Abstract
Gallic acid (GA), a key intermediate in the synthesis of plant hydrolysable tannins, is also a primary anti-inflammatory, cardio-protective agent found in wine, tea, and cocoa. In this publication, we reveal the identity of a gene and encoded protein essential for GA synthesis. Although it has long been recognized that plants, bacteria, and fungi synthesize and accumulate GA, the pathway leading to its synthesis was largely unknown. Here we provide evidence that shikimate dehydrogenase (SDH), a shikimate pathway enzyme essential for aromatic amino acid synthesis, is also required for GA production. Escherichia coli (E. coli) aroE mutants lacking a functional SDH can be complemented with the plant enzyme such that they grew on media lacking aromatic amino acids and produced GA in vitro. Transgenic Nicotiana tabacum lines expressing a Juglans regia SDH exhibited a 500% increase in GA accumulation. The J. regia and E. coli SDH was purified via overexpression in E. coli and used to measure substrate and cofactor kinetics, following reduction of NADP+ to NADPH. Reversed-phase liquid chromatography coupled to electrospray mass spectrometry (RP-LC/ESI–MS) was used to quantify and validate GA production through dehydrogenation of 3-dehydroshikimate (3-DHS) by purified E. coli and J. regia SDH when shikimic acid (SA) or 3-DHS were used as substrates and NADP+ as cofactor. Finally, we show that purified E. coli and J. regia SDH produced GA in vitro.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-011-9739-3) contains supplementary material, which is available to authorized users.
Keywords: AroE, Gallic acid, Juglans regia, Escherichia coli, Shikimate dehydrogenase, Walnut, Tannins
Introduction
Gallic acid (GA) is an essential precursor for many plant secondary metabolites, particularly gallo- and ellagitannins (Gross 1982; Grundhoefer et al. 2001; Haslam 1998). In addition, GA is an active component responsible for reducing coronary arterial disease (CAD) and arterial thrombosis in people who consume red wine, green tea, and fruit (Appeldoorn et al. 2005). Plants and fungi produce high concentrations of GA (Werner et al. 1997). In recent times, an early intermediate of the shikimate pathway, 3-DHS, was identified as a potential precursor for GA synthesis in plants (Werner et al. 1997, 1999, 2004). DHS and GA synthesis using a recombinant E. coli were found in a fed–batch fermentor where glucose availability, oxygenation, and solution pH was controlled, but the authors were not able to obtain a cell-free GA-producing system (Li et al. 1999).
Sequence identities of the gene(s) encoding protein(s) responsible for GA production have not been reported from either plants or bacteria. Two potential pathways for GA synthesis have been postulated: (1) direct oxidation of 3-DHS to GA and (2) dehydration of 3-DHS to protocatechuic acid (PCA) followed by hydroxylation of PCA (Fig. 1; Kambourakis et al. 2000; Li and Frost 1999). Several studies have attempted to validate these competing hypotheses, but clear biochemical/genetic evidence supporting either hypothesis has remained elusive (Werner et al. 1997, 1999, 2004; Chandran and Frost 2001). However, retrobiosynthetic NMR studies with 13C-labeled glucose (Werner et al. 1997, 1999) and oxygen isotope ratio mass spectrometry (Werner et al. 2004) demonstrated that GA is synthesized entirely or predominantly by dehydrogenation of DHS.
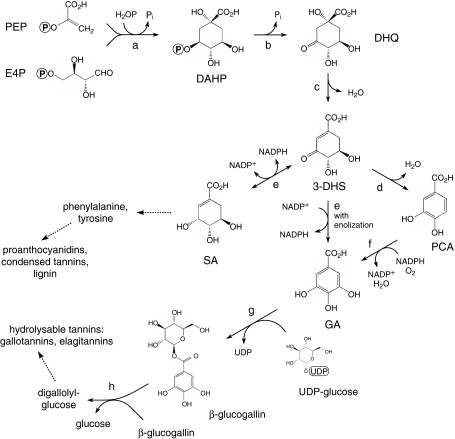
Fig. 1.
Synthesis of gallic and shikimic acids in plants. Enzymes: (a) DAHP synthase (AroF); (b) DHQ synthase (AroB); (c) DHQ dehydratase (AroD); (d) DHS dehydratase (AroZ); (e) shikimate dehydrogenase (AroE); (f) PCA hydroxylase (hypothetical enzyme); (g) UDP-glucose: gallate 1-O-galloyltransferase (Li et al. 1999; Michel et al. 2003); (h) 2-O-gallolyltransferse (Haslam 1994; Michel et al. 2003). Abbreviations of biosynthetic intermediates: PEP phosphoenolpyruvic acid, E4P D-erythrose-4-phosphate, DAHP 3-deoxy-D-arabino-heptulosonic acid-7-phosphate, DHQ 3-dehydroquinic acid, 3-DHS 3-dehydroshikimic acid, PCA protocatechuic acid, SA shikimic acid, GA gallic acid, β-glucogallin 1-O- galloyl-β-D- glucose
In this study, we cloned the gene responsible for GA synthesis from plants (Arabidopsis thaliana and Juglans regia) and E. coli. Interestingly, the corresponding protein had been previously characterized as shikimate dehydrogenase (SDH, EC 1.1.1.25), a key enzyme in the shikimate pathway. SDH had been shown previously to catalyze NADPH-dependent reduction of 3-DHS to shikimic acid (SA), which is ultimately used to produce the aromatic amino acids l-tyrosine, l-tryptophan, and l-phenylalanine (Fig. 1) (Padyana and Burley 2003; Vogan 2003; Ye et al. 2003; Singh and Christendat 2006). Here, we use genetic complementation of E. coli and biochemical analysis to demonstrate that SDH also catalyzes not only the NADP+-dependent dehydrogenation of SA to 3-DHS, but also the dehydrogenation of 3-DHS to GA. Understanding the unique, dual roles of SDH will allow selective modification and increase of GA synthesis, improving the nutritional value of crop plants.
