Abstract
A 12 kb haplotype upstream of the key signaling protein gene, AKT1, has been associated with insulin resistance and metabolic syndrome (Devaney et al. 2010). The region contains the first exon and promoter sequences of AKT1, but also includes the complete transcript unit for a highly conserved yet uncharacterized zinc finger-containing protein (ZBTB42). One of the component SNPs of the 12 kb haplotype metabolic syndrome haplotype changes a conserved amino acid in the predicted ZBTB42 protein, increasing the potential significance of the ZBTB42 transcript unit for contributing to disease risk. Using RT-PCR of human and mouse cells, we verified that the two exon ZBTB42 was expressed and correctly spliced in human skeletal muscle, and murine C2C12 cells. Production of peptide antibodies showed the expected protein in human (47 kD) and mouse (49 kD) immunoblots, and murine tissue distribution showed strongest expression in muscle and ovary. Immunostaining showed nuclear localization of the ZBTB42 protein in human muscle. Confocal imaging analyses of murine muscle showed ZBTB42 distributed in the nucleoplasm, with particular enrichment in nuclei underlying the neuromuscular junctions. The genetic association data of metabolic syndrome, coupled with the molecular characterization of the ZBTB42 transcript unit and encoded protein presented here, suggests that ZBTB42 may be involved in metabolic syndrome phenotypes.
Introduction
We recently reported the discovery of three SNPs present in high LD in highly conserved regions within and upstream of AKT1 (Devaney et al. 2010). The H1 haplotype is composed of the three ancestral alleles at the three loci. As expected this is the predominant haplotype in most populations. The H2 haplotype is composed of the three derived alleles at the loci. We have genotyped individuals from multiple American populations and found high frequencies of H2, ranging from 10 to 35.6%. The lack of intermediate haplotypes (a mix of alleles from H1 and H2) suggests that the H2 haplotype may confer some advantage to the individuals carrying it enforcing its inheritance as a haplotype block.
Due to its role as a mediator of the insulin response pathway as well as a regulator of muscle hypertrophy and muscle atrophy (Bodine et al. 2001; Elghazi et al. 2006; Nader 2005; Zdychova and Komers 2005), we genotyped the H1 and H2 haplotype tagging SNP rs1130214 in four population-based cohorts as a candidate gene for metabolic risk phenotypes (Devaney et al. 2010). These included the FAMUSS study of college-aged individuals (mean age 23.7 years) who participated in supervised resistance training sessions on their non-dominant arm (Thompson et al. 2004); the SHS, a group of 2,134 middle aged (mean age 55.5 years) Native Americans from eight different populations within the United States (Lee et al. 1990); the Health ABC Study of older individuals (mean age 73 years) with the goal of studying the effects of age on a number of health indicators including cardiovascular health and development of metabolic syndrome and T2D (Visser et al. 1999); and the STRRIDE Study, which was developed to study the effect of aerobic exercise on individuals expressing the endophenotypes of metabolic syndrome (Slentz et al. 2004). We found significant associations with all four cohorts with H2: lower fasting glucose levels in young females (FAMUSS), lower BMI and higher LDL levels in middle-aged females (SHS), lower 2 h fasting glucose levels and lower fasting insulin in middle-aged males (SHS); lower fasting glucose levels in older males (Health ABC) and higher Sg levels in middle aged European Americans (STRRIDE) (Devaney et al. 2010). H2 was strongly associated with overall risk of developing metabolic syndrome in older subjects of the Health ABC study. H2 conferred a 40% risk reduction for the development of metabolic syndrome.
The 12 kb haplotype contained three component SNPs, with the common haplotype (H1) showing the ancestral allele at each position. These SNPs were found to have functional relevance to gene expression; promoter assays containing either allele has shown a strong tissue-specific effect on gene expression (Harmon et al. 2010). One of the SNPs of the H1/H2 haplotype is present 12 kb upstream of AKT1 in the putative coding region of an uncharacterized gene, zinc finger and BTB domain containing 42 (ZBTB42). This transcript is predicted to contain 2 exons, and is highly conserved through multiple species down to the stickleback fish. However, there has been very little EST evidence in support of gene expression and splicing of this transcript unit, and the putative protein product has not been characterized. Our interest in the AKT1 H2 haplotype and its strong phenotypic associations in variable populations drew our attention to the characterization of this gene and protein.
