Introduction
The meeting held at the Irsee Convent in Southern Germany was organized by Hans-Henning Arnold (Braunschweig), Renate Renkawitz-Pohl (Marburg), Anna Starzinski-Powitz (Frankfurt) and Bodo Christ (Freiburg). A total of 150 participants, 47 of whom were invited speakers, created an atmosphere of intense progress in a multifaceted field of research. This comprised somite patterning and determination of cell lineages as a paradigm of muscle precursor cell specification, cardiac myogenesis, signals involved in myogenesis, differentiation and the control of muscle-specific gene expression, as well as the molecular genetics of myopathies and repair processes. Eric Olson (Dallas, TX) gave the keynote address on the transcriptional control of myogenesis.
More signals and complexity: the somites
The paraxial mesoderm is the source of cells that can differentiate into skeletal muscle. It is the primary structure in the vertebrate body showing segmentation when the somites form in a periodic sequence of cranio-caudal subdivisions from the presomitic mesoderm in mice and the segmental plate in birds. Three years ago, Pourquié’s group (Palmeirim et al., 1997) presented the first experimental evidence for a periodic molecular event prefiguring the synchronous formation of subsequent somite pairs. They described the oscillating expression of c-hairy1 in the segmental plate prior to somite formation. Such an oscillation had been postulated in theoretical models of somitogenesis. While an ortholog of c-hairy1 in mouse has not been found yet, new oscillating genes have been described and attempts are being made to arrange them in a sequence of events. The most prominent gene with oscillating expression is lunatic fringe, an ortholog of Drosophila fringe, which modulates the Delta-Notch pathway (O.Pourquié, Marseille, France; R.Johnson, Houston, TX). Mice homozygous for a targeted deletion in lunatic fringe lack caudal somites, and markers of a cranio-caudal polarization such as uncx 4.1 are expressed irregularly (Evrard et al., 1998; Pourquie, 1999). It appears that lunatic fringe might act downstream of c-hairy1, because it requires protein synthesis for its dynamic expression, while c-hairy1 expression is unaffected by cycloheximide treatment.
Two sets of bHLH-transcription factors were also shown to be expressed during vertebrate somitogenesis: one includes cMeso1 and cMeso2 in birds (Buchberger et al., 1998) and the other consists of MesP1 and MesP2 in mice (Saga et al., 1997). cMeso1 has an important role in somite formation since interference with cMeso1 transcript expression by antisense oligonucleotides results in attenuation of somite formation and caudal shortening of the trunk. Interestingly, cMeso1 and cMeso2 differ in function: cMeso1 has a transcription activation domain at the C-terminus and a repressor domain at the N-terminus, whereas cMeso2 only has an N-terminal repressor domain. Like cMeso1 and 2, MesP1 and MesP2 are located in close neighborhood on the same chromosome. Double-deficient mice completely lack paraxial mesoderm. MesP2 functions during normal segmentation and during the establishment of rostro-caudal polarity of the somites by regulating the expression of Dll (Delta-like). MesP2 appears to be regulated by the molecular clock. MesP and cMeso genes share some homology; however, whether they are true orthologs remains to be shown. Genes distantly related to MesP and c-Meso are Mespo in Xenopus (Joseph and Cassetta, 1999) and mesogenin in mouse, which are both expressed in the segmental plate. B.Wold (Pasadena, CA) reported that overexpression of mesogenin leads to loss of skeletal muscle differentiation, suggesting an inhibitory function of Mesogenin in this process.
A complex event in somite patterning is the formation of the myotome. Using fluorescent vital dyes, C.Ordahl (San Francisco, CA) and co-workers labeled the dermomyotome to investigate the origin and dynamics of early epaxial and hypaxial muscle formation. They found that precursor cells of the myotome are located in the dorso-medial and ventro-lateral lips of the dermomyotome. From there, they become dislocated to the central portion of the myotome as a consequence of the rapid expansion of the dermomyotome in a dorso-ventral direction. The first myotome cells to form are the ones giving rise to the epaxial myotome (Kaehn et al., 1988). The hypaxial portion of the myotome forms basically in the same way, but with an initial lag phase, from ventro-lateral precursor cells. The birth sequence of continuously forming myotome cells is later reflected by the more superficial location of the early arising cells in the central portion of the myotome and the deeper location of later formed cells.
