Abstract
The [PSI+] prion can be induced by overproduction of the complete Sup35 protein, but only in strains carrying the non-Mendelian [PIN+] determinant. Here we demonstrate that just as [psi –] strains can exist as [PIN+] and [pin–] variants, [PSI+] can also exist in the presence or absence of [PIN+]. [PSI+] and [PIN+] tend to be cured together, but can be lost separately. [PSI+]-related phenotypes are not affected by [PIN+]. Thus, [PIN+] is required for the de novo formation of [PSI+], not for [PSI+] propagation. Although [PSI+] induction is shown to require [PIN+] even when the only overexpressed region of Sup35p is the prion domain, two altered prion domain fragments circumventing the [PIN+] requirement are characterized. Finally, in strains cured of [PIN+], prolonged incubation facilitates the reappearance of [PIN+]. Newly appearing [PIN+] elements are often unstable but become stable in some mitotic progeny. Such reversibility of curing, together with our previous demonstration that the inheritance of [PIN+] is non-Mendelian, supports the hypothesis that [PIN+] is a prion. Models for [PIN+] action, which explain these findings, are discussed.
Keywords: nonsense suppression/[PIN+]/prion/[PSI+]/SUP35
Introduction
Prions are conformational variants of particular proteins that can propagate their conformation to conventionally folded variants. The first hypothesized prion was the causative agent of transmissible spongiform encephalopathies in mammals—PrPSc, an abnormal conformation of the mammalian protein PrPC (for reviews, see Horwich and Weissman, 1997; Prusiner et al., 1998). Now the list of transmissible protein conformations includes [PSI+] (Sup35pPSI+) and [URE3] (Ure2pURE3) in yeast and [Het-s] in Podospora anserina (Wickner, 1994; Coustou et al., 1997; for reviews, see Cox, 1994; Lindquist, 1997; Wickner, 1997; Liebman and Derkatch, 1999; Wickner and Chernoff, 1999). The following genetic criteria for fungal prions were proposed (Wickner et al., 1995, 1999): (i) a non-Mendelian pattern of inheritance; (ii) reversible curing; (iii) induction of the prion by overproduction of the normal form of the protein; and (iv) phenotypic resemblance between strains bearing the prion and strains carrying mutations that inactivate the prion-encoding gene. The first criterion is shared by viruses, plasmids and cytoplasmic organelles. Moreover, like prions, these elements can usually be cured by guanidine hydrochloride (GuHCl). However, prions are the only non-Mendelian elements that have been shown to reappear after curing. Thus, reversibly curable non-Mendelian genetic factors are likely to be prions. The third criterion is met by [PSI+], [URE3] and [Het-s], while the fourth is met only by [PSI+] and [URE3], which inactivate normal cellular functions of Sup35p and Ure2p, but not by the [Het-s] prion, which appears to have a function of its own.
[PSI+] is a prion variant of the yeast Sup35p (for a review, see Liebman and Derkatch, 1999). [PSI+] is characterized by the formation of proteinase K-resistant Sup35p aggregates (Patino et al., 1996; Paushkin et al., 1996) that cause a reduction of translation termination efficiency and can be scored as nonsense suppression because Sup35p is the translation termination factor eRF3. Analyses of nonsense suppression levels in isogenic [PSI+] derivatives demonstrated that there are weak [PSI+] variants characterized by a low level of nonsense suppression and strong [PSI+] variants characterized by a high level of nonsense suppression, and the efficiency of nonsense suppression correlates with the level of Sup35p aggregation (Derkatch et al., 1996; Zhou et al., 1999). These [PSI+] variants are analogous to the mammalian prion strains and are presumed to carry either alternative prion conformations of Sup35p or different types of [PSI+] aggregates.
Sup35p consists of three major domains. The C-terminal part (Sup35C) starting from amino acid 254 is highly conserved and essential for viability (Ter-Avanesyan et al., 1993). It contains the G-domain (Kushnirov et al., 1988; Wilson and Culbertson, 1988) and sequence(s) required for interacting with eRF1 (Paushkin et al., 1997a; Ebihara and Nakamura, 1999; Eurwilaichitr et al., 1999). The Sup35Cp homolog from Xenopus laevis is sufficient for GTP binding, and promotion of the stop codon and eRF1-dependent release of fMet from fMet-tRNA (Zhouravleva et al., 1995; Frolova et al., 1996). The N-terminal domain (Sup35N) ending at amino acid 123 and the internal M-domain (Sup35M) are not conserved and are not essential for the termination process. However, Sup35N is required for the maintenance of [PSI+] (Ter-Avanesyan et al., 1994) and, when overproduced, induces the de novo appearance of [PSI+] (Derkatch et al., 1996; Patino et al., 1996), as does the complete Sup35p (Chernoff et al., 1993). The presence of Sup35N is crucial and sufficient for Sup35p–Sup35p binding in a [PSI+]-dependent manner (Paushkin et al., 1996, 1997a), and the Sup35Np fragment from a [PSI+] strain can convert soluble Sup35p to proteinase K resistance in vitro or be converted to proteinase K resistance in the reverse experiment (Paushkin et al., 1997b). Mutational studies have defined two Sup35N regions, the glutamine/ asparagine-rich stretch and the oligopeptide repeats, which are involved in [PSI+] biogenesis and maintenance, although the roles of these regions may be different (Doel et al., 1994; DePace et al., 1998; Kochneva-Pervukhova et al., 1998a; Derkatch et al., 1999; Liu and Lindquist, 1999).
Purified Sup35p (or Sup35Np) can form amyloid-like fibers in vitro, and the growth of these fibers can be accelerated by seeding with pre-formed fibers as well as by adding lysates of [PSI+] cells (Glover et al., 1997; King et al., 1997). These fibers can be considered to be an in vitro model of [PSI+] behavior. However, additional factors are apparently involved in [PSI+] genesis and maintenance in vivo. For example, the Hsp104p chaperone is required for [PSI+] maintenance but cures [PSI+] when present at a high concentration without the simultaneous overexpression of Ssa1p, a chaperone from the Hsp70p family (Chernoff et al., 1995; Newnam et al., 1999). Also, a non-Mendelian factor [PIN+] has to be present in yeast strains for efficient induction of the de novo appearance of [PSI+] by Sup35p overproduction (Derkatch et al., 1997).
Here we show that [PIN+] is not needed for the propagation of [PSI+] but is required for the de novo formation of [PSI+], because [PSI+] derivatives can be [pin–] and neither [PSI+] stability nor any [PSI+]-related phenotype is affected by [PIN+]. The presence of Sup35M and Sup35C in the [PSI+]-inducing constructs is not crucial for such [PIN+]-dependence, and exceptions to [PIN+]-dependence were observed only in the presence of Sup35p fragments carrying the RVDLQACKLMIQYQRK extension. Finally, we show that curing of [PIN+] is reversible. This, together with our previous observations that [PIN+] is characterized by a non-Mendelian pattern of inheritance and depends upon Hsp104p for its maintenance, strengthens the hypothesis that [PIN+] is a prion.
Results
[PSI+] derivatives obtained in a [pin–] strain remain [pin–]
We have shown previously that the induction of the de novo appearance of [PSI+] by overproduction of the complete Sup35p is observed only in the presence of [PIN+] (Derkatch et al., 1997). Thus, [PIN+] is required for either the maintenance or the induction of [PSI+]. Surprisingly, one of the Sup35Np-encoding plasmids, pEMBL-ΔBalext (previously called pEMBL-ΔBal2), was able to induce [PSI+] when introduced into [pin–] cells (Derkatch et al., 1997). This plasmid encodes a truncated protein containing the first 154 amino acids of Sup35p and a 17 amino acid tail from the vector (see below). Either this Sup35p fragment can seed [PSI+] in the absence of [PIN+], and the induced [PSI+]s remain [pin–] and do not require [PIN+] for further propagation, or [PSI+] induction occurs in conjunction with the de novo appearance of [PIN+].
To distinguish between these possibilities, isogenic weak and strong [PSI+] derivatives were obtained in [PIN+] and [pin–] versions of strain 74-D694 transformed with pEMBL-ΔBalext or the plasmid expressing the complete Sup35p, pEMBL-SUP35 (Table I). These [PSI+] derivatives were then exposed to excess Hsp104p, which causes the loss of [PSI+] (Chernoff et al., 1995) but does not affect [PIN+] (Derkatch et al., 1997), by transforming them with pYS-GAL104. Usually the Pin status of six [psi–] colonies arising from the progeny of three pYS-GAL104 transformants was analyzed for each [PSI+] derivative. The [psi–] progeny of all 15 [PSI+] variants obtained by expression of the pEMBL-ΔBalext encoded Sup35p fragment in a [pin–] derivative were shown to be [pin–]. In contrast, the progeny of the control [PSI+] variants obtained in a [PIN+] version of 74-D694 were usually [PIN+] (Table I).