We describe the characterization of ZBTB42 as a member of the C2H2 zinc finger protein family. Zinc finger proteins are classified by the presence of zinc finger domains, which bind to target DNA sequences and regulate transcription. ZBTB42 is expressed in skeletal muscle, and localized to the myofiber nuclei.
Materials and methods
Amplification of ZBTB42 in human and mouse cDNA
Total RNA was extracted from 50 to 100 mg of human skeletal muscle using TRIzol reagent (Invitrogen Corporation, Carlsbad, CA, USA) and cleaned using RNeasy RNA cleanup kit (Qiagen, Valencia, CA, USA). Complementary DNA was reverse transcribed from 1 ug of mRNA using a cDNA synthesis kit and oligo(dT) primers according to the manufacturer’s protocol (Invitrogen Corporation, Carlsbad, CA, USA).
PCR was performed using a forward and reverse custom primer designed to cover the first intron in human ZBTB42 causing genomic and cDNA amplicons to be different in size (5’-CGGCGACCGGAGAGGAGCTC-3′, 5′-CTTCAACGACAGGTCCAGGG-3′). Accuprime Taq polymerase (Invitrogen Corporation, Carlsbad, CA, USA) was added to the template along with buffer and the ZBTB42-specific primers. The samples were then heat activated at 94°C and cycled between denaturation (94°C for 30 s), annealing (60°C for 30 s) and extension (68°C for 1 min) 30 times. PCR products were size-separated by gel electrophoresis using a 1% agarose gel and were visualized through ethidium bromide chelation of the DNA under UV.
Custom primers (5′-GCAGTGTCTACTTCCATCTC-3′, 5′-CACTTGACTGCTCTGCAACC-3′) were designed against the mouse homolog to ZBTB42 (mZbtb42) for PCR amplification using a panel of cDNAs from various mouse tissues (Clontech, Mountain View, CA, USA). Gel electrophoresis was used to verify expression and the size of the product.
DNA sequencing
Specific primer pairs encompassing the ZBTB42 gene were used to sequence the amplified product described above (5′-CGGCGACCGGAGAGGAGCTC-3′, 5′-CTTCAACGACAGGTCCAGGG-3′). Sequencing was done using the BigDye Terminator version 3.1 Cycle Sequencing Kit and the 3130xl Genetic Analyzer according to the manufacturer’s instructions (Applied Biosystems). Nucleotide changes were identified by aligning sequences generated from both primers (forward and reverse) for comparison with corresponding sequences available in Genbank using SEQUENCHER 4.0.2 analysis software.
Identification of putative ORF and functional domains
The sequence of the coding region of ZBTB42 from the Genome Browser (University of California Santa Cruz) was input into ORFinder (NCBI) to identify the longest open reading frame. The predicted protein sequence was then input into ScanProsite (Swiss Institute of Bioinformatics) to scan the amino acid sequence for any recognizable databased protein domains.
Antibody production
Unique peptide sequences were filtered from the ORF of ZBTB42. Final peptide selection was based on values of antigenicity, hydrophilicity and surface probability (GRLQEKDRSLDPGN, DLSLKSGPRQERVH). Custom polyclonal antibodies were produced in two rabbits against the two unique peptides (for a total of four antibodies) through Invitrogen’s custom antibody service. The immune sera were affinity purified using immobilized peptides. The antibody is available upon request.
Western blot
Protein was extracted from C2C12 cells; (mouse myoblasts grown in 40 cm flasks in Dulbecco’s Modified Eagle’s Medium (Hyclone, Logan, UT, USA) with 10% FBS and 1% penicillin and streptomycin). Radioimmuno precipitation assay (RIPA) cell lysis and extraction buffer (containing DTT, and the protease inhibitors pepstatin, PMSF, antipain, and leupeptin), was used to harvest protein from C2C12 cells.
Protein from human skeletal cells was harvested from muscle sections. Muscle biopsies were sectioned using a cryostat at −20°C. RIPA lysis buffer was used to lyse the cells and the proteins were then isolated using centrifugation at 13,000 rpm for 15 min. A 2-month-old female black/6 mouse was sacrificed and tissues were harvested. Tissues were homogenized in RIPA lysis buffer using a tabletop homogenizer. Total protein extract was then isolated using centrifugation.
