Abstract
Researchers in medical imaging have multiple challenges for storing, indexing, maintaining viability, and sharing their data. Addressing all these concerns requires a constellation of tools, but not all of them need to be local to the site. In particular, the data storage challenges faced by researchers can begin to require professional information technology skills. With limited human resources and funds, the medical imaging researcher may be better served with an outsourcing strategy for some management aspects. This paper outlines an approach to manage the main objectives faced by medical imaging scientists whose work includes processing and data mining on non-standard file formats, and relating those files to the their DICOM standard descendents. The capacity of the approach scales as the researcher’s need grows by leveraging the on-demand provisioning ability of cloud computing.
Key words: Imaging informatics, information storage and retrieval, internet technology
Introduction
The information technology (IT) management complexity facing researchers in medical imaging is increasing relentlessly, driven by government regulations, grant rules, and technical advances. It is not reasonable or sustainable to expect researchers to be capable of doing National Institutes of Health level research and be IT experts as well. Outsourcing is needed. Our task is to consider how this can be achieved.
Researchers in medical imaging face a multitude of challenges:
acquiring data sets, sometimes in DICOM format but often in other “raw” formats
storing the data (and perhaps anonymizing or encrypting it)
possibly sharing data while still protecting patient privacy
indexing the relationships between standard files (i.e., DICOM) and their proprietary ancestral raw files
and maintaining data viability
The management complexity of the above can overwhelm academic researchers who have no desire to become IT professionals and yet may have no alternative if their employer has insufficient IT resources to support research efforts. Further, if an academic medical center even has a data center, space constraints often drive out non-clinical servers and storage—often leaving researchers with their data in unsecured areas with insufficient fault tolerance or back up power. Even if the group has a secured room for their data systems, they may not be comfortable, or fiscally able, to grow their storage infrastructure on a moment’s notice if a grant gets funded.
Methods
Storage
A recent paper suggests that IT professionals will increasingly rely on disk for archive needs as opposed to tape or optical disk.1 However, it is more likely that storage consumers will soon neither know nor care how their data is stored; rather, it will simply be available when needed. In the past few years, a collection of multiple technologies have given rise collectively to the concept of “cloud computing”.2 In cloud computing, a client does not own storage and servers at their physical site, rather they lease capacity (either computational, storage, or both) from providers over the Internet, trusting that said providers can scale without limit and have high reliability. By doing so, a client is freed from the responsibility of maintaining large numbers of servers and storage arrays, but is increasingly dependent on the reliability of a fast and large bandwidth network connection to their cloud provider.
The cloud may not be a panacea for clinical data, however; data needed for immediate patient care should not be subject to the risks of a slow (or broken) network. But, it certainly is reasonable to consider placing research data (or patient data after the patient leaves the medical center) on a remote site until it is needed later. As long as the need can be predicted, it is a simple matter to recall it during overnight pre-fetch operations to a local cache.
There are several key advantages of cloud storage to the researcher:
Costs are predictable and expensed (paid in monthly installments) rather than all at once.
Capacity can be grown as soon as a grant is awarded.
If the grant runs out, the research can just halt payments to the provider and the storage is released.
There are also some new complications once the data leave the physical boundaries of the medical center. First, patient privacy must be protected. This is most simply accomplished by encrypting the data files before transmission to the cloud.3 The simplicity stems from the fact that encryption algorithms exist that are symmetric (what they do they can undo) and can work on any data without knowing what it is.5 The encryption process can be carried out with site-owned appliances, or even open-source software such as the Clinical Trials Processor.4 An even more likely scenario though would be to perform the encryption with the products suggested by the cloud storage vendor being used. The only caveat is that the site must recall the key(s) to decipher the files when they are recalled for use. Second, the site must have a reliable high bandwidth network link to the cloud provider. This requirement may entail new monthly expenses to the site’s internet service provider.
Table 1 lists several cloud storage providers with their current pricing models (as of November 2009). Some companies were included even though they have limited offerings; this was done to spare the reader any confusion over the absence of what some would assume to be major vendors in the field.
Table 1.