Table I. [PSI+] derivatives can be [pin–].
| Derivative used for [PSI+] induction | Inducing Sup35p construct | No. of [PSI+]s induced |
|||
|---|---|---|---|---|---|
| Weak |
Strong |
||||
| [PIN+] | [pin–] | [PIN+] | [pin–] | ||
| [pin–] | truncated | 0 | 7 | 0 | 8 |
| [PIN+] | wild type | 7 | 0 | 6 | 0 |
| [PIN+] | truncated | 9 + 1a | 2b | 5 | 1 |
[PSI+]s were induced in 74-D694 derivatives with pEMBL-SUP35 (wild type) and pEMBL-ΔBalext (truncated). Hsp104p overproduction was used to eliminate [PSI+] from these strains prior to the determination of their Pin status. This was accomplished by transforming [PSI+] derivatives with pYS-GAL104 and inducing HSP104 overexpression. Following colony purification, red [psi –] colonies were assayed for Pin by a cross to a tester [psi –][pin–] strain overproducing the complete Sup35p. Only when the 74-D694 [psi –] derivatives used in the cross carried [PIN+] were the resulting diploids [PIN+], i.e. overproduction of Sup35p induced the appearance of [PSI+], scored as growth on adenineless media.
aBoth [PIN+] and [pin–] progeny were obtained from this [PSI+] derivative.
bThese [PSI+] derivatives also generated [pin–] progeny when [PSI+] was lost spontaneously or on 1 mM GuHCl, proving that they were indeed [pin–]. The appearance of occasional [PSI+][pin–] derivatives among [PSI+]s induced in this experiment reflects the inherent instability of [PIN+] and the ability ofpEMBL-ΔBalext to induce [PSI+] in a [pin–] background. Note that when this experiment was repeated using two freshly isolated colonies of a [PIN+] derivative, none of 12 [PSI+] derivatives induced gave rise to any [pin–] progeny.
Similar results were obtained when the Pin status of [PSI+] derivatives described above was re-determined after [PSI+]s were lost spontaneously or upon treatment with 1 mM GuHCl (data not shown), which apparently cures prions by a mechanism different from that of excess Hsp104p (Eaglestone et al., 1999b). Weak [PSI+]s were chosen for these experiments because they give rise to spontaneous [psi–] colonies at a frequency higher than 10–3 (thus outnumbering any Mendelian mutants with [psi–]-like phenotypes), and growth on 1 mM GuHCl frequently eliminates weak [PSI+]s without eliminating [PIN+] (Derkatch et al., 1997).
Thus, [PSI+] derivatives can be [pin–] as well as [PIN+] and a pEMBL-ΔBalext-encoded protein fragment can induce [PSI+] in a [pin–] strain without the appearance of [PIN+]. Interestingly, we find (data not shown) that [PSI+] strain 783/4c (Eaglestone et al., 1999a), derived from strains used by B.S.Cox and co-workers in the classic [PSI+] studies (for reviews, see Cox et al., 1988; Cox, 1994), is [pin–]. This may be relevant in interpreting some early [PSI+] data.
[PSI+] and [PIN+] tend to be retained or lost together upon growth on GuHCl-containing medium
The above results suggest that [PIN+] is required only at the initial step of Sup35p conformational change and/or [PSI+] seed formation, and that, once induced, [PSI+] does not require [PIN+] for its maintenance. The possibility remains, however, that [PSI+] variants induced by the complete Sup35p or its fragments through a [PIN+]-dependent pathway will be dependent on [PIN+] for their further propagation. To examine this possibility, weak and strong [PSI+][PIN+] 74-D694 variants induced in the presence of [PIN+] were grown on 5 mM GuHCl in an attempt to cure [PIN+] but retain [PSI+]. Cultures were then colony purified on YPD. About 30% of the progeny were non-sectored white (or, in the case of weak [PSI+], pink), indicative of the retention of a mitotically stable [PSI+]. The Pin status of these [PSI+] clones was determined (see above and Materials and methods) and, in agreement with the hypothesis that [PIN+] is not needed for [PSI+] propagation, [pin–]s were detected.
A quantitative analysis of the progeny of [PSI+][PIN+] strains grown on 5 mM GuHCl was performed to test whether [PIN+] and [PSI+] are cured independently (Table II, see also Materials and methods). The frequency of [PSI+][PIN+] and [psi–][pin–] derivatives was higher than expected and the frequency of [psi–][PIN+] and [PSI+][pin–] derivatives was lower than expected, if [PSI+] and [PIN+] were lost independently. The data indicate that even though each of the factors can be cured separately, [PIN+] and [PSI+] tend to be eliminated or retained together.
Table II. [PIN+] and [PSI+] tend to be lost or retained together on GuHCl-containing media.
| Progeny | Strong [PSI+][PIN+] |
Weak [PSI+][PIN+] |
||
|---|---|---|---|---|
| Observed | Expected | Observed | Expected | |
| [PSI+][PIN+] | 63 | 28 | 43 | 19 |
| [psi –][PIN+] | 17 | 52 | 22 | 46 |
| [PSI+][pin–] | 41 | 76 | 42 | 66 |
| [psi –][pin–] | 174 | 139 | 190 | 166 |
| Total | 295 | 295 | 297 | 297 |
The data from three independent subclones, which gave similar results, are combined (see Materials and methods). Strong [PSI+][PIN+] or weak [PSI+][PIN+] 74-D694 derivatives were grown on 5 mM GuHCl for 7.4 generations. The expected values were calculated using the observed frequency of the loss/retention of [PSI+] and the observed frequency of the loss/retention of [PIN+] assuming that the loss of [PSI+] and [PIN+] is independent. For example, the expected value for [psi –][PIN+] is the product of the observed frequency of the loss of [PSI+] and the observed frequency of the retention of [PIN+] times the total number of derivatives analyzed. The difference between the observed and the expected proportions is statistically significant (P <0.005). In control experiments, where cells were grown on YPD, strong [PSI+] was 99.81% stable, weak [PSI+] was 99.55% stable (12 240 and 12 673 colonies counted, respectively) and [PIN+] was 100% stable (26 and 29 progeny analyzed from strong and weak [PSI+][PIN+] derivatives, respectively). Data suggestive of co-loss of [PSI+] and [PIN+] were also obtained in two other experiments, when [PSI+][PIN+] 74-D694 derivatives were grown on 5 mM GuHCl for 6.9 generations (36 [PSI+][PIN+], five [psi –][PIN+], 16 [PSI+][pin–] and 32 [psi –][pin–] derivatives of the strong [PSI+][PIN+] and 25 [PSI+][PIN+], 11 [psi –][PIN+], 15 [PSI+][pin–] and 23 [psi –][pin–] derivatives of the weak [PSI+][PIN+] were scored). Shorter incubation on 5 mM GuHCl (approximately five cell generations) resulted in [PSI+] becoming cryptic (cultures were red on this medium but [PSI+] was not cured).
A ‘protection’ hypothesis postulating that [PIN+] protects [PSI+] from being cured, or [PSI+] protects [PIN+], was tested. In the first experiment, isogenic [PSI+][pin–] and [PSI+][PIN+] 74-D694 derivatives were grown on 5 mM GuHCl for 7.4 cell generations and then plated to YPD. The fraction of [psi–] colonies was estimated from ∼12 000 colonies from each culture. The observation that strong [PSI+] was eliminated in 64.7 and 61.7% of the cells in [PIN+] and [pin–] derivatives, respectively, and weak [PSI+] was eliminated in 71.4 and 72.7% of the cells of [PIN+] and [pin–] derivatives, respectively, excluded the possibility that the presence of [PIN+] protects [PSI+] from being eliminated. In an analogous experiment that involved an isogenic [psi–][PIN+] and the same [PSI+][PIN+] 74-D694 derivatives, the fraction of [pin–] colonies was estimated among 300 colonies from each culture. Curing of [PIN+] was estimated at 79.5% in the [psi–] derivative, and at 72.9 and 78.1% in the strong and weak [PSI+] derivatives, respectively. These differences were shown to be not statistically significant (P >0.25) using a contingency χ2 test (Herskowitz, 1977). Thus, the presence of [PSI+] does not protect [PIN+] from being eliminated.