Experimental procedures
Protein extraction from wild-type E. coli and plant species
Eight different plant species were used in this study (Table 1). Mature leaf tissue of Arabidopsis thaliana, Lycopersicon esculentum, Malus domestica, Medicago sativa, Nicotiana benthamiana L., Nicotiana tabacum L., and Oryza sativa, bract tissue of Gossypium arboretum and Gossypium hirsutum, and juvenile seed coat tissue from Juglans regia were flash frozen in liquid N2 and stored at −80°C. One gram of each sample was homogenized in 2 mL cold extraction buffer (2% insoluble polyvinylpolypyrrolidone, 0.05 M Tris, 0.007 M citric acid monohydrate, 0.006 M cysteine HCl monohydrate, 0.006 M ascorbic acid, and 0.001 M polyethylene glycol 8000; pH 8.3). The slurry was centrifuged 25 min at 13,000×g and 4°C.
Table 1.
Bacterial strains, plasmids, and plant material
| Strain/plasmid/tissue | Genotype | Source/Ref |
|---|---|---|
| Strains | ||
| Top10 | lacZΔM15 recA1 | Invitrogen |
| Top10F′ | lacZΔM15 recA1 F′(lacIq) | Invitrogen |
| AB2834 | aroE353− | CGSCa |
| 38F11 | TAIRb | |
| Ec.AroE.2834 | pEc.AroE.pA in AB2834 | This study |
| At.SDH.2834 | pAt.SDH.pB in AB2834 | This study |
| Jr.SDH.2834 | pJr.SDH.pB in AB2834 | This study |
| Plasmids | ||
| pEc.AroE | Apr, Cmr, AroE in pCR®2.1-TOPO | This study |
| pEc.AroE.pA | Apr, AroE in pTrcHisA | This study |
| pAt.38F11 | Apr, Cmr, SDH in pCR®2.1-TOPO | This study |
| pAt.SDH.pB | Apr, SDH in pTrcHisB | This study |
| pJr.SDH | Apr, Cmr, SDH in pCR®2.1-TOPO | This study |
| pJr.SDH.pB | Apr, SDH in pTrcHisB | This study |
| Plant material | ||
| Apple | M. domestica | |
| Arabidopsis | A. thaliana | |
| Cotton (diploid) | G. arboretum | |
| Cotton (triploid) | G. hirsutum | |
| Tomato | L. esculentum | |
| Tobacco | N. benthamiana L. | |
| Tobacco | N. tabacum L. | |
| Walnut | J. regia | |
| Rice | O. sativa | |
| Alfalfa | M. sativa | |
aCGSC- E. coli Genetic Stock Center, http://cgsc.biology.yale.edu/cgsc.html
bTAIR-Arabidopsis Information Resource Center, http://www.arabidopsis.org/
Soluble proteins were isolated from wild type E. coli and E. coli AB2834, which lacks a functional SDH, using the CelLytic B Bacterial Cell Lysis/Extraction reagent according to the manufacturer’s instructions (Sigma–Aldrich, St. Louis, MO).
Purification of SDHs from transformed E. coli cells
Escherichia coli cells were transformed with protein expression vectors containing the coding sequences for SDH from J. regia, A. thaliana and E. coli (AroE). All transformed cell lines were grown in 200 mL Luria–Bertani (LB) broth containing 100 μg/mL ampicillin and 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). The cultures were grown for 24 h at 37°C and 220 rpm and proteins were extracted using the CelLytic system according to the manufacturer’s instructions. SDH proteins were purified on a precharged nickel Sepharose column under native conditions (Histidine GraviTrap, GE Healthcare/Amersham, Piscataway, NJ).
Protein activity assay
Plant and bacterial soluble proteins were separated on a native polyacrylamide TBE gel (Walker 1994) for 1.5 h at 110 V and 4°C. Following separation, the gel was stained for SDH activity using a staining solution of 20 mg NADP+, 21 mg SA, 20 mg 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT), and 1.5 mg phenazine methosulfate (PMS) in 60 mL 100 mM Tris buffer pH 9.0 (Diaz et al. 1997, 2001). The position of the bands from each sample was measured and recorded. A second, duplicate gel was cut into eight sections: gel slice 1 from the first 0–0.5 cm; slice 2, 0.5–1.0 cm; slice 3, 1.0–1.6 cm; slice 4, 1.6–2.2 cm; slice 5, 2.2–3.0 cm; slice 6, 3.0–3.5 cm; slice 7, 3.5–4.2 cm; and slice 8, 4.2–6.0 cm. Each gel slice was diced into ~1 mm2 pieces and incubated in an assay mixture containing 0.5 mL 50 mM Tris pH 8.5, 5 mM SA, and 5 mM NADP+ (Sigma–Aldrich, St. Louis, MO) at 28°C for 20 h. Heat-denatured J. regia protein and J. regia protein incubated in the absence of SA were included as negative controls. Gallic acid production from each gel slice was measured using rhodanine (Fluka AG, Germany) as described (Sharma and Dawra 2000). The blank contained 0.5 mL assay mixture and 1 cm2 polyacrylamide gel.
Enzyme kinetics assays
The enzymatic activity of purified SDH from E. coli AroE.2834 and J. regia SDH.2834 were assayed at 20°C by monitoring reduction of NADP+ to NADPH spectrophotometrically at 340 nm using an extinction coefficient of 6.18 mM−1 cm−1 in the presence of SA. Saturation kinetics studies were carried out at pH 9.0
To measure the kinetics of SA oxidation, the assay mixture (total volume 200 μL) consisted of 100 mM Tris–HCl, pH 9.0, 4 mM NADP+, and variable SA concentration. To measure the kinetics of NADP+, the assay mixture consisted of 100 mM Tris–HCl, pH 9.0, 4 mM SA, and variable NADP+ concentration. To measure enzyme activity, 0.2 ng E. coli AroE or J. regia SDH dissolved in 10 μL buffer was added to the assay mixture. The enzyme concentrations were chosen to follow the initial reaction rate. The absorbance at 340 nm was measured for 10 min against a blank consisting of the assay mixture without enzyme. To test possible inhibition by NAD+, enzyme activity was assayed using a mixture of 100 mM Tris–HCl, pH 9.0, 4 mM SA, 200 μM NADP+, and 20 mM NAD+. Each measurement was taken in triplicate. Kinetic parameters were calculated using the Lineweaver–Burk equation.
