Abstract
Purpose
MicroRNAs (miRNAs) have emerged as critical regulators of many cellular pathways. Ionizing radiation (IR) exposure causes DNA damage and induces premature senescence. However, the role of miRNAs in IR-induced senescence has not been well defined. Thus, the purpose of this study is to identify and characterize senescence-associated miRNAs (SA-miRNAs) and to investigate the role of SA-miRNAs in IR-induced senescence.
Methods and Materials
In human lung fibroblasts (WI-38 cells), premature senescence was induced either by IR or busulfan (BU) treatment, and replicative senescence was accomplished by serial passaging. MiRNA microarrays were used to identify SA-miRNAs and real-time RT-PCR validated the expression profiles of SA-miRNAs in various senescent cells. The role of SA-miRNAs in IR-induced senescence was characterized by knockdown of miRNA expression with anti-miRNA oligonucleotides (AMOs) or by miRNA overexpression through the transfection of pre-miRNA mimics.
Results
We identified 8 SA-miRNAs, 4 of which were up-regulated (miR-152, -410, -431, and -493) and 4 which were down-regulated (miR-155, -20a, -25, and -15a), that are differentially expressed in both prematurely (induced by IR or BU) and replicatively senescent WI-38 cells. Validation of the expression of these SA-miRNAs indicated that down-regulation of miR-155, -20a, -25, and -15a is a characteristic miRNA expression of cellular senescence. Functional analyses revealed that knockdown of miR-155 or -20a, but not miR-25 or -15a, markedly enhanced IR-induced senescence, whereas ectopic overexpression of miR-155 or miR-20a significantly inhibited senescence induction. Furthermore, our studies indicate that miR-155 modulates IR-induced senescence by acting downstream of the p53 and p38 MAPK pathways and in part via regulating tumor protein 53-induced nuclear protein 1 (TP53INP1) expression.
Conclusion
Our results suggest that SA-miRNAs are involved in the regulation of IR-induced senescence, so targeting these miRNAs may be a novel approach for modulating cellular response to radiation exposure.
Keywords: Ionizing radiation, Senescence, MicroRNAs, p53, p38 MAPK
INTRODUCTION
Cellular senescence is characterized by an irreversible cell cycle arrest that can be triggered by many types of intrinsic and extrinsic stresses. The two major categories of cellular senescence are replicative senescence and stress-induced premature senescence (SIPS). Replicative senescence was first described by Hayflick and Moorehead in human fibroblasts after cells underwent extensive replication as a consequence of serial culture passages (1). Similarly, cells undergo SIPS when exposure to a variety of stresses, including ionizing radiation (IR), oxidative stress, and oncogenic stress (2–3). However, cells undergoing SIPS are morphologically indistinguishable from replicatively senescent cells and exhibit many of the characteristics ascribed to replicatively senescent cells, such as increased senescence associated β-galactosidase (SA-β-gal) activity and elevated expression of p16 (4). Deregulation of senescence may contribute to cancer development and progression (5). Thus, advances in our knowledge of the regulation of cellular senescence may lead to a better understanding of the mechanisms of the aging process and carcinogenesis.
MiRNAs are a new class of small non-coding RNAs that function as gene regulators by base-pairing to complementary sites on their target mRNAs. Previous studies implicate miRNAs in senescence and aging, which includes the observation that overexpression of miRNA lin-4 results in a modest increase in the adult lifespan in C. elegans (6). A role for miRNAs in cellular senescence is also supported by the observation that depletion of Dicer induces senescence through the activation of the p53 pathway (7). Interestingly, recent studies indicated that distinctive miRNA expression profiles were associated with quiescence, oxidative stress-induced or replicatively senescent fibroblasts (8–9). These results suggest that miRNA expression profiles might be dependent upon the status of cell cycle arrest and the types of senescence. However, a characteristic miRNA expression profile that is specifically associated with senescence, but not with quiescence or cell transformation, has not been identified.
We and others have demonstrated that IR exposure induces premature senescence in a variety of cells and tissues both in vitro and in vivo (10–11). It has been shown that IR exposure significantly alters miRNA expression patterns in various normal and cancer cells (12–13). However, the role of miRNAs in IR-induced premature senescence is largely unknown. In the present study, we investigated SA-miRNAs and found that down-regulation of miR-155, -20a, -25, and -15a is a characteristic miRNA expression of cellular senescence. We also show that knockdown of miR-155 or -20a, but not miR-25 or -15a, with anti-miRNA oligonucleotides (AMOs) markedly enhances IR-induced senescence, whereas ectopic overexpression of miR-155 or miR-20a by transfection with pre-miRNA mimics significantly inhibits IR-induced senescence. For the first time, our studies demonstrate that miR-155 and miR-20a are involved in modulating IR-induced senescence. These results suggest that targeting of SA-miRNAs may represent a novel approach to modulate IR-induced cellular senescence.
MATERIALS AND METHODS
Cell Culture and Senescence Induction
WI-38 cells (human embryonic lung diploid fibroblasts) obtained from ATCC (Manassas, VA) were maintained and cultured as previously described (14). Induction of cellular senescence in WI-38 cells by IR exposure or BU treatment was performed as previously described (14–15).
Senescence-associated β-galactosidase (SA-β-gal) Staining and BrdU Incorporation Assay
SA-β-gal staining and BrdU incorporation assays were used to determine cellular senescence as previously described (14–15).