The signals that specify somitic compartments are now more or less well known, but the signaling cascades and downstream events remain to be elucidated in detail. As far as muscle development is concerned, the muscle precursors are specified by signals from the neural tube and the ectoderm. B.Christ (Freiburg, Germany) reported on the upregulation of MyoD by Wnt1 in the chick embryo. The effect of the other important signal, SHH, differs with regard to epaxial and hypaxial muscle. SHH drives epaxial muscle cells of compartmentalized avian somites into differentiation, whereas it seems to maintain hypaxial-derived muscle in the limb in the proliferative state (Amthor et al., 1999). A.Borycki (Philadelphia, PA) coupled Wnt and SHH signaling in the somites by showing that Wnt1 is able to selectively activate gli genes, which are components of the SHH signal transduction pathway.
From somite to limb
Muscle forms not only at the site where precursor cells originate, but also at distant sites after their migration. One such site is the limb bud. The signal leading to de-epithelialization of the dermomyotomes at limb bud level is scatter factor/hepatocyte growth factor (SF/HGF). An additional function for SF/HGF was reported, which explains its persisting expression in developing limb buds: SF/HGF keeps myogenic precursor cells motile while suppressing myoD expression (Scaal et al., 1999). SF/HGF itself is induced by signals from the apical ectodermal ridge (AER) that can be mimicked by FGF2, and inhibited by signals from the zone of polarizing activity (ZPA), possibly BMP2.
Another key regulator of myogenic precursor cell migration into the limb buds is the homeobox gene lbx1, which is expressed by migrating cells. Mice targeted with a disrupted lbx1 gene present with a distinct limb muscle defect: in forelimbs, distal muscle groups are absent only on the extensor side, while in hindlimbs muscles are completely lacking (Schäfer and Braun, 1999). Hence, there seem to be differences in the development of individual muscle groups, a finding which is also reflected by results obtained after Mox2 targeting (P.Rigby, London, UK). Mox2 is expressed in emigrating muscle precursor cells where it follows the expression of Mox1 in the lateral dermomyotome. In knockout mice, an overall reduction in muscle mass is notable with specific muscles absent, especially in the forelimbs. The phenotype can be traced back to early stages of muscle formation and could be due to an incorrect splitting of muscle primordia (Mankoo et al., 1999).
Mending muscles
Muscle repair or regeneration has a program of gene expression that has many similarities to muscle development. A class of quiescent muscle precursor cells called satellite cells, which are situated under the basal lamina, reconstitute skeletal muscle. Until recently, electron microscopy was the only way to identify these cells. However, in the last few years it has become accepted that M-Cadherin is a molecular marker of the quiescent satellite cell (T.Partridge, London, UK).
By isolating single fast muscle fibers from two different strains of mice, myf5nlacZ and 3F-nlacZ-2E (developed in M.Buckingham’s laboratory), T.Partridge elegantly demonstrated that satellite cells express, in addition to M-Cadherin, very low levels of the myf5 transgene, myoD and cd34. Furthermore, in this meeting, evidence was presented (M.Rudnicki, Hamilton, Canada; R.Cooper, Paris, France) for the existence of two different pathways for satellite cell activation: one via myoD and the other via myf5 (Cooper et al., 1999). The satellite cells that express myf5 but not myoD show an increased propensity for cell renewal, and representational difference analysis is currently being used (M.Rudnicki) to identify genes that are involved in the regulation of myoblast identity and satellite cell activation.
What exactly triggers satellite cell activation? A.Starzinski-Powitz (Frankfurt, Germany) has previously proposed that disruption of the M-Cadherin contact, which exists between satellite cells, and the myofiber could be the mechanical trigger that results in satellite cell activation. In this session, J.Anderson (Winnipeg, Canada) showed that NO is also involved in satellite activation and cell fusion. In the present model, satellite cell activation is characterized by satellite cell hypertrophy, decreased adhesion to the myotubes and co-localization of c-Met and SF/HGF. All of these properties were demonstrated to be dependent on nitric oxide synthase activity resulting in loss of M-Cadherin’s adhesion properties and satellite cell activation.
Any potential mechanical or chemical (NO) signal received by the satellite cell during activation has to be transmitted to the nucleus. It has been suggested that M-Cadherin might be involved in this process since it forms a complex with the catenins, which could form a signal transduction network to transmit information to the nucleus. A.Starzinski-Powitz’s group has recently used a yeast two-hybrid system to identify proteins other than the classical catenins that interact with the cytoplasmic domain of M-Cadherin. A very interesting candidate gene, ARVCF, an armadillo protein that is deleted in velo-cardio-facial syndrome, has been isolated and shown to be a novel binding partner of the cadherins. The exogenously expressed protein is usually localized at the cell membrane, but was found in the nucleus of some but not all skeletal and cardiac muscle cells.