For the 3-DHS and GA assay, 0.1 μg purified E. coli AroE or purified J. regia SDH was incubated at 30°C in the presence of 100 mM Tris pH 9.0 with 4 mM SA and 2 mM NADP+ or 4 mM 3-DHS and 2 mM NADP+ (total volume 200 μL). After 20 h, the reactions were stopped by heat denaturation and 3-DHS and GA from the reaction mixtures were assayed by reversed-phase liquid chromatography coupled to electrospray mass spectrometry (RP–LC/ESI–MS) using a modification of a described method (Zou and Tolstikov 2008). An Onyx Monolithic C-18 reversed-phase silica-based column was used for separation (100 × 3 mm; Phenomenex, Torrance, CA). Analytical liquid chromatography was performed using 0.1% formic acid in methanol as the mobile phase at flow rates of 0.7 mL/min at 30°C. After 0.5 min at 0% methanol, a gradient to 100% methanol was concluded over 18 min, then the flow continued at 100% methanol to 25 min. Following a column wash, the run concluded with 100% methanol at 30 min. The column was equilibrated with the starting buffer for at least 5 min before injection of the next sample. The injection volume was 5 μL. Pure 3-DHS, GA and SA were used as external standards for retention time indexing and calibration curve building.
The entire effluent from the HPLC column was directed into the electrospray ionization source (ESI) of a LTQ linear ion trap mass spectrometer (ThermoFinnigan) operated under Xcalibur software (V1.4; ThermoFinnigan). The electrospray voltage was 5 kV. Nitrogen sheath and auxiliary gas flow were 60 and 20 arbitrary units, respectively. The ion transfer capillary temperature was 350°C. Typical ion gauge pressure was 0.90 × 10−5. Full scan spectra were acquired from 150 to 2,000 amu at unit mass resolution with maximum injection time set to 200 ms in one micro scan (Zou and Tolstikov 2008).
Protein sequencing
Proteins were separated on a 10% Tris polyacrylamide gel (READY GEL, BioRad, Hercules, CA). Bands were excised, washed in sterile water, and dried. Following rehydration in 10 mM dithiothreitol and 100 mM ammonium bicarbonate, the gel was incubated 1 h at 55°C. The rehydration solution was replaced with 55 mM iodoacetamide and 100 mM NH4HCO3, the gel was incubated 45 min in the dark and dehydrated in the presence of acetonitrile. Tryptic digestion was performed as described (Lee et al. 2003). The peptide mixture was dissolved in 5% formic acid and loaded onto a micro-column containing POROS R2 resin (Perceptive Biosystems, Framingham, MA). The peptides were eluted onto a gold-coated nanospray ES capillary using a solution of 50% methanol and 5% formic acid (Protana, Odense, Denmark). Peptides from the capillary were sprayed onto a hybrid nanospray/ESI–Quadrupole–TOF–MS and MS/MS mass spectrometer (QSTAR) according to the manufacturer’s instructions (Applied Biosystems Inc., Foster City, CA). QSTAR instrument calibration was performed with a standard peptide mixture yielding mass accuracies of 5 ppm or better. MS/MS fragmentation data were used for manual sequencing.
Cloning SDH from bacteria and plants
To clone SDH from bacteria, the nucleic acid sequence of AroE from E. coli K-12 was obtained from GenBank (acc. AE000406). The 5′ primer (5′Aro: 5′-GGATCCGAAACCTATGCTGTTTTTGGTAATCCG-3′) and the 3′ primer (3′Aro.pA:5′-CTGCAGTCACGCGGACAATTCCTCCTGCAATTGCT-3′) were used for polymerase chain reaction (PCR)-based cloning. TOP10F′ inoculum was used as the template (Invitrogen, Carlsbad, CA). All amplifications were performed on a GeneAmp PCR System 9700 thermal cycler (PE Applied Biosystems, Foster City, CA). The PCR conditions were as follows: 1 cycle of 94°C for 1 min followed by 25 cycles of 94°C for 1 min, 60°C for 1 min, 68°C for 2 min, followed by a final extension of 68°C for 10 min.
To clone SDH from A. thaliana, a full-length cDNA, designated 38F11, was obtained from the Arabidopsis Information Resource Center (TAIR, DNA Stock Center, Columbus, OH). PCR-based cloning was performed. The sequence of the 5′primer (5′Arab.pB) was 5′-CGCGGATCCGAGCTTGATCTGTGCTCCAGTGATGGCGGATTCAATAGAC-3′ and the 3′ primer (3′Arab.pB) was 5′-CGCCTGCAGTCAGTACTTTGACATTATTTGCCAGTAGAGTTCCTTTGGAGCGGG-3′. The PCR conditions were as follows: 25 cycles of 94°C for 1 min, 60°C for 2 min, 68°C for 4 min, followed by a final extension of 68°C for 10 min.
SDH from walnut was obtained from juvenile seed coat of J. regia cv. Tulare. Tissue was dissected away from the embryo, flash frozen in liquid N2, and stored at −80°C until use. mRNA isolated from the seed coat was converted into cDNA using the ProSTAR First-Strand RT–PCR Kit (Stratagene, La Jolla, CA). Degenerate primers were used to amplify the shikimate dehydrogenase message. The sequence of the 5′ primer (5′ShDe-1) was 5′-CAC(C/T)TACAG(A/G)CC(A/C)A(A/C)(A/T)TGGGAAG(A/T/G)GG(C/T)CA(A/G)TATG(A/C)(A/T)GGTGATGA-3′. The sequence of the 3′ primer (3′ShDe-2) was 5′ GC(A/C)CCCTT(C/T)TC(C/T)TTTGC(A/C)CC(A/G)TAAGCAA(T/G)(A/T)GCCTTGCC-3′. The conditions for PCR amplification were as follows: 30 cycles of 94°C for 1 min, 52°C for 1 min, 68°C for 2 min, followed by a final extension of 68°C for 10 min.