[PIN+] does not affect [PSI+]-related phenotypes
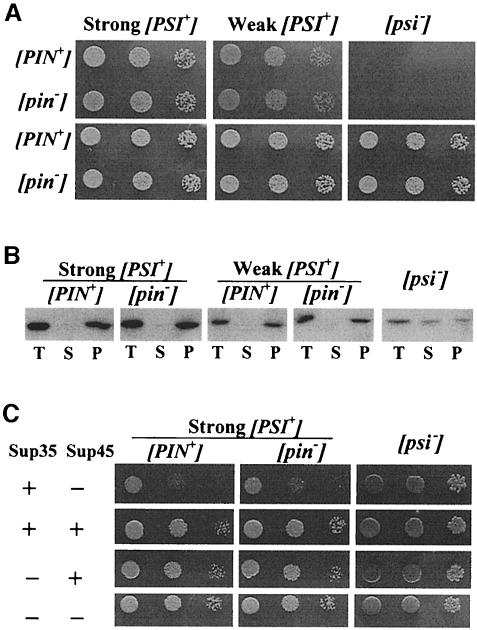
The demonstration of the existence of [PSI+][pin–] as well as [PSI+][PIN+] strains raises the question of whether [PSI+]-related phenotypes are affected by the absence of [PIN+]. No differences in the following phenotypes were detected in isogenic weak and strong [PSI+][PIN+] and [PSI+][pin–] derivatives of 74-D694: (i) the level of nonsense suppression (Figure 1A); (ii) the level of Sup35p aggregation (Figure 1B); (iii) mitotic stability of [PSI+] in the absence and presence of GuHCl (previous section); (iv) growth inhibition by overproduction of wild-type Sup35p (Figure 1C); (v) rescue of this growth inhibition by simultaneous Sup45p overproduction (Figure 1C); (vi) lethality of [PSI+] haploids carrying the sup35-2 allele (data not shown); or (vii) [PSI+] stability and suppression efficiency in the presence of the PNM2 allele of SUP35. PNM2 cured all [PSI+]s in [pin–] 783/4c but did not cure [PSI+] in 74-D694 [PSI+] derivatives whether or not they were [PIN+], suggesting that [PIN+] does not determine the ‘Psi no more’ phenotype of the PNM2 allele. Also, as previously reported for [PSI+][PIN+] strains (Derkatch et al., 1999), PNM2 overexpression increased and reduced, respectively, the suppressor phenotypes of weak and strong [PSI+][pin–] derivatives of 74-D694. The fact that [PIN+] does not affect [PSI+]-related phenotypes supports the hypothesis that [PIN+] affects [PSI+] appearance but not propagation.
Fig. 1. [PIN+] does not affect various [PSI+]-related phenotypes in the isogenic [PSI+][pin–] and [PSI+][PIN+] derivatives of 74-D697. (A) [PIN+] does not affect nonsense suppression caused by [PSI+]. Ten-fold serial dilutions of the indicated cultures were spotted onSD-Ade (top panels) and SD (bottom panels) and incubated at 20°C. Growth on SD-Ade is due to the suppression of the ade1-14 nonsense allele. The SD medium is the growth control. (B) [PIN+] does not affect the levels of Sup35p aggregation. Cells from logarithmically growing cultures of the derivatives indicated were resuspended in buffer (25 mM Tris–HCl pH 7.5, 10 mM MgCl2, 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol) and broken with glass beads. Cell extracts containing 200 µg of protein normalized to 2 µg/µl were centrifuged (30 min, 18 000 g, 4°C), and pellet (P) and supernatant (S) fractions were used in Western blot analysis. T (total) is the ‘no centrifugation’ control. Polyclonal mouse anti-Sup35p antibody M12 (kindly supplied by V.Prapapanich) was used for immunodetection using Western-Star™ reagents and the Tropix protocol (Bronstein et al., 1994). (C) [PIN+] does not affect growth inhibition caused by overproduction of Sup35p in [PSI+] derivatives as well as the rescue of this growth inhibition by simultaneous overproduction of Sup45p. Ten-fold serial dilutions of cultures of co-transformants carrying either pGAL::SUP35 or YCp50 and either YEp13-SUP45 or YEp13 are shown. Incubation was on SGal-Ura,Leu at 30°C for 4 days. + shows that indicated protein is overproduced, – stands for the lack of overproduction.
[PSI+] induction usually remains [PIN+]-dependent even when the inducing constructs lack the Sup35p M and C domains
We have demonstrated previously that overproduction of the complete Sup35p or its pEMBL-ΔBcl-encoded fragment containing the N and M domains cannot cause the de novo appearance of [PSI+] in [pin–] derivatives, whereas the pEMBL-ΔBalext-encoded fragment containing just the N and the beginning of the M domain can (Derkatch et al., 1997). If this [PIN+]-independence were due to the absence of downstream domains in the inducing Sup35p fragment (e.g. if the functional form of the Pin protein, Pinpin–, interacts with Sup35M or Sup35C to inhibit the formation of [PSI+] seeds; see Derkatch et al., 1997), other Sup35p fragments lacking the M and C domains should also induce [PSI+] in a [PIN+]-independent manner. Alternatively, if the [PIN+]-independence of the pEMBL-ΔBalext-encoded fragment is not due to the lack of downstream domains, other Sup35 fragments lacking M and C generally should remain [PIN+]-dependent. This could be, for example, if the Pin protein exerts its effect through the N-terminus of the [PSI+]-seeding fragment. Then, dependence on [PIN+] would disappear only if certain modifications (e.g. extensions present in genetically engineered Sup35p fragments) alter the conformation of Sup35N and/or its ability to take on the [PSI+] shape.
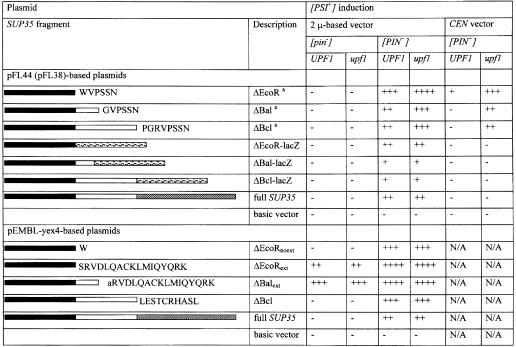
Single- and multi-copy plasmids encoding the complete Sup35p or three N-terminal fragments extended either by different short sequences or by a LacZ fragment were tested for their ability to induce [PSI+] (see Materials and methods; Figure 2). If the presence of the M and C domains is required to make [PSI+] induction [PIN+]-dependent, then [PSI+] is likely to be induced in [pin–] strains by the multicopy plasmids lacking these domains (pFL44-ΔBal, pFL44-ΔEcoR, pFL44-ΔBal-lacZ or pFL44-ΔEcoR-lacZ) but not by the plasmids containing them (pFL44-SUP35, pFL44-ΔBcl or pFL44-ΔBcl-lacZ). The data show that none of the plasmids caused the de novo appearance of [PSI+] in a [pin–] background frequently enough to be detected in a patch test (Figure 2). Despite this, expression of Sup35p fragments was not reduced significantly in [pin–] derivatives (Figure 3), and all multicopy plasmids induced the de novo appearance of [PSI+] in [psi–][PIN+] strains, confirming that the prion domains were functional (Figure 2). Moreover, even the low level of expression of the constructs bearing the N-terminal Sup35p fragments (not fused to LacZ or the Sup35p C-terminus) from single-copy plasmids caused efficient [PSI+] induction in a [PIN+] background (Figure 2). These results show that the absence of downstream Sup35p domains is not sufficient for the relief of [PIN+]-dependence for [PSI+] induction.
Fig. 2. Sequences attached to Sup35N determine whether it can induce [PSI+] in a [PIN+]-independent manner. The results of patch tests in which [PSI+] de novo appearance was detected as growth or papillation on adenineless media are presented. Filled square, open square and shaded square indicate N, M and C domains of Sup35p, respectively; hatched square indicates LacZ extension; the amino acid sequences of shorter extensions are shown in upper case letters. In the ΔBalext construct, the first amino acid in the extension, shown as a lower case ‘a’, is the same as normally found at position 155 in Sup35p. It is included in the extension to show the comparison with the other ΔBal constructs, which include only amino acids 1–154 of Sup35p. ++++ and +++ stand for different levels of growth on adenineless media after 4 days of incubation at 30°C. ++ and + stand for very intensive and intensive papillation after 12 days of incubation in 30°C. – indicates the absence of growth or intensive papillation on adenineless media. aThe absence of a yeast transcription termination signal immediately downstream of the polylinker in pFL44 and pFL38 makes it possible that the mRNAs encoding the ΔEcoR, ΔBal and ΔBcl Sup35p fragments may be extended considerably beyond the stop codons located within the polylinker and may therefore be subject to nonsense-mediated mRNA decay (NMD). Thus, all experiments were repeated using an isogenic Δupf1 strain lacking NMD. Note that control experiments showed that Δupf1 had no effect on the efficiency with which the full-length Sup35p induces [PSI+] (see also Derkatch et al., 1996). Furthermore, there were only minor increases noted even in experiments employing the ΔEcoR, ΔBal and ΔBcl constructs (except for experiments with CEN vectors).