Mice homozygous for a null deletion of α7β1 integrin, described by U.Mayer (München, Germany), are viable and fertile, indicating that α7β1 integrin is not essential for myogenesis. However, a progressive muscular dystrophy appeared with age, indicating that α7β1 integrin is essential to maintain muscle integrity. Ultrastructural studies have confirmed that α7β1 integrin is the major adhesion molecule connecting the muscle fiber to the tendon and is essential to maintain the mechanical stability of the myotendinous junction. N.Rosenthal (Boston, MA) discussed the role of insulin-like growth factor-1 (IGF-1) in muscle differentiation and regeneration. Transgenic animals harboring a transgene that express a muscle-specific isoform of IGF-1 under the control of the MLC1/3 promoter displayed hypertrophied fast muscle fibers. Transgenic animals showed increased expression of calcineurin and GATA-2, which is a novel marker of hypertrophied skeletal muscle. IGF-1 might be utilized in the future for the treatment of muscle atrophy and degeneration (Musaro et al., 1999).
In the session on the genetics of myopathies there were five completely different presentations. M.Fiszman (Paris, France) gave a very complete analysis of the mutations in structural contractile proteins that result in cardiac hypertrophy (familial hypertrophic cardiomyopathy). Mutations in nuclear membrane proteins, although ubiquitously expressed, can provoke both cardiomyopathies as well as muscular dystrophies (Emery-Dreyfuss Muscular Dystrophy). H.Jockusch (Bielefeld, Germany) described a novel system in which fibroblasts that had been previously transfected with myoD were injected under the kidney capsule where they formed ‘mini muscles’. This could provide a possible tool for both the analysis and therapy of muscle disease. The knock out of the DP71 gene (U.Nudel, D.Yaffe, Rehovot, Israel), which is the most abundant and ubiquitous non-muscle product of the dystrophin gene, revealed no pathological effect, as did the experimental deletion of the CAPN3 (calpain) gene (J.Beckmann, Paris, France). In humans, a defect in the calpain gene results in type 2A limb girdle muscular dystrophy. This gene is now thought to be involved in the regulation of cell survival since there is a deregulation of the NFκB pathway resulting in apoptosis of the myofibers. Finally, T.Cooper (Houston, TX) reported on a novel pathological mechanism for myotonic dystrophy (DM). This disease is caused by a CUG expansion in the 3′ untranslated region of the DM gene (Philips et al., 1998). Alternative splicing of the human troponin T pre-mRNA is regulated by a CUG-binding protein and, interestingly, aberrant splicing of troponin T was observed in DM striated muscle and in normal cells expressing transcripts that contain CUG repeats.
Gene regulation in skeletal muscle
Even though mouse mutants in either MyoD or myf5 develop apparently normal muscles, there is growing evidence for distinct functions of the four different members of the MyoD family. M.Rudnicki (Hamilton, Canada) showed that myf5 and MyoD activate overlapping but non-identical different sets of target genes, indicating functional differences. Similarly, in embryoid bodies extra copies of MyoD cannot substitute for Myogenin in myoblast fusion (W.Klein, Houston, TX). S.Tapscott (Seattle, WA) reported that MyoD and Myf5 are distinguishable from Myogenin because they preferentially activate endogenous genes while the latter activates transient reporters more efficiently. This difference is explained by the ability of MyoD to remodel chromatin structure and could be mapped to two domains within the MyoD protein that are also found in Myf5, but not in Myogenin (Gerber et al., 1997).
Acetylation of histones is now considered to be a major mechanism in gene regulation. The acetyltransferase p300 directly interacts with the transactivation domain of MyoD and might be an important bridge between MyoD, MEF2 and the RNA polymerase. The differentiation inhibitory activity of the adenoviral E1A protein is correlated with its ability to disrupt the complex of MyoD with p300 and yet another acetyltransferase, the p300/CBP-associating factor PCAF (Hamamori et al., 1999). At least for PCAF, it was shown that acetyltransferase activity is required for its synergistic effect on MyoD. The first target for this activity is the MyoD gene itself, which is acetylated at multiple sites, several of which are conserved within the MRF family (L.Kedes, Los Angeles, CA). In Xenopus, histone acetylation by p300 and PCAF seems to initiate the phase of ‘muscle competence’, which is confined to a period of ∼90 min from the end-blastula to the mid-gastrula stage (Steinbach et al., 1998). Their enzymatic activity is required for MyoD expression, facilitating stable MyoD autocatalysis. The competence period ends with a change from maternal histones to somatic H1 linker histones, which silences the transcriptional responsiveness of the MyoD gene to growth factor signaling (R.Rupp, Tübingen, Germany).