Total protein in each sample was quantified using the colorimetric assay RC DC Protein Assay (BioRad, Hercules, CA, USA) and BSA standards from 0 mg/ml to 10 mg/ml. 20 μg of protein was loaded for each sample in a NuPAGE® Novex 4–12% Bis–Tris Gel (Invitrogen, Carlsbad, CA, USA). The proteins were then transferred to a nitrocellulose membrane, blocked with 5% milk and incubated overnight with our custom antibody AX-1#3 against peptide DLSLKSGPRQERVH. Washes were done with 5% milk in TBST. The membrane was then incubated with HRP-tagged goat anti-rabbit secondary antibodies. ECL detection reagents (Amersham pharmacia biotech) were used to detect the HRP and the membrane was exposed to film.
Immunohistochemistry
Muscle sections were cut 10 μm thick laterally at the belly of frozen human skeletal muscle biopsies in a cryostat set at −20°C. The sections were arranged on Superfrost glass slides and allowed to air-dry overnight. The sections were blocked with 5% horse serum and then incubated with our custom antibody AX-1#3 (from rabbit) and anti-merosin (from mouse) for 2 h. The excess primary antibodies were removed by three five-minute washes with PBS. The sections were incubated with the secondary antibodies: anti-rabbit Cy3 and anti-mouse Cy2. The sections were treated with the nuclear stain DAPI. Sections were covered and visualized on an apotome microscope as individual stains and merged images.
For confocal microscopy, mouse gastrocnemius muscle was fixed in situ with 2% paraformaldehyde at the time of dissection and stored in 10% sucrose/PBS at 4°C. Gastrocnemius muscle was teased apart into small fiber bundles and incubated in block and permeabilization buffer (0.5% TritonX®-100, 0.1% Tween 20, 2% BSA, 20% goat-horse serum blend in PBS) overnight at 4°C. Fibers were incubated in custom affinity purified antibody AX-1#3 (1:500 dilution in antibody buffer: 0.2% TritonX®-100, 0.1% Tween 20, 2% BSA in PBS) for 2 h with gentle mixing. Fibers were centrifuged at 4,000×g for 1 min then washed with 0.1% Tween-20/PBS, repeating this cycle three times. Fibers were incubated with secondary antibody (Alexa Fluor® 555 F(ab′)2 fragment of goat anti-rabbit IgG) (H+L)(1:500 in antibody buffer) (Invitrogen, Carlsbad, CA, USA) for 2 h and washed three times as stated above. Fibers were counterstained with Alexa Fluor® 488 α-bungarotoxin conjugate and DAPI to label neuromuscular junctions and nuclei respectively and washed three more times (Invitrogen, Carlsbad, CA, USA). Fibers were left in wash buffer overnight at 4°C with gentle mixing then mounted in Fluoromount-G (Southern Biotech, Birmingham, Alabama) the following day. Confocal laser-scanning microscopy was performed using a Carl Zeiss LSM 510 laser scanning microscope coupled to Zen LE microscopy software (Carl Zeiss, Germany).
Results
ZBTB42 is expressed as a transcript in human skeletal muscle cDNA
ZBTB42 is predicted to contain two exons and a single intervening intron according to limited EST evidence on the UCSC genome browser. To determine if ZBTB42 is transcribed into a full length mRNA in vivo, we attempted to amplify the transcript using RT-PCR with ZBTB42-specific primers in human skeletal muscle. The PCR primers were designed against the two exons; the forward primer in exon 1, the reverse primer in exon 2. The primers cover intron 1, therefore allowing us to differentiate cDNA (introns spliced out) from genomic DNA (containing introns), which was included as a control. The expected size products between the two primers are 600 bps for cDNA and 1,500 bps for genomic DNA. The genomic sample showed one band at 1,500 bp corresponding to the non-spliced genomic locus. The cDNA showed a band at 600 bp, corresponding to the predicted size between the primers. There are other larger bands that were amplified in the cDNA sample, including one at 1,500 bp, which is probably genomic DNA contamination from the mRNA isolation and cDNA synthesis steps (Fig. 1). This indicated that the EST ZBTB42 exists as an mRNA transcript within human skeletal muscle.
Fig. 1.