Each PCR product described above was ligated to the pCR2.1-TOPO vector (TA TOPO Cloning Kit, Invitrogen, Carlsbad, CA). Plasmid DNA from selected colonies was isolated (QIAprep Spin Miniprep Kit, Qiagen, Valencia, CA), sequenced (Davis Sequencing, Davis, CA), and designated pEC.AroE, pAt.38F11, and pJr.SDH for the E. coli, A. thaliana, and J. regia clones, respectively (Table 1). DNA was digested with the restriction endonucleases BamHI and PstI for E. coli and A. thaliana and with BamHI and HindIII for J. regia, then ligated to the appropriate pTrcHis expression vector (TrcHis A, B, and C, Version F, Invitrogen, Carlsbad, CA).
Functional complementation in E.coli mutants
The E. coli strain AB2834, which lacks a functional SDH, was obtained from the E. coli Genetic Stock Center, Yale University. Electrochemically competent AB2834 cells were generated and transformed with each expression vector construct as described (Ausubel 1998). The transformants were grown in SOC media for 1 h at 37°C, transferred to M63 minimal medium broth (100 mM KH2PO4, 15 mM (NH4)2SO4, 90 nM FeSO4, 71 mM KOH, 22 mM glucose, 0.001% thiamine, 1 mM MgSO4, and 100 μg/mL ampicillin), grown for 3 h at 37°C, and finally plated on M63 minimal medium agar plates and incubated for 36 h at 37°C. Colonies were selected and grown overnight in 5 mL LB containing 100 μg/mL ampicillin.
Vector construction for Agrobacterium-mediated plant transformation
A full-length transit peptide from the Silene pratensis ferredoxin precursor, designated pTP12, was kindly provided by K. Inoue (University of California, Davis) (Smeekens et al. 1985). PCR-based cloning was performed. The sequence of the 5′primer (PTPXho2) was 5′- CGCCTCGAGATGGCTTCTACACTCTCTACCCTCTCGGTGAGCG -3′ and that of the 3′ primer (PTPBam2) was 5′- CGCGGATCCACCTTGTATGTGGCCATTGCAGTCACTCTGCC -3′. The PCR conditions were as for J. regia SDH, but followed by a final extension at 68°C for 4 min. The PCR product was ligated to the pCR2.1-TOPO and screened as above; the correct clone was labeled pTP.PTP. Clone pTP.PTP was ligated into an intermediary cassette containing a CaMV35S promoter sequence and octopine synthase terminator to generate pDU04.1706. The SDH gene from pJr.SDH was then ligated to the transit peptide to generate pDU04.2327. This expression cassette was then ligated into the binary plasmid pDU99.2215 (Escobar et al. 2001) to generate pDU04.2601 (Supplementary Fig. 1). As a negative control, the transit peptide from pDU04.1706 was ligated to pDU99.2215 to generate pPTP. The resultant binary vectors were transformed into the disarmed A. tumefaciens strain EHA105:pCH32 (Hamilton 1997) by electroporation.
Plant Transformation
The Ralph M. Parsons Foundation Plant Transformation Facility (University of California, Davis) transformed pDU04.2601 and pPTP into N. tabacum cv SR1. Transformants were selected on kanamycin plates and screened for β-glucuronidase (GUS) activity (Jefferson 1987). Transformations with these plasmids were confirmed through PCR. The 5′ primer corresponded to the CaMV35S promoter and the 3′ primer (3′Stop) was 5′- AAGCTTCTAGTTATTGGCCATAACTTTTCTAAATAGCTCC-3′.
Overexpression of J. regia SDH in N. tabacum
Four to five leaves were collected from each 4-week-old transformed experimental or control line. Total GA was extracted from the fresh tissue and measured in triplicate using reverse-phase HPLC as described above.
Results
Co-localization of shikimic acid and gallic acid production in wild type plants
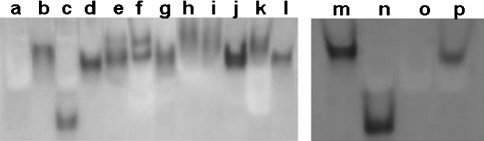
Soluble protein isolated from wild type E. coli and plants (J. regia, A. thaliana, M. sativa, N. tabacum, M. domestica, G. arboretum, G. hirsutum, O. sativa, L. esculentum, and N. benthamiana) exhibited functional SDH activity using SA as the substrate (Fig. 2). Soluble protein from N. tabacum exhibited two SDH activity bands, consistent with prior reports of polymorphic forms of SDH (Fig 2f, Colich et al. 2009). As expected, E. coli SDH migrated on the gel approximately twice as fast as the plant proteins because it lacks the plant dehydroquinase domain (Ye et al. 2003; Singh and Christendat 2006; Haslam 1994; Michel et al. 2003). Heat-denatured J. regia protein was included as a negative control and did not exhibit SDH activity. 3.5 μg protein extract from J. regia, 3.1 μg from wild type E. coli, 7.1 μg from A. thaliana, 9.1 μg from M. sativa, 9.7 μg from N. tabacum L, 6.2 μg from M. domestica, 8.6 μg from G. arboretum, 8.7 μg from G. hirsutum, 10.1 μg from O. sativa., 11.6 μg from L. esculentum, and 5.8 μg from N. benthamiana L. yielded approximately equal SDH activity, as evidenced by band intensity on the gel. Of the J. regia, E. coli, and A. thaliana SDH expressed in E. coli AB2834, 13.1 μg, 3.1 μg, and 6.0 μg protein, respectively, was required to detect approximately equal enzyme activity (Fig. 2).
Fig. 2.