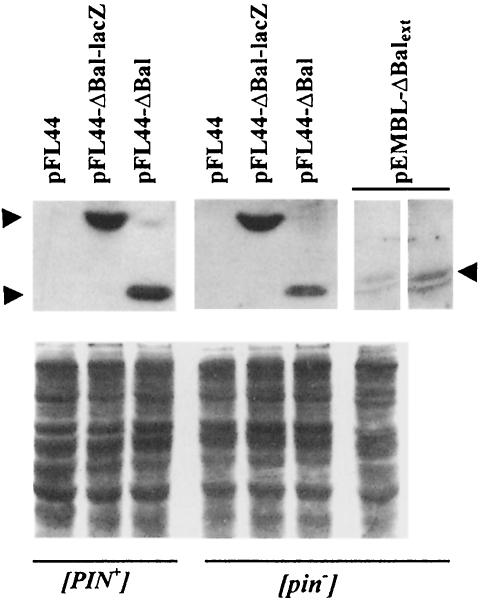
Fig. 3. Comparison of expression of Sup35p fragments in [PIN+] and [pin–] derivatives of 74-D694. The indicated [psi–] derivatives of the Δupf1 mutant of 74-D694 were transformed with the plasmids listed. Cells from logarithmically growing cultures were resuspended in buffer (50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, 0.1 mM dithiothreitol, 100 µg/ml RNase A, 100 µg/ml cycloheximide, 2 µg/ml pepstatin A, 2 µg/ml phenylmethylsulfonyl fluoride, 1 µg/ml benzamidine, 10 µg/ml leupeptin) and broken with glass beads. Cell extracts containing 120 µg of protein were used in Western blot analysis (top panel). Antipeptide antibody against amino acids 137–151 of Sup35p (Patino et al., 1996; kindly supplied by M.Patino and S.Lindquist) was used for immunodetection using Western-Star™ reagents and the Tropix protocol (Bronstein et al., 1994). All lanes are from the same gel. Lanes 1–7 were exposed for the same amount of time, whereas lane 8 shows a longer exposure of lane 7. Arrows indicate the positions of the proteins encoded by pFL44-ΔBal-lacZ (top left), pFL44-ΔBal (bottom left) and pEMBL-ΔBalext (right). The amount of protein loaded was normalized by Coomassie Blue staining (bottom panel).
A comparison of constructs that do or do not induce [PSI+] in [pin–] strains indicates that the extensions attached to the construct, rather than the length of the N-terminal Sup35p fragment, determines the [PIN+]-dependence (see Figure 2). Indeed, the proteins encoded by pFL44-ΔBal, pEMBL-ΔBalext, pFL44-ΔBal-lacZ, pFL44-SUP35 and pEMBL-SUP35 differ only by sequences (which are six, 17, 129, 531 and 531 amino acids long, respectively) downstream of the 154 amino acid Sup35 piece. Western analysis shows that the 17 amino acid extension present in the inducing pEMBL-ΔBalext construct does not increase the protein expression or stability (Figure 3). On the contrary, the level of the pEMBL-ΔBalext-encoded fragment was significantly lower than the level of the fragments encoded by pFL44-ΔBal, pFL44-ΔBal-lacZ, pFL44-SUP35 and pEMBL-SUP35 (the last is in agreement with Kochneva-Pervukhova et al., 1998b).
To see if the presence of an analogous 17 amino acid extension on a different fragment containing the prion domain could also facilitate the appearance of [PSI+] in [pin–] derivatives, the pEMBL-ΔEcoRext plasmid was utilized. Indeed, the de novo appearance of [PSI+] was observed in [pin–] derivatives when transformed with pEMBL-ΔEcoRext but not when transformed with the control pEMBL-ΔEcoRnoext plasmid (Figure 2). Thus, [PIN+]-dependence is relieved in cells expressing two different Sup35Np constructs with the RVDLQACKL MIQYQRK extension.
Extended incubation allows for the induction of the de novo appearance of [PSI+] in [pin–] derivatives: [PIN+] reappearance
According to the prion model, the loss of a prion should be reversible as long as the gene encoding the prion protein is expressed. Thus, if [PIN+] is a prion, it should reappear spontaneously at a low frequency. Since prolonged incubation at low temperature apparently increases the spontaneous appearance of prions (see Discussion), we used these conditions in an attempt to increase the frequency of spontaneous [PIN+] appearance to a detectable level. To detect the de novo appearance of [PIN+], we selected for the appearance of [PSI+] in a [psi–][pin–] strain overproducing Sup35p and screened these [PSI+]s for the presence of [PIN+].
Transformants of the [psi–][pin–] 74-D694 derivative with the pGAL::SUP35 plasmid were incubated on SD-Ura for 1 month at 4°C prior to replica plating to SGal-Ura, where Sup35p is overproduced, and then to SD-Ade and SEt-Ade, which select for [PSI+] as well as for other Ade+ revertants. The slight increase in the number of the Ade+ colonies following prolonged incubation and GAL::SUP35 induction relative to the control ‘no induction’ experiments was due to the de novo appearance of [PSI+]s (Table III, A). Importantly, the majority of these [PSI+]s were shown to be [PIN+] and the remainder appear to have gone through a transient [PIN+] state (see below). Because the [psi–][pin–] 74-D694 derivative used in this experiment was obtained by GuHCl curing of [PIN+] from the [psi–][PIN+] 74-D694 strain (see Materials and methods), this result confirms that the curing of [PIN+] is reversible.
Table III. The effects of prolonged incubation on [PSI+] induction and [PIN+] de novo appearance.
| Plasmid | 4°C incubation | Incubation on SGal-Ura | No. of colonies |
|||
|---|---|---|---|---|---|---|
| Ade+ | [PSI+] | Confirmed as [PSI+][PIN+] | ||||
| A | pGAL::SUP35 | 1 month | immediately after 4°C | 106 | 53 | 28 |
| pGAL::SUP35 | 1 month | none | 25 | 0 | NA | |
| YCp50 | 1 month | immediately after 4°C | 42 | 0 | NA | |
| YCp50 | 1 month | none | 23 | 0 | NA | |
| B | pGAL::SUP35 | 1 month | delayed after 4°C | 30 | 3 | 2 |
| YCp50 | 1 month | delayed after 4°C | 21 | 0 | NA | |
| C | pGAL::SUP35 | 1 month | during 4°C | 34 | 22 | 6a |
| YCp50 | 1 month | during 4°C | 11 | 0 | NA | |
| D | pGAL::SUP35 | 7 days | immediately after 4°C | 30 | 5 | 1 |
| pGAL::SUP35 | 7 days | none | 36 | 0 | NA | |
| E | pGAL::SUP35 | 4 days | immediately after 4°C | 124 | 7 | 1a |
| pGAL::SUP35 | 4 days | none | 69 | 0 | NA | |
| F | pGAL::SUP35 | none | yes | 174 | 1b | 0 |
(A, D and E) Like-size patches on SD-Ura of nine (A), three (D) and seven (E) transformants with each plasmid were incubated at 4°C for the time period listed. After the 4°C incubation, yeast were replica plated first to SGal-Ura (if ‘immediately after 4°C’ is indicated) or to SD-Ura (if ‘none’ is indicated), and then toSD-Ade and SEt-Ade to select for Ade+ colonies. (B) Same as above except that after the 4°C incubation, four transformants were first replica plated to SD-Ura, then to SGal-Ura and only then to SD-Ade and SEt-Ade. (C) Like-size patches on SGal-Ura of two transformants with each plasmid were incubated at 4°C for 1 month and then replica plated to SD-Ade and SEt-Ade. (F) Combined control data in which 4°C incubation was omitted for transformants used in A–E. When comparing data between sections with different letters, the proportion of [PSI+]s among Ade+ clones and not the absolute number of Ade+ and [PSI+] colonies best indicates the efficiency of [PSI+] induction because the number of transformants and the size of patches varied between these experiments.
aPin status was determined in 19 of 22 and in and five of seven [PSI+] derivatives, respectively.
bThe transformant that gave rise to this [PSI+] was also used in experiment E where it did not give rise to any [PSI+]s.
Several results suggest that the newly induced [PIN+]s are unstable. First, 10 of 28 [PSI+][PIN+] derivatives, when cured of [PSI+], gave rise to [psi–][pin–] as well as [psi–][PIN+] progeny. Indeed, when pYS-GAL104 was introduced into the [PSI+][PIN+] derivatives to cure [PSI+], both [psi–][PIN+] and [psi–][pin–] clones could occasionally be obtained from a single transformant. Secondly, when Sup35p overproduction was induced after several generations of growth at 30°C following the 4°C incubation (delayed induction, Table III, B), the proportion of [PSI+] colonies among Ade+ colonies was reduced from 50 to 10%, indicating a reduction in the de novo appearance of [PSI+], possibly because some cells lost [PIN+] prior to the Sup35p overexpression. Likewise, when yeast overexpressing Sup35p on SGal-Ura medium were incubated at 4°C for a prolonged period (Table III, C), the proportion of [PSI+]s among Ade+ colonies was increased to 65%. Finally, newly generated [PSI+] derivatives that at first appeared to be [pin–] have apparently lost [PIN+] following [PSI+] induction. In these cases, although no [PIN+] was found among the first six clones tested, [PIN+] was detected when several additional clones were analyzed (data not shown).
The detection of [PSI+] derivatives carrying stable and unstable [PIN+]s could be explained if [PIN+] were generally unstable following its de novo appearance but is stabilized in some clones. To test the possibility of [PIN+] stabilization, four [PSI+] derivatives were colony purified, and for each [PSI+] the stability of [PIN+] was analyzed in three colonies (Table IV). In support of the stabilization hypothesis, two [PSI+] derivatives that were prone to lose [PIN+] at high frequency each gave rise to one colony with a stable [PIN+].
