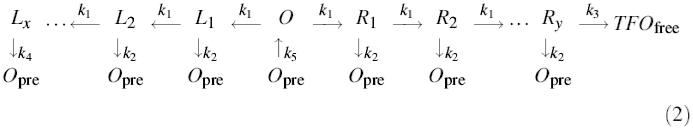Abstract
The type I restriction enzyme EcoR124I cleaves DNA following extensive linear translocation dependent upon ATP hydrolysis. Using protein-directed displacement of a DNA triplex, we have determined the kinetics of one-dimensional motion without the necessity of measuring DNA or ATP hydrolysis. The triplex was pre-formed specifically on linear DNA, 4370 bp from an EcoR124I site, and then incubated with endonuclease. Upon ATP addition, a distinct lag phase was observed before the triplex-forming oligonucleotide was displaced with exponential kinetics. As the distance between type I and triplex sites was shortened, the lag time decreased whilst the displacement reaction remained exponential. This is indicative of processive DNA translocation followed by collision with the triplex and oligonucleotide displacement. A linear relationship between lag duration and inter-site distance gives a translocation velocity of 400 ± 32 bp/s at 20°C. Furthermore, the data can only be explained by bi-directional translocation. An endonuclease with only one of the two HsdR subunits responsible for motion could still catalyse translocation. The reaction is less processive, but can ‘reset’ in either direction whenever the DNA is released.
Keywords: DNA translocation/DNA triplex/motor protein/restriction–modification
Introduction
Protein motion along DNA is essential for the repair (Jiricny, 1998), recombination (George et al., 1999), replication (Hiasa and Marians, 1999), transcription (Landick, 1999) and restriction (Bickle, 1993) of genetic material. Enzymes that catalyse directed linear steps along polymers differ from other ‘molecular machines’ in that they generate both motion and force, i.e. they are true motors. This has been investigated most thoroughly for the classical ‘motor proteins’, kinesin and myosin. Although it is clear that DNA translocation also fits these criteria (Szczelkun, 2000), the dynamics of the nucleic acid enzymes remain inadequately characterized. A rigorous evaluation of any DNA-bound motor must first provide answers to some fundamental questions: what is the translocation rate; what is the direction of movement relative to binding-site polarity; and how often does an enzyme stall on, or release the DNA? In this study, these questions will be addressed with the type I restriction enzyme EcoR124I.
Type I restriction endonucleases recognize specific asymmetric DNA sequences (e.g. Figure 1A). However, DNA cleavage is dependent upon the free energy of ATP hydrolysis and occurs at random non-specific loci from 50 to >10 000 bp distant (Horiuchi and Zinder, 1972; Szczelkun et al., 1997). It was first suggested >25 years ago that the link between binding and cleavage may be due to protein motion along DNA (Shulman, 1974). Electron microscopy of endonucleases bound to DNA in the presence of ATP revealed loops of DNA (Rosamund et al., 1979; Yuan et al., 1980), and a general model was proposed in which an enzyme remains bound to its recognition site whilst simultaneously pulling adjacent non-specific DNA past itself (Yuan et al., 1980). Studier and Bandyopadhyay (1988) developed the model by demonstrating that DNA cleavage follows the collision of two converging EcoKI enzymes, midway between each pair of sites. Their results suggested that despite the asymmetry of the recognition sequence, movement occurs in both ‘leftward’ and ‘rightward’ directions simultaneously. This was termed bi-directional translocation. More recent experiments using atomic force microscopy have confirmed the presence of the expected multiple DNA loops (Ellis et al., 1999), but more importantly have permitted direct observation of temporal changes in the size of the loops, indicative of translocation. An unambiguous demonstration of linear communication in vitro was provided by examining cleavage of DNA catenanes (Szczelkun et al., 1996). Of two singly interlinked DNA circles, only the ring carrying an EcoR124I recognition site could be cleaved—the connected ring remained intact despite its physical proximity. This proved that DNA recognition and cleavage must be coupled by one-dimensional translocation. Furthermore, motion generates a decrease in twist behind and an increase in twist ahead of the complex (Szczelkun et al., 1996; Janscak and Bickle, 2000), indicating that translocation follows the helical path of DNA. Molineux and co-workers have analysed DNA translocation elegantly in vivo (Davies et al., 1999a; Garcia and Molineux, 1999). Introduction of an EcoKI site into the leader sequence of a modified T7 phage genome allowed endonuclease translocation to internalize the DNA into bacteria. By following the progressive methylation of Sau3AI restriction sites, an EcoKI translocation velocity of ∼100 bp/s was estimated (Garcia and Molineux, 1999). An accurate determination of translocation rate is fundamental to the description of molecular motor function. Consequently, a direct assay of linear motion would be an invaluable tool in the study of type I enzymes.
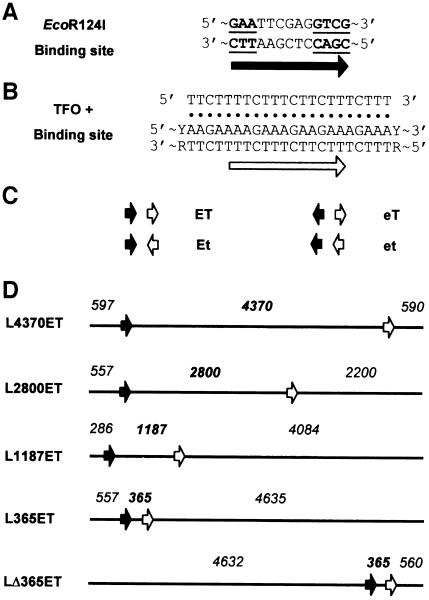
Fig. 1. Triple helix substrates. (A) EcoR124I recognition sequence. Conserved nucleotides are underlined. (B) TFO and TFO-binding site sequences. Hoogsten base pairs between the TFO and purine-rich strand of the duplex are indicated (filled circles). Arbitrary site orientations are defined by the black (EcoR124I) and white (triplex) arrows. (C) Substrates are designated a two-letter code depending on the relative orientations of the EcoR124I (E or e) and TFO (T or t) sites. Upper case characters indicate that the site runs left to right on the substrate as drawn, lower case characters indicate the reverse. (D) Examples of linear substrates (5557 bp) used in the TFO displacement assay. DNA is drawn as a solid line with arrows as in (C). Numbers indicate base pairs from a DNA end to the G of GAA in the EcoR124I site, to the T⋅AT triplet at the 5′ end of the triplex, and to the other DNA end. Substrates are named L (for linear), followed by inter-site distance and relative site orientation (from C). Cleavage of pMDS21 with NcoI or ScaI produced L365ET or LΔ365ET, respectively; cleavage of pMDS22 with NcoI or ScaI produced L4370ET or L1187ET, respectively; and cleavage of pMDS28 with ScaI produced L2800ET.
