Abstract
The TCR α enhancer (Eα) has served as a paradigm for studying how enhancers organize trans-activators into nucleo-protein complexes thought to recruit and synergistically stimulate the transcriptional machinery. Little is known, however, of either the extent or dynamics of Eα occupancy by nuclear factors during T cell development. Using dimethyl sulfate (DMS) in vivo footprinting, we demonstrate extensive Eα occupancy, encompassing both previously identified and novel sites, not only in T cells representing a developmental stage where Eα is known to be active (CD4+CD8+–DP cells), but surprisingly, also in cells at an earlier developmental stage where Eα is not active (CD4–CD8––DN cells). Partial occupancy was also established in B-lymphoid but not non-lymphoid cells. In vivo DNase I footprinting, however, implied developmentally induced changes in nucleo-protein complex topography. Stage-specific differences in factor composition at Eα sequences were also suggested by EMSA analysis. These results, which indicate that alterations in the structure of a pre-assembled nucleo-protein complex correlate with the onset of Eα activity, may exemplify one mechanism by which enhancers can rapidly respond to incoming stimuli.
Keywords: in vivo footprint/nucleo-protein complex/TCRα enhancer
Introduction
Cellular differentiation requires the coordinated utilization of complex programs of gene expression in response to specific signaling cues. To orchestrate the spatio-temporal control of these programs, eukaryotic cells utilize a relatively limited number of trans-acting factors in different combinations at discrete cis-acting DNA regulatory elements, such as enhancers. The prevailing view is that enhancers ensure cooperativity and synergy between trans-activators by organizing them into tightly clustered nucleo-protein complexes, known as ‘enhanceosomes’ (reviewed in Carey, 1998). Pioneering studies on the interferon β (IFN-β) and T cell receptor (TCR) α (Εα) gene enhancers have shown that these complexes exhibit a distinct three-dimensional topology, dependent upon architectural proteins that distort the DNA helix and facilitate interactions between remotely bound factors (Falvo et al., 1995; Giese et al., 1995). These structures are assumed to provide complementary surfaces to recruit molecular machines, such as those involved in chromatin remodeling and DNA transcription, or recombination (Grosschedl, 1995; Maniatis et al., 1998).
The molecular basis for the regulation of Εα function has been the focus of intense study. This element, located at the 3′ end of the TCRα locus, was originally identified based on its ability to stimulate reporter gene expression following transfection into T lymphoid cells and was narrowed down to a 230 bp fragment in the mouse and a 116 bp fragment in the human (Ho et al., 1989; Winoto and Baltimore, 1989). Complementary analyses by many laboratories have shown that the highly homologous murine and human Eα (shown in Figure 1A) possess overlapping binding sites for a number of transcription factors (Leiden, 1993; Kuo and Leiden, 1999; and references therein). Notably, the human core enhancer of 116 bp, which was determined to be sufficient for transcriptional activation in vitro (Ho et al., 1989) and, subsequently, for transcriptional activation and V(D)J recombination in vivo (see below), contains four major sites that bind, respectively, the transcription factors ATF/CREB, LEF-1 or TCF-1 [two closely related members of the ‘high mobility group’ (HMG) box family of DNA-binding proteins], CBF/PEBP2 and Ets-1. All four sites, in their correct helical phasing, are required for enhancer activity (Giese et al., 1995; Mayall et al., 1997; Roberts et al., 1997). In vitro studies, using either naked DNA or chromatinized templates, suggest the stepwise assembly of these four factors onto the Εα core, with a pivotal role for the architectural HMG-domain factor in organizing the multiprotein complex (Giese et al., 1995; Mayall et al., 1997). Transgenic studies, however, have suggested that factor binding to the Εα core in vivo is all or none, supporting a model for the highly cooperative assembly of the nucleo-protein complex (Hernandez-Munain et al., 1998). In addition to these factors, other trans-activators have been shown to interact with Eα in vitro, including SP-1, at the 5′ end of the core (Hernandez-Munain et al., 1998), and, within the 3′ flanking region, GATA-3 and members of the basic helix–loop–helix (bHLH) transcription factor family (Marine and Winoto, 1991; Bernard et al., 1998).
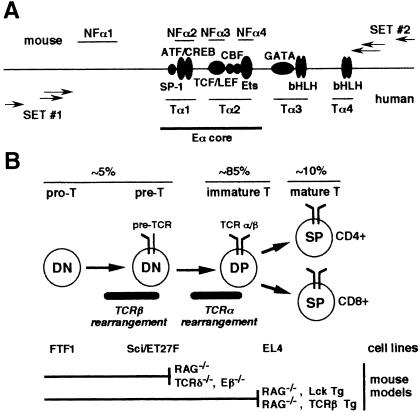
Fig. 1. (A) Comparative representation of mouse/human Eα organization. Previously identified nuclear factor binding sites are shown for the mouse (NFα1 to NFα4) and human (Tα1 to Tα4). Known transcription factor binding motifs of both the murine and human sequences are represented by gray circles and ovals. The 116 bp Eα core fragment is indicated. Arrows indicate the relative positions of primer sets 1 and 2 used in LM–PCR to analyze the bottom and top strands, respectively. (B) Schematic representation of T cell differentiation in the mouse thymus. The principal thymocyte cell stages with respect to the expression of the CD4 and CD8 markers and TCR complexes are schematized. The developmental stage of the T cell lines and T cell developmental blocks observed in the thymi of the mouse models used in this study are indicated.
A more complete understanding of the regulation of Eα function will require knowledge of how enhanceosome formation is controlled within the context of T cell differentiation. During T cell development in the thymus (reviewed in Kisielow and von Boehmer, 1995; also see Figure 1B), the onset of TCR gene expression and V(D)J recombination are both tightly regulated. TCRα gene expression and recombination are delayed until the CD4–CD8– double negative (DN) thymocytes, which produce TCRβ chains, become CD4+CD8+ double positive (DP) cells. Expression of a functionally rearranged TCRα gene leads to the formation of a TCR αβ complex and, ultimately, to the selection of αβ-expressing cells into terminally differentiated CD4+ or CD8+ single positive (SP) thymocytes. Accordingly, in the absence of TCRα gene expression, αβ T cell development is arrested at the DP cell stage.
One of the earliest expression events at the TCRα gene locus is the production, at the DN to DP cell stage transition, of the so-called T early α (TEA) germline transcripts that initiate upstream of the TCR-Jα gene segment cluster; TEA transcription is rapidly followed by Vα-to-Jα recombination (Krangel et al., 1998). Several in vivo studies argue that Eα activity is critical for these processes. Using transgenic mice, we have previously demonstrated that a 515 bp fragment containing the mouse Eα element confers tissue- and stage-specificity to germline transcription and V(D)J recombination within a reporter substrate (Capone et al., 1993). Other studies, using a similar strategy to test DNA fragments containing the human enhancer, confirmed these findings (Lauzurica and Krangel, 1994), and further demonstrated that the 116 bp Eα core was sufficient to drive these activities, although with slightly altered temporal specificity (Roberts et al., 1997). Finally, targeted deletion of a 1.6 kb fragment centered on Eα has been shown to result in a drastic reduction of TEA transcription and Vα-to-Jα recombination at the targeted locus and a block of thymocyte differentiation at the DP cell stage (Sleckman et al., 1997).
While in vitro, transgenic and gene targeting studies have provided invaluable insights into the molecular biology and function of Eα, our knowledge is still limited concerning the precise relationship between early T cell development and the molecular cues that trigger Eα activity, in particular with respect to the assembly of nucleo-protein complexes at the endogenous locus. We have examined in vivo factor occupancy of Eα during early T cell development by genomic footprinting of appropriate cell lines and mouse models. Surprisingly, extensive enhancer occupancy appears to be established prior to the onset of Eα activity. Additional results, however, argue that the three-dimensional topology and factor composition of the assembled complex(es) changes during the DN to DP transition, correlating with Eα activation.
Results
Eα occupancy by nuclear factors in TCRα expressing cells
To examine Eα occupancy by nuclear factors at the endogenous TCRα locus, we utilized the technique of in vivo genomic footprinting, which consists of treating cells with the membrane permeable, DNA methylating agent dimethyl sulfate (DMS), followed by ligation-mediated (LM) PCR (for example, Garrity and Wold, 1992). As a source of TCRα expressing T cells, we first used wild-type thymocytes (comprised of mostly TCRαβ+, DP + SP cells). Cultured cells from the TCRαβ+ EL4 thymoma cell line were similarly analyzed. Footprinting was carried out over a region that encompasses not only the core enhancer but also previously in vitro identified transcription factor binding sites within Eα (Figure 1A).