SDH activity from plant and bacterial protein extracts. (a) 3.1 μg protein extract from AB2834, (b) 3.5 μg from J. regia, (c) 3.1 μg E. coli, (d) 7.1 μg from A. thaliana, (e) 9.1 μg from M. sativa, (f) 9.7 μg from N. tabacum, (g) 6.2 μg from M. domestica, (h) 8.6 μg from G. arboretum, (i) 8.7 μg from G. hirsutum, (j) 10.1 μg from O. sativa, (k) 11.6 μg from L. esculentum, and (l) 5.8 μg from N. benthamiana. SDH fror A. thaliana, J. regia, and E. coli expressed in E. coli were also purified. Protein from the following samples are in lanes (m) pJr.SDH.pB expressing J. regia SDH in AB2834 (13.1μg protein), (n) pEc.AroE.pA expressing E. coli SDH in AB2834 (3.1μg protein), (o) AB2834 (3.1μg protein), and (p) pAt.SDH.pB expressing A. thaliana SDH in AB2834 (6.0 μg protein)
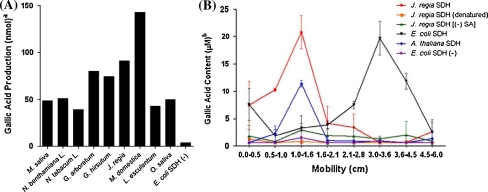
We measured GA synthesis catalyzed by fractionated protein from each sample. We detected SDH activity in a single section of each gel. Gallic acid production at a given SA concentration was highest in the two tree species, J. regia and M. domestica, with herbaceous plants and E. coli producing 33 and 66% less total GA, respectively (Fig. 3a). Heat-denatured J. regia proteins and proteins incubated without SA did not produce GA above background levels (Fig. 3b). For each bacterial and plant sample tested, only protein from the region of the gel containing the SDH band catalyzed GA production.
Fig. 3.
Functional analysis of SDH from various sources. Bands were excised from the gel and tested for gallic acid production. a SDH activity in excised gel bands from the following plant sources: 1) M. sativa; 2) N. benthamiana L; 3) N. tabacum; 4) G. arboretum; 5) G. hirsutum; 6) J. regia; 7) M. domestica; 8) L. esculentum; 9) O. sativa and 10) AB2834. b SDH from E. coli, A. thaliana and J. regia was cloned, transformed into AB2834, and purified on a nickel column. Gallic acid production was measured from each of the following: (black inverted triangle) purified Ec.AroE; (green triangle) purified Jr.SDH [(-) substrate (shikimic acid)]; (orange square) purified Jr. SDH (denatured protein); (red circle ) purified Jr.SDH; (blue diamond) purified At.SDH and (purple diamond) purified protein from E. coli AB2834 (non-transfromed). a aThe same amount of protein (950 ng) was used for all species, the mean values presented are the average of three replicates. b b The values presented are the average ± standard deviation of three replicates
Characterization of aroE mutant strain of E. coli (AB2834)
AB2834 cells of E. coli (aroE) deficient in endogenous SA production required shikimate pathway products and intermediates to grow on minimal medium. Conversely, wild-type E. coli colonies were observed on minimal medium lacking SA after 16 h at 37°C. AB2834 cells required medium with 10−6 M SA, 10−6 M L-Phe, 10−6 M L-Trp, and 10−6 M L-Tyr to exhibit growth similar to wild-type cells.
Proteins isolated from wild type E. coli and mutant E. coli strain AB2834 were separated on a native gel and stained for SDH activity. Detectable shikimate-dependent NADPH formation was generated by 3.1 μg total wild type protein, indicating the presence of a functional, endogenous SDH. Conversely, 44.1 μg AB2834 protein yielded no detectable SDH activity.
Proteins isolated from wild type and AB2834 E. coli were separated on a native gel and their GA concentrations were quantified using the rhodanine stain (Fig. 3B; Salminen 2003). Wild type protein produced 57.5 μmoles GA/μg. Conversely, AB2834 protein did not produce GA above background levels.
Cloning SDH from bacteria and plants
To determine the role of SDH in synthesis of GA, we cloned aroE from E. coli (pEc.AroE) and 3-dehydroquinase/3-shikimate dehydrogenase from A. thaliana (pAt.38F11) and J. regia (pJr.SDH) (Table 1). To eliminate any effects of the transit peptide on protein expression and enzyme activity in E.coli, we deleted the 5′ ends from the plant cDNA sequences. We estimated that the transit peptide included the first 76 amino acids of the immature A. thaliana protein based on its deduced sequence homology with SDH from O. sativa, N. tabacum, and L. esculentum (data not shown). The predicted average hydrophobicity of SDH residues 1-76 from pAt.38F11 was 0.12, compared with 0.04 for the remainder of the protein. Similarly, we generated a truncated walnut SDH that lacked a transit peptide, based on its sequence homology with the predicted transit peptide from A. thaliana. The 5′ ends of pJr.SDH and pAt.38F11 corresponded to approximately the same region of SDH. Because of the absence of targeting sequences in bacteria, pEc.AroE represented the full-length SDH message obtained from GenBank (acc: D90811).
pAt.38F11, pJr.SDH, and pEc.AroE were aligned using the multiple alignment Clustal W parameters (unpublished information). pAt.38F11 and pJr.SDH exhibited 75% sequence homology of both nucleotides and amino acids. The amino acid sequence identity between pEc.AroE and pAt.38F11 or pJr.SDH was 29% and 28%, respectively. AroE-3 exhibited increased homology to the C-terminal ends of the plant proteins. 3-Dehydroquinase from E. coli, encoded by AroD, was 28% homologous to the region immediately following the transit peptide from both A. thaliana and J. regia.
Functional complementation in E. coli
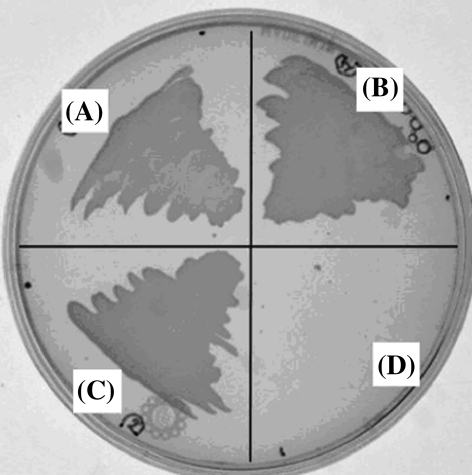
Each sequence encoding heterologous protein was cloned into the expression vector pTrcHis, screened, and designated pEc.AroE.pA, pAt.SDH.pB, and pJr.SDH.pB for the E. coli, A. thaliana, and J. regia SDH, respectively (Table 1). The vectors were electroporated into AB2834, where their expression complemented the mutant phenotype. The bacterial strains Ec.AroE.2834, At.SDH.2834 and Jr.SDH.2834 grew on minimal medium lacking shikimate pathway products and intermediates. Bacterial colonies from these strains were observed after 16 h incubation at 37°C (Fig. 4). Soluble proteins from each mutant line showed SDH activity with SA used as the substrate (Fig. 2).