Considerable progress has been made in investigating the regulation of myf5 expression; however, the case seems to become more and more complex, the closer we look. P.Rigby and D.Summerbell have identified a number of separate enhancers, which are dispersed throughout a 14 kb region spanning both Myf5 and MRF4, but which is still much smaller than the >50 kb required to truly mimic Myf5 expression. Yet, transcriptional control is only part of the story. M.Buckingham (Paris, France) reported that no Myf5 protein could be detected in differentiating neurons, which by means of activation of a Myf5-nlacZ knock-in are clearly identified as myf5-expressing cells. As in the somite, Myf5 transcription in these neurons is Wnt1 responsive. When cultured in vitro, such neurons can display myogenic potential (Daubas et al., 2000). The lack of Myf5 protein could, thus, be due to regulation at the level of mRNA translation or protein stability. As demonstrated by D.Montarras, C.Pinset and their co-workers (Paris, France), Myf5 protein is cell cycle regulated and specifically degraded during mitosis (Lindon et al., 1998). This cell-cycle-specific degradation of Myf5 is independent of the normal turnover and requires a putative specific ‘destruction box’ in the protein.
Drosophila contains ∼30 multinucleated muscle fibers per hemisegment, which are defined by shape, position and insertion into the epidermal attachment sites. This pattern is prefigured by the selection of muscle progenitors, in which Wg (Wingless) and Dpp (Decapentaplegic) as secreted factors play essential roles. Once the cell fate is established, the pattern is maintained by lateral inhibition via Notch signaling. When muscle progenitors are established, they divide and give rise to different muscle precursors, which are characterized by the expression of distinct transcription factors (e.g. Eve, S59, Nau and Kr) activated by the above-mentioned signal transduction cascade (Frasch, 1999; Paululat et al., 1999a). These cells fuse with 1–2 neighboring myoblasts to definite muscle precursor cells. Successive fusion of fusion-competent myoblasts to these precursor cells gives rise to syncytial myofibers. This requires cell recognition, alignment, formation of prefusion complexes and membrane breakdown (recently reviewed by Paululat et al., 1999b). By genetic analysis, several genes could be defined that are essential for the formation of syncytial muscle fibers. R.Renkawitz-Pohl (Marburg, Germany) discussed the possible role of the genes rolling stone and rolling pebbles, both of which are essential for myoblast fusion. The rolling stone gene encodes an integral membrane protein, which is expressed in a subset of developing muscle fibers. For one of the above-mentioned transcription factors, Nautilus, the MyoD homolog, direct binding to the rolling stone promoter was demonstrated. The presumptive rolling pebbles gene is expressed in muscle precursor cells. For surrounding fusion-competent myoblasts, S.Abmayr (Pennsylvania State University) reported the analysis and expression pattern of sticks and stones, which encodes a member of the nephrin class of cell adhesion molecules. Thus, we start to get insight into different components of this cell-type-specific fusion mechanism.
Molecular basis of heart development
In Drosophila, the NK-2 class homeobox gene tinman (tin) is expressed in cardiac and visceral mesodermal progenitors and is essential for their specification. In vertebrates, the tin homolog Nkx2.5 and related genes are expressed in early cardiac and visceral mesodermal progenitors. In his keynote address, E.Olson (Dallas, TX) reported on the analysis of an early cardiogenic function for Nkx2.5. The mouse gene was introduced into the Drosophila germline, and tested for its ability to rescue the tin mutant phenotype. While Tin itself strongly rescued both heart and visceral mesoderm, Nkx2.5 rescued only visceral mesoderm. The cardiogenic domain of Tin was mapped to a unique region at its N-terminus and, when transferred to Nkx2.5, this region conferred a strong ability to rescue heart (Ranganayakulu et al., 1998). A protein interacting with the N-terminus of Tin is the bHLH/leucine zipper protein Tinwoman, which is broadly expressed in mesoderm and partly overlaps with the tin expression domain.
Members of the GATA class of transcription factors are also vital for vertebrate heart formation. The existence of a cardiac GATA factor in Drosophila has been implicated by the analysis of the cardiac enhancer of the D-mef2 gene. R.Schulz (Houston, TX) presented evidence that in Drosophila, pannier encodes the functional homolog of the vertebrate cardiac GATA factors (Gajewski et al., 1999). pannier is expressed in the dorsal mesoderm overlapping with the expression domain of tinman. Like tin, pannier expression is dependent on Dpp signals from the overlying ectoderm. pannier mutant embryos have a dorsal vessel phenotype and overexpression of pannier results in the formation of supernumerary cardiac cells at the expense of other derivatives of the dorsal mesoderm.