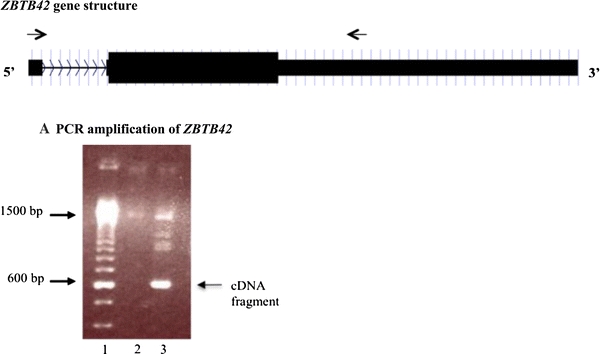
ZBTB42 cDNA is expressed in human skeletal muscle. a shows a schematic of the ZBTB42 gene. The black boxes represent the two exons of ZBTB42. The line with arrow heads is the intronic region and shows the direction of the gene. The black arrows above the gene are the forward and reverse primers used to amplify the transcript. The primers were designed across the intron, in exons 1 and 2, to distinguish between genomic DNA (1,500 bp) and cDNA (600 bp). b shows the amplification of ZBTB42 from human cDNA and genomic DNA. cDNA was reverse transcribed from total RNA isolated from a frozen human skeletal muscle biopsy (lane 3). Genomic DNA (lane 2) was isolated from human skeletal muscle. cDNA was amplified at the correct size of 600 bp. There are additional bands in the cDNA sample including one at 1,500 bp. This may be genomic DNA contamination. PCR products were loaded in a 1% agarose gel for size discrimination. A 100 bp marker was used for size comparison (lane 1)
To confirm that the cDNA PCR product was ZBTB42 and not a non-specific binding event, we eluted the 600 bp product from the gel and sequenced it. The sequence matched the sequence of ZBTB42 from the UCSC genome browser database.
ZBTB42 has C2H2 zinc finger domains and an N-terminal POZ domain
Upon sequence investigation using online protein prediction tools (ORFinder, ScanProsite), the predicted primary structure of the protein translated from ZBTB42 shows four zinc finger domains at its C-terminus, which are highly homologous with other members of the C2H2 class of zinc finger proteins. In addition, there is a POZ domain (named for its prevalence in POxvirus proteins and Zinc finger proteins) at the N-terminus of the protein, which is common to most members of the C2H2 family of proteins (Fig. 2). The predicted size of the protein using the amino acid sequence from ORFinder is 47 kDa. Upon comparison with the mouse homolog, Zbtb42, we observed that the two proteins share 75% amino acid identity at the protein level, with 94% amino acid identity in the zinc finger and POZ domains. These analyses indicated that ZBTB42 would be an expressed protein.
Fig. 2.
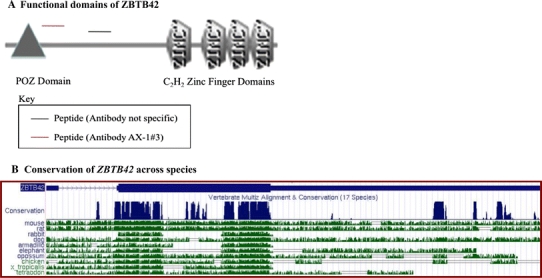
ZBTB42 contains C2H2 zinc finger domains and shares high sequence homology with mouse. ZBTB42 is a member of the C2H2 zinc finger family of proteins. a The four C2H2 zinc finger domains of ZBTB42 at the C-terminal and the POZ domain at the N-terminal. The antibody AX-1#3 was designed against peptide DLSLKSGPRQERVH (black line) present in between the conserved domains. The antibody with non-specific binding (red line) was designed against peptide GRLQEKDRSLDPGN. b The conservation of ZBTB42 (annotated as ZBTB42) across species. ZBTB42 shares the highest level of homology with mouse and rat sequences
ZBTB42 is translated into a protein in human skeletal muscle
To directly identify whether ZBTB42 is an expressed protein in vivo we produced two antibodies against two distinct peptide sequences near the predicted N-terminus (Fig. 2). The peptides were designed to exclude the POZ and zinc finger functional domains due to anticipated cross-reaction with other zinc finger domain and POZ domain-containing proteins (amino acids 189–202 [DLSLKSGPRQERVH] and 116–129 [GRLQEKDRSLDPGN]).
Each of the four antibody preparations was tested by immunoblot using human skeletal muscle lysates as well as lysates from C2C12 mouse myoblasts. The antibody against peptide DLSLKSGPRQERVH from rabbit #3 (AX-1#3) showed the best specificity and sensitivity for the predicted 47 kDa protein in both the human skeletal muscle and mouse myoblasts (Fig. 3). The antibodies raised against GRLQEKDRSLDPGN were non-specific as evidenced by large amount of non-specific binding in the immunoblot analysis of human skeletal muscle lysates and C2C12 lysates. We also observed a second band at 36 kDa in the human protein lysates. This may be a splice variant or reflect non-specific binding of another protein. The immunoblot analysis provided strong evidence that this protein is not only transcribed into an mRNA in vivo but is also translated as a protein in human skeletal muscle.