Table IV. Mitotic stability of [PIN+] in [PSI+][PIN+] derivatives obtained following prolonged incubation at 4°C.
| [PSI+][PIN+] derivative | Colonies | No. [psi–] |
|
|---|---|---|---|
| [PIN+] | [pin–] | ||
| 1 | 1–3 | 18 | 0 |
| 2 | 1 | 6 | 0 |
| 2, 3 | 3a | 9 | |
| 3 | 1 | 6 | 0 |
| 2, 3 | 0 | 12 | |
| 4 | 1 | 2a | 4 |
| 2, 3 | 0 | 10 | |
The [PSI+] derivatives listed were colony purified and the Pin status of three colonies of each was determined. For each colony, two [psi–]s obtained by overexpression of Hsp104p in each of three pYS-GAL104 transformants were examined.
aTwo [psi–][PIN+] derivatives were from a single pYS-GAL104 transformant.
Although we established that the de novo appearance of [PIN+] can be detected only following prolonged incubation at 4°C, the possibility remained that [PIN+] is encoded by Sup35p and is induced by Sup35p overproduction just as the appearance of [PSI+] and [URE3] is induced by overexpression of the normal forms of the prion proteins Sup35p and Ure2p, respectively, and that prolonged incubation only facilitates this process. This possibility was excluded by our finding that [PIN+] reappeared following prolonged 4°C incubation of cells transformed with a SUP35 fragment encoded by pFL44-ΔBcl that was shown previously (Derkatch et al., 1997) not to encode [PIN+]. Indeed, 65 out of 85 Ade+ colonies that appeared in four pFL44-ΔBcl transformants following prolonged incubation were [PSI+], and three of eight [PSI+] derivatives examined were shown definitively to be [PIN+].
Finally, we demonstrated that short-term incubation at 4°C for 4 or 7 days was not sufficient to increase the frequency of [PSI+] or [PIN+] appearance to the same level as the month long incubation. The data suggest that the longer the incubation, the more frequent the appearance of prions (Table III, D and E; data not shown). The possibility that prolonged incubation in the absence of cold shock causes the induction of [PIN+] was also examined. A month long incubation at 30°C resulted in the severe loss of viability, whereas a 7 day incubation at 30°C did not induce [PSI+] (data not shown). However, following a month long 20°C incubation of [psi–][pin–] transformants carrying pGAL::SUP35, yeast were still alive and [PSI+] appearance was detected when Sup35p was overproduced during the prolonged incubation: out of 26 Ade+ colonies, 18 were [PSI+] and at least five were [PIN+].
Discussion
Prion model for [PIN+]
The GuHCl-curable factor [PIN+] was added to the list of provisional prion elements when its non-Mendelian pattern of inheritance and Hsp104p-dependence were established (Derkatch et al., 1997). Here we show that [PIN+] can reappear in derivatives from which it was cured. Such a demonstration of reversible curing, an important genetic criterion for prions (see Introduction), strongly supports the hypothesis that [PIN+] is a prion. In addition, our finding that [PIN+] shares two other properties with prions: (i) frequent de novo appearance caused by prolonged incubation and (ii) instability following de novo induction, strengthens the prion hypothesis for [PIN+].
We show that the de novo appearance of [PIN+] is facilitated by prolonged incubation at 4°C, a condition that also induces the de novo appearance of the other yeast prions, [URE3] (M.Aigle, reproduced and cited in Chernoff et al., 1995) and [PSI+] (our unpublished results including experiments in [PIN+] 74-D694; Y.O.Chernoff, personal communication). The mechanism behind this phenomenon is unknown. Our data exclude the possibility that cold shock induces [PIN+] and indicate that prolonged incubation is the crucial factor. Possibly the shift in the chaperone balance in deep stationary phase causes the appearance of [PIN+]. It is important to stress that even though SUP35 overexpression was essential to detect the de novo appearance of [PIN+] following prolonged incubation, this does not suggest that overexperssion of Sup35 induces the appearance of [PIN+] as would be expected if SUP35 encoded [PIN+]. This is because [PIN+] appearance was also observed when SUP35 fragments encoding only Sup35NMp or Sup35Np, previously shown not to encode [PIN+] (Derkatch et al., 1997), were overexpressed in place of the complete SUP35 (see Results; data not shown). Furthermore, substitution of the complete Saccharomyces cerevisiae SUP35 with the Pichia methanolica SUP35 did not cause the loss of [PIN+] (Chernoff et al., 2000; our unpublished observations).
Another trait shared by [PIN+] and prions is their instability when they are newly induced. We observe that when [PIN+] factors appear de novo they are frequently unstable and segregate both [pin–] and stable [PIN+] clones. We find that such instability and subsequent stabilization is also typical of freshly induced [PSI+] derivatives that form sectored (red/white or red/pink) colonies, which keep producing sectored as well as stable non-sectored (red and white or pink) clones (data not shown). Also, prions induced by the overproduction of N-terminal domains of Sup35p homologs from other yeast species in S.cerevisiae are frequently unstable (Chernoff et al., 2000; Santoso et al., 2000). Possibly, prions must go through a maturation step (Glover et al., 1997) when they first arise before they can propagate efficiently. Alternatively, a certain number of prion seeds may have to accumulate in order to ensure inheritance.
Relationship of [PSI+] and [PIN+]
Considerable experimental data in yeast (see the Saccharomyces Genome Database for references; http://genome-www.stanford.edu/Saccharomyces/) show that usually all effects of gene overexpression disappear when overexpression is discontinued and that phenotypes caused by wild-type gene overdose rarely resemble those of mutations inactivating the same gene. This indicates that few proteins are likely to be able to take on a prion conformation. The discovery of the [PSI+]–[PIN+] system that appears to involve two prion elements suggests the existence of a prion-based regulation pathway. [PSI+] may be engaged in adaptation to changing environmental conditions. Indeed, the presence of [PSI+] was associated with increased survival at high temperature or on ethanol in some strains of S.cerevisiae (Eaglestone et al., 1999a), and the amino acid composition of the prion domain and the ability to prionize is conserved in Sup35p (Chernoff et al., 2000; Santoso et al., 2000). However, under many conditions, the presence of [PSI+] is likely to be disadvantageous because [PSI+] partially inactivates an essential translation termination factor. Indeed, Sup35p aggregation (i.e. Sup35pPSI+ state) is rare among strains of various yeast species (Chernoff et al., 2000). We propose that [PIN+] may modulate the frequency of spontaneous [PSI+] appearance. For example, [PIN+] could appear under certain conditions allowing for the appearance of [PSI+]. Prolonged incubation at low temperature could be one such condition. [PIN+] could then be retained in some, but lost in other progeny of the [PSI+] derivative without causing any phenotypic change. The two types of cells would provide the population with variation that will become important upon the loss of [PSI+]. Those retaining [PIN+] will acquire [PSI+] more easily if environmental changes become repetitious, while those without [PIN+] will resist induction of the harmful [PSI+].
We observed that while [PSI+] and [PIN+] can be maintained/cured independently, they tend to be retained or lost together during growth on GuHCl. One explanation is that a state of competence for prion loss is established in some but not other cells during growth on GuHCl. Another possibility is that GuHCl inhibits the propagation of prion seeds, causing them to be diluted at each division. This model was proposed to explain [PSI+] loss caused by the presence of PNM2 (McCready et al., 1977) or by growth on GuHCl (Eaglestone et al., 1999b), and to predict that ∼60 randomly distributing [PSI+] particles are present in [PSI+] cells. Likewise, GuHCl curing of [PIN+] (Derkatch et al., 1997) may result from inhibition of [PIN+] propagation. According to this dilution model, different prions with a similar number of seeds should be cured simultaneously. During the first few divisions, all cells would retain some seeds for each prion. Interestingly, [PSI+] is cryptic in such cells (see Table II footnote) possibly because newly synthesized Sup35p is not aggregated. As the number of cell divisions on GuHCl increases, the number of [PSI+] and [PIN+] prion seeds per cell would be reduced, until the chance of losing all seeds increases dramatically, causing efficient curing of both prions. Thus, on the GuHCl plate, cells that divided more would tend to lose both prions, while cells with fewer cell divisions would retain both prions.
[PSI+] appearance in the presence and absence of [PIN+]
Our data indicate that [PIN+] is not involved in [PSI+] maintenance or propagation and thus cannot affect the ability of Sup35p to join [PSI+] aggregates. However, [PIN+] has to be present in the cell for [PSI+] to appear de novo. Because most Sup35Np fragments lacking the M and C domains still induce [PSI+] only in the presence of [PIN+], we suggest that [PIN+] exerts its effect on [PSI+] induction through Sup35N. These results are compatible either with the hypothesis that Pinpin– inhibits [PSI+] seeding or with the hypothesis that PinPIN+ makes such seeding possible. In the first case, Pinpin– could inhibit the completion of [PSI+] seed formation by recognizing rare self-interacting Sup35p molecules. Inhibition of [PSI+] induction by a direct interaction between Pinpin– and individual native Sup35p molecules is unlikely because extreme overproduction of Sup35p fails to overcome the inhibition caused by [pin–] (Derkatch et al., 1997), but does overcome the inhibitory effect of excess Sup45p (Derkatch et al., 1998) that appears to bind to individual Sup35p molecules (Stansfield et al., 1995; Zhouravleva et al., 1995). In the second case, the proposed [PIN+] prion aggregates could help to seed the formation of [PSI+]. Since prions generally promote self-aggregation, we expand this idea to propose that certain prions may be able to seed the formation of other prions and that this is how the [PIN+]-dependent pathway of [PSI+] induction works.