Signals in heart induction
M.Frasch (New York, USA) reported on the analysis of signaling components and cis-acting elements controlling tinman and bagpipe (bap) expression. The dorsal mesoderm enhancers of tin and bap contain both Mad/Medea and Tin binding sites (Xu et al., 1998). While the Mad binding sites mediate Dpp-dependent signaling, the Tin binding sites are important for mesoderm-specific induction of tin and bap. Furthermore, overlap of wg and dpp expression domains promotes induction of heart progenitors, while Wg counteracts induction of bap, which is thus restricted to segmental visceral mesoderm progenitors. The observed Wg effects are mediated by the Wg-dependent activation of the forkhead gene sloppy paired (slp) in striped mesodermal domains. Ectopic slp on its own results in a complete loss of bap expression and, if combined with ectopic wg, in ectopic heart progenitors. A.Michelson (Boston, MA) discussed the combinatorial activities of Wg, Dpp, receptor tyrosine kinase (RTK) and Notch (N) signaling pathways in the specification of particular muscle and cardiac progenitors in the Drosophila embryo. Whereas Wg and Dpp cooperate with the Ras/MAPK cascade in this developmental process, N antagonizes RTK signaling by suppressing MAPK activation. Moreover, evidence was presented that Wg, Dpp and Ras signals converge at the transcriptional level, thereby providing a molecular mechanism for signal integration during cardiogenesis and myogenesis in this model organism. Like Dpp in Drosophila, BMP2 is required in vertebrates for cardiac specification. In the chick, BMP2 is expressed in the pharyngeal endoderm underlying the precardiac mesoderm between stages 4 and 8. T.Brand (Braunschweig, Germany) showed that blocking of BMP signaling by implanting noggin-expressing cells into the heart field of embryos at stages 4–6 results in complete loss of Nkx2.5 expression, while both Nkx2.5 expression and tubular heart formation were impaired at stages 7 and 8 (Schlange et al., 2000). Interestingly, R.Bodmer (Ann Arbor, MI) observed that in Drosophila Dpp is required (in combination with Wg) during multiple developmental stages for a stepwise restriction of tin expression to the cardiac mesoderm and for formation of the dorsal vessel. As R.Schwartz (Houston, TX) reported, one of the inhibitory Smads, cSmad6 is abundantly expressed in the chick heart-forming regions and is induced by BMP and lost after Noggin treatment (Yamada et al., 1999). cSmad6, which is co-expressed with cSmad1, might modulate BMP signaling in the heart-forming regions by keeping a balance between constitutively expressed cSmad1 and ligand-induced inhibitory cSmad6.
The tubular heart
The helix–loop–helix protein MesP1 is expressed in the mesodermal cell lineage during gastrulation. As Y.Saga (Tokyo, Japan) reported, disruption of the MesP1 gene leads to failure of heart tube formation and the two heart tubes remain unfused (Saga et al., 1999). In the MesP1-deficient mice, a delayed migration of cardiac precursor cells from the primitive streak is probably the cause for the phenotype. Double null embryos for MesP1 and MesP2 displayed a complete lack of non-axial mesoderm. Interestingly, when introduced into wild-type embryos, cells lacking both MesP1 and MesP2 populated any tissue of the embryo except the heart. Thus, in addition to their function in early somite formation, MesP proteins are also required for early heart development. R.Harvey (Sydney, Australia) discussed the significance of Nkx2.5 function in the vertebrate heart. In contrast to the complete loss of dorsal vessel formation in tinman mutant embryos, null embryos for Nkx2.5 display a block of specification of the ventricles, as judged by the lack of expression of several genes and transgene markers. By use of a differential expression technique, a novel Nkx2.5-dependent clone named chisel has been isolated. Interestingly, chisel is absent from the primary myocardium at the tubular heart stage, but becomes expressed in those regions that will become the atrial appendages and ventricular outer curvature. Transfection of skeletal muscle cultures with chisel appears to have a pro-myogenic activity; however, further work is required to define the precise function of Chisel in vertebrate heart development.