Fig. 3.
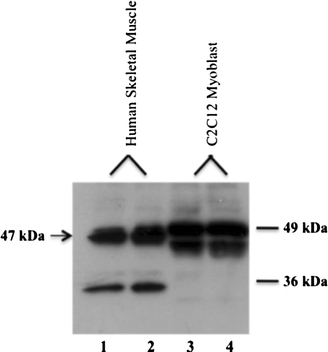
ZBTB42 protein is expressed in human skeletal muscle and a murine cell line C2C12 myotubes. Immunoblot analysis with AX-1#3 shows ZBTB42 protein expression (47 kDa) in lysates isolated from two human skeletal muscle samples sectioned from frozen biopsies (lanes 1, 2) and mZbtb42 protein expression from two cultures of murine cell line C2C12 myotubes (lanes 3, 4) (47 kDa). 20 μg of protein was loaded for each sample. A second band of 36 kDa is present in human skeletal muscle, but is absent from the C2C12. There is a double band in the C2C12 samples. This may be a splice variant or post-translational modification
mZbtb42 protein is differentially expressed in a panel of mouse tissues
A panel of murine tissues was tested by immunoblot using our AX-1#3 antibody, and a loading control (vinculin) (Fig. 4). The murine orthologue of ZBTB42, Zbtb42, was detected at the expected 47 kDa molecular weight, and showed highest expression in skeletal muscle and ovary, as well as C2C12 cell line myotubes. Low expression was seen in brain, lung, spleen, liver and heart. The kidney and intestines do not express mZbtb42. The liver and skeletal muscle showed strong expression of a smaller protein product (36 kDa), which may be a splice variant of mZbtb42 specific to these tissues.
Fig. 4.
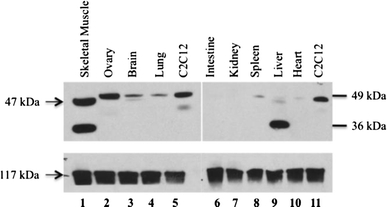
mZbtb42 is variably expressed in mouse tissues with strong expression in skeletal muscle. Immunoblot analysis of lysates from a panel of mouse tissues shows that mZbtb42 is highly expressed variably across tissues. 20 μg of protein was loaded for each tissue. High expression is seen in skeletal muscle and ovary (lanes 1, 2). Brain, lung, spleen, liver and heart show low expression (lanes 3, 4, 8, 9, 10) while mouse intestines and kidneys show no expression of mZbtb42 (lanes 6, 7). The skeletal muscle (lane 1) and liver (lane 9) show strong expression of a smaller molecular weight product (36 kDa). This may be a splice variant specific to those tissues, or a cross-reactive protein. Lysates from C2C12 cells were loaded as positive controls (lanes 5, 11). Total vinculin is shown as a loading control
ZBTB42 localizes to the nucleus in skeletal muscle, with enrichment at neuromuscular junction nuclei
Zinc finger proteins typically localize to the nuclei of cells and function as transcription factors when the zinc finger domains bind to DNA targets to initiate transcription (Iuchi 2001). To evaluate cellular localization we co-stained human skeletal muscle cross sections with an antibody against ZBTB42 (AX-1#3), and an antibody against merosin, which is a muscle cell membrane protein (Fig. 5). ZBTB42 protein was localized to the nuclei in skeletal muscle myofibers and interstitial (perimysial) connective tissue cells. The addition of 4′,6-diamidino-2-phenylindole (DAPI), a nuclear stain, verified that the ZBTB42 protein was present in all nuclei (Fig. 6).
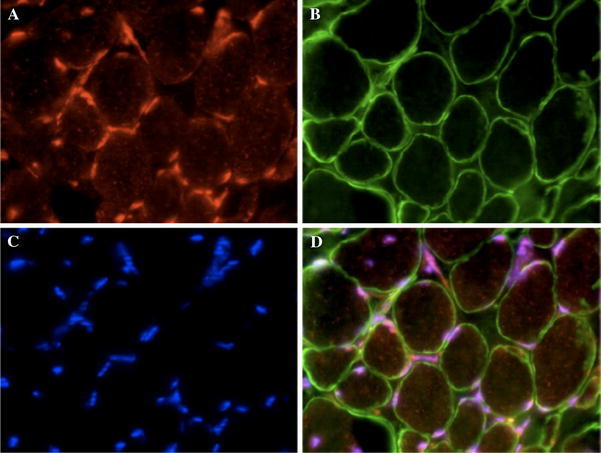
Fig. 5.