The discovery that [PSI+] colonies appear frequently in [pin–] strains carrying the pEMBL-ΔBalext or pEMBL-ΔEcoRext plasmids without the induction of [PIN+] indicates that [PSI+] can appear independently of [PIN+]. Both inducing plasmids encode proteins containing the [PSI+] prion domain at their N-termini and the RVDLQACKLMIQYQRK sequence at their C-termini, but in one construct these termini are separated by a piece of Sup35M which might cause the observed increase in the efficiency of the [PIN+]-independent [PSI+] induction (Figure 2). Apparently, the truncation of Sup35p is not the cause of the [PIN+] independence, since other constructs expressing analogously truncated Sup35 proteins do not allow for efficient [PSI+] induction in [pin–], even though they can induce [PSI+] much more efficiently than complete Sup35p in [PIN+] backgrounds (Figure 2). Thus, we propose that [PIN+]-dependence is reversed due to the presence of the RVDLQACKLMIQYQRK ‘tail’. However, it appears that in addition to using a [PIN+]-independent [PSI+] induction pathway, the ‘tailed’ Sup35p fragments are also converted into [PSI+] through a [PIN+]-dependent pathway. This is because such fragments induce [PSI+] better than the analogous fragments not carrying the RVDLQACKLMIQYQRK ‘tail’ even in [PIN+] cells (e.g. compare ΔEcoRnoext and ΔEcoRext in [PIN+] columns of Figure 2), and the efficiency of [PSI+] induction by the ‘tailed’ proteins is increased by [PIN+] (e.g. compare [PIN+] and [pin–] columns for ΔEcoRext in Figure 2).
One possibility is that the ‘tail’ retards Sup35Np folding, thus shifting the balance from the stable native state to a meta-stable intermediate conformation prone to both degradation and [PSI+] seeding. Slow folding could be an intrinsic property of the ‘tailed’ Sup35Np fragment, or a consequence of its altered interaction with chaperones. Indeed, it appears that the RVDLQACKL MIQYQRK-‘tailed’ proteins are unstable because their level is very low relative to the level of complete and other truncated Sup35p constructs expressed from similar plasmids, even when the nonsense-mediated mRNA degradation pathway is disrupted (Kochneva-Pervukhova et al., 1998b; Figure 3). Because unfolded or partially folded proteins are prone to form aggregates that are usually highly disordered but occasionally are nucleated and extended into ordered fibrils, like amyloid (see Dobson and Karplus, 1999), we propose that the meta-stable conformation of Sup35Np is prone to form [PSI+] seeds through a [PIN+]-independent pathway. Indeed, Dobson and Karplus (1999) argue that the rapid folding of proteins is one of the major roles of chaperones and an important evolutionary development to minimize aggregation. A strong tendency of the ‘tailed’ Sup35Np to aggregate could allow it to form [PSI+] seeds in the absence of PinPIN+ crystallization centers. Alternatively, if it is Pinpin– that plays the active role in [PSI+] induction, the lack of interaction between partially unfolded Sup35Np and the Pinpin– inhibitor may cause [PIN+]-independence.
Materials and methods
Yeast strains
Derivatives of strain 74-D694 (MATa ade1-14UGA trp1-289 his3-Δ200 ura3-52 leu2-3,112) were used in [PSI+] induction experiments and to compare [PSI+]-related phenotypes. The original derivative was [psi–][PIN+] (Chernoff et al., 1995; Derkatch et al., 1996). Previously used weak (21) and strong (8) [PSI+][PIN+] derivatives were induced by overproduction of the complete Sup35p (Derkatch et al., 1997, 1998, 1999). Unless specifically mentioned, these [PSI+] derivatives were used throughout this study. [pin–] variants of [psi–] and [PSI+] derivatives were obtained on 5 mM GuHCl. [psi–][PIN+] and [psi–][pin–] derivatives of the Δupf1 mutant of 74-D694 (Derkatch et al., 1996) were used to overcome the degradation of messages with premature stop codons encoding Sup35p fragments. [psi–][PIN+] and [psi–][pin–] derivatives of strain 64-D697 (MATα ade1-14UGA trp1-289 lys9-A21 ura3-52 leu2-3,112; Derkatch et al., 1997) were used to determine the Pin status of 74-D694 derivatives (see below). [PSI+] 783/4c (MATa ade2-1UAA leu2-3,112 ura3-1 his3-11,15 SUQ5; kindly provided by C.R.Nierras) is a [PSI+] strain used by B.S.Cox (Eaglestone et al., 1999a).
Plasmids
The pGAL::SUP35 plasmid (Derkatch et al., 1996) is a YCp50-based (Rose et al., 1987) centromeric URA3 vector that contains the promoterless SUP35 gene under the control of the inducible GAL1 (GAL) promoter.
pEMBL-SUP35, pEMBL-ΔBcl, pEMBL-ΔBal2 (here called pEMBL-ΔBalext) and pEMBL-ΔEcoR (here called pEMBL-ΔEcoRext) are pEMBL-yex4-based (Cesarini and Murray, 1987) 2µ plasmids containing either the complete SUP35 gene (pEMBL-SUP35) or SUP35 deletion alleles controlled by the original SUP35 promoter (Ter-Avanesyan et al., 1993) that were used previously to study the induction of [PSI+] in [pin–] and/or [PIN+] backgrounds (Derkatch et al., 1996, 1997; Kochneva-Pervukhova et al., 19998b). pEMBL-ΔEcoRnoext was constructed by inserting the PvuII–EcoRV fragment containing the SUP35 promoter and the first 112 codons into the SmaI site of pEMBL-yex4 followed by BamHI digestion, Klenow fill-in and religation. The URA3 marker was used to select for transformants and to maintain the plasmids.
Four new series of vectors bearing SUP35 or its fragments were constructed on the base of URA3-bearing centromeric and 2µ plasmids pFL38 and pFL44 (Bonneaud et al., 1991), respectively. pFL38-SUP35 contains the complete SUP35 gene on a PvuII–EcoRI fragment of yeast DNA inserted into pFL38 digested with SmaI and EcoRI. pFL38-ΔBal-lacZ and pFL38-ΔEcoR-lacZ were constructed by inserting the PvuII–BalI and PvuII–EcoRV fragments of yeast DNA containing the SUP35 promoter and the first 154 and 112 codons, respectively, into the SmaI site of pFL38. pFL38-ΔBcl-lacZ carries the truncated SUP35 ending at codon 239 on a PvuII–BclI fragment of chromosomal DNA. To construct this plasmid, the pYCH-U2 vector (Chernoff et al., 1992) was digested with BamHI and BclI, and self-ligated. The resulting construct was linearized with EcoRI, filled-in with Klenow and self-ligated, the ligation product (pID24) was digested with PvuII and the smaller fragment was inserted into the pFL38 plasmid cut with PvuII and SmaI. In all three plasmids, pFL38-ΔBcl-lacZ, pFL38-ΔBal-lacZ and pFL38-ΔEcoR-lacZ, the 129 amino acid segment of the lacZ gene present in the pFL vectors was fused to the 3′ termini of the SUP35 fragments. pFL38-ΔBcl, pFL38-ΔBal and pFL38-ΔEcoR were constructed by digesting the pFL38-ΔBcl-lacZ, pFL38-ΔBal-lacZ and pFL38-ΔEcoR-lacZ plasmids with AvaI, EcoRI and EcoRI, respectively, and religating following Klenow fill-in. pFL44-SUP35, pFL44-ΔBcl-lacZ, pFL44-ΔBal-lacZ, pFL44-ΔEcoR-lacZ, pFL44-ΔBcl, pFL44-ΔBal and pFL44-ΔEcoR are identical to the respective pFL38-based constructs except that they are 2µ based.
The pYS-GAL104 plasmid (Lindquist and Kim, 1996) is a centromeric URA3 vector containing the HSP104 gene under the control of the GAL promoter. The YEp13-SUP45 plasmid (Chernoff et al., 1992) contains the 4.9 kb BamHI–HindIII insert bearing a functional SUP45 fragment under the original promoter cloned into a 2µ LEU2 vector, YEp13 (Broach and Hicks, 1980).
All DNA manipulations were according to standard protocols (Sambrook et al., 1989). Sequencing used to confirm the structure of plasmids was performed by Lark Inc. (University of Florida) and by the University of Chicago Cancer Research Center DNA Sequencing Facility.