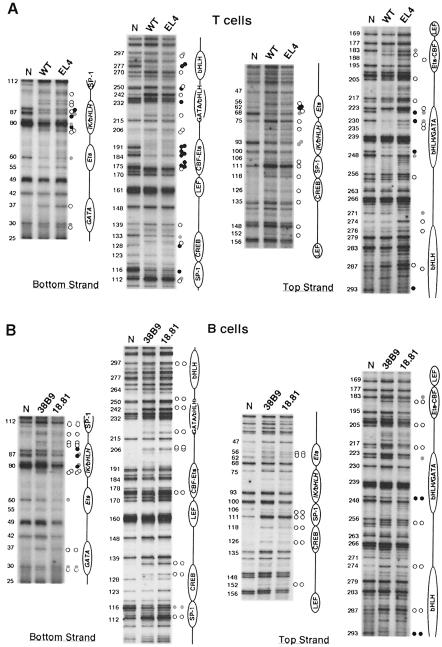
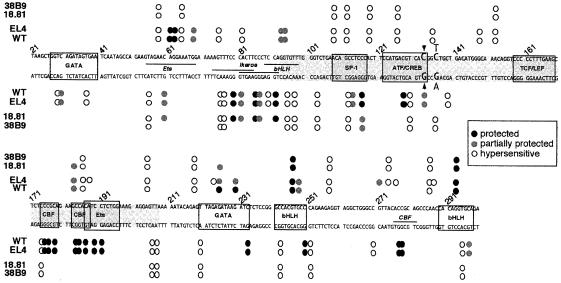
A comparison between the pattern generated from in vitro DMS treated (‘naked’) DNA, which shows all G residues, and those from in vivo treated wild-type thymocytes and EL4 cells revealed a complex pattern of protected and hypersensitive sites, indicative of widespread protein binding throughout Eα for both types of cells (Figure 2A; summarized in Figure 3). As predicted, strong footprints were detected within the minimal core enhancer. Moving 5′ to 3′, occupancy of the ATF/CREB site was visualized on the bottom strand by the protection of one G (nt 128) along with a hypersensitive A (nt 130) and, on the top strand, by a hypersensitive G (nt 126). Additional evidence for occupancy comes from a footprint at the 3′ border of this site, consisting of a partially protected G on the bottom strand (nt 133) and a hypersensitive G on the top strand (nt 135). Generally, direct evidence of factor binding at the TCF/LEF site was not observed (but also see below), most likely because DMS methylates residues within the major groove whereas the corresponding site-specific HMG-domain proteins bind within the minor groove (van de Wetering and Clevers, 1992). However, on the top strand, a hypersensitive G (nt 148) and A (nt 152) immediately 5′ of this site may actually reflect factor binding. Finally, occupancy at the CBF–Ets composite motif was visualized on the bottom strand by a single hypersensitive G followed by a series of protected Gs (nts 175–191) and, on the top strand, by a weakly protected G (nt 183) followed by a hypersensitive A three nucleotides later (nt 186). Such an extensive in vivo footprint appears to be characteristic of CBF and Ets occupancy as it has been observed for similar motifs, for example within the TCRβ gene enhancer (Eβ) (Tripathi et al., 2000). Of note, we also observed a footprint within a GC rich region immediately upstream of the ATF/CREB site, visualized on the bottom strand by a hypersensitive and two protected Gs (nt 112, 113 and 116, respectively) and, on the top strand, by an intensely hypersensitive G (nt 111) flanked by hypersensitive As (nt 108 and 118). The corresponding region in the human Eα core has previously been demonstrated to contain an SP-1 binding site (Hernandez-Munain et al., 1998).
Fig. 2. Eα occupancy in wild-type thymocytes, the EL4 cell line (A) and the 38.B9 and 18.81 B cell lines (B). In vivo occupancy was determined by DMS genomic footprinting and LM–PCR. Analytical gels were run for different times to optimize band resolution for distinct regions. The numbers to the left of each panel represent the nucleotide positions based on the published murine sequence (Winoto and Baltimore, 1989), commencing at the 5′ Pvu II restriction site. The known transcription factor binding sites (in plain text) as well as the new identified potential binding sites (in italics) are indicated to the right of each panel. Filled and open circles represent protected and hypersensitive nucleotides, respectively. Gray circles represent partial protections. For (A), circles in the left column correspond to footprints found in common between wild-type thymocytes and EL4 cells; circles in the right column (top strand) are footprints specific to EL4 cells. For (B), circles in the left and right columns correspond to footprints for 38B9 and 18.81 cells, respectively. ‘N’: naked, in vitro methylated genomic DNA. In (A), C57BL/J6 wild-type thymocytes and naked DNA were used. In (B), naked DNA was from Balb/c mice.
Fig. 3. Summary of the in vivo DMS genomic footprints at Eα in T and B lymphoid cells. The analyzed sequence is shown. Protected and hypersensitive sites are indicated for each cell type, as in Figure 2. Previously described and new putative DNA binding sites are indicated by boxes and horizontal lines, respectively. The nucleotide sequence corresponding to the Eα core is shadowed. The nucleotides within the ATF/CREB site involved in the polymorphism between the various cells used in this study are shown in enlarged type letters, with the boldface nucleotides corresponding to the C57BL/6J sequence. The site of the polymorphic deletion is indicated by an arrowhead.
Footprints were also visualized outside of the Eα core. On the 5′ side, we observed hypersensitive and protected Gs on the bottom strand (nt 29, 30 and 37) within a region encompassing the murine NF-α1 site (Figures 1, 2A and 3). We also obtained evidence of nuclear factor binding within the region between the NF-α1 and SP-1 motifs forming two distinct sites (centered at nt 61 and 85, respectively), which have not been previously recognized. Among the latter two sites, one appears as a complex that is best visualized on the top strand by a series of protected Gs (nt 61, 62 and 68) and a hypersensitive G and A (nt 57 and 65). The second complex is mainly seen on the bottom strand as a series of protected Gs and hypersensitive Gs and As spanning nt 76–97. Analysis for sequence homology with previously identified nuclear factor binding motifs indicated that NF-α1 contains a putative GATA binding site, while the footprints centered at nt 61 and 85 correspond with a putative Ets and two overlapping Ikaros and one bHLH binding motif, respectively. Hereafter, we will refer to these sites by the names of the factors that putatively bind to them.
On the 3′ side of the Eα core, footprints were mainly detected in two regions that correspond to the Tα3 and Tα4 sites previously identified in the human sequences (Ho et al., 1989), as well as in the region between these two elements. Tα3 consists of identified binding sites for the GATA-3 and bHLH factors, while Tα4 overlaps with a known bHLH motif (Marine and Winoto, 1991; Bernard et al., 1998). The region between Tα3 and Tα4, which also demonstrates a footprint, contains a putative binding site for CBF. The Tα3 GATA and bHLH sites are well visualized on the bottom strand by protected and hypersensitive Gs (nt 233 and 250; 220, 243 and 251, respectively) and, on the top strand, by a number of protected Gs (nt 225, 230, 246 and 248). The 3′ CBF and Tα4 bHLH sites show up on the bottom strand by the strong protection of two Gs (CBF, nt 277 and 278) and, on the top strand, by two hypersensitive and one protected G (bHLH, nt 287, 290 and 293). In addition, there were a small number of relatively weak footprints located in regions devoid of homology with known DNA binding motifs and generally found in non-lymphoid cells as well (see below). The principle example consists of three clustered hypersensitive As on the bottom and top strands (nt 206, 207 and 204, respectively), which we consider unlikely to be functionally relevant. Finally, although footprints in wild-type primary thymocytes and EL4 cells were well conserved throughout almost the entire length of Eα (Figures 2A and 3), several differences were observed at the 3′ end, on the top strand (e.g. nt 188, 232, 235, 271 and 276). These differences, which may suggest changes in factor binding at the underlying sites (notably, Tα3/Tα4 bHLH and 3′ CBF) between the two cell types, have not been investigated further in this study.