The type I endonucleases consist of three subunits: HsdS (DNA recognition), HsdM (DNA methylation) and HsdR (DNA and ATP hydrolysis), with the stoichiometry R2M2S1 (Dryden et al., 1997). Which part of the complex acts as the motor? The HsdR amino acid sequences contain motifs previously identified in helicases, the so-called DEAD-box motifs (Gorbalenya and Koonin, 1991), and also have predicted secondary structure homology to the PcrA and Rep helicases (Davies et al., 1999a). Mutations in these motifs affect ATPase activity (Davies et al., 1998), impair DNA restriction (Webb et al., 1996) and prevent internalization of phage DNA (Davies et al., 1999b), suggesting a DNA translocation mechanism akin to that suggested for the DNA helicases (Soultanas and Wigley, 2000). Changes in DNA topology during translocation are also indicative of strand separation (Janscak and Bickle, 2000), but a helicase-like DNA unwinding activity has yet to be demonstrated directly for type I enzymes (Szczelkun, 2000). Moreover, it has yet to be established whether HsdR subunits contact DNA with a defined polarity (i.e. 5′–3′ or 3′–5′) or even if the strands need to be separated at all. A full understanding of DNA translocation by EcoR214I would not only pertain to type I enzymes, but would generate valuable information about DEAD-box proteins in general, as well as other DNA-based motors such as the methyl-directed mismatch repair enzymes for which an equivalent bi-directional translocation model has been proposed (Allen et al., 1997).
To evaluate protein motion on DNA, the displacement of a DNA triplex was assayed. Triplexes are potential therapeutic gene regulators (Vasquez and Wilson, 1998), produced when a triplex-forming oligonucleotide (TFO) binds specifically to duplex DNA (Gowers and Fox, 1999). Triplex formation within transcription initiation regions is highly effective at blocking RNA polymerases (Kim and Miller, 1995). However, the effect on transcription elongation of a TFO binding downstream of a promoter depends on triplex stability. Both phage and Escherichia coli polymerases can read-through a weakly associated TFO (Skoog and Maher, 1993). However, augmenting the binding efficiency using phosphorothioates or covalently attached intercalating moieties increases the chance of inhibiting motion (Xodo et al., 1994; Giovannangeli et al., 1996). In contrast, DNA polymerase elongation is inhibited readily by triplexes (Hacia et al., 1994). The most effective unmodified TFOs are pyrimidine rich (CT), occupy the major groove parallel to a purine-rich strand and utilize Hoogsteen hydrogen bonds to form T⋅AT and C+⋅GC triplets (Gowers and Fox, 1999). Sub-nanomolar equilibrium dissociation constants are attainable (Maher et al., 1990; Plum et al., 1990), but protonation of the N3 position of cytosine demands a low pH. However, Kopel et al. (1996) demonstrated that a CT-rich TFO bound to a GA sequence in acid conditions and subsequently diluted into neutral buffer remained associated indefinitely. Using this strategy, we have evaluated the EcoR124I-directed displacement of a stable triplex in vitro under ‘standard’ reaction conditions.
A triplex was formed on either leftward or rightward sides of an EcoR124I recognition site. DNA translocation resulted in a lag phase followed by TFO displacement. By measuring lag time as a function of distance translocated, a translocation speed was calculated. The data also indicate that translocation is both processive and bi-directional. Using conditions that produce endonucleases with a single HsdR subunit, unidirectional motion independent of site asymmetry could still be recorded. The reaction was less processive than with a full complement of HsdR and correspondingly slower. However, the enzyme could ‘reset’ and resume translocation in either direction if it released the DNA during motion or at a DNA end.
Results and discussion
Triplex formation and dissociation
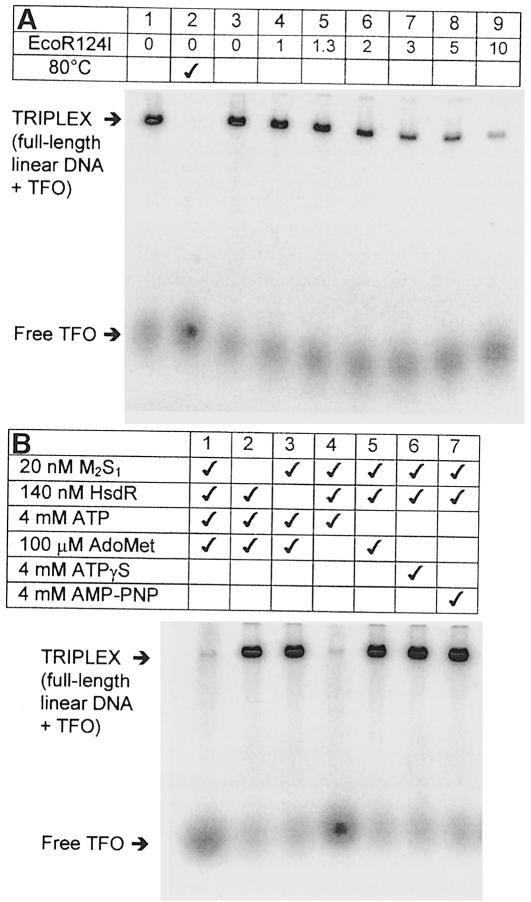
The aim of this study was to investigate the effect of a DNA triplex ‘roadblock’ on translocation by the type I restriction endonuclease EcoR124I. Linear DNA substrates were generated containing both EcoR124I- and TFO-binding sites (Figure 1A–D). Since bimolecular association rate constants for CT-rich TFOs are ∼1000/M/s (Maher et al., 1990; Paes and Fox, 1997), the 32P-labelled TFO was incubated with its target DNA overnight. Triplex formation was assessed by agarose gel electrophoresis (Materials and methods). After binding at pH 5.5, the triplex could be diluted into EcoR124I reaction buffer R (pH 8.0; Materials and methods) without the labelled TFO dissociating (Figure 2A, lane 1). The triplex was stable for at least 12 h at 20°C (data not shown). However, once displaced by heating to 80°C, the TFO did not re-associate when cooled to 20°C (lane 2). Similarly, removing Mg2+ ions from the buffer completely dissociated the triplex (not shown). Neither ATP nor the methyl donor S-adenosyl methionine (AdoMet) affected the binding. These results confirm the observations of Kopel et al. (1996) that a CT-based triplex is stable close to neutral pH if it is pre-bound in acid conditions. The simplest explanation is that the protonated form of cytosine in a C+⋅GC triplet is stabilized at neutral pH by Mg2+ ions bound to the DNA (Singleton and Dervan, 1993). Consequently, the off-rate at pH 8.0 remains as slow as it would be in acid conditions (Maher et al., 1990; Paes and Fox, 1997). Once dissociated, however, the cytosines are deprotonated rapidly, C+⋅GC triplets cannot form and the on-rate becomes so slow that rebinding is negligible. These distinct non-equilibrium ‘on’ and ‘off’ states make this triplex an ideal probe for protein–DNA interactions.
Fig. 2. TFO binding and displacement. Reactions were examined by agarose gel electrophoresis of the 32P-labelled TFO (Materials and methods). Bound and free TFO are indicated. (A) TFO (50 nM) and linear DNA were incubated at pH 5.5 overnight. Lane 1, 5 nM L365ET was incubated in buffer R (Materials and methods) for 25°C for 1 h; lane 2, 5 nM L365ET was heated to 80°C for 10 min in buffer R, cooled to 20°C and then incubated for a further hour; lanes 3–9, 5 nM L365ET was incubated with varying nanomolar concentrations of EcoR124I in buffer R as indicated, ATP added to 4 mM and the reaction incubated for a further hour at 20°C. (B) L365ET (5 nM) was pre-incubated with MTase (M2S1), HsdR and/or AdoMet in buffer R as shown. Where indicated ATP, ATPγS or AMP-PNP were also added to 4 mM. Reactions were allowed to proceed for 1 h at 20°C.