Western blotting Analysis
Western blotting analysis was performed as previously described (14). Rabbit polyclonal antibody against TP53INP1 and mouse monoclonal antibody against β-actin were obtained from Santa Cruz Biotechnology.
MiRNA Microarrays
Total RNA was extracted with TRIzol solution (Invitrogen, Carlsbad, CA) and quantified using a Nanodrop spectrophotometer. Five micrograms of total RNA from each sample was reverse transcripted using biotin end-labeled random octamer oligonucleotide primers. Hybridization of biotin-labeled complementary DNA was carried out with the Ohio State University miRNA microarray chips (OSU_CCC version 3.0). The hybridized chips were processed and scanned on an Axon 4000B microarray scanner (Axon Instruments, Sunnyvale, CA). Raw data were normalized and analyzed using Genespring 7.2 software (ZcomSilicon Genetics, Redwood City, CA).
Real-time Reverse Transcription-PCR Analysis of MiRNAs
Total RNAs were extracted from senescent or control WI-38 cells using a mirVana™ miRNA Isolation Kit (Ambion, Austin, TX) following the manufacturer's protocol. RNA samples were reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit and a set of stem loop miRNA-specific primers (Applied Biosystems, Foster City, CA). Expression levels of individual SA-miRNAs were normalized to endogenous U6 miRNA and analyzed using the CT method (10).
Retrovirus-mediated Gene Delivery into WI-38 Cells
Retroviruses were generated by co-transfection of HEK 293T cells with pBabe-H-Ras, VSV-G and gag/pol using FuGen 6 (Roche, Mannheim, Germany). The pBabe-puro empty vector was used as control. WI-38 cells were infected with retrovirus expressing H-Ras in the presence of polybrene (8mg/ml, Sigma, St. Louis, MO). Two days after virus infection, cells were selected using puromycin (2μg/ml, Invitrogen) for seven days. Senescence was determined by SA-β-gal staining and BrdU incorporation assays as previously described (14).
Transfection of Cells with MiRNAs
To overexpress miRNAs in WI-38 cells, we reverse transfected the cells with Pre-miR miRNA precursors using the siPORT NeoFX Transfection Agent (Ambion, Austin, TX) according to the manufacturer's protocol. Pre-miR negative miRNA control #1 (Ambion, Austin, TX) was used as control.
MiRNA Knockdown
The knockdown of miRNAs in WI-38 cells was achieved by reverse transfection with Anti-miRNA antisense 2'-o-methyl oligoribonucleotides using the siPORT NeoFX Transfection Agent according to the manufacturer's protocol.
SiRNA and p38 Inhibitor Treatment
WI38 cells were transfected with 100 nM scrambled oligonucleotides (Scr) or p53 small interference RNA (siGENOME SMART pool reagent for Human TP53) (Dharmacon, Chicago, IL) using Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions (15). Two days after initial transfection, WI38 cells were irradiated with 5 Gy using a Cs-137 irradiator. For studies involved in p38 MAPK inhibitor, WI-38 cells were treated with SB 203580 (Calbiochem, San Diego, CA), one hour before 5 Gy irradiation.
Statistical Analysis
Data were analyzed by analysis of variance (ANOVA). Differences were considered significant at P < 0.05. All analyses were carried out with the GraphPad Prism program from GraphPad Software, Inc. (San Diego, CA).
RESULTS
Identification of SA-miRNAs
To indentify SA-miRNAs and elucidate their roles in regulating cellular senescence, we induced cellular senescence in WI-38 cells by serial passaging, exposure to IR, or treatment with the alkylating agent BU as previously described (14–15). After 35 passages (about 60 population doublings), WI38 cells entered replicative senescence. Similarly, 12 days after IR or BU treatment, WI-38 cells became permanently arrested and exhibited characteristics ascribed to replicative senescence, such as increased SA-β-gal activity and inability to incorporate BrdU (Fig. 1A).
Fig. 1. Identification of SA-miRNAs.
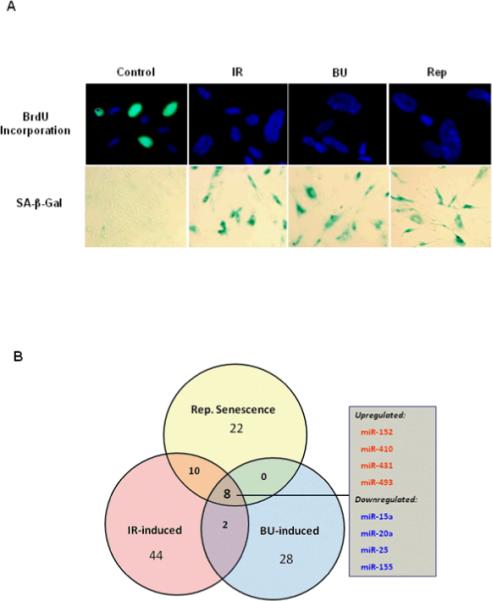
A. Induction of senescence in WI-38 cells was performed by serial passaging, exposure to 10 Gy IR, or treatment with the alkylating agent busulfan (BU, 120 μM) as previously described (14–15). Cellular senescence was determined by SA-β-gal staining and BrdU incorporation assays. B. Using miRNA microarrays and bioinformatics analyses, we identified eight SA-miRNAs that are differentially expressed in both prematurely (induced by either IR or BU treatment) and replicatively senescent cells.