ZBTB42 localizes to the nuclei of human skeletal muscle fibers. Immunofluorescence analysis with AX-1#3 (visualized by Cy3-conjugated secondary antibody) of human skeletal muscle sections (10 μm) shows ZBTB42 expression exclusive to the nuclei. Sections are cut laterally through the muscle resulting in cross-sectional view of the muscle cells. Muscle cell nuclei are peripherally localized. The membrane is stained with anti-merosin (visualized by Cy2-conjugated secondary antibody). Images were taken on an apotome microscope at 100× magnification
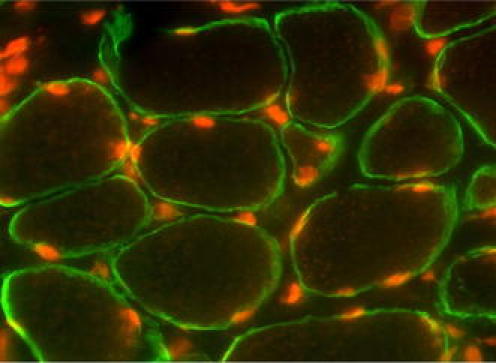
Fig. 6.
ZBTB42 localizes to the nuclei of human skeletal muscle fibers and merges with a nuclear stain. Immunofluorescence analysis of a triple-stained muscle section shows nuclear localization of ZBTB42 in skeletal muscle fibers. Sections are cut laterally through the muscle resulting in cross-sectional view of the muscle cells. Muscle cell nuclei are peripherally localized. a AX-1#3 stain of ZBTB42 in human skeletal muscle shows staining exclusive to the nuclei (Cy3-conjugated secondary antibody). b Anti-merosin staining shows expression of the membrane protein, merosin (Cy2-conjugated secondary antibody). c DAPI nuclear stain. d Merge of all three stains shows perfect alignment of DAPI and ZBTB42 expression in muscle cell nuclei. Images were taken on an apotome microscope at 40× magnification
To determine the localization within nuclei, the ZBTB42 protein was studied using confocal microscopy in murine skeletal fixed and teased myofibers (Fig. 7). The ZBTB42 protein was seen localized to myofiber nuclei superimposed on the DNA chromatin (DAPI), suggesting a localization within the nucleoplasm (Fig. 7a). Myofibers have distinct nuclear domains within a syncytium, with myonuclei underlying the neuromuscular junction showing specialized function. Identification of neuromuscular junctions (NMJs) was done using biochemical staining for acetylcholine receptors with labeled bungarotoxin, and ZBTB42 co-localized with NMJs (Fig. 7b). This analysis showed ZBTB42 to be highly enriched in subsynaptic nuclei at the NMJ, again within the nucleoplasm (Fig. 7b, c).
Fig. 7.
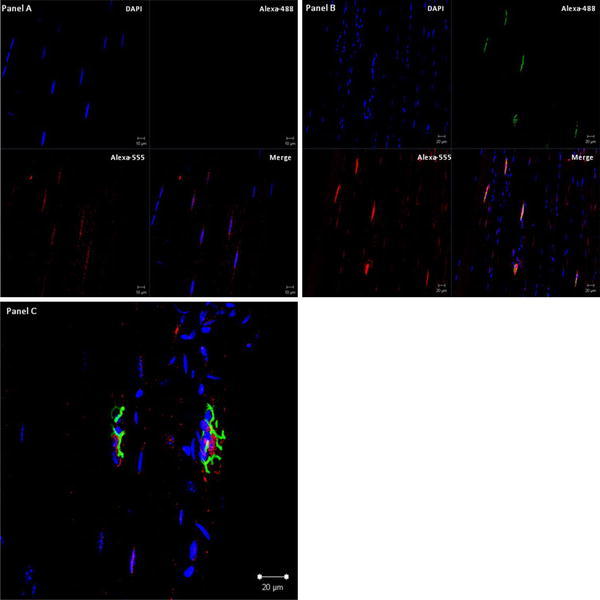
ZBTB42 localizes to the nuclei of mouse skeletal muscle fibers, and is enriched at the neuromuscular junction. Shown is mouse gastrocnemius skeletal muscle fiber bundles stained for immunofluorescence of ZBTB42 protein (Alexa Fluor® 555; AX-1#3 antibody), nuclear DNA (DAPI), and neuromuscular junctions (Alexa Fluor® 488 α-bungarotoxin conjugate). a Extra-synaptic myofiber nuclei (absence of Alexa Fluor® 488 α-bungarotoxin signal). b Synaptic myofiber nuclei (presence of Alexa Fluor® 488 α-bungarotoxin signal). c High resolution superimposed image of the ZBTB42 protein (red), nuclear DNA (blue) and bungarotoxin post-synaptic myofiber membrane (green). ZBTB42 protein signal is seen in most myofiber nuclei within the nucleoplasm. The protein was highly enriched in nuclei underlying the neuromuscular junction
Discussion
We recently reported strong associations between a three SNP haplotype encompassing 12 kb upstream and inclusive of the first exon of the AKT1 gene, and containing the ZBTB42 gene. One of the strongly linked SNPs, rs10141867, was in the coding sequence of the ZBTB42 transcript. We show that ZBTB42 is transcribed into a spliced mRNA in human skeletal muscle through RT-PCR. We further demonstrated that this transcript is translated into a 47 kD protein in vivo in human skeletal muscle immunoblots using a custom designed antibody directed against a unique peptide on the N-terminus of the predicted open reading frame (ORF) of ZBTB42.