MEF2 and cardiac hypertrophy
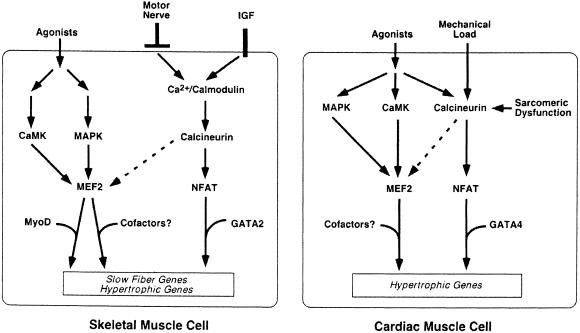
MEF2 proteins are targets for various signal transduction pathways both in skeletal and cardiac muscle (Figure 1). L.Megeney (Ottawa, Canada) reported that MEF2 is specifically phosphorylated by p38 MAP kinase, resulting in enhanced transcriptional activity (Kolodziejczyk et al., 1999). This phosphorylation is instrumental in both physiological and pathophysiological hypertrophy. Importantly, transgenic animals overexpressing a dominant-negative form of MEF2C display attenuated postnatal growth of the myocardium, suggesting that MEF2 is a nodal point for both physiological and hypertrophy signals. E.Olson (Dallas, TX) reported that both MAP kinases and CaM kinases can phosphorylate MEF2 within the MADS box. CaM kinase/MEF2 might constitute a secondary hypertrophy pathway besides the previously identified calcineurin/NFAT pathway (Molkentin et al., 1998). A multimerized MEF2 binding in front of lacZ faithfully imitates MEF2 activity in the embryo; however, no activity was visible in the adult heart despite the presence of MEF2 protein, suggesting the presence of an inhibitor in the adult (Naya et al., 1999). Using the two-hybrid system, an inhibitor for MEF2 has been isolated and found to be a member of a novel subfamily of histone deacetylases (HDAC).
Fig. 1. Signal transduction pathways regulating fiber-type specific gene expression in skeletal muscle and activation of the hypertrophic gene expression program in both striated muscle types. In both tissues, prolonged Ca2+ transients, triggered by mechanical load, agonists or nerve activity among other signals, activate the phosphatase calcineurin, which controls nuclear accumulation of NFAT. NFAT together with GATA2 and GATA4 in skeletal and cardiac muscle, respectively, activate gene expression. MEF2 proteins are targets of an alternative signal transduction pathway mediated by CaM kinase (CaMK) and p38 MAPK. Cell-type-specific cofactors interact with MEF2 to activate the hypertrophic gene expression program independent of the calcineurin/NFAT pathway.
Telling left from right
While in recent years we have learned a lot about signaling molecules involved in setting up the embryonic left–right (L–R) axis, there is a complete lack of information on how this embryonic axis is converted into organ- specific asymmetric morphogenesis (Harvey, 1998). Unfortunately, in Drosophila the heart lacks any overt L–R asymmetry; however, the hindgut displays asymmetric morphogenesis. R.Bodmer reported on initial attempts to define the signaling pathway involved in setting up L–R asymmetry in Drosophila, and overexpression of Dpp and Wg in conjunction with Dpp resulted in randomized hindgut turning. T.Brand reported on the asymmetric expression of the bagpipe homolog NKX3.2 in chick and mouse stomach development. This gene, like Pitx2, is strongly expressed in the left lateral plate mesoderm (Schneider et al., 1999). Misexpression of left-sided signals on the right side results in upregulation of NKX3.2 contralateral to its normal expression in left lateral plate mesoderm (LPM). Surprisingly, in the mouse, NKX3.2 expression is also asymmetric; however, in contrast to chick, it is expressed in right LPM.
Relatively little is also known about the molecular control of anterior–posterior polarity of the tubular heart. As F.Stockdale (Stanford University, CA) pointed out, many genes with a chamber-restricted expression pattern are initially expressed throughout the tubular heart and only after looping chamber-specific expression patterns emerge. In the avian embryo, the sMyHC3 gene encodes an atrial-specific myosin heavy chain gene that is among the first to show chamber-restricted expression. Recently, the promoter of the sMyHC3 gene has been characterized. Only two elements, a GATA motif and a vitamin D receptor-like motif (VDRE), control atrial-specific expression. While the GATA element is sufficient to drive expression in both chambers, atrial specificity is achieved by inhibition of ventricular expression, which is mediated by the iroquois class homeobox gene Irx4 (Bao et al., 1999).
Outlook
The field of muscle development has gained immensely from the identification of many signaling molecules and transcription factors, which are involved in setting up the cardiac and skeletal muscle lineages. While different sets of transcription factors are utilized in these two forms of striated muscle, it now becomes apparent that some of the signal transduction pathways and target genes regulating cardiac and skeletal muscle hypertrophy and fiber type transitions are surprisingly similar (Figure 1). Recently, a new perspective has been opened by the finding that both organ systems can recruit bone-marrow-derived stem cells to differentiate into cardiac and skeletal muscle cells, which is an important avenue with enormous biomedical impact (Bittner et al., 1999; Gussoni et al., 1999). From the recent knock-out data of the lbx-1 and Mox-2 genes, it becomes apparent that different limb muscles are probably generated by utilizing different genetic programs, which most likely are very complex (Kardon, 1998). Similarly, in the heart, different segments are defined by the expression of distinct sets of transcription factors and we are only beginning to understand the molecular complexity of these process (Schwartz and Olson, 1999). Thus, while we have learned a lot about muscle development in the past, there is much to be learned in the future.