Media and cultivation procedures
Standard yeast media, cultivation procedures and genetic methods were used (Sherman et al., 1986; Rose et al., 1990). Unless specifically mentioned, yeast were grown on organic complete glucose medium (YPD) and at 30°C. YPD containing 1 or 5 mM GuHCl was used to cure prions (Tuite et al., 1981). Transformants were grown on synthetic glucose media selective for plasmid maintenance (e.g. SD-Ura). Synthetic media selective for plasmid maintenance and containing 2% galactose as the single carbon source (e.g. SGal-Ura) were used for the induction of the GAL promoter in transformants bearing pYS-GAL104 or pGAL::SUP35. Synthetic media lacking adenine and containing glucose or ethanol (2%) as the single carbon source (SD-Ade or SEt-Ade) were used for the selection of [PSI+] derivatives and for suppression analyses.
[PSI+]-related phenotypes
Suppression of the ade1-14 and ade2-1 nonsense mutations was estimated from growth at 20 and/or 30°C on SD-Ade and SEt-Ade and from the color on YPD, because, in addition to adenine auxotrophy, ade1 and ade2 mutations cause the accumulation of red pigment. The better the growth on adenineless media and the lighter the color on YPD, the stronger the suppression. Weak [PSI+] derivatives are pink on YPD and grow poorly on adenineless media (growth can be detected only at 20°C or on SEt-Ade). Strong [PSI+] derivatives are white on YPD and grow well on both SD-Ade and SEt-Ade. [psi–] derivatives are red and do not grow on adenineless media.
Analysis of Sup35p aggregation was according to Patino et al. (1996). Growth inhibition caused by Sup35p overproduction in [PSI+] derivatives and the rescue of this growth inhibition by Sup45p overproduction were scored exactly as in Derkatch et al. (1998). The incompatibility of [PSI+] variants with the sup35-2 nonsense allele was analyzed as described previously (Liebman and All-Robyn, 1984; Zhou et al., 1999) by crossing the [PSI+] derivatives of 74-D694 with the sup35-2-bearing strain, SL555-10C. The stability and suppressor phenotypes of weak and strong [PSI+]s overexpressing the PNM2 allele of SUP35 were analyzed in transformants carrying the YEp-PNM2 plasmid exactly as described in Derkatch et al. (1999).
[PSI+] curing
HSP104 overexpression was used to cure [PSI+] without affecting the Pin status of the cells. [PSI+] derivatives were transformed with pYS-GAL104. Three transformants of each [PSI+] derivative were replica plated to SGal-Ura and then colony purified on YPD. Usually >50% of the colonies were red, indicative of [PSI+] loss, and some were also unable to grow on SD-Ura, indicative of plasmid loss. Two Ura– [psi–] derivatives from each transformant were selected for further experimentation.
A ‘curing test’ on 5 mM GuHCl was used to demonstrate that the presence of [PSI+] was responsible for the Ade+ phenotype. Ade+ papillae were picked from adenineless media, colony purified on YPD, and two colonies were patched on YPD and replica plated twice onto 5 mM GuHCl. Patches that were white or pink on YPD but turned red on 5 mM GuHCl medium were colony purified on YPD. The presence of a significant proportion (usually >50%) of red colonies after GuHCl was indicative of a curable [PSI+] suppressor. Parallel replica plating on YPD and subsequent colony purification were used to confirm the relative stability of the Ade+ phenotype in the absence of GuHCl.
When indicated, 1 mM GuHCl was used to eliminate weak [PSI+]s. Independent clones were grown on 1 mM GuHCl for ∼14 cell generations and then colony purified on YPD. Approximately 10% of the progeny were [psi–] (red). One [psi–] colony from each culture was selected for further analysis. Spontaneous [psi–] derivatives were selected analogously except that YPD was used throughout the experiment.
[PSI+] induction
The de novo induction of [PSI+] by overproduction of Sup35p and its fragments was performed as previously described (Derkatch et al., 1996). Transformants were grown in patches on SD-Ura (or on SGal-Ura, if pGAL::SUP35 was used) for ∼14 cell generations. Patches were then replica plated onto SD-Ade and SEt-Ade. Growth or intensive papillation on adenineless media was indicative of [PSI+] induction. Curing of several [PSI+] isolates on 5 mM GuHCl was used to confirm the presence of [PSI+]. Experiments testing low-level [PSI+] induction, where the number of Ade+ colonies did not increase significantly, are described in the last section of Materials and methods.
Determination of the Pin status of [psi–] and [PSI+] derivatives
To determine the Pin status of [psi–] derivatives, they were crossed to [psi–][pin–] 64-D697-bearing pGAL::SUP35. Diploids selected on SD-Ura,Lys,His were replica plated twice to SGal-Ura,Lys,His (to induce Sup35p overproduction) and then to SD-Ade and SEt-Ade (to score for the appearance of [PSI+]). Derivatives that produced diploids that grew or heavily papillated on adenineless media in this experiment, but not in the control experiment where incubations were on non-inducing SD-Ura,Lys,His, were scored as [PIN+]. Derivatives that in a cross with [psi–][pin–] 64-D694 produced diploids that remained Ade– following Sup35p overproduction were scored as [pin–], but only after confirming that these derivatives produced [PSI+]-inducing diploids when crossed to [psi–][PIN+] 64-D694. The validity of this test was confirmed by transforming a group of derivatives defined as [PIN+] or [pin–] with pGAL::SUP35 and pEMBL-SUP35 and testing their Pin status directly (e.g. all spontaneous and 1 mM GuHCl-induced [psi–] derivatives described in the first section of Results were tested in both ways). [PSI+] derivatives were cured of [PSI+] by HSP104 overexpression (see above) prior to determination of their Pin status.
Quantitative analysis of [PSI+] and [PIN+] curing
Three fresh colonies of the indicated 74-D694 derivatives were picked from YPD and suspended in water. Then 106 cells were spotted onto 5 mM GuHCl (three plates, one spot per plate) and incubated for 7.4 cell generations (unless otherwise stated). Cells from each plate were then washed off and spread on three YPD plates at ∼300 cells per plate. The [psi–] (red), [PSI+] (white or pink) and Pet– (small beige) colonies were counted. The Psi status of Pet– colonies was determined on SD-Ade. The samples chosen for Pin determination maintained the proportion of [psi–], [PSI+] and Pet– colonies in the whole culture. YPD was used instead of 5 mM GuHCl in control experiments.
Experiments involving prolonged incubations
pGAL::SUP35 transformants of [psi–][pin–] 74-D694 were patched on SD-Ura (each patch was approximately one-fifth of the plate), incubated at 30°C for 2 days, replica plated to SD-Ura, grown at 30°C for 2 days, transferred to 4°C and kept there for 1 month. Following this prolonged incubation, cells were replica plated to SGal-Ura and then to SD-Ade and SEt-Ade. All Ade+ colonies were subjected to the ‘curing test’ to check if the Ade+ phenotype was due to the appearance of [PSI+] (see above), and the Pin status of the newly induced [PSI+] derivatives was determined (see above). Control experiments involved ‘no incubation’, in which the prolonged incubation at 4°C was omitted, ‘no induction’, in which yeast were transferred to the non-inducing SD-Ura medium instead of to SGal-Ura, and ‘no overproduction’, in which control vector, YCp50, was substituted for GAL::SUP35. The conclusion that the appearance of [PSI+] was induced in an experiment was made only if significantly more colonies were Ade+ due to the presence of [PSI+] in the experimental data compared with the control. Variations of the standard experiment are described in Results (see also Table III footnotes).
Acknowledgments
Acknowledgements
We thank V.Prapapanich for kindly providing the M12 anti-Sup35p antibody, for expert assistance with protein analysis and for participation in experiments. We also thank J.Moore and C.Phillips for help with experiments. We are grateful to M.D.Ter-Avanesyan, M.Nierras, M.F.Tuite, M.Patino and S.Lindquist for providing plasmids, a strain and an antibody, to Y.O.Chernoff for permission to refer to his unpublished observations, and to Y.O.Chernoff, M.F.Tuite, C.Jeffery and R.B.Wickner for helpful discussions. This work was partially supported by grants from National Institutes of Health (GM56350), the Alzheimer’s Association, the Civilian Research and Development Foundation (RB1-238) and RFBR (99-04-49601).