As shown in Table 1, using a strict cutoff of p<0.01 for significance, our miRNA microarray data revealed that 22, 44, and 28 miRNAs are differentially expressed in replicatively (Rep), IR- and BU-induced prematurely senescent cells, respectively, compared to control WI-38 cells. These results indicate that miRNA expression profiles of senescent cells are stress type- and senescence type-dependent. Subsequent bioinformatic analyses identified that a set of 8 miRNAs, including 4 up-regulated miRNAs (miR-152, -410, -431, and 493) and 4 down-regulated miRNAs (miR-15a, -20a, -25, and -155), are differentially altered in both replicatively senescent and stress-induced senescent cells (Fig. 1B).
Table 1.
miRNA microarray demonstrating shared miRNA expression among replicative and stress-induced senescence
| Replicative Senescence |
IR-induced Senescence |
BU-induced Senescence |
||||
|---|---|---|---|---|---|---|
| Gene Name | Fold Change | p value | Fold Change | p value | Fold change | p-value |
| Up-Regulated | ||||||
| miR-516-35p | – | – | – | – | 2.63 | 1.27E-03 |
| miR-496 | 3.89 | 1.64E-03 | 5.49 | 1.77E-04 | – | – |
| miR-494 | – | – | 2.54 | 5.67E-03 | – | – |
| miR-493 | 2.07 | 8.51E-03 | 3.51 | 8.78E-04 | 3.05 | 1.50E-03 |
| miR-486 | – | – | – | – | 2.18 | 1.01E-04 |
| miR-433 | – | – | – | – | 1.86 | 8.60E-03 |
| miR-432 | 5.07 | 1.26E-03 | 4.33 | 3.32E-04 | – | – |
| miR-431 | 3.14 | 3.97E-03 | 2.79 | 5.75E-03 | 3.73 | 7.77E-04 |
| miR-410 | 2.60 | 8.36E-03 | 3.37 | 7.20E-04 | 3.83 | 3.21E-03 |
| miR-382 | – | – | 2.64 | 6.17E-03 | – | – |
| miR-380-3p | – | – | 4.04 | 7.70E-04 | – | – |
| miR-379 | – | – | 3.56 | 7.56E-03 | – | – |
| miR-376a | – | – | – | – | 2.30 | 5.10E-03 |
| miR-376b | – | – | 2.12 | 9.32E-03 | 2.27 | 1.66E-03 |
| miR-340 | – | – | – | – | 1.38 | 9.15E-03 |
| miR-337 | – | – | 2.61 | 2.68E-03 | – | – |
| miR-323 | – | – | 2.34 | 9.51E-03 | – | – |
| miR-296 | 2.56 | 2.53E-03 | – | – | – | – |
| miR-219 | – | – | 2.17 | 8.11E-03 | – | – |
| miR-190 | – | – | – | – | 128.20 | 8.24E-03 |
| miR-152 | 2.42 | 4.31E-03 | 2.22 | 7.56E-03 | 3.15 | 4.31E-03 |
| miR-134 | 2.37 | 7.58E-03 | – | – | 2.35 | 9.11E-03 |
| miR-129 | – | – | – | – | 1.49 | 3.72E-03 |
| miR-128a | – | – | – | – | 3.27 | 1.09E-03 |
| miR-127 | _ | – | – | – | 2.25 | 5.03E-05 |
| miR-123 | – | – | – | – | 1.32 | 1.09E.03 |
| miR-106a | – | – | – | – | 1.47 | 3.60E-03 |
| miR-103 | – | – | – | – | 1.39 | 9.64E-03 |
| miR-101b | – | – | 2.91 | 9.31E-03 | – | – |
| miR-34a | – | – | – | – | 1.68 | 1.97E-03 |
| miR-30a | – | – | 2.38 | 6.28E-03 | 1.95 | 4.79E-03 |
| miR-30c | – | – | – | – | 1.49 | 7.43E-03 |
| miR-29b | – | – | – | – | 1.49 | 7.55E-03 |
| miR-27 | – | – | 3.03 | 8.17E-03 | – | – |
| miR-22 | – | – | – | – | 1.83 | 8.27E-03 |
| Down-Regulated | ||||||
| miR-483 | – | – | – | – | 0.59 | 7.32E-03 |
| miR-450-2b | – | – | 0.03 | 4.04E-04 | – | – |
| miR-424-2 | 0.24 | 1.21E-02 | 0.19 | 2.53E-04 | – | – |
| miR-321 | – | – | 0.44 | 7.14E-03 | – | – |
| miR-224 | – | – | 038 | 8.26E-03 | – | – |
| miR-218a | – | – | 0.20 | 9.74E-04 | – | – |
| miR-217b | 0.14 | 6.07E-03 | 0.12 | 7.25E-04 | – | – |
| miR-195 | – | – | 0.39 | 8.21E-03 | – | – |
| miR-155 | 0.27 | 2.14E-03 | 0.10 | 1.61E-05 | 0.08 | 1.75E-03 |
| miR-145 | 0.45 | 7.48E-03 | 0.39 | 3.25E-03 | – | – |
| miR-143 | – | – | 0.25 | 2.48E-04 | – | – |
| miR-123b | – | – | 0.26 | 8.87E-03 | – | – |
| miR-135b | – | – | 0.26 | 3.14E-03 | – | – |
| miR-106a | 0.35 | 5.37E-03 | 0.19 | 2.56E-04 | – | – |
| miR-106b | 0.39 | 8.18E-03 | 0.25 | 5.88E-03 | – | – |
| miR-93a | 0.39 | 6.06E-03 | 0.26 | 6.10E-04 | – | – |
| miR-92-1a | – | – | – | – | 0.36 | 3.65E-03 |
| miR-92-1b | 0.39 | 6.72E-03 | 0.22 | 5.13E-04 | – | – |
| miR-32 | – | – | 0.34 | 3.98E-03 | – | – |
| miR-30c-1 | – | – | – | – | 0.36 | 3.65E-03 |