Fig. 4.
Complementation of AB2834 with A. thaliana, J. regia, and E. coli SDH clones. The samples are as follows: a pJr.SDH.pB expressing J. regia SDH in AB2834, b pEc.AroE.pA expressing E. coli SDH in AB2834, c pAt.SDH.pB expressing A. thaliana SDH in AB2834, and d non-transformed AB2834
The heterologous proteins from E. coli, Ec.AroE.2834, At.SDH.2834, and Jr.SDH.2834 contained a 6xHis tag at their N-terminus to facilitate binding to a nickel column. Purified SDH from these lines was digested with trypsin. The resulting peptides were sequenced using ESI–MS–MS. The amino acid sequences exhibited perfect homology to regions of the corresponding translated SDH clones.
We measured GA production by the three purified proteins. Fractionated Ec.AroE.2835, At.SDH2834, and Jr.SDH.2834 SDH produced 14.6, 23.2, and 17.0 times more GA, respectively, than denatured controls (Fig. 3b). Across all three samples, GA extended into the gel slice immediately preceding that with SDH activity.
Kinetic properties of purified SDH from Ec.AroE.2834 and Jr.SDH.2834
Kinetic properties of E. coli SDH purified from E. coli (Ec.AroE.2834) were determined. The kinetic parameters for SA were Km = 102 μM, Vmax = 3.36 nmol min−1 and k cat = 8,750 s−1 (Lineweaver–Burk method, R2 = 0.99). The kinetic parameters for NADP+ were Km = 347 μM, Vmax = 4.6 nmol min−1 (Lineweaver–Burk method, R2 = 0.96).
We also determined the kinetic properties of J. regia SDH purified from E. coli (Jr.SDH.2834). The kinetic parameters for SA were Km = 860 μM, Vmax = 14.9 nmol min−1 and k cat = 137,833 s−1 (Lineweaver–Burk method, R2 = 0.93). The Km for SA was similar to previously reported values for the SDH enzyme in other plant species (Singh and Christendat 2006; Ossipov et al. 2000; Balinsky et al. 1971). The kinetic parameters for NADP+ were Km = 860 μM, Vmax = 10.7 nmol min−1 (Lineweaver–Burk method, R2 = 0.94). E. coli and J. regia SDH oxidized SA using NADP+ as a cofactor but displayed no activity in the presence of NAD+. E. coli and J. regia SDH activity was not significantly altered when both cofactors were in the same assay mixture, which agrees with published reports that NAD+ is not a competitive inhibitor of NADP+ or that AroE does not bind NAD+ (Michel et al. 2003).
Gallate and 3-dehydroshikimate analysis
3-DHS and GA production by enzymatic action of E. coli (Ec.AroE.2834) and J. regia (Jr.SDH.2834) SDH expressed in E. coli was quantified by RP-LC/ESI–MS. 3-DHS was produced at about the same concentration for both E. coli and J. regia SDH. However, GA was produced at highest concentrations by E. coli SDH (Table 2).
Table 2.
3-Dehydroshikimate (3-DHS) and gallic acid (GA) production by the action of E. coli (Ec.AroE.2834) and J. regia (Jr.SDH.2834) purified shikimate dehydrogenase expressed in E. coli
| Enzyme | Substrate (4 mM) | Cofactor (2 mM) | 3-DHSa (μM) | GAa (μΜ) |
|---|---|---|---|---|
| E. coli AroE | SA | NADP+ | 40.57 ± 2.15 | 94.03 ± 1.99 |
| J. regia SDH | SA | NADP+ | 39.87 ± 0.70 | 14.03 ± 0.90 |
| J. regia SDH | 3-DHS | NADP+ | 19.35 ± 0.60 | 25.06 ± 0.20 |
Shikimic acid or 3-DHS and NADP+ were used as substrate and cofactor, respectively. Quantification was done by RP-LC/ESI–MS
aThe values presented are the average ± standard deviation of three replicates
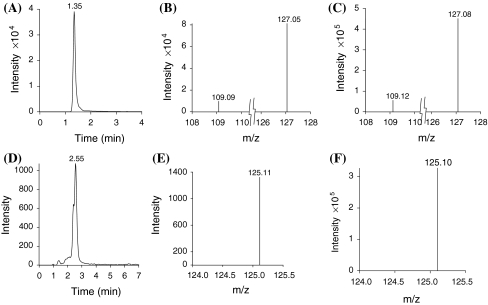
Peaks corresponding to the retention times for 3-DHS (1.35 min, Fig. 5a) and GA (~2.55 min, Fig. 5D) were detected by RP-LC from the same reaction mixture resulting from the J. regia (Jr.SDH.2834) purified SDH activity. Gallic acid and 3-DHS peaks were not observed from negative controls lacking enzyme or substrate. 3-DHS (Fig. 5b) and GA (Fig. 5e) peaks from the same reaction mixture and from pure 3-DHS (Fig. 5c) and pure GA corresponded to the ESI–MS fragmentation pattern (Fig. 5f).
Fig. 5.