TFO displacement by EcoR124I motion
The effect of EcoR124I on triplex stability was assayed by varying the concentration of endonuclease and analysing the products 1 h after addition of 4 mM ATP (Figure 2A, lanes 3–9). Unlike equivalent observations with RNA and DNA polymerases, EcoR124I readily displaced the tightly bound TFO. Under the conditions used, no rebinding of the free TFO was observed. As the concentration of EcoR124I was increased, the percentage of free TFO also increased, until at a 2-fold molar excess of enzyme over full-length linear DNA, >95% free TFO had been generated (average value quantified from three repeat experiments; see Materials and methods). The relative orientation of the TFO had no effect on this process (not shown). The requirement for a 2-fold molar excess of enzyme over DNA for maximum activity does not represent an absolute stoichiometry, but probably results from dissociation of the enzyme complex at nanomolar protein concentrations (Janscak et al., 1998). Data from DNA cleavage reactions indicate that type I enzymes cannot turn over (Bickle, 1993). Similarly, our data show that endonuclease–DNA complexes do not dissociate appreciably once translocation has started; TFO displacement did not increase if the reactions were incubated for longer periods (not shown). TFO displacement was not seen with methyltransferase (MTase) or HsdR alone (Figure 2B, lanes 1–3), in the absence of ATP (lane 5) or with non-hydrolysable ATP analogues ATPγS or AMP-PNP (lanes 6 and 7). Excluding AdoMet from the reaction had no apparent effect (lane 4). Similarly, DNA cleavage by EcoR124I is unaffected by the absence of AdoMet (Szczelkun et al., 1996, 1997). Although AdoMet is unstable in aqueous solution, it may be stable indefinitely when bound tightly to a protein. Since we cannot rule out co-purification of AdoMet with EcoR124I, the effect of cofactor binding on translocation could not be assessed. Overall, these results suggest that DNA translocation must be responsible for TFO displacement.
The full-length linear DNA remained intact during these reactions (not shown), confirming that DNA cleavage is not a prerequisite of translocation (Davies et al., 1999a,b; Janscak et al., 1999b). However, a feature of type I enzyme activity is that DNA is cleaved wherever an endonuclease stalls, e.g. upon collision of converging enzymes (Studier and Bandyopadhyay, 1988) or upon collision of a single enzyme with an immobile Holliday junction (Janscak et al., 1999a). The same is not true upon collision with a triplex. Is a simple pause in translocation sufficient to generate DNA cleavage or is a significant build-up in mechanical load required? Morris and Raney (1999) have shown that DNA helicases can displace streptavidin from biotin-labelled oligonucleotides. The force required to disrupt this interaction has been estimated at ∼250 pN, far more energy than that available to a motor protein. However, the force required to break a non-covalent bond correlates with activation enthalpy and not overall free energy. Consequently, the application of a force, even a weak one, results in a shift in thermal fluctuations in free energy to the extent that the dissociation rate of the bond is increased considerably (Morris and Raney, 1999). Therefore, the increase in force associated with TFO displacement may not be as great as that required for DNA cleavage. Similarly, displacement of Lac repressor by EcoR124II did not result in cleavage at the operator site (Dreier et al., 1996). Moreover, if type I enzymes unwind DNA as they translocate (Davies et al., 1999a; Janscak and Bickle, 2000), then displacement of a bound ligand may be accelerated by disruption of the underlying duplex DNA.
Kinetics of DNA translocation by EcoR124I
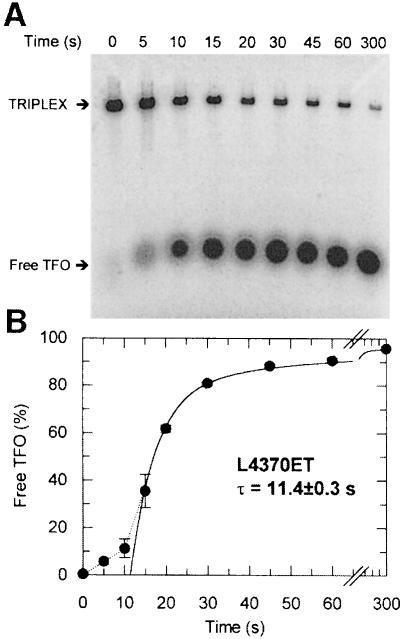
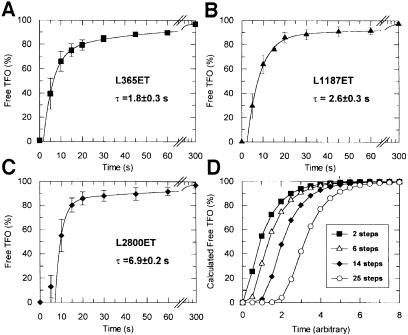
To analyse the kinetics of TFO displacement, samples were withdrawn from reactions containing fixed concentrations of EcoR124I at sequential time points after addition of ATP. These were analysed by agarose gel electrophoresis (Figure 3A) and the percentage of free TFO ascertained by densitometry (Materials and methods). The kinetic profile for L4370ET is shown (Figure 3B). Initially, there is little dissociation of the triplex, but after 10 s there is a rapid rise in free TFO, reaching ∼90% by 1 min, followed by a 10- to 20-fold slower phase reaching >95% by 5 min. To analyse the kinetics further, we measured TFO displacement on a series of linear substrates with varying distances between the EcoR124I and triplex sites (Figure 4A–C). As the distance between the sites was lengthened, the lag phase increased whilst the rates of the exponential phases remained equivalent.
Fig. 3. Kinetics of TFO displacement on L4370ET. (A) L4370ET (5 nM) was pre-incubated in buffer R with 20 nM EcoR124I for 5 min, and the reaction initiated with ATP. Aliquots were removed and the reaction quenched at the time points shown, and analysed as described (Materials and methods). Triplex and free TFO are indicated. (B) TFO displacement in terms of the percentage of free TFO liberated at each time point (filled circles). Error bars represent the standard deviation from the mean of three independent experiments. The solid line is a least-squares fit to equation (3) (Materials and methods). The estimated lag time (τ) is taken from the intercept with the abscissa. A dotted line emphasizes the lag phase.
Fig. 4. Kinetics of TFO displacement as a function of distance. (A–C) L365ET, L1187ET or L2800ET (5 nM) were pre-incubated in buffer R with 20 nM EcoR124I for 5 min, and the reactions initiated with ATP. Aliquots were removed and the reactions quenched at the time points shown, and analysed as in Figure 3. TFO displacement is shown in terms of the percentage of free TFO liberated at each time point. Error bars represent the standard deviation from the mean of at least three independent experiments. The solid lines are least-squares fits to equation (3) (Materials and methods). The resulting lag times (τ) are shown. (D) Theoretical TFO displacement profiles. The percentage of free TFO was calculated by numerical integration using equation (1) (Materials and methods). In each case, k2 was an order of magnitude slower than k1 (absolute values are not defined; see main text). The percentage of displaced TFO is plotted for reactions where y = 2, 6, 14 or 25.