| miR-29b | – | – | 0.39 | 6.73E-03 | – | – |
| miR-25 | 0.39 | 8.67E-03 | 0.26 | 3.88E-04 | 0.29 | 6.01E-03 |
| miR-20a | 0.25 | 2.41E-03 | 0.20 | 1.19E-04 | 0.15 | 5.38E-03 |
| miR-20b | – | – | 0.23 | 8.39E-04 | – | – |
| miR-19b | – | – | 0.25 | 2.20E-03 | – | – |
| miR-17 | 0.36 | 6.68E-03 | 0.14 | 1.50E-03 | – | – |
| miR-16b | – | – | 0.39 | 4.10E-03 | – | – |
| miR-16-1 | – | – | 0.27 | 7.70E-04 | – | – |
| miR-15a | 0.39 | 3.04E-03 | 0.33 | 1.21E-03 | 0.35 | 9.56E-03 |
| miR-15b | 0.34 | 6.07E-03 | 0.23 | 4.15E-04 | – | – |
| miR-7 | 0.26 | 1.78E-02 | – | – | – | – |
Validation of SA-miRNA Expression Profiles in Senescent Cells
To validate the microarray results, we profiled the expression signature of SA-miRNAs in senescent versus control WI-38 cells. As shown in Figures 2A & 2B, the results confirmed that 7 of the 8 SA-miRNAs (all but miR-431) were differentially expressed in BU- and IR-induced senescent cells as well as replicatively senescent cells (p<0.01). To determine if alterations in miRNA expression might be time-dependent, we examined the timing kinetics of SA-miRNA expression after IR exposure. The results showed that changes in both up- and down-regulated SA-miRNA expression peaked at day 7 and was sustained for at least 14 days after irradiation (Figs. 2C & 2D).
Fig. 2. Confirmation of differential expression of SA-miRNAs in senescent cells.
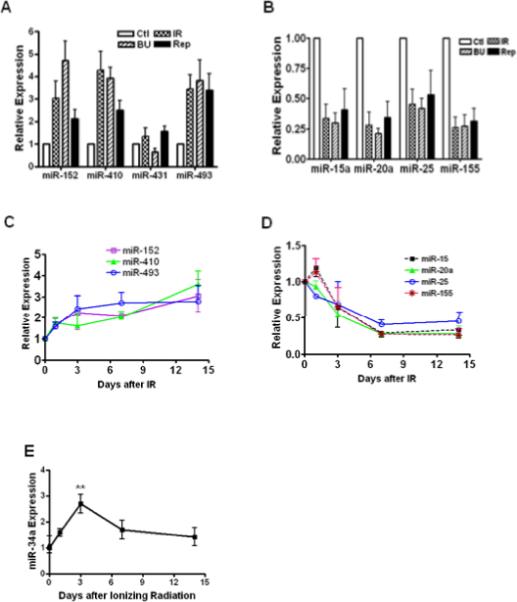
TaqMan MicroRNA Assays were used to determine the expression levels of SA-miRNAs in senescent WI-38 cells. The levels of up-regulated (A) and down-regulated (B) SA-miRNAs in replicatively senescent cells (Rep) and prematurely senescent cells (induced by IR and BU) were normalized to the control cells and graphed. Kinetic changes of up-regulated (C) and down-regulated (D) SA-miRNAs versus time after IR were plotted. Kinetic changes of miR-34a expression were determined (E). Data are presented as fold changes compared with control (Mean ± SE, n = 4). **, p < 0.01 vs. control.
Because miR-34a is reported to be involved in regulating senescence in some tumor cells (16), we compared the kinetic changes of miR-34a expression with that of SA-miRNAs in WI-38 cells after exposure to IR. The data showed that increases in miR-34a expression peaked 3 days after IR and then subsided to an insignificant level by day 7 (Fig. 2E).
Down-regulation of miR-155, -20a, -25, and -15a is Characteristic of Senescence, But Not Quiescence and Transformation
Next, we investigated whether the expression of SA-miRNAs is specific to senescence, but not to quiescence and transformation of WI-38 cells. We compared the SA-miRNA expression of quiescent and SV40 transformed cells with that of senescent cells as shown in Fig. 2. We induced quiescence of WI-38 cells by serum starvation as previously described (17) and the quiescent cells had negative Ki67 staining (data not shown). We observed that miR-152 was markedly increased in quiescent WI-38 cells (Fig. 3A), suggesting that up-regulation of miR-152 is not only associated with senescence, but also associated with quiescence. Our data also showed that miR-493 and miR-410 were markedly increased in SV40 transformed WI-38 cells (Fig. 3C). Collectively, these data suggest that increased levels of miR-152, -410 and -493 are not specifically associated with cellular senescence, as their expression levels are also altered in quiescent or transformed cells. In contrast, down-regulation of miR-155, -25, -20a and -15a appears to be a specific miRNA expression signature of senescence (Fig. 2), as no significant changes of these 4 down-regulated SA-miRNAs were observed in both quiescent (Fig. 3B) and SV40 transformed WI-38 cells (Fig. 3D). These results demonstrate that only the 4 down-regulated SA-RNAs are specifically associated with senescence.