Acknowledgments
Acknowledgements
We thank all colleagues who have allowed us to report on their partly unpublished data and for their helpful suggestions. Eric Olson is gratefully acknowledged for preparing the summarizing figure.
References
- Amthor H., Christ,B. and Patel,K. (1999) A molecular mechanism enabling continuous embryonic muscle growth—a balance between proliferation and differentiation. Development, 126, 1041–1053. [DOI] [PubMed] [Google Scholar]
- Bao Z.Z., Bruneau,B.G., Seidman,J.G., Seidman,C.E. and Cepko,C.L. (1999) Regulation of chamber-specific gene expression in the developing heart by Irx4. Science, 283, 1161–1164. [DOI] [PubMed] [Google Scholar]
- Bittner R.E. et al. (1999) Recruitment of bone-marrow-derived cells by skeletal and cardiac muscle in adult dystrophic mdx mice. Anat. Embryol. (Berl.), 199, 391–396. [DOI] [PubMed] [Google Scholar]
- Buchberger A., Seidl,K., Klein,C., Eberhardt,H. and Arnold,H.H. (1998) cMeso-1, a novel bHLH transcription factor, is involved in somite formation in chicken embryos. Dev. Biol., 199, 201–215. [DOI] [PubMed] [Google Scholar]
- Cooper R.N., Tajbakhsh,S., Mouly,V., Cossu,G., Buckingham,M. and Butler-Browne,G.S. (1999) In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell Sci., 112, 2895–2901. [DOI] [PubMed] [Google Scholar]
- Daubas P., Tajbakhsh,S., Hadchouel,J., Primig,M. and Buckingham,M. (2000) Myf5 is a novel early axonal marker in the mouse brain and is subjected to post-transcriptional regulation in neurons. Development, 127, 319–331. [DOI] [PubMed] [Google Scholar]
- Evrard Y.A., Lun,Y., Aulehla,A., Gan,L. and Johnson,R.L. (1998) lunatic fringe is an essential mediator of somite segmentation and patterning. Nature, 394, 377–381. [DOI] [PubMed] [Google Scholar]
- Frasch M. (1999) Controls in patterning and diversification of somatic muscles during Drosophila embryogenesis. Curr. Opin. Genet. Dev., 9, 522–529. [DOI] [PubMed] [Google Scholar]
- Gajewski K., Fossett,N., Molkentin,J.D., Kim,Y., Choi,C.Y. and Schulz,R.A. (1999) The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila.Development, 126, 5679–5688. [DOI] [PubMed] [Google Scholar]
- Gerber A.N., Klesert,T.R., Bergstrom,D.A. and Tapscott,S.J. (1997) Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev., 11, 436–450. [DOI] [PubMed] [Google Scholar]
- Gussoni E., Soneoka,Y., Strickland,C.D., Buzney,E.A., Khan,M.K., Flint,A.F., Kunkel,L.M. and Mulligan,R.C. (1999) Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature, 401, 390–394. [DOI] [PubMed] [Google Scholar]
- Hamamori Y., Sartorelli,V., Ogryzko,V., Puri,P.L., Wu,H.Y., Wang,J.Y., Nakatani,Y. and Kedes,L. (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell, 96, 405–413. [DOI] [PubMed] [Google Scholar]
- Harvey R. (1998) Links in the left/right axial pathway. Cell, 94, 273–276. [DOI] [PubMed] [Google Scholar]
- Joseph E.M. and Cassetta,L.A. (1999) Mespo: a novel basic helix–loop–helix gene expressed in the presomitic mesoderm and posterior tailbud of Xenopus embryos. Mech. Dev., 82, 191–194. [DOI] [PubMed] [Google Scholar]
- Kaehn K., Jacob,H.J., Christ,B., Hinrichsen,K. and Poelmann,R.E. (1988) The onset of myotome formation in the chick. Anat. Embryol., 177, 191–201. [DOI] [PubMed] [Google Scholar]
- Kardon G. (1998) Muscle and tendon morphogenesis in the avian hind limb. Development, 125, 4019–4032. [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk S.M., Wang,L., Balazsi,K., DeRepentigny,Y., Kothary,R. and Megeney,L.A. (1999) MEF2 is upregulated during cardiac hypertrophy and is required for normal post-natal growth of the myocardium. Curr. Biol., 9, 1203–1206. [DOI] [PubMed] [Google Scholar]