If motion on DNA is considered as a series of reactions governed by single macroscopic rate constants, then translocation will generate a succession of highly populated intermediate states (Ali and Lohman, 1997). If the rate constants are equal, then the culmination of such reactions will be a lag phase, the length of which is diagnostic of the number of steps (Gutfreund, 1995). The unwinding of short DNA duplexes by UvrD helicase has been analysed in this manner (Ali and Lohman, 1997). In our experiments, however, TFO displacement is not part of the translocation mechanism and is rate limiting with respect to the preceding steps. Consequently, the same fitting procedure could not be applied. Instead, we evaluated the effect of altering the number of steps prior to TFO displacement by modelling theoretical reactions using equation (1) (Materials and methods). This model considers translocation as a series of unimolecular reactions as described above, except that the final step, TFO displacement, is slower than the preceding translocation steps. Although motion is only considered in one direction, bi-directional motion can still be accommodated (see below). The resulting series of kinetic profiles (Figure 4D) illustrate two important features of the model. First, as the number of steps is increased from two to 25, a linearly increasing lag phase is observed. Secondly, the ensuing TFO displacement follows an exponential function, the rate of which is independent of the number of steps. Because TFO displacement is rate limiting, the model cannot distinguish between a large number of fast steps and a small number of slow steps (not shown). Furthermore, each ‘step’ in reality comprises multiple reactions associated with ATP binding and hydrolysis, physical protein motion along DNA and release of ADP and inorganic phosphate. Nevertheless, the pattern of kinetic profiles in Figures 3 and 4A–C matches the predictions of the model.
If our data can be represented by equation (1) (albeit with a larger number of steps), there should be a linear relationship between lag time and inter-site distance, where the gradient gives the average lag time per base pair (or bp/s in reciprocal form). To evaluate the lag times, the exponential phases of each reaction were fitted to equation (3) (Figures 3B and 4A–C). The rates and relative proportions of the two exponential phases did not vary significantly with distance (not shown). Variations in the profiles would be indicative of an increased frequency of events that slow translocation. Therefore, stalling (Uptain et al., 1997), back-stepping (Komissarova and Kashlev, 1997) or protein–DNA dissociation events during DNA translocation must be rare. In other words, translocation by EcoR124I has a high processivity; thousands of base pairs can be transported before an enzyme dissociates or stalls. The slower exponential phase most probably results from a subset of the enzyme population that moves less efficiently. This heterogeneity may be due to a fraction of R1M2S1 species being present in the R2M2S1 reactions (see below). Figure 5 shows the combined lag times from Figures 3 and 4. A linear relationship between lag time and inter-site distance is observed, justifying the contraction of multiple kinetic events to a single unimolecular rate constant. The increase in free TFO must be due to DNA translocation via a discrete number of rapid steps, collision with the triplex and finally rate-limiting TFO displacement.
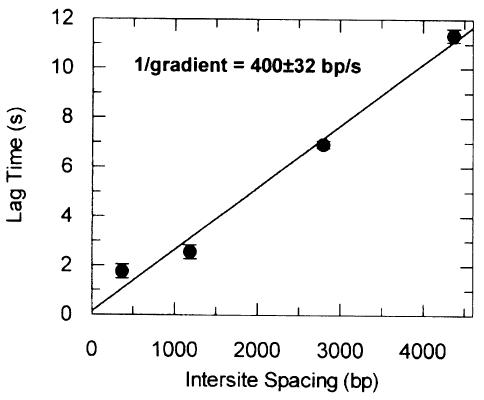
Fig. 5. Velocity of DNA translocation. Lag times from Figures 3 and 4 are plotted as a function of distance between EcoR124I and triplex sites (from Figure 1D). Error bars are the standard deviation from the mean of the intercepts. The solid line represents a least-squares linear fit to the data (Materials and methods). The velocity (reciprocal gradient) is shown as bp/s.
The reciprocal gradient of the best-fit line in Figure 5 gives a translocation velocity of 400 ± 32 bp/s at 20°C. It should be noted, however, that the inter-site distances defined in Figure 1D are not the exact lengths of DNA translocated, as the true starting position is unknown. DNA footprinting of EcoR124I has not revealed where HsdR binds (Mernagh et al., 1998), but it almost certainly occurs outside the recognition sequence (Powell et al., 1998). Although we can discount long-range random DNA looping events (Szczelkun et al., 1996), we cannot discount local DNA wrapping around the protein such that translocation starts at an appreciable distance from the site (i.e. tens of base pairs). If this were the case, then the x-axis intercept of the line in Figure 5 would specify this distance. Because of errors in evaluation of lag time, however, we cannot state precisely where translocation starts. Note that the difference in rate that this uncertainty generates is less than the observed standard error.
Bi-directional translocation by EcoR124I
According to the bi-directional model of Studier and Bandyopadhyay (1988), the relative orientation of an EcoR124I site should have little or no effect on TFO displacement kinetics. Conversely, a sequence-dependent unidirectional model as described for EcoBI (Rosamund et al., 1979) would result in motion in one direction exclusively. However, similar kinetic profiles to that for L4370ET were obtained with the substrates L4370Et and L4370eT (data not shown), in which the relative orientations of EcoR124I and triplex sites were inverted. Similarly, DNA cleavage shows no dependence on site orientation (Szczelkun et al., 1996, 1997). Therefore, translocation cannot be constrained rigidly by site asymmetry. However, a unidirectional model could still be accommodated by our results if DNA binding resulted in, on average, 50% of enzymes going leftward and 50% going rightward, and if, when an enzyme reached a DNA end, it could dissociate, randomly rebind and restart translocation. We term this last process ‘resetting’. This alternative unidirectional scheme is described by equation (2) (Materials and methods). Enzymes travel down either a leftward (L) or rightward (R) pathway (because motion is highly processive, we will assume that the dissociation rate, k2, is zero). Any enzyme moving down the L pathway eventually will reach the end of the DNA and ‘step off’. Translocation can then reset, governed by a unimolecular rate constant k5. There is no memory of the antecedent pathway, e.g. an enzyme that starts down the L pathway will dissociate, reset and restart down either L or R pathways with equal probability. Given that TFO displacement rapidly approaches 95% (Figures 3 and 4), the resetting process cannot be rate limiting. However, if resetting was fast, then equation (2) could produce kinetic profiles indistinguishable from those illustrated in Figure 4 (not shown).
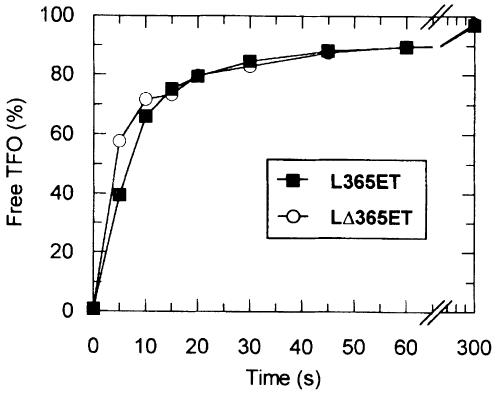
To differentiate between uni- and bi-directional models, we compared TFO displacement kinetics on L365ET and LΔ365ET (Figure 6). The distance between EcoR124I- and triplex-binding sites is the same on each substrate (365 bp), but their context is different (Figure 1D). Enzymes moving leftward as drawn on LΔ365ET would have to travel 4632 bp before they could reset, compared with only 557 bp on L365ET (Figure 1D). Even if resetting was fast, the kinetic profile on LΔ365ET would be markedly biphasic compared with that on L365ET. However, little difference can be seen between the profiles (Figure 6). Furthermore, TFO displacement on circular DNA shows a similar displacement profile to that on linear DNA (M.D.Szczelkun, unpublished data). Collectively, the data indicate that EcoR124I is capable of translocating DNA from both sides of its site simultaneously, independently of recognition sequence asymmetry. Therefore, it should also be noted that the translocation rate refers to motion in one direction only—twice the amount of DNA is actually transported per second by each enzyme.