The mouse homolog mZbtb42 is homologous to hZBTB42, sharing 94% amino acid identity in the functional domains and an overall amino acid identity of 75%. We found that the mZbtb42 protein is variably expressed in mouse tissues, with skeletal muscle and ovaries showing the strongest expression. Both human and mouse immunoblots showed some heterogeneity of protein size, with possible doublets of similar molecular weight, and also a strong 36 kD smaller signal in murine muscle and liver (Fig. 4). Further work is required to determine if the 36 kD protein represents alternative processing of the mZbtb42 protein, or a cross-reactive protein specific to those tissues.
The predicted ORF and putative functional domains were identified (virtual characterization) with the web tools ORFinder (NCBI) and ScanProsite. The functional domains of ZBTB42 are most similar to members of the C2H2 zinc finger protein family. There are different families of zinc finger proteins characterized by domains at the C-terminus that contain the amino acids cysteine and histidine in a specific order to create a unique secondary structure stabilized by a zinc ion bond. The C2H2 zinc finger proteins have two cysteine and two histidine residues organized CX2-4CX12HX2-6H. The C2H2 domains bind to the DNA through a short alpha-helix domain and initiate transcription of target genes (Chang et al. 1996; Iuchi 2001). Depending on the number of zinc finger domains, they are able to bind to different sequences of DNA, RNA and, in some instances, proteins (Chang et al. 1996; Iuchi 2001). At the N-terminal of the ZBTB42ORF there is a POZ domain. POZ domains are named for their presence in a large number of POxvirus proteins and Zinc finger proteins (Albagli et al. 1995; Bardwell and Treisman 1994; Li et al. 1999). POZ domains function as binding mediators either by aiding homodimerization or heterodimerization (Bardwell and Treisman 1994; Chen et al. 1995; Huynh and Bardwell 1998). In the case of transcriptional regulators, such as zinc finger proteins that contain POZ domains, these domains mediate transcriptional repression through recruitment of co-repressor proteins or by altering the binding efficiency between the zinc finger domains and the target DNA (Chang et al. 1996).
The virtual evidence from ORFinder and ScanProsite described above, by way of sequence analysis and a protein domain database query is significant but the true characterization of a protein must be done through studies of expression and other functional tests. Zinc finger proteins localize to the nuclei of cells where they regulate the transcription of target genes. Immunohistochemistry of human skeletal muscle sections showed sensitive and specific binding of ZBTB42 in the nuclei of human muscle fibers. The staining of ZBTB42 co-localized with the nuclear stain DAPI. Confocal localizations within murine muscle confirmed nuclear localization, with further localization within the nucleoplasm (co-localization with DAPI nucleic acid stain) (Fig. 6). There are distinct nuclear domains within the syncytial muscle fibers, with nuclei underlying the neuromuscular junction (NMJ) having distinct transcriptional programs. We identified sub-synaptic NMJ nuclei using biochemical staining with labeled bungarotoxin, and showed that mZbtb42 protein was highly enriched within the nucleoplasm of NMJ nuclei (Fig. 7). These date bolster our hypothesis that this protein has active zinc finger domains and may function as a novel muscle-relevant transcription factor in vivo.