Reversed-phase liquid chromatography coupled to electrospray mass spectrometry (RP-LC/ESI–MS analysis of 3-dehydroshikimate (3-DHS) and gallic acid (GA) produced by dehydrogenation of 3-dehydroshikimate (3-DHS) by the action of J. regia SDH expressed in E. coli (Jr.SDH.2834) when shikimic acid was used as a substrate a 3-DHS formed in the reaction mixture b MS spectrum for 3-DHS from the reaction products, c MS spectrum of free 3-DHS standard, d GA formed in the reaction mixture, e MS spectrum for GA from reaction products, and f MS spectrum of free GA standard
In planta validation via transgenic expression of J. regia SDH in N. tabacum
Juglans regia SDH was expressed under the control of the CaMV35S promoter. Seven independently transformed lines containing the SDH gene from p.Jr.SDH from pDU04.2601 were identified and designated JrSkD016, JrSkD017, JrSkD023, JrSkD036, JrSkD038, JrSkD042, and JrSkD043. N. tabacum transformed with pPTP, the SDH sequence lacking a transit peptide, was used as a negative control. Two independent control lines were identified and designated PTP014 and PTP018. We did not detect any visible phenotypic differences between non-transformed N. tabacum, the controls and experimental transformed lines. Mild acid hydrolysis was used to liberate GA from hydrolysable tannins and galloyl-glucose molecules. There was no significant difference in GA accumulation between non-transformed and pPTP lines (Table 3). Six of the seven experimental lines exhibited increased GA accumulation: JrSkD016, JrSkD017, JrSkD023, JrSkD036, JrSkD038, and JrSkD043 produced 540, 178, 237, 290, 470, and 150% more GA, respectively, than the average from PTP014 and PTP018. JrSD042 exhibited 35% less GA.
Table 3.
Gallic acid production by four week old transformed experimental and control tobacco lines
| Plant Line | Gallic Acid (pmol/mg FW)a |
|---|---|
| JrSkD016 | 309 ± 1 |
| JrSkD017 | 102 ± 2 |
| JrSkD023 | 135 ± 3 |
| JrSkD036 | 165 ± 1 |
| JrSkD038 | 270 ± 3 |
| JrSkD042 | 37 ± 1 |
| JrSkD043 | 88 ± 1 |
| PTP014 | 49 ± 1 |
| PTP018 | 65 ± 1 |
| Non-transformed | 38 ± 1 |
JrSkD samples were transformed with J. regia SDH. PTP samples lacked J. regia SDH. The non-transformed control is wild type N. tabacum. Quantification was done by RP-LC
aThe values presented are the average ± standard deviation of two replicates
Discussion
Although previous studies examined the biosynthetic route leading to GA formation, the gene responsible for its synthesis has not yet been reported (Kambourakis et al. 2000; Haslam 1994; Conn and Swain 1961; Werner et al. 1999). Here, we demonstrate that a shikimate pathway enzyme, SDH, is directly responsible for GA production in both plants and bacteria. We obtained an E. coli cell line, AB2834, known to contain a non-functional SDH (Li et al. 1999). In characterizing its mutant phenotype, we determined that the cells were: (1) not able to convert 3-DHS to SA and vice versa and (2) completely lacked the ability to synthesize GA in vitro. This led us to postulate that SDH was directly and/or indirectly responsible for both SA and GA production.
We cloned SDH from J. regia, a species that produces very elevated concentrations of GA. To conclusively demonstrate that SDH is responsible for GA production, we performed over-expression analysis of J. regia SDH in N. tabacum. Significantly, our transformed lines accumulated over 500% more GA than controls. To verify these results, we cloned and purified SDH from J. regia (Jr.SDH.2834). The purified protein synthesized GA in vitro, confirming that SDH is directly responsible for GA production in planta.
Previous studies examining synthesis of GA in plants were based on the underlying assumption that a mono-functional enzyme is responsible for GA formation (Ossipov et al. 2003; Kambourakis et al. 2000; Werner et al. 1999). For example, an enzyme capable of synthesizing GA from 3-DHS was partially purified from Betula pubescens and designated dehydroshikimate dehydrogenase (DSkDG) (Ossipov et al. 2003). DSkDG synthesized GA from 3-DHS, which had been produced from SA. Our results suggest that GA synthesis is catalyzed by a bi-functional SDH, instead of by a mono-functional enzyme such as DSkDG.
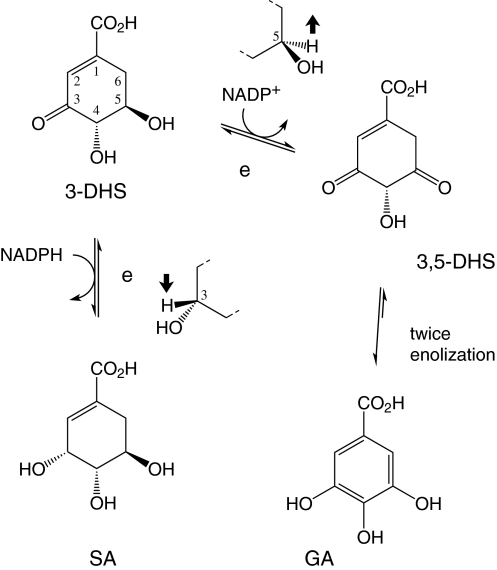
Retrobiosynthetic NMR studies with 13C-labeled glucose (Werner et al. 1997, 1999) and oxygen isotope ratio mass spectrometry (Werner et al. 2004) demonstrated that GA is synthesized entirely or predominantly by dehydrogenation of DHS. Our enzyme activity analysis for measuring GA production demonstrates that 3-DHS, the fifth intermediate of the shikimic acid pathway, is the substrate required for GA production. We demonstrated that SDH converted SA to 3-DHS (Fig. 5a) and that the 3-DHS produced was subsequently used as the precursor for GA (Fig. 5d). SDH had previously been shown to catalyze the NADPH-dependent reduction of 3-DHS to SA, which requires reduction of the carbonyl group on C-3 of the 3-DHS ring structure (Fig. 1 and 6) (Li et al. 1999; Chandran et al. 2003; Li and Frost 1999). Here, the cofactor NADPH and substrate 3-DHS must be oriented in such a way as to form a key that matches the enzyme’s lock for the enzymatic reaction to proceed. Based on the structures of 3-DHS and SA, NADPH must be oriented properly in the enzyme-cofactor-substrate complex. Then, while H− is delivered to C-3 from the top, an H+ is available to immediately bond with the forming O− (bottom left with respect to the 3-DHS structure in Fig. 6). For the reverse reaction, oxidation of SA to 3-DHS, NADP+ extracts the H− from SA, and the H+ linked with O goes back to form 3-DHS. The experimental results suggest that SDH can also catalyze a second oxidation with another substrate (i.e, 3-DHS).