Fig. 3. Expression of various SA-miRNAs in quiescent and transformed cells.
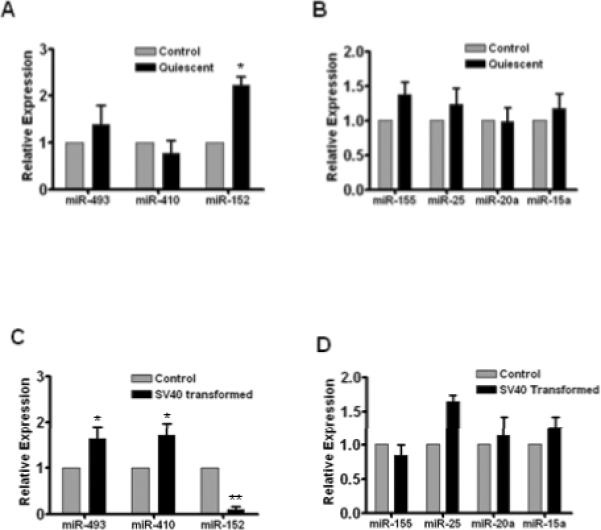
TaqMan MicroRNA Assays were used to profile the expression patterns of SA-miRNAs in serum starvation-induced quiescent WI-38 cells (A & B) and in SV40 transformed WI-38 cells (C & D). Expression levels of SA-miRNAs were normalized to the control cells. Data are presented as fold changes compared with control (Mean ± SE, n = 3). *, p < 0.05 vs. control; **, p < 0.01 vs. control.
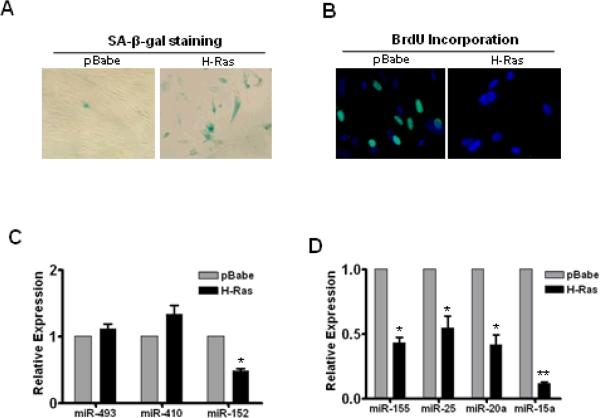
SA-miRNA Expression Profile in Ras-induced Senescence
Oncogenic stress-induced senescence represents an important type of cellular senescence, which significantly contributes to tumor prevention (18). Our next goal was to determine if oncogenic stress-induced senescence has the same miRNA expression profile as manifested in replicative and IR- and BU-induced senescence (Fig. 2). Ten days after the introduction of H-Ras, WI-38 cells become senescent as measured by increased SA-β-gal staining and an inability to incorporate BrdU (Figs. 4A & 4B).
Fig. 4. Expression profiles of SA-miRNAs in Ras-induced senescent WI-38 cells.
H-Ras was introduced into WI-38 cells by retroviral gene delivery system to induce senescence. Cellular senescence was verified by SA-β-gal staining (A) and BrdU incorporation assays (B). TaqMan MicroRNA Assays were used to assess the up-regulation (C) and down-regulation (D) of SA-miRNAs in Ras-induced senescent cells. Data are presented as fold changes normalized to pBabe control (Mean ± SE, n = 3). *, p < 0.05 vs. control; **, p < 0.01 vs. control.
The SA-miRNA profiling data showed that no significant changes were observed in the levels of miR-493 and miR-410 in Ras-transduced senescent cells versus pBabe-transduced cells (Fig. 4C). In contrast, all of the four down-regulated SA-miRNAs (miR-155, -25, -20a and -15a) were significantly decreased in Ras-induced senescent WI-38 cells (Fig. 4D). Interestingly, despite being up-regulated in both prematurely and replicatively senescent WI-38 cells (Fig. 2A), miR-152 was markedly reduced in Ras-induced senescent cells (Fig. 4C), suggesting that miR-152 levels can be up- or down-regulated in senescent cells depending upon the senescence type.
SA-miRNAs Regulate IR-induced Cellular Senescence
Next, we investigated whether any of the 4 down-regulated SA-miRNAs might play a role in regulating IR-induced premature senescence. AMOs (also known as antagomirs) is an effective tool to knock down miRNAs (19). Thus, we performed a loss-of-function (LOF) analysis of SA-miRNAs by transfecting WI-38 cells with AMOs against each of the 4 individual SA-miRNAs to evaluate their effects on IR-induced senescence. Our data indicate that SA-β-gal positive cells are significantly increased in cells transfected with miR-155 or miR-20a AMOs, but not with AMOs of other down-regulated SA-miRNAs, suggesting that knockdown of miR-155 or miR-20a enhances IR-induced senescence in WI-38 cells (Figs. 5A & 5B). The ability of miR-155 and miR-20a AMOs to enhance IR-induced senescence was further confirmed by BrdU incorporation assays, which demonstrate that miR-155 and miR-20a AMO-treated cells have a reduced capacity to incorporate BrdU (Fig. 5C).
Fig. 5. Effect of SA-miRNAs on IR-induced cellular senescence.