In summary, wild-type thymocytes not only demonstrate occupancy of transcription factor binding motifs within the core enhancer, but also 5′ and 3′ of this element. Overall, our footprinting profiles compare favorably with those recently reported by Hernandez-Munain and colleagues, who have also analyzed a homologous DNA region of human and mouse Eα in vivo (Hernandez-Munain et al., 1998, 1999). The more extensive profile that we describe 5′ of the Eα core results from the use of a different set of oligonucleotide primers, allowing examination of the upstream NFα1 site and intervening sequences. The latter region, notably, contains strongly occupied sites (e.g. Ets and Ikaros) not previously recognized.
Eα occupancy by nuclear factors in B cells
In order to determine whether Eα occupancy is restricted or not to T lineage cells, we also examined the footprints of chromosomal DNA in B lymphoid and non-lymphoid cell lines. These included the 38B9 pro-B and 18.81 pre-B cell lines and the NIH 3T3 fibroblast and J774 macrophage lines. As predicted, both the NIH 3T3 and J774 cells exhibited a pattern largely unchanged from that of naked DNA, with the exception of several weakly hypersensitive nucleotides (including the site at nt 204–207 mentioned above), which were also found in all of the other cells analyzed (data not shown).
Surprisingly, however, the B lymphoid cells exhibited in vivo footprints at several Eα sites, which varied slightly depending on the cell line studied but were close to those found in T cells (Figures 2B and 3). Thus, 38B9 cells exhibited partial footprints at the 5′ Ets and core ATF/CREB motifs, as well as complete footprints at the core SP-1 and 3′ bHLH motifs. The footprint pattern for 18.81 cells was even closer to that for T cells, sharing most of the same changes as those found for 38B9 cells, as well as additional footprints similar to those found in wild-type thymocytes and EL4 cells (e.g. the 5′-Ets, -Ikaros and -bHLH sites). Interestingly, while the Eα core flanking sequences were mostly occupied in the B cell lines, the CBF–Ets composite motif within the core itself was largely unbound. We conclude that, even though the complete pattern of Eα occupancy is T-cell specific, this element also demonstrates unexpectedly extensive factor binding in cells of B-lymphoid origin.
Eα occupancy in DN thymocytes
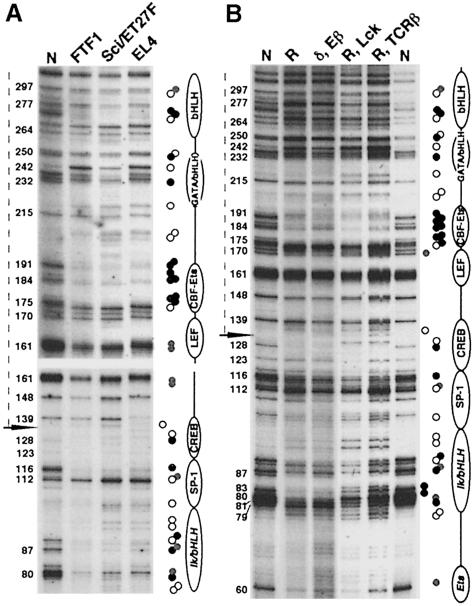
In light of the fact that the TCRα gene is not expressed in B cells [nor the 38B9 and 18.81 cell lines (Leiden, 1993); our unpublished results], our data raise the possibility that Eα occupancy may, at least in part, be established prior to the onset of enhancer activity. Therefore, we examined the Eα footprints for cell lines representative of early T cell development at a stage prior to TCRα locus activation. As schematized in Figure 1B, the FTF1 cell line has a pro-T cell phenotype with its TCR genes unrearranged (Pelkonen et al., 1987), whereas the Sci/ET27F line has been recognized as a pre-T cell (Groettrup et al., 1992). Footprints obtained with these cell lines were compared with those of TCRαβ+ EL4 T cells. Surprisingly, we observed essentially the same footprints for all three cell lines at all of the major factor binding sites [Figure 4A, only the results on the bottom strand are shown; in this particular experiment, note the partial protection of two Gs adjacent to LEF/TCF (nt 160, 161) suggesting that this site is also occupied]. The only significant difference between EL4 and the other two cell lines was a 1 bp upshift in the EL4 lane (compared with the naked DNA sample also), visible from nt 139 onward, as well as a protection at nt 139. These differences, however, are due to a genetic polymorphism between the EL4 cell line (of C57BL/6J origin) and the pro-/pre-T cells and naked DNA used in this experiment (of Balb/c origin), including a 1 bp deletion within the ATF/CREB motif (for details, see Materials and methods; Figure 3). The protection at nt 139 is also a result of this polymorphism, as naked DNA that does not have the deletion shows a reduced intensity of this band (e.g. Figure 2A). These results, notably the footprint pattern observed in FTF1 pro-T cells, strongly suggest that the nuclear factor-binding site occupancy of Eα is established early during T cell development.
Fig. 4. Eα occupancy in DN versus DP thymocytes. Bottom strand in vivo footprint analysis of (A) the developmentally staged FTF1, Sci/ET27F and EL4 T cell lines and (B) thymocytes from the various T cell developmental model mice (R: RAG–/–; δ, Eβ: TCRδ–/–, Eβ–/–; R, Lck: RAG–/–, Lck Tg; R, TCRβ: RAG–/–, TCRβ Tg). Legends are as in Figure 2. Circles in the right column represent the general footprint pattern; differences found between the DN (R or δ, Eβ) and mostly DP (R, Lck or R, TCRβ) cells are shown in the left column. The position of the polymorphic deletion at nt 133 is indicated by the horizontal arrow; the dotted line to the left of each panel indicates the Eα region where footprints may be up shifted. The R and δ, Eβ mice were on a 129/Ola genetic background; the R, Lck and R, TCRβ mice were on a mixed 129/Ola × C57BL/6J background. Naked DNA was from Balb/c mice.
To extend our analysis further, we examined Eα occupancy using primary thymocytes from several engineered mouse models that, due to a specific block in early or late T cell development, have their thymuses highly enriched in either DN or DP cell populations (i.e. the RAG–/– and TCRδ–/–, Eβ–/– mice or the RAG–/–, TCRβ Tg and RAG–/–, Lck Tg mice, respectively, shown in Figure 1B; also see Fischer and Malissen, 1998 and references therein). We used two different models, each for the DN or DP cell stage block, to ensure that our observations were not based on the peculiarity of one mouse line. Strikingly, as observed for the developmentally staged T cells, the in vivo footprints of thymocytes derived from the various mouse models were essentially identical to each other (and to those of wild-type thymocytes) for both the bottom and top strands throughout Eα (Figure 4B and data not shown). Furthermore, the intensity of the footprints at most of the sites was similar between the two developmental stages, suggesting that these sites are occupied to the same degree. Two exceptions were (i) at the 5′ Ikaros site (nt 80 and 83), and (ii) 3′ of the TCF/LEF site within the core (nt 170), where the corresponding Gs were protected in the DN mouse models only. In addition, the footprints for the DP mouse models appear as doublets 3′ of nt 133 due to the fact that they both carry the Eα polymorphic deletion on only one allele, whereas the DN mouse models (and the naked DNA) used here carry the mutation on both (Figure 4B, legend). In independent experiments, we have performed RT–PCR analysis of TCRα germline transcription for the developmental mouse models used in this study (TEA and germline Cα transcription were investigated) to confirm that, as expected, TCRα gene expression is activated in thymocytes from the RAG–/–, TCRβ Tg and RAG–/–, Lck Tg mice only (data not shown). Altogether, the above results led us to conclude that (i) essentially all of the nuclear factor binding sites of Eα are occupied in DN cells (even though the TCRα locus is transcriptionally silent at this stage) and (ii) aside from the two exceptions mentioned above, no major modification in Eα occupancy can be detected in vivo by DMS footprinting as thymocytes progress from the DN to the DP cell stage.