- Lindon C., Montarras,D. and Pinset,C. (1998) Cell cycle-regulated expression of the muscle determination factor Myf5 in proliferating myoblasts. J. Cell Biol., 140, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankoo B.S., Collins,N.S., Ashby,P., Grigorieva,E., Pevny,L.H., Candia,A., Wright,C.V., Rigby,P.W. and Pachnis,V. (1999) Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature, 400, 69–73. [DOI] [PubMed] [Google Scholar]
- Molkentin J.D., Lu,J.R., Antos,C.L., Markham,B., Richardson,J., Robbins,J., Grant,S.R. and Olson,E.N. (1998) A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell, 93, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musaro A., McCullagh,K.J., Naya,F.J., Olson,E.N. and Rosenthal,N. (1999) IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature, 400, 581–585. [DOI] [PubMed] [Google Scholar]
- Naya F.J., Wu,C., Richardson,J.A., Overbeek,P. and Olson,E.N. (1999) Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development, 126, 2045–2052. [DOI] [PubMed] [Google Scholar]
- Palmeirim I., Henrique,D., Ish-Horowicz,D. and Pourquie,O. (1997) Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell, 91, 639–648. [DOI] [PubMed] [Google Scholar]
- Paululat A., Breuer,S. and Renkawitz-Pohl,R. (1999a) Determination and development of the larval muscle pattern in Drosophila melanogaster.Cell Tissue Res., 296, 151–160. [DOI] [PubMed] [Google Scholar]
- Paululat A., Holz,A. and Renkawitz-Pohl,R. (1999b) Essential genes for myoblast fusion in Drosophila embryogenesis. Mech. Dev., 83, 17–26. [DOI] [PubMed] [Google Scholar]
- Philips A.V., Timchenko,L.T. and Cooper,T.A. (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science, 280, 737–741. [DOI] [PubMed] [Google Scholar]
- Pourquie O. (1999) Notch around the clock. Curr. Opin. Genet. Dev., 9, 559–565. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G., Elliott,D.A., Harvey,R.P. and Olson,E.N. (1998) Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development, 125, 3037–3048. [DOI] [PubMed] [Google Scholar]
- Saga Y., Hata,N., Koseki,H. and Taketo,M.M. (1997) Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev., 11, 1827–1839. [DOI] [PubMed] [Google Scholar]
- Saga Y., Miyagawa-Tomita,S., Takagi,A., Kitajima,S., Miyazaki,J. and Inoue,T. (1999) MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development, 126, 3437–3447. [DOI] [PubMed] [Google Scholar]
- Scaal M., Bonafede,A., Dathe,V., Sachs,M., Cann,G., Christ,B. and Brand-Saberi,B. (1999) SF/HGF is a mediator between limb patterning and muscle development. Development, 126, 4885–4893. [DOI] [PubMed] [Google Scholar]
- Schäfer K. and Braun,T. (1999) Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nature Genet., 23, 213–216. [DOI] [PubMed] [Google Scholar]
- Schlange T., Andrée,B., Arnold,H.-H. and Brand,T. (2000) BMP2 is required for early heart development during a distinct time period. Mech. Dev., 91, 259–270. [DOI] [PubMed] [Google Scholar]
- Schneider A., Mijalski,T., Schlange,T., Dai,W., Overbeek,P., Arnold,H.H. and Brand,T. (1999) The homeobox gene NKX3.2 is a target of left–right signalling and is expressed on opposite sides in chick and mouse embryos. Curr. Biol., 9, 911–914. [DOI] [PubMed] [Google Scholar]
- Schwartz R.J. and Olson,E.N. (1999) Building the heart piece by piece: modularity of cis-elements regulating Nkx2–5 transcription. Development, 126, 4187–4192. [DOI] [PubMed] [Google Scholar]
- Steinbach O.C., Ulshofer,A., Authaler,A. and Rupp,R.A. (1998) Temporal restriction of MyoD induction and autocatalysis during Xenopus mesoderm formation. Dev. Biol., 202, 280–292. [DOI] [PubMed] [Google Scholar]
- Xu X., Yin,Z., Hudson,J.B., Ferguson,E.L. and Frasch,M. (1998) Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev., 12, 2354–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Szendro,P.I., Prokscha,A., Schwartz,R.J. and Eichele,G. (1999) Evidence for a role of Smad6 in chick cardiac development. Dev. Biol., 215, 48–61. [DOI] [PubMed] [Google Scholar]