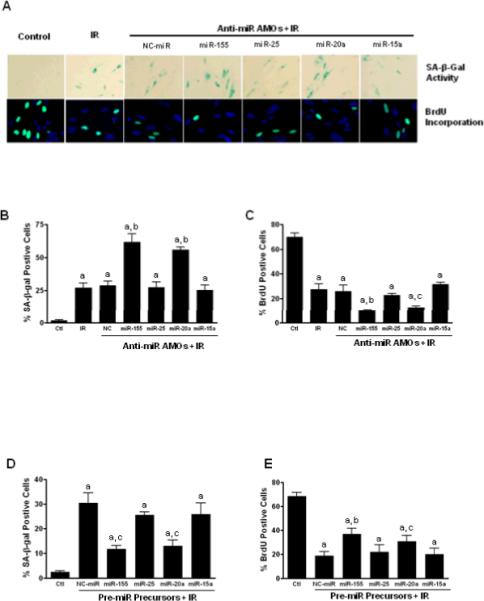
WI-38 cells were transfected with specific AMOs against individual SA-miRNAs (50nM) or negative control miRNAs (NC). Two days after transfection, cells were irradiated (5Gy) to induce cellular senescence. A. upper panel, representative pictures of IR-induced senescent cells showing SA-β-gal positive staining (Blue); lower panel, representative pictures of proliferating cells that are positive for BrdU staining (Green). B. Quantification of senescent cells was determined by SA-β-gal staining. Results are presented as mean ± SE (n = 4). C. Seven days after irradiation, cells were pulsed with BrdU (10 nM) for 24 hours and percentage of BrdU positive cells were counted and plotted. D & E. WI-38 cells were transfected with pre-miR™ precursors (50nM.) or negative control pre-miRNAs (NC-miR). Two days after transfection, cells were irradiated (5Gy) to induce senescence. Senescent cells were determined by SA-β-gal staining (D) and cell proliferation was determined by BrdU incorporation assays (E). Data are presented as mean ± SE (n = 3). a, P < 0.001 vs. control (Ctl); b, P < 0.01 vs. NC + IR; c, P < 0.05 vs. NC + IR.
We also performed gain-of-function (GOF) analysis of SA-miRNAs by transfecting the cells with pre-miRNA precursors to determine if exogenous overexpression of SA-miRNAs influences IR-induced senescence. As shown in Fig. 5D, the percentage of SA-β-gal positive cells decreased in WI-38 cells transfected with miR-155 or miR-20a precursors, whereas the percentage of BrdU positive cells dramatically increased (Fig. 5E) in WI-38 cells transfected with miR-155 or -20a precursors. These results demonstrate that overexpression of miR-155 or -20a inhibits IR-induced senescence.
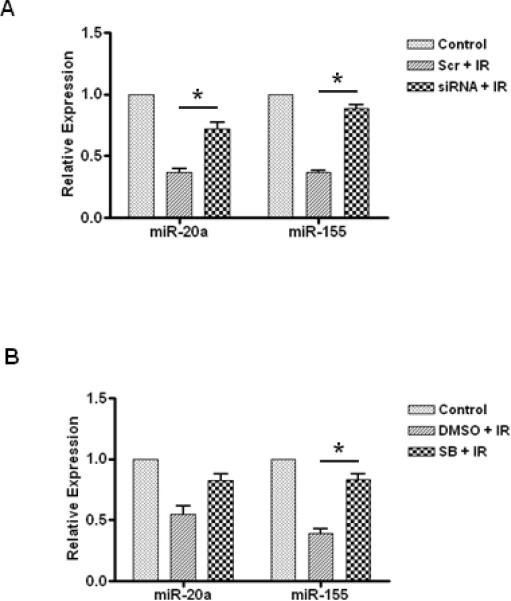
Inhibition of p53 Attenuates IR-induced Down-regulation of SA-miRNAs
Because tumor suppressor p53 is a key regulator of DNA damage responses (DDR) and IR-induced cell cycle arrest (20), we investigated if the expression of SA-miRNAs is regulated by p53 in irradiated WI-38 cells. We employed an RNAi technique to knockdown p53 expression and observed that p53 knockdown significantly attenuates IR-induced down-regulation of miR-155 and miR-20a in irradiated WI-38 cells (Fig. 6A). These data suggest that p53 may contribute to the regulation of IR-induced senescence partially by regulating miR-155 and miR-20a expression.
Fig. 6. Regulation of SA-miRNA expression by p53 and p38 MAPK.
A. WI38 cells were doubly transfected (48 hr prior to and 3 days post IR) with either 100 nM p53 siRNA (siRNA) or 100 nM control scramble oligonucleotides (Scr). Seven days post irradiation, total RNA was extracted and SA-miRNA expression levels were determined by real-time RT-PCR. B. WI-38 cells were treated with SB 203580 (5μM), one hour before 5 Gy of irradiation, and with fresh inhibitor added every other day. Seven days post IR, SA-miRNA expression levels in irradiated cells were determined by real-time RT-PCR and normalized to those of control cells. Data are presented as mean ± SE (n = 3). *, P < 0.05.
To determine if the p38 MAPK pathway is implicated in the regulation of SA-miRNAs, we used SB203580 to pharmacologically inhibit the p38 MAPK pathway and performed SA-miRNA expression profiling of irradiated WI-38 cells. As shown in Fig. 6B, our data show that inhibition of the p38 MAPK partially restores IR-induced down-regulation of miR-155 but not miR-20a, suggesting that p38 MAPK may regulate miR-155 expression.