Structural changes at Eα during early T cell development
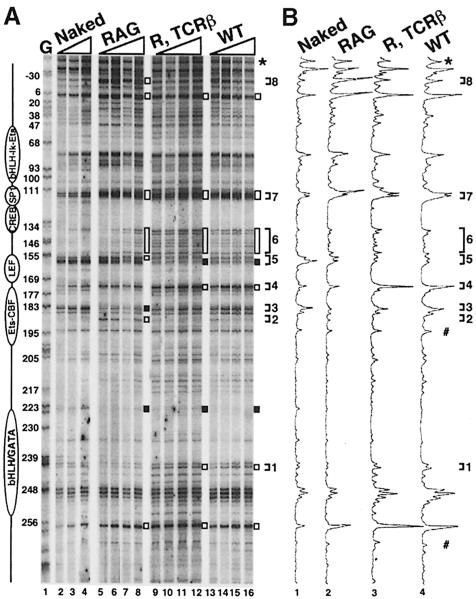
Because significant differences in the DNA–protein interactions at Eα were not observed between DN and DP thymocytes following DMS treatment, we suspected that a change(s) in the overall conformation of the pre-assembled nucleo-protein complex(es) may correlate with the onset of enhancer activation. To test this hypothesis, we subjected thymocytes from the RAG–/–, RAG–/–, TCRβ Tg and wild-type mice to limited DNase I digestion followed by LM–PCR. Differences in sensitivity to DNase I-mediated cleavage can reflect changes in protein–DNA interactions and/or chromatin structure that are not detectable by the DMS footprint technique, in particular those that affect DNA bending (Rigaud et al., 1991).
Representative results are shown in Figure 5. As expected from our previous findings, DNase I footprinting of RAG–/– thymocytes (DN cells) resulted in a series of protected and hypersensitive regions compared with naked DNA (Figure 5A, lanes 2–4 and 5–8; 5B, 1st and 2nd histograms). Within the core enhancer, these included regions of increased sensitivity within the Sp-1 motif and between the ATF/CREB and TCF/LEF motifs, as well as protected and hypersensitive sites within the CBF–Ets motifs. In addition, a site of increased sensitivity was found 5′ of the TCF/LEF motif. Outside of the core, protected and hypersensitive sites were observed flanking the 3′ Tα3 GATA and bHLH motifs. Moreover, two hypersensitive sites were also found at the 5′ end of Eα, within sequences not previously described to interact with nuclear factors.
Fig. 5. In vivo DNase I genomic footprinting analysis of Eα. (A) Thymocytes from the mice indicated, as well as wild-type genomic DNA (Naked) were treated with increasing amounts of DNase I and subjected to LM–PCR (top strand analysis). Lane G is a display of guanine residues of in vitro DMS treated DNA. All mice and DNA are of C57BL/6J origin. DNase I-hypersensitive and protected regions are indicated by open and filled bars, respectively. The differences in DNase I sensitivity detected between the RAG and RAG, TCRβ or wild-type thymocytes are indicated by brackets numbered 1–8 (see details in the text). The star indicates the band used to equilibrate for peak intensity in (B). Utilization of a reference point in the intermediate or lower regions of the gel (#) gave consistent results. (B) Histogram analysis of the LM–PCR products shown in (A) [histograms 1–4 correspond to, respectively, lanes 3, 7, 11 and 15 in (A)].
Strikingly, while retaining a majority of the same protected and/or hypersensitive sites, the DNase I footprints of the RAG–/–, TCRβ Tg and wild-type thymocytes (both mostly DP cells) demonstrated additional changes in sensitivity, most of them within the Eα core (Figure 5A, lanes 9–12 and 13–16; 5B, 3rd and 4th histograms; sites 1–8). The most dramatic difference in terms of increased sensitivity in the DP samples consists of one prominent band at the border between the TCF/LEF and CBF–Ets motifs (site 4; note that hypersensitivity at this site is not detected in the RAG–/– sample). At the same time, a site on the 5′ edge of the TCF/LEF motif becomes protected (site 5). Significantly, using DNase I in vitro footprinting, a related profile (consisting of the association of a DNase protection at the TCF/LEF site and increased sensitivity 3′ of it) has been previously described upon LEF-1 binding to its cognate Eα motif in chromatinized templates (Mayall et al., 1997). Furthermore, we also noted increased sensitivity within the ATF/CREB-to-TCF/LEF linking region (site 6; most evident at the lowest DNase I concentrations, compare lanes 5–7 with lanes 9–11 and 13–15), as well as a change in the relative intensity of the hypersensitive bands within the SP-1 motif (site 7). However, in contrast to the RAG–/– samples, no specific footprint (i.e. relative to naked DNA) was observed at the downstream CBF–Ets motifs, and one Eα 5′ flanking hypersensitive site (site 8) also disappeared. Overall, such opposite changes in sensitivity between the DN and DP samples render it unlikely that they are attributable to variations in the DNase I titration and, rather, suggest that they correspond to differences in factor binding and/or conformational changes within the enhanceosome. This interpretation is further supported by our additional findings that, using the same DNA samples, less striking differences were found in parallel analyses of Eβ, an enhancer known to be active at both the DN and DP cell stages (data not shown). These results, therefore, while confirming the extent of Eα occupancy (including at the TCF/LEF motif), strongly support the presence of developmentally induced changes in the higher order architecture of a pre-assembled DNA–protein complex(es) as thymocytes progress from the DN to DP cell stage, which may, indeed, reflect Eα activation for TCRα locus expression and recombination.
Electrophoretic mobility shift assay (EMSA) analysis of Eα motifs
Two unanticipated findings can be drawn from our footprint analyses. First, Eα occupancy in vivo is more widespread than expected, and includes sequences not previously shown to interact with nuclear proteins. Secondly, while protein binding motifs at Eα are occupied throughout thymocyte ontogeny, cell stage-specific structural changes are observed that suggest qualitative and/or quantitative differences in factor composition at some sites. To lend additional support to these issues, we subjected Eα motifs to EMSA analysis using specific oligonucleotides (listed in Table I) and nuclear extracts (NE) from either RAG–/– or wild-type thymocytes.
Table I. Oligonucleotide probes used in EMSAs.
| Probe | Nucleotide sequencea | Nt position/reference |
|---|---|---|
| Eα 5′-Ets | 5′-GAAGTAGAACAGGAAATGGAAA-3′ | Eα 51–72 |
| Eα 5′-Ik | 5′-GAAAAAGTTTCCCACTTCCCTCCAG-3′ | Eα 69–93 |
| Eα ATF/CREB | 5′-TCCATGACGTCACGGCTGCT-3′ | Eα 121–140 |
| Eα LEF/CBF/Ets | 5′-ACAGGTCCCCCTTTGAAGCTCTCCCGCAGAAGCCACATCCTCTGGAA-3′ | Eα 152–199 |
| Eα CBF-Ets | 5′-TCTCCCGCAGAAGCCACATCCTCTGGAA-3′ | Eα 161–199 |
| EBS-a | 5′-GATAACAGGAAGTGGTTGTA-3′ | Bosselut et al. (1993) |
| EBS-z | 5′-GATAACACCAAGTGGTTGTA-3′ | Bosselut et al. (1993) |
| IK-BS4 | 5′-TCAGCTTTTGGGAATGTATTCCCTGTCA-3′ | Molnár and Georgopoulos (1994) |
| IK-BS5 | 5′-TCAGCTTTTGAGAATACCCTGTCA-3′ | Molnár and Georgopoulos (1994) |
| CREB-BS | 5′-CGCCTTGAATGACGTCAAGGC-3′ | Plet et al. (1997) |
aSequences are in the 5′ to 3′ orientation; only the top strand is indicated. Transcription factor binding motifs are underlined.