Fig. 6. Uni- and bi-directional translocation by EcoR124I. The percentage of TFO liberated as a function of time is shown for L365ET and LΔ365ET. Reactions were performed and analysed as in Figures 3 and 4.
Unidirectional translocation and resetting by EcoR124I with a single HsdR subunit
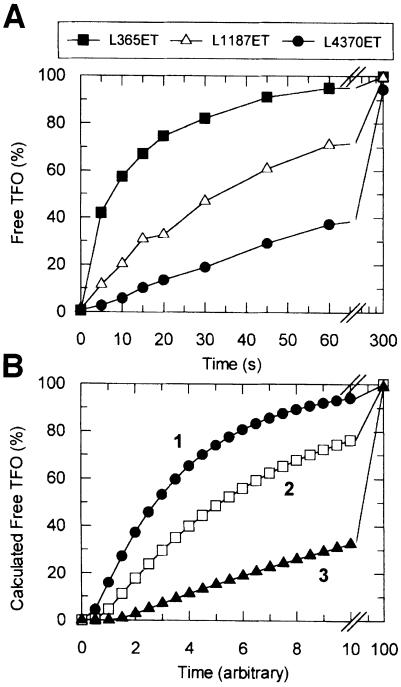
Since EcoR124I can translocate DNA in two directions, how do the HsdR subunits generate motion? Do the subunits act independently, a single HsdR translocating DNA on each side of the site, or do they communicate directly to provide added processivity and/or synchronicity? To assess these possibilities, we examined TFO displacement kinetics as a function of endonuclease stoichiometry. Since the equilibrium dissociation constant for the first HsdR subunit of R2M2S1 is >1000-fold lower than the second (Janscak et al., 1998), mixing equal nanomolar concentrations of HsdR and MTase produces the R1M2S1 form exclusively (<0.01% R2M2S1 will be present assuming equivalent sites). This form of the enzyme cannot cleave linear or plasmid DNA (not shown), whereas a DNA-activated ATP hydrolysis rate approximately one-quarter that of R2M2S1 has been measured (Janscak et al., 1998). TFO displacement by R1M2S1 was observed on L365ET, L1187ET and L4370ET (Figure 7A), and on L365eT (not shown). Despite there being only one HsdR subunit, in each case >95% free TFO was generated in 5 min, independently of the relative site orientations. However, as the inter-site distance was increased (Figure 7A), the shapes of the resulting profiles varied markedly compared with those expected from equation (1) (Figure 4D). The exponential phases became more extended with distance, indicating an increasing frequency of events that slow translocation. To explain these observations, we will consider a random unidirectional model, introducing dissociation events during motion. Although a bi-directional model can also accommodate these results, there is no evidence from studies of DNA helicases that a monomeric enzyme can bind and transport two DNA duplexes simultaneously (Soultanas and Wigley, 2000).
Fig. 7. DNA translocation by EcoR124I with one HsdR subunit. (A) DNA (5 nM; as indicated) was pre-incubated with 20 nM M2S1 and 20 nM HsdR in buffer R at 20°C. Reactions started with ATP were analysed as in Figures 3 and 4. The percentage of liberated TFO is indicated as a function of time. (B) The theoretical percentage of TFOfree was calculated by numerical integration using equation (2) (Materials and methods). Simulation 1 (filled circles), x = 4, y = 4; simulation 2 (open squares), x = 4, y = 8; simulation 3 (filled triangles), x = 4, y = 16. For each simulation, k5>>k1 = k4>k2>k3 (absolute values are not defined; see main text).
To evaluate a unidirectional model, we simulated equation (2) (Materials and methods), varying the number of R steps and keeping the number of L steps equal (Figure 7B). Forward motion (k1) was defined as 10 times faster than the dissociation rate (k2) and 10 times slower than the resetting rate (k5). If dissociation were more frequent, then translocation would be less processive, to the extent that free TFO would not approach 100% over a reasonable time scale (not shown). A slower resetting rate would have the same result. Conversely, if there were no dissociation events and resetting only occurred at DNA ends, then the exponential reactions would be less extended than observed. As the number of R steps is increased, the cumulative occurrence of dissociation events results in a corresponding decrease in the apparent translocation rate (Figure 7B). Nevertheless, due to resetting, free TFO reaches >95% in each case. Thus, the distribution of kinetic profiles is graphically equivalent to that in Figure 7A. However, the model can neither determine the number of L or R steps directly nor estimate the relative values of k1, k2 or k5. Furthermore, the profiles illustrated assume that upon reaching a DNA end, an enzyme steps off at the same rate as any previous forward step (Figure 7B). An alternative model, in which enzymes stall at ends and dissociate more slowly, results in an equivalent distribution of curves (not shown). Never theless, the R1M2S1 data (Figure 7A) can only be justified by a model that incorporates both dissociation during motion and resetting.
In their previous study, Janscak et al. (1998) could not distinguish between an endonuclease assembly model with two non-equivalent HsdR-binding sites, which would result in only one type of R1M2S1 complex, and an assembly model with two equivalent HsdR sites, which would result in two distinct R1M2S1 complexes. Since our data indicate no preference for recognition site asymmetry, binding of the first HsdR to the MTase must result in an equal distribution between leftward and rightward translocation states. As the alignment of HsdS is fixed on DNA by its TRD sequences (Powell et al., 1998), the HsdR-binding sites must therefore be equivalent. The weaker binding of the second HsdR subunit could be due to physical occlusion by the first subunit or a conformational change resulting in negative cooperativity (Janscak et al., 1998).
Conclusions
Although a linear relationship between specific binding and non-specific cleavage of DNA by the type I restriction enzymes has been investigated thoroughly (see Szczelkun, 2000), a direct in vitro assay of protein motion had not been described. In this study, EcoR124I-directed TFO displacement allowed direct measurement of translocation velocity. Furthermore, the kinetics discriminated clearly between uni- and bi-directional translocation schemes, depending upon endonuclease stoichiometry. The R2M2S1 species translocates DNA bi-directionally, each side of its site moving at a speed of 400 ± 32 bp/s at 20°C. Unidirectional models can be discounted. Additional events that could slow translocation, such as sequence-dependent pauses or dissociation from the DNA, occur infrequently. Consequently, DNA translocation by EcoR124I is highly processive. This explains how DNA >1000 bp from the recognition site can be cleaved efficiently (Janscak et al., 1996; Szczelkun et al., 1997). However, we have yet to determine the exact number of steps that give rise to the kinetics observed. As a result, we cannot define a step size, i.e. the number of base pairs translocated per physical step. The R1M2S1 species cannot cleave DNA but still generated translocation, independently of site asymmetry. However, the reaction was less processive than that catalysed by the R2M2S1 form, as dissociation events during motion are more frequent. Therefore, the presence of two HsdR subunits must stabilize the translocating complex such that dissociation is rare. A similar increase in processivity with subunit complexity has been observed for the PcrA helicase (Soultanas et al., 1999). Nevertheless, the HsdR subunits appear to act independently. Alongside the subunit assembly of the type I enzymes (Dryden et al., 1997; Janscak et al., 1998), and the pseudo-symmetrical domain organization (Dryden et al., 1995; Powell et al., 1998), this suggests that translocation on each ‘side’ of the recognition site is undertaken by a single HsdR.