A Comparison of the Price Structures and Functionality of Several Cloud Storage Providers
| Vendor | Product Information | $/TB/year | $/TB In | $/TB Out | Security | Admin | Responsiveness |
|---|---|---|---|---|---|---|---|
| Amazon S3 | http://aws.amazon.com/s3/ | $1,440 (best) | $100 | $100 (best) | HIPAA Compliant Encryption | Via software API | 1-Day provisioning. No uptime SLA |
| $1,800 (worst) | $170 (worst) | ||||||
| EMC Atmos | http://www.emccis.com/ | Not publically published | Beta testing | ||||
| http://googleblog.blogspot.com/2009/11/twice-storage-for-quarter-of-price.html | $4,000 | No encryption, assume photos/email only | Web-based upload and indexing tools | ||||
| Max 16 TB | |||||||
| IBM | No public cloud storage at this time | ||||||
| Microsoft Azure | http://www.microsoft.com/windowsazure/pricing/ | $1,800 | $100 | $150 | Encryption available | .NET-based user interface application | 1-Day provisioning. No published uptime SLA |
| Mozy | http://mozy.com/pro/pricing | $6,000 | None listed | None listed | Encrypted upload and storage | Client side user application | 1-Day provisioning. No uptime SLA |
| Nirvanix | http://www.nirvanix.com/products-services/index.aspx | $2,169 | $180 | $180 | Optional encrypted upload and storage | Management portal, special healthcare data tools | 99.9% uptime, 1-day provisioning |
| Zetta | http://www.zetta.net/products.php | $3,000 (worst) | None listed | None listed | Encrypted upload and storage | 99.9% uptime, 1-day provisioning |
A comparison of the pricing models and functionality of several cloud storage providers
Also noted are their user interfaces and application programming interface (APIs) and service level agreements (SLA) terms for uptime. Of note are some major vendors that have limited or no offering as of this writing
Data Sharing
As previously mentioned, not all data of interest to the medical imaging researcher are in DICOM format; if it were, research storage needs could be met easily with an enterprise DICOM archive.6 However, the CT investigator may also need access to the projection sinograms, the MR researcher to the K-space Fourier signals, and the US researcher to the transducer polar or rectilinear data to name just a few possibilities. It is likely that each of these data are in a proprietary file format known only to the vendor. This reality compels a difficult choice on the investigator as will be seen.
As detailed in the “Storage” section, encryption offers a clean solution to offsite storage as long as there is no need to process the data offsite. The situation is acceptable for a single site storing data offsite for later recall and research processing at the original patient care site (pursuant to Internal Review Board regulations at the site). However, a multi-center research trial that requires data processing at remote medical centers presents a much more complex challenge. In this case, the data cannot be encrypted without also preventing image processing. The answer is to anonymize the file(s) before they leave the performing site, and substitute a study identifier that can link back to the original patient demographics if required.
Anonymization of DICOM files is a well-understood procedure; numerous tools exist that can assist with it in a HIPAA compliant manner.4,7 This then allows offsite researchers to process unencrypted DICOM files while still preserving patient privacy. However, it is unlikely that the patient scanning site will have the software tools to open and anonymize any data elements in the ancillary raw data files (i.e., the CT projection sinograms). This leaves open the temptation to transfer the raw files unencrypted and endure the possibility that a site with another (for example) CT scanner of the appropriate make and model could open the data files and learn the patient identity. This is a vexing issue for which there is likely no alternative except for tools that would be provided by the vendor. Of course, such tools would likely have to change over time as the vendor’s proprietary files change, which leads to issues in maintaining the data’s viability over time.
Indexing
A further concern is building a semantic association among data files that are related. For example, it may be the case in CT or MR that a single raw data file can be used to reconstruct DICOM image files for a single image, series or multiple series. So while a single DICOM image may have a single antecedent, the parent raw data file may have numerous descendants. The researcher is thus challenged with how to relate a given clinical image back to the raw file (assuming it is saved) to perform further analysis. Figure 1 shows a simplified view of the data flow that represents this concept, and one conceptual way to deal with it. In this figure, the related DICOM and raw files are both sent from the scanner, but proprietary “raw” files are stored to a simple file server, while the DICOM image files are stored to both a DICOM archive and forwarded to a parser (MIRTH, http://www.mirthcorp.com) and database (PostGreSQL, http://www.postgresql.org).
Fig 1.
A high level view of the key image file components, showing the relationships between the raw files, their DICOM descendent files, and the MIRTH parsing engine creating a database to cross-reference the two data sets for subsequent data mining.
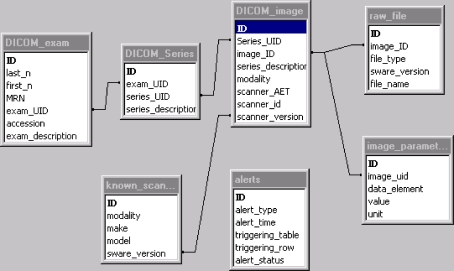
An early database schema used in our lab illustrates the relationship among the DICOM and raw files, and a strategy to deal with it (figure generated from Microsoft Access, Microsoft Corp., Redmond, WA, USA). Note the two tables on the bottom of the figure: “known scanners” and “alerts”; the former acts as a “knowledge base” of scanners that the database knows about, and the second records events when the database is exposed to scanner software versions that are unknown. For known scanners, the upper tables come into effect: one can search on patients, exams types, scanner types, or even image reconstruction parameters (see table “image_parameters” that encodes slice thickness, reconstruction filters, pulse sequence, image projections, etc.). This schema permits, for example, locating all images done on CT Scanner X using dose optimizer method Z and relates the found DICOM images back to the raw data file(s) that produced them.