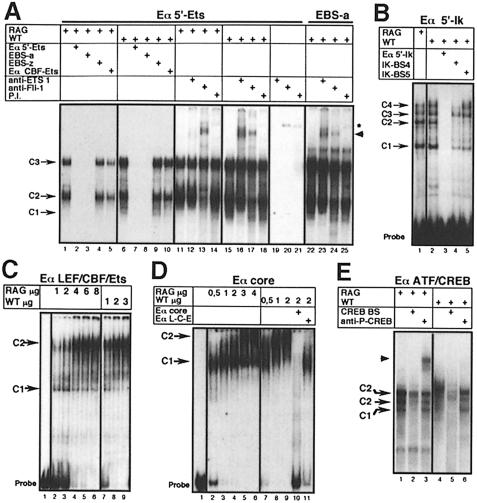
Representative data from such analyses are shown, focusing on the Eα 5′-Ets and -Ikaros motifs (Figure 6A and B) and sites within the Eα core, including the TCF/LEF plus CBF–Ets and ATF/CREB motifs (Figure 6C–E). Significantly, complexes were detected with each probe, several of which were similar when using NE of either RAG–/– or wild-type origin, suggesting that they are composed of identical (or highly related) factors (but also see below). In all cases, complexes were competed by an excess of unlabeled probe and/or oligonucleotides carrying the consensus motif, but not those in which the same motif was mutated (Figure 6 and data not shown), demonstrating that all are specific. Most strikingly, analyses of the novel motifs identified in this study (5′-Ets and -Ikaros), using oligonucleotide competition or supershift analysis with specific antisera, demonstrated that these sites can indeed be bound by a member of the predicted transcription factor family. Thus, the 5′-Ets probe revealed two major complexes (C2 and C3) for both wild-type and RAG–/– NE and an additional, minor one (C1) for wild-type NE, which were all competed by an excess of the Ets-specific competitor EBS-a but not the control EBS-z mutated probe (Figure 6A, lanes 1, 3, 4/6, 8 and 9). Specificity of complex formation at the 5′-Ets site was further emphasized by the fact that an oligonucleotide encompassing the CBF–Ets motifs within the Eα core competed inefficiently (Figure 6A, lanes 5/10). Furthermore, supershift analysis using anti-sera against specific Ets family members argues that C1 contains Ets-1 (Figure 6A, lane 16) whereas C2 contains Fli-1 (lanes 13/17). Note that similar supershifts were observed with both antisera when using the EBS-a probe as a positive control (lanes 22–25), but not when using pre-immune serum (lanes 14/18) or when the NE was omitted (lanes 19–21), indicating that the shifted bands do not result from the stabilization of a non-specific complex(es). Along the same lines, EMSA analysis of the putative Ikaros binding motifs revealed several prominent complexes (C1–C4) using both RAG–/– and wild-type NEs, which were differentially inhibited by an excess of the IK-BS4 oligonucleotide, known to bind all Ikaros isoforms (Figure 6B, lanes 1, 2, 4, and data not shown). However, competition with IK-BS5, which is not bound by Ikaros (see Molnár and Georgopoulos, 1994, for a detailed analysis of IK-BS oligoprobes), indicated that only the C4 complex may actually contain this factor (compare lanes 2, 4 and 5). Of note, the C4 complex appears more abundant with wild-type than RAG–/– NE (lanes 1/2), a finding that is reminiscent of the different DMS protection profiles observed at the corresponding site in DP versus DN thymocytes.
Fig. 6. EMSA analysis of Eα factor binding sites. Radiolabeled probes corresponding to different Eα regions (indicated at the top of each panel) were used together with RAG–/– (RAG) or wild-type thymocyte nuclear extracts (NE), as indicated. Unlabelled oligonucleotides used for competition analyses (A, B, D and E), as well as anti-sera used for supershift analyses (A and E), are also indicated. P.I., pre-immune rabbit serum. The position of the major complexes observed with the individual probe are indicated by arrows (C#; see details in the text). Supershifts are indicated by arrowheads. In (A), lanes 1–10 and 11–25 correspond to two separate experiments; a longer exposure is presented for lanes 11–14 to visualize the shifted bands better. The star indicates a non-specific complex that can be formed in the absence of NE. Probes and oligonucleotide competitors are defined in Table I.
Our DNase I studies suggest that the major changes in enhanceosome structure between DN and DP cells occur within the Eα core itself. Consistent with this, standard concentrations of NE (0.5–2 µg/ml of proteins) resulted in a marked difference in the bandshift pattern for complexes generated from either RAG–/– or wild-type NEs using a probe that includes the TCF/LEF and CBF–Ets motifs (Figure 6C). Whereas several distinct complexes were formed with RAG–/– NE, among which a complex of intermediate mobility (C1) predominates, essentially one large, slow migrating complex (C2) was detected with wild-type NE (lanes 2, 3/7 and 8). These profiles may indicate increased stability and/or synergy in the formation of the wild type-derived complex, possibly in relation to the presence of additional, identical or novel factor(s). Indeed, a complex similar to C2 was observed when using higher concentrations of RAG–/– NE (Figure 6C, lanes 4–6). The same phenomenon was reproduced upon analysis of the entire Eα core (Figure 6D; lanes 2–6/7–9). Moreover, the low-mobility Eα core–wild-type NE complex was competed by an excess of unlabeled LEF/CBF/Ets probe, leading to a faster migrating complex similar to the one observed at a low concentration of RAG–/– NE (Figure 6D, lane 11). These results are unlikely to be due to a difference in factor extractability as highly related complexes were found when using 1 µg of either NE in EMSA analysis of the CBF–Ets site from Eβ (data not shown). Altogether, these data lead us to conclude that there is a difference, between the wild-type and RAG–/– NEs, in the concentration of the factors that interact with the core motifs in vitro. This difference may correlate, at least in part, with the changes in DNase I sensitivity observed by in vivo footprinting of Eα in DP versus DN thymocytes.
Another possibility that could also account for the stage-specific activation of Eα would be post-translational modifications of pre-bound factors. One likely candidate would be CREB as activity of this factor is known to correlate with its phosphorylation state (Shaywitz and Greenberg, 1999). Indeed, using an oligonucleotide that includes the ATF/CREB motif and antisera specific for either CREB or phospho-CREB, we found that while both NEs resulted in complexes that could be supershifted by the CREB anti-serum (data not shown), only those formed using the RAG–/– NE could be supershifted in the presence of phospho-CREB antibodies (Figure 6E). These results support a model in which the in vivo complex organized at the ATF/CREB binding site of Eα may actually contain phosphorylated CREB in DN cells but, predominantly, the non-phosphorylated form of this factor in DP cells. As discussed further below, such a change may also contribute to stage-specific alterations in the architecture of the nucleo-protein complex(es) assembled at the TCRα enhancer.
Discussion
Eα shows an extensive pattern of nuclear factor occupancy in T lymphocytes
We demonstrate widespread DMS protection in vivo throughout a ∼300 bp region of Eα in T cells derived from wild-type and T-cell developmental mouse models. Not surprisingly, the occupied sites included those within the core enhancer that are sufficient to drive V(D)J recombination and/or transcription in transgenic and/or cell transfection systems (the ATF/CREB, TCF/LEF and CBF–Ets sites; see Giese et al., 1995; Mayall et al., 1997; Roberts et al., 1997). However, we also detected additional factor binding both within (the CG-rich SP-1 consensus site) and outside the core enhancer, including at sites not previously identified in Eα (detailed below). In total, up to 15 sites may be engaged, suggesting that a very large nucleo-protein complex(es) is organized at this element.
In contrast, DMS-protection studies of enhancers at the immunoglobulin light (IgL) κ or TCRβ loci have demonstrated a limited extent of nuclear factor occupancy in developing B or T lymphocytes, far less than the complex profiles described in vitro (Roque et al., 1996; Shaffer et al., 1997; Tripathi et al., 2000). How then can one explain such extensive occupancy in the case of Eα? Ultimately, the multimerization of interacting interfaces is thought to confer transcriptional synergy to the bound factors, hence increasing enhancer strength (Carey, 1998). Extensive in vivo occupancy of Eα may, at least in part, be related to the large size and complex structure of the TCRα locus (Koop and Hood, 1994) and the need for Eα to act over long distances (e.g. Eα-dependent TEA transcripts initiate >70 kb upstream).
Eα regions outside the core sequences may also be required for the fine control of enhancer function by holding in check the potential for premature activation of Eα directed by core-interacting factors (Roberts et al., 1997). The cis-elements that are responsible for this suppressive effect are currently unknown, but may include, for example, the 3′-bHLH (E-box) and associated GATA and/or CBF motifs (see Figure 3). The bHLH proteins, notably, make up a large family of transcription factors that play both positive and negative regulatory roles during cell differentiation in a large number of cell types, including B and T cells (Bain and Murre, 1998). Intriguingly, the resulting effect of bHLH binding onto Eα could be transcriptional silencing (Bernard et al., 1998). In addition, however, the new binding sites that we have identified in the 5′ Eα region may also be worth considering. They include a site for Ets and two overlapping sites for Ikaros factors, the latter associated with an E-box motif. The expression of both the Ets and Ikaros family of transcription factors is critical to T cell development (Clevers and Grosschedl, 1996) and, within the T cell lineage, is subject to precise developmental regulation (Morgan et al., 1997; Kelley et al., 1998; Anderson et al., 1999). Moreover, recent studies indicate that, in lymphoid cells, Ikaros proteins preferentially associate with silenced genes at centromeric heterochromatin (Brown et al., 1997; Hahm et al., 1998) and regulate the formation of chromatin remodeling complexes (Kim et al., 1999). Taking these facts into consideration, it is noteworthy that, whereas our in vivo DMS analyses revealed strong footprints over the Ikaros (and Ets) sites, they were not recognized in previous in vitro studies.