Despite dissociating from the DNA during motion, the R1M2S1 complex could restart translocation via resetting. This process must allow for a change in the direction of motion. One possibility is that HsdR completely dissociates from both the MTase and DNA. The free subunit could then rebind to the DNA-bound MTase at either position. However, the concentration of free HsdR would be low in this case and binding would be correspondingly slow (even if it occurred with a bi-molecular association rate constant close to the diffusion limit; Gutfreund, 1995). Alternatively, the HsdR subunit may release the MTase but not DNA. Translocation could not continue, but the local concentration of the proteins would remain artificially high and reassociation could be faster. A third scenario is that HsdR releases the DNA but remains part of an R1M2S1 complex; the HsdR subunit would then have to redistribute between the equal affinity binding sites. This would only be feasible if the sites were on the same ‘face’ of the MTase. Conversely, we do not believe that resetting plays a role in DNA translocation by the R2M2S1 species. There is a possibility that when one of the two HsdR subunits reaches a DNA end it could dissociate from both the DNA and enzyme. An R1M2S1 complex would result, and translocation in the opposite direction would become noticeably less processive. However, the kinetics do not support this (Figures 3, 4 and 5). The added stability of the R2M2S1 complex may therefore result in stalling at DNA ends, as described for the PcrA DNA helicase (Dillingham et al., 2000).
A comparison of the translocation velocity determined in this study with rates determined in other studies is dependent upon the different reaction conditions used. The experiments of Garcia and Molineux (1999) suggest an in vivo EcoKI translocation rate of ∼100 bp/s. This may differ from our rate not only because of ionic factors, but because their internalization assay relies on pulling DNA through a bacterial pore out of a phage capsid; this may represent an increased load that slows translocation. Comparisons with other DNA motor proteins reveal a range of velocities in vitro: elongation rates vary dramatically among RNA polymerases and correlate with subunit complexity (Uptain et al., 1997): single subunit bacteriophage RNA polymerases translocate at 200–400 nucleotides per second (nt/s); bacterial RNA polymerases translocate at an intermediate rate of 10–40 nt/s; and yeast RNA polymerase II and III elongate at 20–30 nt/s. For DNA helicases, unwinding rates over long distances have only been reported for a few enzymes. For instance, RecBCD unwinds DNA at 470 ± 80 bp/s at 25°C and at ∼900 bp/s at 37°C (Roman and Kowalczykowski, 1989). On the other hand, single strand translocation rates for PriA (Lee and Marians, 1990) and PcrA (Dillingham et al., 2000) are 90 and 50 nt/s, respectively. The rate of DNA translocation by EcoR124I is thus one of the fastest measured so far.
Materials and methods
Proteins
EcoR124I MTase was purified as described previously (Taylor et al., 1992). Purified HsdR was generously provided by Pavel Janscak (Biozentrum, Switzerland). Protein concentrations were calculated from absorbance at 280 nm using molar extinction coefficients derived from amino acid sequences (MTase, 160 400/M/cm; HsdR, 91 900/M/cm). Unless otherwise stated, endonuclease was formed by mixing M2S1 with a 7 molar excess of hsdR in buffer R (50 mM Tris–HCl pH 8.0, 10 mM MgCl2, 1.0 mM dithiothreitol, 0.2 mM AdoMet). The final concentration of endonuclease was taken as the input concentration of M2S1. The reconstituted enzyme had DNA cleavage activity equivalent to that previously published (Szczelkun et al., 1997). For experiments with one HsdR, endonuclease was prepared with equimolar concentrations of M2S1 and HsdR. All other enzymes were from New England Biolabs (Massachusetts, USA).
DNA
Unless stated otherwise, all DNA manipulations were carried out using standard procedures (Sambrook et al., 1989). Six oligonucleotides were supplied desalted by Cruachem (Glasgow, UK) and annealed as below to produce duplexes possessing either an EcoR124I site (single underlined) or a TFO-binding site (double underlined):

Plasmid pMDS20 was created by introducing an EcoR124I site in the BamHI of pNEO (Pharmacia, Sweden) as outlined previously (Szczelkun et al., 1996), whilst pMDS21 was created by cloning duplex 1 into the HindIII site of pNEO. The EcoR124I-binding site has the same spacer and flanking sequences as used previously (Szczelkun et al., 1996, 1997). Cloning of duplex 2 into the BglI site of pMDS20 or pMDS21 produced pMDS22 and pMDS23, respectively. Cloning duplex 3 into the BspEI site of pMDS20 produced pMDS28. Alternative orientations of the EcoR124I and TFO sites were selected by sequencing (L.Hall, Bristol). Plasmids were either used directly as supercoiled substrates or were first digested with an appropriate type II restriction endonuclease, which cleaved the DNA once to produce full-length linear substrates (Figure 1D). The 22 bp CT-rich TFO sequence (Figure 1B) was designed according to observations by Gowers and Fox (1997, 1999). It was supplied desalted by Cruachem, 5′ end-labelled with 32P using T4 polynucleotide kinase and purified by ultrafiltration (Bio-Spin, Bio-Rad).
Triplex formation and analysis
Equimolar concentrations (50 nM) of linear DNA and 32P-labelled TFO were mixed in buffer MM (25 mM MES, pH 5.5, 10 mM MgCl2) at 57°C for 15 min and left to cool to room temperature overnight. The resulting triplex was diluted 1/10 into buffer R before use. To analyse the proportion of bound and free TFO, reactions were quenched using GSMB buffer [15% (w/v) glucose, 3% (w/v) SDS, 250 mM MOPS pH 5.5, 0.4 mg/ml bromophenol blue] and analysed in 1% (w/v) agarose gels (40 mM Tris-acetate, 5.0 mM sodium acetate, 1.0 mM MgCl2 pH 5.5) at 10 V/cm for 2 h at 4°C. Gels were fixed in 5% (v/v) acetic acid, 50% (v/v) methanol for at least 1 h and air-dried overnight between sheets of cellophane. Dry gels were scanned in a Molecular Dynamics PhosphorImager, and the image analysed with ImageQuant software (MD) to determine the volume of each ‘band’, taking into account background readings. The fraction of free TFO in each sample was calculated as volumefree TFO/(volumefree TFO + volumetriplex). Triplexes were not observed at or above pH 6.0 at 20°C using nanomolar concentrations of linear DNA and TFO (not shown). However, full occupancy at this concentration range and temperature was observed at pH 5.5 (see main text). Attempts to ascertain an equilibrium dissociation constant for the triplex were hampered because at the detection limit of the assay the TFO was still tightly bound, indicating a sub-nanomolar Kd value.
EcoR124I translocation reactions
For a standard reaction, 5 nM triplex (see above) was pre-incubated with the appropriate concentration of EcoR124I at 20°C in buffer R for 5 min. Unless otherwise stated, reactions were started by the addition of ATP to a final concentration of 4 mM, and aliquots quenched by the addition of 0.25 vols of GSMB buffer. Optimal reaction times were determined empirically (not shown). Resulting samples were analysed as above. DNA cleavage was analysed as described elsewhere (Szczelkun et al., 1996).