TP53INP1 is a Target of miR-155 Involved in IR-induced Senescence
To characterize miR-155 targets, we have identified TP53INP1 as a predicted target of miR-155 using two complementary bioinformatic miRNA target prediction tools (TargetScan & miRana). As shown in Fig. 7A, the seed sequence of miR-155 matches the 3'-UTR sequence of TP53INP1, suggesting that TP53INP1 might be a target of miR-155. Western blotting data indicated that overexpression of miR-155 significantly decreases TP53INP1 expression, whereas transfection of negative control miRNA (NC) did not alter the expression of TP53INP1 (Fig. 7B). These results experimentally demonstrate that TP53INP1 is a target of miR-155. Next, we performed LOF analysis of TP53INP1 using siRNA to determine the role of TP53INP1 in IR-induced senescence. As shown in Fig. 7D, cells transfected with TP53INP1 siRNA had significantly reduced IR-induced cellular senescence, whereas scramble control (Scr) RNA transfection did not affect IR-induced cellular senescence. These results suggest that miR-155 may regulate IR-induced senescence by modulating its target gene, TP53INP1 expression.
Fig. 7. TP53INP1 is a target of miR-155.
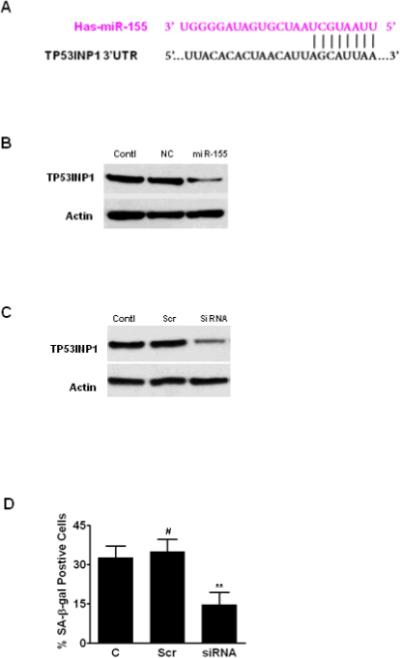
A. Schematic showing the predicted miR-155 binding site of TP53INP1 3'-UTR. B. WI-38 cells were transfected with pre-miR precursors to overexpress miR-155 or NC as control. Western blotting was used to assess the expression of TP53INP1 48 hr after transfection. C. WI-38 cells were transfected with 100 nM TP53INP1 siRNA or 100 nM scrambled RNA (Scr) in 6-well plates. Downregulation of TP53INP1 by siRNA was confirmed by Western blotting. D. WI-38 cells were irradiated (5Gy) at 48 hours after siRNA transfection. Senescence was determined by SA-β-gal staining and data are presented as mean ± SE (n = 3). #, P > 0.05 vs. irradiation control (C); **, P < 0.01 vs. Scr + irradiation.
DISCUSSION
In this study, we demonstrated that the down-regulation of miR-155, -25, -20a, and -15a is a characteristic miRNA expression signature of cellular senescence, but not quiescence and transformation. Consistent with our findings, a recent study indicated that down-regulation of miR-155, -20a and -15a was also associated with oxidative stress-induced senescence in human diploid fibroblasts (9). Interestingly, it has been shown that miR-17, -19b, -20a, and -106a are down-regulated in senescent cells (21). Our miRNA array data also show that miR-106a and –miR-17 are down-regulated in both IR-induced senescence and replicative senescence but not BU-induced senescence; however, miR-19b is down-regulated only in IR-induced senescence (Table 1). These results suggest that down-regulation of miR-17, -19b, and -106a in senescent cells might be senescence type specific.
Because miR-152 is significantly increased in quiescent cells and miR-493 and miR-410 are elevated in SV40 transformed cells, it is likely that these up-regulated SA-miRNAs may not be specific biomarkers of cellular senescence. However, we cannot exclude the possibility that, to some extent, they may be involved in the regulation of senescence. Further studies are required to test this possibility. Surprisingly, we found that miR-152 was significantly down-regulated in SV40 transformed cells and in Ras-induced senescent cells, indicating that miR-155 may mediate temporary cell cycle arrest, particularly cell quiescence.
MiR-34a has been shown to be a transcriptional target of p53, and exogenous overexpression of miR-34a induces senescence in tumor cells (16). In this study, we found that increases in miR-34a expression peaked 3 days after IR and then subsided to an insignificant level by day 7, suggesting that miR-34a may play an important role in initiating senescence induction but may not be indispensible for the maintenance of cellular senescence. In contrast, our time-course studies revealed that a significant down-regulation of miR-155, −25, −20a, and −15a expression occurred about 3 days after IR and continued to day 7 and 14 when the irradiated cells became permanently arrested, which suggests that, unlike miR-34a, the SA-miRNAs identified in our study may play a critical role both in the initiation and maintenance of cellular senescence.
To the best of our knowledge, this is the first study demonstrating that knockdown of endogenous miR-155 or miR-20a with AMOs enhances IR-induced senescence, whereas exogenous overexpression of miR-155 or miR-20a attenuates senescence. These results suggest that intervention of SA-miRNA expression may represent a novel tool to modulate the cellular responses of radiation effects. However, it remains to be determined if manipulation of these SA-miRNAs can alter the process of replicative senescence. Interestingly, it has been shown that inhibition of miR-20a causes a significant increase in DNA double-stranded breaks and G1 arrest by targeting the transcription factor E2F1 (22).