To populate the database, various MIRTH processing “channels” were constructed that take actions based on modality and other filters. Hence, one can define a CT channel or an MR channel and copy the contents of specific image tags to the database defined in Figure 1. What cannot be automated via this process is the association of a given raw file(s) to the derived DICOM file(s). In this example, this linkage has to be performed manually by completing the “file_name” element in the “raw_file” table. Hence, ongoing database support is required (Fig. 2).
Fig 2.
One potential database schema for relating DICOM image objects to the proprietary “raw” files that created them. The table “known_scanners” forms a knowledge base of scanner and software versions that allow linking a specific DICOM image object to the raw file(s) that created it. The table “DICOM_image” then links to one or more “image_parameter” tables to record values of interest to the researcher (slice thickness, reconstruction kernel, pulse sequence, etc.). If the database encounters an unknown scanner model, an entry is made in the “alerts” table so that the knowledge base can be expanded.
Maintaining Viability
As alluded to in the “Data Sharing” section, there are several issues involved in maintaining data viability over time, not so much with standard DICOM files but more likely with proprietary file formats that the vendor may use. Consider an example: A researcher saves the raw file from a CT for subsequent data processing, possibly using different reconstruction kernels. As long as the original CT scanner is in-house, the raw file can likely be reprocessed. But if the scanner is upgraded, there is no guarantee that the new version will be able to read the old file—and this likelihood is assured if the new CT is from a different vendor.
A possible method to address this issue is to persuade the vendor to license the software emulators (under non-disclosure agreements of course) that they use at IHE Connectathons.8 If one attends a Connectathon or views the picture in the reference, they will see attendees poring over numerous laptop computers. Many of those laptops are running modality emulator software (MR, CT, US, etc.). It is a simple matter to save each emulator as a virtual machine and in this way maintain the ability over time to process legacy raw files.9
Discussion and Conclusions
This paper has outlined an approach to manage the main objectives faced by medical imaging scientists: storing growing amounts of data in a scalable way, protecting patient information in the offsite data, relating non-standard raw file formats to their DICOM file descendents, and maintaining data viability of the non-standard files over time.
The capacity of the approach scales as the researcher’s need grows by leveraging the on-demand provisioning ability of cloud computing. Furthermore, with the exceptions of anonymization and other processing on proprietary “raw” files, the approach is based on open tools and methods, and a scheme was proposed to overcome the raw file exception with low vendor effort.
A topic we have not addressed explicitly is the possibility that the researcher may also require high-performance compute clusters to perform image processing on massive data sets or perform data mining pattern searches across large numbers of exams. Such needs also align well with clouds, and many of the vendors that provide cloud storage also can provide cloud-based compute clusters that leverage virtualization methods to dynamically grow capacity. This is an enticing approach because it keeps data sets and compute resources near each other for optimal efficiency across high bandwidth data center networks.
Cloud computing is in its infancy, as evidenced by the still relatively low penetration of major vendors into this market (as shown in Table 1). Yet, the drivers for it are compelling, and it will no doubt become an indispensible tool for medical researchers as their storage and computational demands grow with few limits in sight.
References
- 1.Nagy PG. The future of PACS. Med Phys. 2007;34(7):2676–2682. doi: 10.1118/1.2743097. [DOI] [PubMed] [Google Scholar]
- 2.He R, Niu J, Yuan M, Hu J: A novel cloud based model of pervasive computing. Computer and information technology, 2004. CIT '04. The Fourth International Conference on 14–16 Sept. 2004, pp 693–700
- 3.Langer SG, Stewart BK. Computer security: A primer. J Digit Imaging. 1999;12(3):114–113. doi: 10.1007/BF03168630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CTP: http://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor. Last viewed April 2010
- 5.Schneier B. Applied Cryptography. 2. New York: Wiley; 1996. pp. 28–29. [Google Scholar]
- 6.Langer S. Issues surrounding PACS archiving to external, third-party DICOM archives. J Digit Imaging. 2009;22(1):48–52. doi: 10.1007/s10278-008-9125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez DR, Carpenter T, Hemert JI, Wardlaw J: An open-source toolkit for medical imaging de-identification. Eur Radiol doi:10.1007/s00330-010-1745-3 [DOI] [PubMed]
- 8.IHE: http://www.ihe.net/north_america/connectathon2010.cfm. Last viewed November 2009
- 9.Langer S, Charboneau N, French T: DCMTB: A virtual appliance DICOM Toolbox. J Digit Imaging 2009. PMID:19705204. doi:10.1007/s10278-009-9230-8 [DOI] [PMC free article] [PubMed]