Eα occupancy is established early during lymphoid cell differentiation
We observe Eα occupancy in thymocytes arrested at an early DN developmental stage when the TCRα locus is not expressed, in agreement with another recently published report (Hernandez-Munain et al., 1999). Furthermore, we demonstrate the generalizability of this finding as Eα occupancy was observed in DN thymocytes independent of the nature of the developmental block, as well as in DN T cell lines. Importantly, we detected occupancy in FTF1 pro-T cells, suggesting that Eα is accessible for factor binding at the time of, or soon after, the commitment of lymphoid precursors to the T cell lineage. Interestingly, we also observed Eα binding in two immature B cell lines, although the sites that demonstrated occupancy are mainly those known to bind ubiquitous factors (such as ATF/CREB or SP-1) or factors critical for B cell development (such as the bHLH proteins). The lymphoid specificity of the footprinting profiles is emphasized by the fact that we observed no factor binding in a macrophage line, although bi-potential precursors for B cells and macrophages have been described (Cumano et al., 1992). It is believed that specific nuclear factors (master regulators) are able to bind their target sites within nucleosomal packaged DNA and reorganize the surrounding chromatin to permit additional factor binding. We propose that, as lymphoid precursors commit to the T cell lineage, expression and binding of a specific factor(s) onto Eα results in the local disruption of repressive chromatin structure and establishment of enhancer occupancy. B cell lineage commitment may also lead to Eα occupancy due to the expression of a distinct, but overlapping set of factor(s) (e.g. bHLH proteins) and the presence of cognate motifs within this element. From the comparison between the T and B cell footprints at the Eα core that, respectively, show full occupancy or occupancy of only the SP-1 and ATF/CREB sites, it is tempting to speculate that one T cell specific regulator may be LEF-1 (or TCF-1), which has been shown to nucleate the CBF/Ets protein complex on chromatin-assembled Eα templates independently of ATF/CREB binding (Mayall et al., 1997). This scenario is also consistent with the finding that TCF-1 and LEF-1 are expressed early during T cell development even though their expression peaks during the DN to DP transition (Verbeek et al., 1995).
Another interesting point regarding occupancy of the core ATF/CREB, but not CBF–Ets (and most likely TCF/LEF) sites in B cells, is the fact that Eα core occupancy does not appear to require simultaneous binding of all the core factors, as previously suggested (Hernandez-Munain et al., 1998). Although all-or-none binding at the four ATF/CREB, TCF/LEF and CBF–Ets sites may be required within the context of the minimal enhancer, it may not be necessary within the context of the full-length element at the endogenous TCRα locus.
Structural changes at Eα correlate with enhancer activity
TCRα gene expression is restricted to DP and latter stage T cells. Transgenic studies demonstrate that Eα is sufficient to activate TCRα transcription and recombination at the correct developmental stage, i.e. the DN to DP cell-transition (Krangel et al., 1998). Thus, the nucleo-protein complex(es) organized onto Eα must be different between these two thymic subpopulations. Whereas DMS footprinting revealed no major difference at Eα between DN and DP cells arguing for similar, extensive factor occupancy of cis-acting motifs, DNase I footprinting demonstrated more widespread changes in the latter, centered over the core motifs and intervening sequences. Because digestion by DNase I is especially sensitive to protein-induced wrapping, looping or bending of DNA (Mymryk et al., 1997), these results most likely imply a critical change(s) in the topographical architecture of the Eα nucleo-protein complex(es) as thymocytes develop from DN to DP cells, which correlates with activation of Eα function.
The specific nucleo-protein change(s) involved in Eα activation are still unclear. However, our study provides a first glimpse into these processes. These might include stage-specific differences in nuclear factor composition at specific Eα motifs involving a related family member or protein isoform (for example at the 5′-Ikaros sites, which demonstrate DMS protection changes in DP versus DN cells) (Figure 4B) and/or post-translational changes of a pre-existing factor(s) (e.g. a modification in the phosphorylation state, as observed for CREB) (Figure 6E). Factors such as Ikaros or CREB could provide the backbone for the assembly of a higher-order complex that includes factors that do not, themselves, contact DNA but that may either modulate chromatin structure or bridge the already bound factor(s) to modulate their activity differentially (Kim et al., 1999; Kuo and Leiden, 1999). In this respect, although CREB phosphorylation has generally been correlated with transcriptional activation (i.e. contrary to our own results, see Mayall et al., 1997; Shaywitz and Greenberg, 1999), recent evidence also points toward the possibility of the stimulation of the repressive function of certain nuclear factors, depending on the nucleo-protein context (Blobel, 2000). It is also noteworthy that Eα activation by phospho-CREB as defined in vitro by Mayall et al. (1997) most probably corresponds to a phenomenon that may take place in differentiated, mature T cells upon TCR stimulation (Mayall et al., 1997; Kuo and Leiden, 1999 and references therein), emphasizing the more complex nature of the in vivo situation. In addition to these potential mechanisms, however, the fact that the strongest DNase I hypersensitive/protected sites in DP cells are formed in the regions flanking the TCF/LEF motif (Figure 5), together with the EMSA patterns generated using probes comprised of the TCF/LEF and Ets–CBF sites (Figure 6C and D), strongly suggest that differences in the availability of the corresponding factors (and/or putative non-DNA binding partners) are involved, a notion that is consistent with the defects in expression and rearrangement of the TCRα locus in double knock-out Lef1–/–Tcf1–/– thymocytes (Okamura et al., 1998). Recent studies have brought to light several non-DNA binding proteins that could potentially be considered in this context, including ALY and TLE/Groucho, respectively, a coactivator and corepressors that interact with both LEF-1 and CBF (Bruhn et al., 1997; Levanon et al., 1998). Finally, other non-mutually-exclusive mechanisms could also be at play, including for example the involvement of an additional cis-regulatory element(s) and/or a change in the positioning of the TCRα locus in the nucleus during the DN to DP cell transition.
Previous studies, which recognized that transcription factor binding at the Eα core is highly coordinated and inter-dependent, anticipated a single-step mechanism for enhancer occupancy and activation (Giese et al., 1995; Mayall et al., 1997; Hernandez-Munain et al., 1998). Our results, however, imply a novel mode whereby the initial binding of nuclear factors (Eα occupancy) is temporally distinct from formation of the final three-dimensional complex specifically required for enhancer function (Eα activity). This apparent contradiction may stem largely from differences in the nature of the systems studied (e.g. in vitro versus in vivo, transgenic versus endogenous). Another enhancer assembly model that has been extensively analyzed is that of the IFN-β enhanceosome, an element that, in response to viral infection, recruits and organizes the transcription factors NF-κB, ATF-2/c-Jun, IRFs and the HMG protein HMG I(Y) (Maniatis et al., 1998). As recently shown, IFN-β enhanceosome formation appears to proceed through two steps by which HMG I(Y) first facilitates binding of NF-κB and ATF-2/c-Jun by inducing a bend in the DNA and then, via protein–protein contacts, aids in the formation of the final complex (Yie et al., 1999). This mechanism, however, is different from what we observe at Eα as there is no distinct temporal separation between the two phases of complex assembly.