As noted above, the ZBTB42 transcript unit is contained within a 12 kb haplotype associated with insulin resistance and metabolic syndrome (Devaney et al. 2010). The component polymorphisms in this haplotype show strong activity in modulating transcriptional reporter assays, and haplotype-dependent variable transcriptional modulation of AKT1, or ZBTB42, or both provides a potential molecular mechanism for modulating insulin resistance (a muscle phenotype), and metabolic syndrome (Harmon et al. 2010). AKT1 has clear functional roles in muscle remodeling that are likely relevant to insulin resistance and metabolic syndrome. The data presented here on ZBTB42 suggests that this novel protein may also have a hitherto undiscovered role in muscle and metabolic syndrome phenotypes.
Acknowledgments
Supported by grants from the National Institutes of Health (2RO1 AR55100; National Center for Medical Rehabilitation Research 5R24HD050846).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein–protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein–protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zollman S, Couderc JL, Laski FA. The BTB domain of bric a brac mediates dimerization in vitro. Mol Cell Biol. 1995;15:3424–3429. doi: 10.1128/mcb.15.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Devaney JM, Gordish-Dressman H, Harmon BT, Bradbury MK, Devaney SA, Harris TB, Thompson PD, Clarkson PM, Price TB, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Seip RL, Seo J, Kim BH, Tosi LL, Garcia M, Li R, Zmuda JM, Delmonico MJ, Lindsay RS, Howard BV, Kraus WE, Hoffman EP (2010) AKT1 polymorphisms are associated with risk for metabolic syndrome. Hum Genet (in press) PubMed PMID: 21061022 [DOI] [PMC free article] [PubMed]
- Elghazi L, Balcazar N, Bernal-Mizrachi E. Emerging role of protein kinase B/Akt signaling in pancreatic beta-cell mass and function. Int J Biochem Cell Biol. 2006;38:157–163. doi: 10.1016/j.biocel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Harmon BT, Devaney SA, Gordish-Dressman H, Reeves EK, Zhao P, Devaney JM, Hoffman EP. Functional characterization of a haplotype in the AKT1 gene associated with glucose homeostasis and metabolic syndrome. Hum Genet. 2010;128:635–645. doi: 10.1007/s00439-010-0891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, Cucchiara AJ, Savage PJ, Howard BV. The strong heart study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–1155. doi: 10.1093/oxfordjournals.aje.a115757. [DOI] [PubMed] [Google Scholar]
- Li X, Peng H, Schultz DC, Lopez-Guisa JM, Rauscher FJ, 3rd, Marmorstein R. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 1999;59:5275–5282. [PubMed] [Google Scholar]
- Nader GA. Molecular determinants of skeletal muscle mass: getting the “AKT” together. Int J Biochem Cell Biol. 2005;37:1985–1996. doi: 10.1016/j.biocel.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Slentz CA, Duscha BD, Johnson JL, Ketchum K, Aiken LB, Samsa GP, Houmard JA, Bales CW, Kraus WE. Effects of the amount of exercise on body weight, body composition, and measures of central obesity: STRRIDE—a randomized controlled study. Arch Intern Med. 2004;164:31–39. doi: 10.1001/archinte.164.1.31. [DOI] [PubMed] [Google Scholar]
- Thompson MNPD, Seip R, Price T, Clarkson P, Angelopoulos T, Gordon P, Pescatello L, Visich P, Zoeller R, Devaney JM, Gordish H, Bilbie S, Hoffman EP. Functional polymorphisms associated with human muscle size and strength. Med Sci Sports Exerc. 2004;36:1132–1139. doi: 10.1249/01.MSS.0000132274.26612.23. [DOI] [PubMed] [Google Scholar]
- Visser M, Fuerst T, Lang T, Salamone L, Harris TB. Validity of fan-beam dual-energy X-ray absorptiometry for measuring fat-free mass and leg muscle mass. Health, aging, and body composition study—dual-energy X-ray absorptiometry and body composition working group. J Appl Physiol. 1999;87:1513–1520. doi: 10.1152/jappl.1999.87.4.1513. [DOI] [PubMed] [Google Scholar]
- Zdychova J, Komers R. Emerging role of Akt kinase/protein kinase B signaling in pathophysiology of diabetes and its complications. Physiol Res. 2005;54:1–16. doi: 10.33549/physiolres.930582. [DOI] [PubMed] [Google Scholar]