Kinetic models for DNA translocation
Theoretical TFO displacement kinetics were simulated by numerical integration in Scientist (MicroMath Software, Salt Lake City, UT). Models were constructed from a series of consecutive reactions governed by unimolecular rate constants as described by Ali and Lohman (1997). Equation (1) describes one branch of a bi-directional model, where only those molecules that travel from the origin (O) towards the triplex are considered.
Each step (R1, R2, etc.) is governed by the same unimolecular rate constant, k1. Subsequent TFO displacement is governed by k2. Equation (2) describes a two-branch scheme for unidirectional motion.
This scheme has a number of features different from equation (1): (i) motion begins at an origin (O), but then proceeds down either the L (leftward) or R (rightward) pathway; (ii) each step can be governed by two unimolecular rate constants—a forward translocation rate (k1) and a dissociation rate (k2); (iii) only those molecules that travel down the R pathway can reach the triplex, generating free TFO with the rate constant k3; (iv) those molecules that travel down the L pathway eventually reach a DNA end (Lx): at this stage, the enzyme can dissociate (k4); and (v) any molecule that releases the DNA, either at an end or during motion, produces Opre. This form of the enzyme can then restart translocation at the origin, governed by a unimolecular rate constant, k5. For both models, the number of steps and the rate constants were varied as indicated in the main text. The percentage of final product was analysed in GraFit (Erithacus Software, Slough, UK).
Kinetic data analysis
Kinetic data were fitted to linear or exponential functions in GraFit. Residual data analysis of single exponential fits of TFO displacement showed non-random errors indicative of a higher order exponential relationship (data not shown). Consequently, displacement data were fitted using a double exponential increase with offset function:
TFOfree = A1⋅[1 – e–k1(t – offset)] + A2⋅[1 – e–k2(t – offset)] (3)
where TFOfree is the percentage of liberated oligonucleotide, A1 and A2 are the amplitudes of the first and second phases, respectively, k1 and k2 are the rates of the first and second phases, respectively, and offset is the x-axis intercept (or lag time).
Acknowledgments
Acknowledgements
We thank Dr Darren Gowers for valuable advice on triplex design, Dr Pavel Janscak for kindly donating purified HsdR, and Graham Davies, Dr David Dryden, Professor Freddie Gutfreund, Professor Steve Halford, Professor Noreen Murray and Dr Nigel Savery for discussions. This work was supported by grants from the Wellcome Trust to M.D.S. and K.F. M.D.S is a Wellcome Trust Research Career Development Fellow.
References
- Ali J.A. and Lohman,T.M. (1997) Kinetic measurement of the step size of DNA unwinding by the Escherichia coli UvrD helicase. Science, 275, 377–380. [DOI] [PubMed] [Google Scholar]
- Allen D.J., Makhov,A., Grilley,M., Taylor,J., Thresher,R., Modrich,P. and Griffith,J.D. (1997) MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J., 16, 4467–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle T.A. (1993) The ATP-dependent restriction enzymes. In Linn,S.M., Lloyd,R.S. and Roberts,R.J. (eds), Nucleases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 35–88. [Google Scholar]
- Davies G.P., Powell,L.M., Webb,J.L., Cooper,L.P. and Murray,N.E. (1998) EcoKI with an amino acid substitution in any one of seven DEAD-box motifs has impaired ATPase and endonuclease activities. Nucleic Acids Res., 26, 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G.P., Martin,I., Sturrock,S.S., Cronshaw,A., Murray,N.E. and Dryden,D.T.F. (1999a) On the structure and operation of type I DNA restriction enzymes. J. Mol. Biol., 290, 565–579. [DOI] [PubMed] [Google Scholar]
- Davies G.P., Kemp,P., Molineux,I.J. and Murray,N.E. (1999b) The DNA translocation and ATPase activities of restriction-deficient mutants of EcoKI. J. Mol. Biol., 292, 787–796. [DOI] [PubMed] [Google Scholar]
- Dillingham M.S., Wigley,D.B. and Webb,M.R. (2000) Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry, 39, 205–212. [DOI] [PubMed] [Google Scholar]
- Dreier J., MacWilliams,M.P. and Bickle,T.A. (1996) DNA cleavage by the type IC restriction modification system EcoR124II. J. Mol. Biol., 264, 722–733. [DOI] [PubMed] [Google Scholar]
- Dryden D.T.F., Sturrock,S.S. and Winter,M. (1995) Structural modelling of a type I DNA methyltransferase. Nature Struct. Biol., 2, 632–635. [DOI] [PubMed] [Google Scholar]
- Dryden D.T.F., Cooper,L.P., Thorpe,P.H. and Byron,O. (1997) The in vitro assembly of the EcoKI type I DNA restriction/modification enzyme and its in vivo implications. Biochemistry, 36, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Ellis D.J., Dryden,D.T.F., Berge,T., Edwardson,J.M. and Henderson, R.M. (1999) Direct observation of DNA translocation and cleavage by the EcoKI endonuclease using atomic force microscopy. Nature Struct. Biol., 6, 15–17. [DOI] [PubMed] [Google Scholar]
- Garcia L.R. and Molineux,I.J. (1999) Translocation and specific cleavage of bacteriophage T7 in vivo by EcoKI. Proc. Natl Acad. Sci. USA, 96, 12430–12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George H., Mezard,C., Stasiak,A. and West,S.C. (1999) Helicase-defective RuvB(D113E) promotes RuvAB-mediated branch migration in vitro. J. Mol. Biol., 293, 505–519. [DOI] [PubMed] [Google Scholar]
- Giovannangeli C., Perrouault,L., Escude,C., Thuong,N. and Helene,C. (1996) Specific inhibition of in vitro transcription elongation by triplex-forming oligonucleotide–intercalator conjugates targeted to HIV proviral DNA. Biochemistry, 35, 10539–10548. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E. and Koonin,E.V. (1991) Endonuclease (R) subunits of type I and type III restriction enzymes contain a helicase-like domain. FEBS Lett., 291, 277–281. [DOI] [PubMed] [Google Scholar]
- Gowers D.M. and Fox,K.R. (1997) DNA triple helix formation at oligopurine sites containing multiple contiguous pyrimidines. Nucleic Acids Res., 25, 3787–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers D.M. and Fox,K.R. (1999) Towards mixed sequence recognition by triple helix formation. Nucleic Acids Res., 27, 1569–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutfreund H. (1995) Kinetics for the Life Sciences: Receptors, Transmitters and Catalysts. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Hacia J.G., Dervan,P.B. and Wold,B.J. (1994) Inhibition of Klenow fragment DNA-polymerase on double-helical templates by oligonucleotide-directed triple-helix formation. Biochemistry, 33, 6192–6200. [DOI] [PubMed] [Google Scholar]
- Hiasa H. and Marians,K.J. (1999) Initiation of bidirectional replication at the chromosomal origin is directed by the interaction between helicase and primase. J. Biol. Chem., 274, 27244–27248. [DOI] [PubMed] [Google Scholar]