While temporal separation between assembly and final activation by the Eα nucleo-protein complex(es) is intriguing, it may represent a relatively common mechanism by which enhancers function during the course of hematopoiesis. Thus, occupation of Eα by nuclear factors in early αβ T cell precursors may serve an important function by disrupting chromatin in this region and creating an epigenetic mark that is transmitted to the cell progeny. As these precursors differentiate, going through periods of cell proliferation and DNA replication (Kisielow and von Boehmer, 1995), the enhancer would remain ‘primed’ for activation by the binding of a complex containing one or a few additional factors and/or post-translational modifications of factors already expressed, despite the requirement for a large number of protein–DNA interactions. Moreover, as T cell precursors commit to either the αβ or γδ lineages, enhancer ‘priming’ may be a molecular mechanism required for subsequent transcriptional control by Eα of the linked TCRα or δ genes in mature αβ or γδ T cells, respectively (Sleckman et al., 1997; Monroe et al., 1999). By analogy, during hematopoietic cell differentiation, maintenance of an epigenetic memory at cis-regulatory elements would allow for rapid responsiveness in the context of a recent model that proposes the stepwise exchange of a limited number of factors within DNA-independent, pre-assembled complexes (Sieweke and Graf, 1998).
Materials and methods
Mice
The mice used in this study included wild-type C57BL/6J and Balb/c mice, single knock-out RAG-1 deficient (RAG–/–) mice (Spanopoulou et al., 1994), RAG–/– mice expressing either a TCR β transgene, P14 TCR-β (Pircher et al., 1989) (RAG–/–, TCRβ Tg), or a transgene encoding a constitutively active form of the tyrosine kinase p56lck (Abraham et al., 1991) (RAG–/–, Lck Tg), and double knock-out TCRδ deficient, TCRβ enhancer-deleted (TCRδ–/–, Eβ–/–) mice (Hempel et al., 1998). Mice were maintained in a specific pathogen-free breeding facility and were sacrificed for analysis between 4 and 6 weeks of age.
In vivo genomic footprinting assays
DMS and DNase I treatment of thymocytes and cell lines, preparation of genomic DNA from the DMS-treated and untreated cells, DMS and DNase I in vitro treatment for the naked DNA control, LM–PCR, polyacrylamide gel analysis of the amplified products, and analyses of nucleotide sequence homologies were performed as described previously (Rigaud et al., 1991; Garrity and Wold, 1992; Tripathi et al., 2000). For DNase I treatment, cells were first permeabilized using NP-40 (0.2%), then treated by increasing amounts of enzyme (2.5, 5, 10 and 20 U/ml). Two sets of specific primers were designed to analyze footprint patterns between nt 19 to 300 (nucleotide position according to Winoto and Baltimore, 1989) on the bottom strand (set 1: 5′-CAGACACACAAGG CATAGG-3′; 5′-CCTCCAAGAATCACACAGACAGCTG-3′; 5′-TCA CACAGACAGCTGCACCCTGAAATGGG-3′) and nt 288–3 on the top strand (set 2: 5′-GTGAATGGGTGTTACCACC-3′; 5′-CAATGTGC TCTCCGTGGCCTTCTTT-3′; 5′-TCTCCGTGGCCTTCTTTTCTGC ACCTGTG-3′). At least two independent experiments were performed to analyze Eα profiles for every cell type. For histogram analysis, gels were exposed to a phosphorimager and band intensities quantified using Tina 2.09 software.
During the in vivo footprint analysis, we uncovered an allelic polymorphism within the ATF/CREB binding site of Eα (verified by sequence analyses) with respect to the published sequence, consisting of a 1 nt deletion and a G/C to A/T conversion at nt 133 and 136, respectively. Only alleles from C57BL/6J origin demonstrated the published sequence. Of the cell lines used, all but EL4 had the altered sequence.
EMSA analyses
Preparation of NE, binding reaction and polyacrylamide gel analysis were performed as previously described (Costello et al., 1993). For the CREB-binding studies, the binding buffer was as follows: 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM MgCl2, 10 mM dithiothreitol (DTT), 5% glycerol, 0.05 µg/µl poly(dI–dC). Supershift and competition assays were performed by incubating the reactions for 15 min with the anti-sera or a 100-fold excess of cold competitor before the addition of the labeled probe. The following rabbit polyclonal anti-sera were used: anti-ETS1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); anti-Fli-1 (kindly provided by J.Ghysdael); anti-phospho-CREB (Upstate Biotechnology/Euromedex, Souffelweyersheim, France).
Acknowledgments
Acknowledgements
We thank Drs M.Sieweke for critical reading of this manuscript, R.Palacios for the gift of the FTF1 and SciET27F cell lines, and R.Perlmutter and H.Pircher for the gift of the RAG–/–, Lck Tg mice and the P14 TCRβ Tg mice, respectively. This work was supported by institutional grants, and specific grants from the ‘Association pour la Recherche sur le Cancer‘, the Commission of the European Communities, the ‘Fondation Princesse Grace de Monaco‘, the ‘Ligue Nationale Contre le Cancer‘, and Rhone-Poulenc Pharmaceuticals.
References
- Abraham K.M., Levin,S.D., Marth,J.D., Forbush,K.A. and Perlmutter, R.M. (1991) Delayed thymocyte development induced by augmented expression of p56lck. J. Exp. Med., 173, 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.K., Hernandez-Hoyos,G., Diamond,R.A. and Rothenberg, E.V. (1999) Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development, 126, 3131–3148. [DOI] [PubMed] [Google Scholar]
- Bain G. and Murre,C. (1998) The role of E-proteins in B- and T-lymphocyte development. Semin. Immunol., 10, 143–153. [DOI] [PubMed] [Google Scholar]
- Bernard M., Delabesse,E., Millien,C., Kirsch,I.R., Strominger,J.L. and Macintyre,E.A. (1998) Helix–loop–helix (E2-5, HEB, TAL1 and Id1) protein interaction with the TCRαδ enhancers. Int. Immunol., 10, 1539–1549. [DOI] [PubMed] [Google Scholar]
- Blobel G.A. (2000) CREB-binding protein and p300: Molecular integrators of hematopoietic transcription. Blood, 95, 745–755. [PubMed] [Google Scholar]
- Bosselut R., Levin,J., Adjadj,E. and Ghysdael,J. (1993) A single amino-acid substitution in the Ets domain alters core DNA binding specificity of Ets1 to that of the related transcription factors Elf1 and E74. Nucleic Acids Res., 21, 5184–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Guest,S.S., Smale,S.T., Hahm,K., Merkenschlager,M. and Fisher,A.G. (1997) Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell, 91, 845–854. [DOI] [PubMed] [Google Scholar]
- Bruhn L., Munnerlyn,A. and Grosschedl,R. (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev., 11, 640–653. [DOI] [PubMed] [Google Scholar]
- Capone M., Watrin,F., Fernex,C., Horvat,B., Krippl,B., Scollay,R. and Ferrier,P. (1993) TCRβ and TCRα gene enhancers confer tissue- and stage-specificity on V(D)J recombination events. EMBO J., 12, 4335–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. (1998) The enhanceosome and transcriptional synergy. Cell, 92, 5–8. [DOI] [PubMed] [Google Scholar]
- Clevers H.C. and Grosschedl,R. (1996). Transcriptional control of lymphoid development: lessons from gene targeting. Immunol. Today 17, 336–343. [DOI] [PubMed] [Google Scholar]
- Costello R., Lipcey,C., Algarte,M., Cerdan,C., Baeuerle,P.A., Olive,D. and Imbert,J. (1993) Activation of primary human T-lymphocytes through CD2 plus CD28 adhesion molecules induces long-term nuclear expression of NF-κB. Cell Growth. Differ., 4, 329–339. [PubMed] [Google Scholar]
- Cumano A., Paige,C.J., Iscove,N.N. and Brady,G. (1992) Bipotential precursors of B cells and macrophages in murine fetal liver. Nature, 356, 612–615. [DOI] [PubMed] [Google Scholar]
- Falvo J.V., Thanos,D. and Maniatis,T. (1995) Reversal of intrinsic DNA bends in the IFNβ gene enhancer by transcription factors and the architectural protein HMG I(Y). Cell, 83, 1101–1111. [DOI] [PubMed] [Google Scholar]
- Fischer A. and Malissen,B. (1998) Natural and engineered disorders of lymphocyte development. Science, 280, 237–243. [DOI] [PubMed] [Google Scholar]