References
- Bonneaud N., Ozier-Kalogeropoulos,O., Li,G.Y., Labouesse,M., Minvielle-Sebastia,L. and Lacroute,F. (1991) A family of low and high copy replicative, integrative and single-stranded S.cerevisiae/E.coli shuttle vectors. Yeast, 7, 609–615. [DOI] [PubMed] [Google Scholar]
- Broach J.R. and Hicks,J.B. (1980) Replication and recombination functions associated with the yeast 2 micron circle. Cell, 21, 501–508. [DOI] [PubMed] [Google Scholar]
- Bronstein I., Olesen,C.E.M., Martin,C.S., Schneider,G., Edwards,B., Sparks,A. and Voyta,J.C. (1994) Chemiluminescent detection of DNA and protein with CDP™ and CDP-Star™ 1,2-dioxetane enzyme substrates. In Campbell,A.K. et al. (eds), Bioluminescence and Chemiluminescence: Fundamentals and Applied Aspects. Wiley, New York, NY, pp. 269–272. [Google Scholar]
- Cesarini G. and Murray,A.H. (1987) Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In Setlow,J.K. (ed.), Genetic Engineering: Principles and Methods. Plenum Press, New York, NY, Vol. 4, pp. 135–154. [Google Scholar]
- Chernoff Y.O., Inge-Vechtomov,S.G., Derkach,I.L., Ptyushkina,M.V., Tarunina,O.V., Dagkesamanskaya,A.R. and Ter-Avanesyan,M.D. (1992) Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast, 8, 489–499. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Derkach,I.L. and Inge-Vechtomov,S.G. (1993) Multicopy SUP35 gene induces de novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet., 24, 268–270. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist,S.L., Ono,B.-I., Inge-Vechtomov,S.G. and Liebman,S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science, 268, 880–884. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Galkin,A.P., Lewitin,E., Chernova,T.A., Newman,G.P. and Belenkiy,S.M. (2000) Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol. Microbiol., 35, 865–876. [DOI] [PubMed] [Google Scholar]
- Coustou V., Deleu,C., Saupe,S. and Begueret,J. (1997) The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl Acad. Sci. USA, 94, 9773–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B.S. (1994) Prion-like factors in yeast. Curr. Biol., 4, 744–748. [DOI] [PubMed] [Google Scholar]
- Cox B.S., Tuite,M.F. and McLaughlin,C.S. (1988) The psi factor of yeast: a problem of inheritance. Yeast, 4, 159–178. [DOI] [PubMed] [Google Scholar]
- DePace A.H., Santoso,A., Hillner,P. and Weissman,J.S. (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell, 93, 1241–1252. [DOI] [PubMed] [Google Scholar]
- Derkatch I.L., Chernoff,Y.O., Kushnirov,V.V., Inge-Vechtomov,S.G. and Liebman,S.W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics, 144, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M.E., Zhou,P., Chernoff,Y.O. and Liebman,S.W. (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics, 147, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M.E. and Liebman,S.W. (1998) Overexpression of the SUP45 gene encoding a Sup35-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc. Natl Acad. Sci. USA, 95, 2400–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M.E., Zhou,P. and Liebman,S.W. (1999) The PNM2 mutation in the prion protein domain of SUP35 has distinct effects on different variants of the [PSI+] prion in yeast. Curr. Genet., 35, 59–67. [DOI] [PubMed] [Google Scholar]
- Dobson C.M. and Karplus,M. (1999) The fundamentals of protein folding: bringing together theory and experiment. Curr. Opin. Struct. Biol., 9, 92–101. [DOI] [PubMed] [Google Scholar]
- Doel S.M., McCready,S.J., Nierras,C.R. and Cox,B.S. (1994) The dominant PNM2– mutation that eliminates the ψ factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics, 137, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S.S., Cox,B.S. and Tuite,M.F. (1999a) Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-related mechanism. EMBO J., 18, 1974–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S.S., Ruddock,L.W., Cox,B.S. and Tuite,M.F. (1999b) Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara K. and Nakamura,Y. (1999) C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA, 5, 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eurwilaichitr L., Graves,F., Stansfield,I. and Tuite,M.F. (1999) The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol., 32, 485–496. [DOI] [PubMed] [Google Scholar]
- Frolova L., Le Goff,X., Zhouravleva,G., Davydova,E., Philippe,M. and Kisselev,L. (1996) Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA, 2, 334–341. [PMC free article] [PubMed] [Google Scholar]
- Glover J.R., Kowal,A.S., Schirmer,E.C., Patino,M.M., Liu,J.J. and Lindquist,S. (1997) Self-seeded fibers formed by Sup35, the prion determinant of [PSI+], a heritable prion-like factor of S.cerevisiae. Cell, 89, 811–819. [DOI] [PubMed] [Google Scholar]
- Herskowitz I.H. (1977) Principles of Genetics. Macmillan Publishing Co. and Collier Macmillan Publishers, New York, NY. [Google Scholar]
- Horwich A.L. and Weissman,J.S. (1997) Deadly conformations—protein misfolding in prion disease. Cell, 89, 499–510. [DOI] [PubMed] [Google Scholar]
- King C.Y., Tittmann,P., Gross,H., Gebert,R., Aebi,M. and Wuthrich,K. (1997) Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl Acad. Sci. USA, 94, 6618–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N.V., Paushkin,S.V., Kushnirov,V.V., Cox,B.S., Tuite,M.F. and Ter-Avanesyan,M.D. (1998a) Mechanism of inhibition of Psi+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J., 17, 5805–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N.V., Poznyakovski,A.I., Smirnov,V.N. and Ter-Avanesyan,M.D. (1998b) C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+] determinant in Saccharomyces cerevisiae. Curr. Genet., 34, 146–151. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V., Ter-Avanesyan,M.D., Telckov,M.V., Surguchov,A.P., Smirnov,V.N. and Inge-Vechtomov,S.G. (1988) Nucleotide sequence of the SUP2 (SUP35) gene of Saccharomyces cerevisiae. Gene, 66, 45–54. [DOI] [PubMed] [Google Scholar]
- Liebman S.W. and All-Robyn,J.A. (1984) A non-Mendelian factor, [eta+], causes lethality of yeast omnipotent-suppressor strains. Curr. Genet., 8, 567–573. [DOI] [PubMed] [Google Scholar]
- Liebman S.W. and Derkatch,I.L. (1999) The yeast [PSI+] prion: making sense of nonsense. J. Biol. Chem., 274, 1181–1184. [DOI] [PubMed] [Google Scholar]
- Lindquist S. (1997) Mad cows meet psi-chotic yeast: the expansion of the prion hypothesis. Cell, 89, 495–498. [DOI] [PubMed] [Google Scholar]
- Lindquist S. and Kim,G. (1996) Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc. Natl Acad. Sci. USA, 93, 5301–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.J. and Lindquist,S. (1999) Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature, 400, 573–576. [DOI] [PubMed] [Google Scholar]
- McCready S.J., Cox,B.S. and McLaughlin,C.S. (1977) The extra chromosomal control of nonsense suppression in yeast: an analysis of the elimination of [psi+] in the presence of a nuclear gene PNM–. Mol. Gen. Genet., 150, 265–270. [DOI] [PubMed] [Google Scholar]
- Newnam G.P., Wegrzyn,R.D., Lindquist,S.L. and Chernoff,Y.O. (1999) Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol., 19, 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino M.M., Liu,J., Glover,J.R. and Lindquist,S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1997a) Interaction between yeast Sup45p (eRF1) and Sup35 (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol. Cell. Biol., 17, 2798–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1997b) In vitro propagation of the prion-like state of yeast Sup35 protein. Science, 277, 381–383. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B., Scott,M.R., DeArmond,S.J. and Cohen,F.E. (1998) Prion protein biology. Cell, 93, 337–348. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Novick,P., Thomas,J.H., Botstein,D. and Fink,G.R. (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene, 60, 237–243. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Heiter,P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Santoso A., Chien,P., Osherovich,L.Z. and Weissman,J.S. (2000) Molecular basis of a yeast prion species barrier. Cell, 100, 277–288. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Hicks,J.B. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Stansfield I. et al. (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J., 14, 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Kushnirov,V.V., Dagkesamanskaya,A.R., Didichenko,S.A., Chernoff,Y.O. and Inge-Vechtomov,S.G. (1993) Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol., 7, 683–692. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Dagkesamanskaya,A.R., Kushnirov,V.V. and Smirnov,V.N. (1994) The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae.Genetics, 137, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M.F., Mundy,C.R. and Cox,B.S. (1981) Agents that cause a high efficiency of genetic change from [psi+] to [psi–] in Saccharomyces cerevisiae. Genetics, 98, 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science, 264, 566–569. [DOI] [PubMed] [Google Scholar]
- Wickner R.B. (1997) A new prion controls fungal cell fusion incompatibility. Proc. Natl Acad. Sci. USA, 94, 10012–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B. and Chernoff,Y.O. (1999) Prions of fungi: [URE3], [PSI], and [HET-s] discovered as heritable traits. In Prusiner,S. (ed.), Prion Biology and Diseases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 229–272. [Google Scholar]
- Wickner R.B., Masison,D.C. and Edskes,H.K. (1995) [PSI] and [URE3] as yeast prions. Yeast, 11, 1671–1685. [DOI] [PubMed] [Google Scholar]
- Wickner R.B., Edskes,H.K., Maddelein,M.-L., Taylor,K. and Moriyama,H. (1999) Prions of yeast and fungi: proteins as genetic material. J. Biol. Chem., 274, 555–558. [DOI] [PubMed] [Google Scholar]
- Wilson P.G. and Culbertson,M.R. (1988) SUF12 suppressor protein of yeast. A fusion protein related to the EF-1 family of elongation factors. J. Mol. Biol., 199, 559–573. [DOI] [PubMed] [Google Scholar]
- Zhou P., Derkatch,I.L., Patino,M., Uptain,S., Lindquist,S. and Liebman,S.W. (1999) The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J., 18, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhouravleva G., Frolova,L., LeGoff,X., LeGuellec,R., Inge-Vechtomov,S.G., Kisselev,L. and Philippe,M. (1995) Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J., 14, 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]