- Horiuchi K. and Zinder,N.D. (1972) Cleavage of bacteriophage f1 DNA by the restriction enzyme of Escherichia coli B. Proc. Natl Acad. Sci. USA, 69, 3220–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janscak P. and Bickle,T.A. (2000) DNA supercoiling during ATP-dependent DNA translocation by the type I restriction enzyme EcoAI. J. Mol. Biol., 295, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Janscak P., Adadjieva,A. and Firman,K. (1996) The type I restriction endonuclease R.EcoR124I: over-production and biochemical properties. J. Mol. Biol., 257, 977–991. [DOI] [PubMed] [Google Scholar]
- Janscak P., Dryden,D.T.F. and Firman,K. (1998) Analysis of the subunit assembly of the type IC restriction–modification enzyme EcoR124I. Nucleic Acids Res., 26, 4439–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janscak P., MacWilliams,M.P., Sandmeier,U., Nagaraja,V. and Bickle,T.A. (1999a) DNA translocation blockage, a general mechanism of cleavage site selection by type I restriction enzymes. EMBO J., 18, 2638–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janscak P., Sandmeier,U., and Bickle,T.A. (1999b) Single amino acid substitutions in the HsdR subunit of the type IB restriction enzyme EcoAI uncouple the DNA translocation and DNA activities of the enzyme. Nucleic Acids Res., 27, 2638–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. (1998) Replication errors: cha(lle)nging the genome. EMBO J., 17, 6427–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.G. and Miller,D.M. (1995) Inhibition of in vitro transcription by a triplex-forming oligonucleotide targeted to human C-MYC P2 promoter. Biochemistry, 34, 8165–8171. [DOI] [PubMed] [Google Scholar]
- Komissarova N. and Kashlev,M. (1997) Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA, 94, 1755–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopel V., Pozner,A., Baran,N. and Manor,H. (1996) Unwinding of the third strand of a DNA triple helix, a novel activity of the SV40 large T-antigen helicase. Nucleic Acids Res., 24, 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R. (1999) Transcription—shifting RNA polymerase into overdrive. Science, 284, 598–599. [DOI] [PubMed] [Google Scholar]
- Lee M.S. and Marians,K.J. (1990) Differential ATP requirements distinguish the DNA translocation and DNA unwinding activities of the Escherichia coli PriA protein. J. Biol. Chem., 265, 17078–17083. [PubMed] [Google Scholar]
- Maher L.J. III, Dervan,P.B. and Wold,B.J. (1990) Kinetic-analysis of oligodeoxyribonucleotide-directed triple-helix formation on DNA. Biochemistry, 29, 8820–8826. [DOI] [PubMed] [Google Scholar]
- Mernagh D.R., Janscak,P., Firman,K. and Kneale,G.G. (1998) Protein–protein and protein–DNA interactions in the type I restriction endonuclease EcoR124I. Biol. Chem., 379, 497–503. [DOI] [PubMed] [Google Scholar]
- Morris P.D. and Raney,K.D. (1999) DNA helicases displace streptavidin from biotin-labelled oligonucleotide. Biochemistry, 38, 5164–5171. [DOI] [PubMed] [Google Scholar]
- Paes H.M. and Fox,K.R. (1997) Kinetic studies on the formation of intermolecular triple helices. Nucleic Acids Res., 25, 3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum G.E., Park,Y.W., Singleton,S.F., Dervan,P.B. and Breslauer,K.J. (1990) Thermodynamic characterisation of the stability and the melting behavior of a DNA triplex—a spectroscopic and calorimetric study. Proc. Natl Acad. Sci. USA, 87, 9436–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L.M., Connolly,B.A. and Dryden,D.T.F. (1998) The DNA binding characteristics of the trimeric EcoKI methyltransferase and its partially assembled dimeric form determined by fluorescence polarisation and DNA footprinting. J. Mol. Biol., 283, 947–961. [DOI] [PubMed] [Google Scholar]
- Roman L.J. and Kowalczykowski,S.C. (1989) Characterisation of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry, 28, 2863–2873. [DOI] [PubMed] [Google Scholar]
- Rosamund J., Endlich,B. and Linn,S. (1979) Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J. Mol. Biol., 129, 619–635. [DOI] [PubMed] [Google Scholar]
- Sambrook J.C., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Shulman M.J. (1974) Model for wandering restriction enzymes. Nature, 252, 76–78. [DOI] [PubMed] [Google Scholar]
- Singleton S.F. and Dervan,P.B. (1993) Equilibrium association constants for oligonucleotide-directed triple helix formation at single DNA sites: linkage to cation valence and concentration. Biochemistry, 32, 13171–13179. [DOI] [PubMed] [Google Scholar]
- Skoog J.U. and Maher,L.J. (1993) Relief of triple-helix-mediated promoter inhibition by elongating RNA-polymerases. Nucleic Acids Res., 21, 4055–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P. and Wigley,D.B. (2000) DNA helicases: ‘inching forward’. Curr. Opin. Struct. Biol., 10, 124–128. [DOI] [PubMed] [Google Scholar]
- Soultanas P., Dillingham,M.S., Papadopoulos,F., Phillips,S.E.V., Thomas,C.D. and Wigley,D.B. (1999) Plasmid replication initiator protein RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res., 27, 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W. and Bandyopadhyay,P.K. (1988) Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA, 85, 4677–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun M.D. (2000) How do proteins move along DNA? Lessons from the type I and type III restriction endonucleases. In Banting,G. and Higgins,S.J. (eds), Essays in Biochemistry: Motor Proteins. Portland Press, London, UK, Vol. 35, pp. 131–143. [DOI] [PubMed] [Google Scholar]
- Szczelkun M.D., Dillingham,M.S., Janscak,P., Firman,K. and Halford,S.E. (1996) Repercussions of DNA tracking by the type IC restriction endonuclease EcoR124I on linear, circular and catenated substrates. EMBO J., 15, 6335–6347. [PMC free article] [PubMed] [Google Scholar]
- Szczelkun M.D., Janscak,P., Firman,K. and Halford,S.E. (1997) Selection of non-specific DNA cleavage sites by the type IC restriction endonuclease EcoR124/I. J. Mol. Biol., 271, 112–123. [DOI] [PubMed] [Google Scholar]
- Taylor I., Patel,K., Firman,K. and Kneale,G. (1992) Purification and biochemical characterization of the EcoR124 type I modification methylase. Nucleic Acids Res., 20, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain S.M., Kane,C.M. and Chamberlin,M.J. (1997) Basic mechanisms of transcriptional elongation and its regulation. Annu. Rev. Biochem., 66, 117–172. [DOI] [PubMed] [Google Scholar]
- Vasquez K.M. and Wilson,J.H. (1998) Triplex-directed modification of genes and gene activity. Trends Biochem. Sci., 23, 4–9. [DOI] [PubMed] [Google Scholar]
- Webb J.L., King,G., Ternet,D., Titheradge,A.J.B. and Murray,N.E. (1996) Restriction by EcoKI is enhanced by co-operative interactions between target sequences and is dependent on DEAD box motifs. EMBO J., 15, 2003–2009. [PMC free article] [PubMed] [Google Scholar]
- Xodo L., Alunnifabbroni,M., Manzini,G. and Quadrifoglio,F (1994) Pyrimidine phosphorothioate oligonucleotides form triple-stranded helices and promote transcription inhibition. Nucleic Acids Res., 22, 3322–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R., Hamilton,D.L. and Burckhardt,J. (1980) DNA translocation by the restriction enzyme from E.coli K. Cell, 20, 237–244. [DOI] [PubMed] [Google Scholar]