- Garrity P.A. and Wold,B.J. (1992) Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc. Natl Acad. Sci. USA, 89, 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K., Kingsley,C., Kirshner,J.R. and Grosschedl,R. (1995) Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein–protein interactions. Genes Dev., 9, 995–1008. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Baron,A., Griffiths,G., Palacios,R. and von Boehmer,H. (1992) T cell receptor (TCR) β chain homodimers on the surface of immature but not mature α, γ, δ chain deficient T cell line. EMBO J., 11, 2735–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R. (1995) Higher-order nucleoprotein complexes in tran scription: analogies with site-specific recombination. Curr. Opin. Cell Biol., 7, 362–370. [DOI] [PubMed] [Google Scholar]
- Hahm K., Cobb,B.S., McCarty,A.S., Brown,K.E., Klug,C.A., Lee,R., Akashi,K., Weissman,I.L., Fisher,A.G. and Smale,S.T. (1998) Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev., 12, 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel W.M., Stanhope-Baker,P., Mathieu,N., Huang,F., Schlissel,M.S. and Ferrier,P. (1998) Enhancer control of V(D)J recombination at the TCRβ locus: differential effects on DNA cleavage and joining. Genes Dev., 12, 2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munain C., Roberts,J.L. and Krangel,M.S. (1998) Cooper ation among multiple transcription factors is required for access to minimal T-cell receptor α-enhancer chromatin in vivo. Mol. Cell. Biol., 18, 3223–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munain C., Sleckman,B.P. and Krangel,M.S. (1999) A developmental switch from TCR δ enhancer to TCR α enhancer function during thymocyte maturation. Immunity, 10, 723–733. [DOI] [PubMed] [Google Scholar]
- Ho I-C., Yang,L-H., Morle,G. and Leiden,J.M. (1989) A T-cell-specific transcriptional enhancer element 3′ of Cα in the human T-cell receptor α locus. Proc. Natl Acad. Sci. USA, 86, 6714–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley C.M., Ikeda,T., Koipally,J., Avitahl,N., Wu,L., Georgopoulos,K. and Morgan,B.A. (1998) Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr. Biol., 8, 508–515. [DOI] [PubMed] [Google Scholar]
- Kim J. et al. (1999) Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity, 10, 345–355. [DOI] [PubMed] [Google Scholar]
- Kisielow P. and von Boehmer,H. (1995) Development and selection of T cells: facts and puzzles. Adv. Immunol., 58, 87–208. [DOI] [PubMed] [Google Scholar]
- Koop B.F. and Hood,L. (1994) Striking sequences similarity over almost 100 kilobases of human and mouse T-cell receptor DNA. Nature Genet., 7, 48–53. [DOI] [PubMed] [Google Scholar]
- Krangel M.S., Hernandez-Munain,C., Lauzurica,P., McMurry,M., Roberts,J.L. and Zhong,X.P. (1998) Developmental regulation of V(D)J recombination at the TCR α/δ locus. Immunol. Rev., 165, 131–147. [DOI] [PubMed] [Google Scholar]
- Kuo C.T. and Leiden,J.M. (1999) Transcriptional regulation of T lymphocyte development and function. Annu. Rev. Immunol., 17, 149–197. [DOI] [PubMed] [Google Scholar]
- Lauzurica P. and Krangel,M.S. (1994) Temporal and lineage-specific control of T Cell Receptor α/δ gene rearrangement by T Cell Receptor α and δ enhancers. J. Exp. Med., 179, 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiden J.M. (1993) Transcriptional regulation of T cell receptor genes. Annu. Rev. Immunol., 11, 539–570. [DOI] [PubMed] [Google Scholar]
- Levanon D., Goldstein,R.E., Bernstein,Y., Tang,H., Goldenberg,D., Stifani,S., Paroush,Z. and Groner,Y. (1998) Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl Acad. Sci. USA, 95, 11590–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Falvo,J.V., Kim,T.H., Kim,T.K., Lin,C.H., Parekh,B.S. and Wathelet,M.G. (1998) Structure and function of the interferon-β enhanceosome. Cold Spring Harb. Symp. Quant. Biol., 63, 609–620. [DOI] [PubMed] [Google Scholar]
- Marine J. and Winoto,A. (1991) The human enhancer binding GATA-3 binds to several T-cell receptor regulatory elements. Proc. Natl Acad. Sci. USA, 88, 7284–7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayall T.P., Sheridan,P.L., Montminy,M.R. and Jones,K.A. (1997) Distinct roles for P-CREB and LEF-1 in TCRα enhancer assembly and activation on chromatin templates in vitro. Genes Dev., 11, 887–899. [DOI] [PubMed] [Google Scholar]
- Molnár A. and Georgopoulos,K. (1994) The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol., 14, 8292–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe R.J., Sleckman,B.P., Monroe,B.C., Khor,B., Claypool,S., Ferrini,R., Davidson,L. and Alt,F.W. (1999) Developmental regulation of TCRδ locus accessibility and expression by the TCRδ enhancer. Immunity, 10, 503–513. [DOI] [PubMed] [Google Scholar]
- Morgan B., Sun,L., Avitahl,N., Andrikopoulos,K., Ikeda,T., Gonzales,E., Wu,P., Neben,S. and Georgopoulos,K. (1997) Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J., 16, 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymryk J.S., Fryer,C.J., Jung,L.A. and Archer,T.K. (1997) Analysis of chromatin structure in vivo. Methods Enzymol., 12, 105–114. [DOI] [PubMed] [Google Scholar]
- Okamura R.M., Sigvardsson,M., Galceran,J., Verbeek,S., Clevers,H. and Grosschedl,R. (1998) Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity, 8, 11–20. [DOI] [PubMed] [Google Scholar]
- Pelkonen J., Sideras,P., Rammensee,H.G., Karjalainen,K. and Palacios,R. (1987) Thymocyte clones from 14-day mouse embryos. I. State of T cell receptor genes, surface markers and growth requirements. J. Exp. Med., 166, 1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher H., Bürki,K., Lang,R., Hengartner,H. and Zinkernagel,R.M. (1989) Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature, 342, 559–561. [DOI] [PubMed] [Google Scholar]
- Plet A., Huet,X., Algarte,M., Rech,J., Imbert,J., Philips,A. and Blanchard,J.M. (1997) Relief of cyclin A gene transcriptional inhibition during activation of human primary T lymphocytes via CD2 and CD28 adhesion molecules. Oncogene, 14, 2575–2583. [DOI] [PubMed] [Google Scholar]
- Rigaud G., Roux,J., Pictet,R. and Grange,T. (1991) In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell, 67, 977–986. [DOI] [PubMed] [Google Scholar]
- Roberts J.L., Lauzurica,P. and Krangel,M.S. (1997) Developmental regulation of VDJ recombination by the core fragment of the T cell receptor α enhancer. J. Exp. Med., 185, 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque M.C., Smith,P.A. and Blasquez,V.C. (1996) A developmentally modulated chromatin structure at the mouse immunoglobulin κ 3′ enhancer. Mol. Cell. Biol., 16, 3138–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Peng,A. and Schlissel,M.S. (1997) In vivo occupancy of the κ light chain enhancers in primary pro- and pre-B cells: a model for κ locus activation. Immunity, 6, 131–143. [DOI] [PubMed] [Google Scholar]
- Shaywitz A.J. and Greenberg,M.E. (1999) CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem., 68, 821–861. [DOI] [PubMed] [Google Scholar]
- Sieweke M.H. and Graf,T. (1998) A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev., 8, 545–551. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Bardon,C.G., Ferrini,R., Davidson,L. and Alt,F.W. (1997) Function of the TCRα enhancer in αβ and γδ T cells. Immunity, 7, 505–515. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E. et al. (1994) Functional immunoglobulin transgenes guide ordered B-cell differentiation in RAG-1-deficient mice. Genes Dev., 8, 1030–1042. [DOI] [PubMed] [Google Scholar]
- Tripathi R.K. et al. (2000) Definition of a T-cell receptor β gene core enhancer of V(D)J recombination by transgenic mapping. Mol. Cell. Biol., 20, 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M. and Clevers,H. (1992) Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson–Crick double helix. EMBO J., 11, 3039–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek S. et al. (1995) An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature, 374, 70–74. [DOI] [PubMed] [Google Scholar]
- Winoto A. and Baltimore,D. (1989) A novel, inducible and T cell-specific enhancer located at 3′ end of the T cell receptor α locus. EMBO J., 8, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yie J., Merika,M., Munshi,N., Chen,G. and Thanos,D. (1999) The role of HMG I(Y) in the assembly and function of the IFN-β enhanceosome. EMBO J., 18, 3074–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]