhedrosis-Virus. Journal of Virology. 1978;27(3):754–767. doi: 10.1128/jvi.27.3.754-767.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lyupina YV, Dmitrieva SB, Timokhova AV, Beljelarskaya SN, Zatsepina OG, Evgen'ev MB, Mikhailov VS. An important role of the heat shock response in infected cells for replication of baculoviruses. Virology. 2010;406(2):336–341. doi: 10.1016/j.virol.2010.07.039. [DOI] [PubMed] [Google Scholar]
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 angstrom resolution. Nature. 2009;458(7238):597–U61. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- Makela AR, Oker-Blom C. Baculovirus display: A multifunctional technology for gene delivery and eukaryotic library development. Insect Viruses: Biotechnological Applications. 2006;68:91. doi: 10.1016/S0065-3527(06)68003-2. + [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruniak JE, Summers MD. Autographa-Californica Nuclear Polyhedrosis-Virus Phosphoproteins and Synthesis of Intracellular Proteins after Virus-Infection. Virology. 1981;109(1):25–34. doi: 10.1016/0042-6822(81)90468-2. [DOI] [PubMed] [Google Scholar]
- Massotte D. G protein-coupled receptor overexpression with the baculovirus-insect cell system: a tool for structural and functional studies. Biochimica Et Biophysica Acta-Biomembranes. 2003;1610(1):77–89. doi: 10.1016/s0005-2736(02)00720-4. [DOI] [PubMed] [Google Scholar]
- Mayer MP. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Reviews of Physiology Biochemistry and Pharmacology. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- Negre V, Hotelier T, Volkoff AN, Gimenez S, Cousserans F, Mita K, Sabau X, Rocher J, Lopez-Ferber M, d'Alencon E, Audant P, Sabourault C, Bidegainberry V, Hilliou F, Fournier P. SPODOBASE : an EST database for the lepidopteran crop pest Spodoptera. Bmc Bioinformatics. 2006:7. doi: 10.1186/1471-2105-7-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobiron I, O'Reilly D, Olzewski JA. Autographa californica nucleopolyhedrovirus infection of Spodoptera frugiperda cells: a global analysis of host gene regulation during infection, using a differential display approach. Journal of General Virology. 2003;84:3029–3039. doi: 10.1099/vir.0.19270-0. [DOI] [PubMed] [Google Scholar]
- O'Reilly DR, Miller LK, Luckow V. Baculovirus Expression Vectors: A Laboratory Manual. W. H. Freeman and Company; New York: 1992. [Google Scholar]
- Ochiai M, Ashida M. A pattern-recognition protein for beta-1,3-glucan - The binding domain and the cDNA cloning of beta-1,3-glucan recognition protein from the silkworm, Bombyx mori. Journal of Biological Chemistry. 2000;275(7):4995–5002. doi: 10.1074/jbc.275.7.4995. [DOI] [PubMed] [Google Scholar]
- Ooi BG, Miller LK. Regulation of Host RNA Levels During Baculovirus Infection. Virology. 1988;166(2):515–523. doi: 10.1016/0042-6822(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Popham HJR, Grasela JJ, Goodman CL, McIntosh AH. Baculovirus infection influences host protein expression in two established insect cell lines. Journal of Insect Physiology. 2010;56(9):1237–1245. doi: 10.1016/j.jinsphys.2010.03.024. [DOI] [PubMed] [Google Scholar]
- Quadt I, Mainz D, Mans R, Kremer A, Knebel-Morsdorf D. Baculovirus infection raises the level of TATA-binding protein that colocalizes with viral DNA replication sites. Journal of Virology. 2002;76(21):11123–11127. doi: 10.1128/JVI.76.21.11123-11127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SGF, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK. Crystal structure of the human beta(2) adrenergic G-protein-coupled receptor. Nature. 2007;450(7168):383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Rohrmann GF. Baculovirus Molecular Biology. Nationa; Library of Medicine (US), National Center for Biotechnology Information; Bethesda, MD: 2008. [Google Scholar]
- Roncarati R, Knebel-Morsdorf D. Identification of the early actin-rearrangement-inducing factor gene, arif-1, from Autographa californica multicapsid nuclear polyhedrosis virus. Journal of Virology. 1997;71(10):7933–7941. doi: 10.1128/jvi.71.10.7933-7941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagisaka A, Fujita K, Nakamura Y, Ishibashi J, Noda H, Imanishi S, Mita K, Yamakawa M, Tanaka H. Genome-wide analysis of host gene expression in the silkworm cells infected with Bombyx mori nucleopolyhedrovirus. Virus Research. 2010;147(2):166–175. doi: 10.1016/j.virusres.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Sarramegna V, Talmont R, Demange P, Milon A. Heterologous expression of G-protein-coupled receptors: comparison of expression systems from the standpoint of large-scale production and purification. Cellular and Molecular Life Sciences. 2003;60(8):1529–1546. doi: 10.1007/s00018-003-3168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nature Protocols. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schultz KLW, Friesen PD. Baculovirus DNA Replication-Specific Expression Factors Trigger Apoptosis and Shutoff of Host Protein Synthesis during Infection. Journal of Virology. 2009;83(21):11123–11132. doi: 10.1128/JVI.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu RS, Lee JY, Yuan C, Smith WL. Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers. Biochemistry. 2010;49(33):7069–7079. doi: 10.1021/bi1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GE, Fraser MJ, Summers MD. Molecular Engineering of the Autographa-Californica Nuclear Polyhedrosis-Virus Genome - Deletion Mutations within the Polyhedrin Gene. Journal of Virology. 1983;46(2):584–593. doi: 10.1128/jvi.46.2.584-593.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462(7274):745–U66. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults JT, Oconnell KL, Garcia C, Wong S, Engel AM, Garbers DL, Lowe DG. The Disulfide Linkages And Glycosylation Sites Of The Human Natriuretic Peptide Receptor-C Homodimer. Biochemistry. 1994;33(37):11372–11381. doi: 10.1021/bi00203a036. [DOI] [PubMed] [Google Scholar]
- Summers MD. Milestones leading to the genetic engineering of baculoviruses as expression vector systems and viral pesticides. Insect Viruses: Biotechnological Applications. 2006;68:3–73. doi: 10.1016/S0065-3527(06)68001-9. [DOI] [PubMed] [Google Scholar]
- Summers MD, Smith GE. A Manual of Methods for Baculovirus Vectors and Insect Cell- Culture Procedures. Texas Agricultural Experiment Station Bulletin. 1987;(1555):1–56. [Google Scholar]
- Tani K, Mitsuma T, Hiroaki Y, Kamegawa A, Nishikawa K, Tanimura Y, Fujiyoshi Y. Mechanism of Aquaporin-4's Fast and Highly Selective Water Conduction and Proton Exclusion. Journal of Molecular Biology. 2009;389(4):694–706. doi: 10.1016/j.jmb.2009.04.049. [DOI] [PubMed] [Google Scholar]
- Tate CG, Haase J, Baker C, Boorsma M, Magnani F, Vallis Y, Williams DC. Comparison of seven different heterologous protein expression systems for the production of the serotonin transporter. Biochimica Et Biophysica Acta-Biomembranes. 2003;1610(1):141–153. doi: 10.1016/s0005-2736(02)00719-8. [DOI] [PubMed] [Google Scholar]
- Tate CG, Whiteley E, Betenbaugh MJ. Molecular chaperones improve functional expression of the serotonin (5-hydroxytryptamine) transporter in insect cells. Biochemical Society Transactions. 1999a;27:932–936. doi: 10.1042/bst0270932. [DOI] [PubMed] [Google Scholar]
- Tate CG, Whiteley E, Betenbaugh MJ. Molecular chaperones stimulate the functional expression of the cocaine-sensitive serotonin transporter. Journal of Biological Chemistry. 1999b;274(25):17551–17558. doi: 10.1074/jbc.274.25.17551. [DOI] [PubMed] [Google Scholar]
- van Oers MM. Vaccines for viral and parasitic diseases produced with baculovirus vectors. Insect Viruses: Biotechnological Applications. 2006;68:193. doi: 10.1016/S0065-3527(06)68006-8. + [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oers MM, Doitsidou M, Thomas AAM, de Maagd RA, Vlak JM. Translation of both 5 ′ TOP and non-TOP host mRNAs continues into the late phase of Baculovirus infection. Insect Molecular Biology. 2003;12(1):75–84. doi: 10.1046/j.1365-2583.2003.00389.x. [DOI] [PubMed] [Google Scholar]
- van Oers MM, Thomas AAM, Moormann RJM, Vlak JM. Secretory pathway limits the enhanced expression of classical swine fever virus E2 glycoprotein in insect cells. Journal of Biotechnology. 2001a;86(1):31–38. doi: 10.1016/s0168-1656(00)00403-x. [DOI] [PubMed] [Google Scholar]
- van Oers MM, van der Veken L, Vlak JM, Thomas AAM. Effect of baculovirus infection on the mRNA and protein levels of the Spodoptera frugiperda eukaryotic initiation factor 4E. Insect Molecular Biology. 2001b;10(3):255–264. doi: 10.1046/j.1365-2583.2001.00263.x. [DOI] [PubMed] [Google Scholar]
- Vaughn JL, Goodwin RH, Tompkins GJ, McCawley P. Establishment of 2 cell lines from insect Spodoptera frugiperda (Lepidoptera-Noctuidae) In Vitro-Journal of the Tissue Culture Association. 1977;13(4):213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- Volkman LE, Zaal KJM. Autographa-Californica-M Nuclear Polyhedrosis-Virus - Microtubules and Replication. Virology. 1990;175(1):292–302. doi: 10.1016/0042-6822(90)90211-9. [DOI] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature. 2008;454(7203):486–U2. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH. The progress of membrane protein structure determination. Protein Science. 2004;13(7):1948–1949. doi: 10.1110/ps.04712004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BL, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010a;330(6007):1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YK, Yang Y, Ye S, Jiang YX. Structure of the gating ring from the human large-conductance Ca2+-gated K+ channel. Nature. 2010b;466(7304):393–U148. doi: 10.1038/nature09252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Leonetti MD, Pico AR, Hsiung YC, MacKinnon R. Structure of the Human BK Channel Ca2+-Activation Apparatus at 3.0 angstrom Resolution. Science. 2010;329(5988):182–186. doi: 10.1126/science.1190414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun EY, Goo TW, Kim SW, Choi KH, Hwang JS, Kang SW, Kwon OY. Changes in cellular secretory processing during baculovirus infection. Biotechnology Letters. 2005;27(14):1041–1045. doi: 10.1007/s10529-005-8108-1. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology. 2000;7(1-2):203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.