In contrast, little is known about the targets of miR-155 that are likely involved in the regulation of senescence. To identify such targets of miR-155, we found that TP53INP1 is a target of miR-155 that is likely involved in modulating IR-induced senescence. Our data also show that inhibition of TP53INP1 by siRNA attenuates IR-induced senescence in WI-38 cells. These results suggest that overexpression miR-155 may suppress cellular senescence by down-regulating TP53INP1 expression. Whether TP53INP1 is a direct or secondary target of miR-155 will be determined in our follow-up study using 3'-UTR reporter assays. Our observations are in agreement with the previous studies showing that over expression of TP53INP1 induces DNA damage-induced cell cycle arrest in both p53-dependent and p53-independent manners (23). Because a single miRNA may regulate the expression of multiple target genes (24), it is possible that additional miR-155 targets may be involved in IR-induced senescence.
Given the importance of p53 pathway in regulating DDR and IR-induced cell cycle arrest (20), it is rational to predict that expression of SA-miRNAs are possibly regulated by the p53 pathway. Our results show that inhibition of p53 using RNAi attenuates IR-induced down-regulation of miR-155 and miR-20a in irradiated WI-38 cells, suggesting that SA-miRNAs are likely regulated by the activation of p53 after radiation exposure. In agreement with our findings, a previous study indicated that miR-155 and miR-20a are p53-responsive miRNAs (25). We also found that inhibition of p38 MAPK using a pharmacological inhibitor can partially reverse IR-induced down-regulation of miR-155. Together, these data indicate that the p38 MAPK and p53 pathways may modulate cellular senescence by regulation their downstream targets such as miR-155 expression.
In summary, our studies indicate that down-regulation of miR-155, −20a, −25, and −15a is a characteristic miRNA expression profile of senescence and that SA-miRNAs are implicated in the regulation of IR-induced senescence. Given that induction of senescence has been increasingly recognized as an important mechanism of action for anti-cancer therapy (26); our studies suggest that these SA-miRNAs may have implications in modulating cancer cell response to radiotherapy. In support of this idea, recent studies suggest that manipulation of miRNA expression can sensitize cancer cells to radiation therapy (27). Further studies will be needed to determine if targeting of SA-miRNAs can benefit radiotherapy of cancers by increasing radiation-induced senescence.
ACKNOWLEDGEMENTS
The authors thank Mrs. Jennifer Schulte and Mrs. Aimin Yang for their excellent technical assistance. We also thank Dr. Yi-Te Hsu for critically reviewing the manuscript and Dr. Jennifer Schnellmann for editing the manuscript. This study was partially supported by CA08860, CA122023, ACS IRG-97-219-08, AI067770-05 and NIH RR014516.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Notification The authors declare no conflict of interest.
REFERENCES
- 1.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki M, Boothman DA. Stress-induced premature senescence (SIPS)--influence of SIPS on radiotherapy. J Radiat Res. 2008;49:105–12. doi: 10.1269/jrr.07081. [DOI] [PubMed] [Google Scholar]
- 3.Yaswen P, Campisi J. Oncogene-induced senescence pathways weave an intricate tapestry. Cell. 2007;128:233–4. doi: 10.1016/j.cell.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–22. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Boehm M, Slack F. A developmental timing microRNA and its target regulate lifespan in C. elegans. Science. 2005;310:1954–7. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 7.Mudhasani R, Zhu Z, Hutvagner G, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–63. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maes OC, Sarojini H, Wang E. Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J Cell Physiol. 2009;221:109–19. doi: 10.1002/jcp.21834. [DOI] [PubMed] [Google Scholar]
- 9.Li G, Luna C, Qiu J, et al. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev. 2009;130:731–41. doi: 10.1016/j.mad.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Schulte BA, Larue AC, et al. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 2006;107:358–66. doi: 10.1182/blood-2005-04-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson RB. Ionizing radiation and aging: rejuvenating an old idea. Aging. 2009;1:887–902. doi: 10.18632/aging.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner-Ecker M, Schwager C, Wirkner U, et al. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simone NL, Soule BP, Ly D, et al. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Meng A, Zhou D. Inhibition of phosphatidylinostol 3-kinase uncouples H2O2-induced senescent phenotype and cell cycle arrest in normal human diploid fibroblasts. Exp Cell Res. 2004;298:188–96. doi: 10.1016/j.yexcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Probin V, Wang Y, Bai A, et al. Busulfan selectively induces cellular senescence but not apoptosis in WI38 fibroblasts via a p53-independent but Erk-p38 MAPK-dependent mechanism. J Pharmacol Exp Ther. 2006;319:551–60. doi: 10.1124/jpet.106.107771. [DOI] [PubMed] [Google Scholar]
- 16.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–7. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prieur A, peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–5. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Krutzfeldt J, Kuwajima S, Braich R, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–92. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883–96. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 21.Hackl M, Brunner S, Fortschegger K, et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. 2010;9:291–6. doi: 10.1111/j.1474-9726.2010.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–5. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomasini R, Seux M, Nowak J, et al. TP53INP1 is a novel p73 target gene that induces cell cycle arrest and cell death by modulating p73 transcriptional activity. Oncogene. 2005;24:8093–104. doi: 10.1038/sj.onc.1208951. [DOI] [PubMed] [Google Scholar]
- 24.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Brosh R, Shalgi R, Liran A, et al. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gewirtz DA, Holt SE, Elmore LW. Accelerated senescence: an emerging role in tumor cell response to chemotherapy and radiation. Biochem Pharmacol. 2008;76:947–57. doi: 10.1016/j.bcp.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 27.Weidhaas JB, Babar I, Nallur SM, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]