Fig. 6.
Mechanism of shikimic acid and gallic acid synthesis from 3-dehydroshikimate by SDH. Enzyme: (e) Shikimate dehydrogenase, SDH, (Aro E). Abbreviations of biosynthetic intermediates: SA Shikimic acid, 3-DHS 3-dehydroshikimate, 3,5-DHS 3,5-dehydroshikimate, GA, Gallic acid
It is clear from the above description that the precursor for the reaction to GA should have features similar to SA for this reaction to occur. After flipping the orientation of 3-DHS vertically (Fig. 6), this substance has the necessary requirements for being a suitable substrate. The C-5 of 3-DHS is in a similar position with respect to SDH and NADP+, reminiscent of C-3 in SA of the complex for the dehydrogenation of SA to 3-DHS (Fig. 6). In this new orientation of 3-DHS the OH at C-5 is down and H is up, analogous to OH and H at C-3 of SA. In addition, because of the OH at the C-4 position and COO− at the C-1 position, it is possible that while holding the substrate in the enzyme active site, the enzyme can still perform the oxidation of SA to 3-DHS or the oxidization of 3-DHS to GA.
Given the structural similarities between 3-DHS and SA, it is likely that because SA can be oxidized to 3-DHS, 3-DHS can be oxidized to 3,5-DHS. The intermediate 3,5-DHS immediately and spontaneously converts to GA, as the enolization of 3,5-DHS to GA is energetically favorable. Countering one suggestion, the conversion between two alternate forms of 3-DHS (keto form to enol form) is likely not required for GA production, since the enol form of 3-DHS is energetically highly unfavorable. In the presence of SDH, 3-DHS is converted to the intermediate 3,5-DHS which immediately enolizes to yield GA. The enolization is highly energetically favorable and should therefore occur spontaneously. Interestingly, the proposed SDH reaction mechanism would result in continuous generation of both SA and GA. The NADP-bound enzyme could convert 3-DHS to SA (reduction with NADPH) as well as GA (oxidation with the just-formed NADP+). This cycle could potentially continue as long as the supply of 3-DHS exists, with the result that both SA and GA are produced, albeit not necessarily in equal amounts.
Plants synthesize two major classes of tannins: hydrolysable tannins containing GA residues and structurally distinct, but functionally equivalent, condensed tannins (proanthocyanidins) (Fig. 1; Charlton et al. 1996; Khanbabaee and van Ree 2001). Condensed tannins are synthesized from l-phenylalanine, an end product of the shikimate pathway, as part of plant phenylpropanoid metabolism (Haslam 1994; Khanbabaee and van Ree 2001). The redox state of the cell could potentially control the direction of SDH activity. Therefore, a change in the ratio of intracellular [NADPH]/[NADPH + NADP+] could affect accumulation of the different tannins in plant tissues (Lee et al. 2003). Variability in the affinity of NADP+ binding by SDH, potentially due to structural differences in the enzyme among species, could control the relative proportions of condensed and hydrolysable tannins (Ye et al. 2003).
In plants, SDH is associated with 3-dehydroquinase (DHQ, AroD), an enzyme that synthesizes 3-DHS from dehydroquinate. Dehydroquinate is converted to 3-DHS (DHQ activity) and 3-DHS is subsequently used to produce SA (SDH activity) (Fig. 1; Vogan 2003; Ye et al. 2003; Michel et al. 2003). The N- and C- terminal domains of the plant protein exhibit sequence homology to the bacterial DHQ (AroD) and SDH (AroE) proteins, respectively. As we have now demonstrated that purified SDH (AroE) from E. coli and J. regia synthesize GA in vitro, we propose that the C-terminal, AroE/SDH domain of the plant enzyme is the region of the protein responsible for GA production. Further expression studies using fragments of the plant gene(s) will be performed to verify the activity of each individual domain in GA production. Because of the importance of GA as an antioxidant in foods, controlling its production and accumulation in plants could significantly increase the nutritional value of many crop species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Schematic of binary vectors pDU04.2601 and pPTP. CaMV35S 5’, 35S promoter from cauliflower mosaic virus; TP, transit peptide from silene ferrodoxin; SDH cDNA, shikimate dehydrogenase cDNA from J. regia cv. Tulare; 3’ocs, terminator sequence from octopine synthase gene; ubi3-5’, 5’ regulatory sequence from the potato ubi3 gene; nos3’, 3’ regulatory sequence from the nopaline synthase genes; uidA, β-glucuronidase, GUS scorable marker gene; mas5’/mas3’, 5’ and 3’ regulatory sequences from the mannopine synthase gene, nptII, neomycin phosphotransferase gene encoding resistance to kanamycin, a selectable marker gene. (DOC 61 kb)
Acknowledgments
This work has been supported by the United States Task Force for Aflatoxin Reduction/Elimination and the Walnut Marketing Board of California. RMM would like to acknowledge the Stuke Fellowship of the Department of Plant Sciences and the NIH Biomolecular Training grant of the UC Davis campus. Finally, we would like to thank Prof. Eric E. Conn for his insightful comments and suggestions, J. Frost for providing us with purified 3-DHS, and K. Inoue for providing the ferredoxin transit peptide from Silene pratensis.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CAD
Coronary arterial disease
- DHS
Dehydroshikimate
- DSkDG
Dehydroshikimate dehydrogenase
- ESI
Electrospray ionization
- GA
Gallic acid
- GUS
β-glucuronidase
- IPTG
Isopropyl-β-D-thiogalactopyranoside
- LB
Luria–Bertani
- LTQ
Linear ion trap
- MS
Mass spectrometry
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- PCA
Protocatechuic acid
- PCR
Polymerase chain reaction
- PMS
Phenazine methosulfate
- RT
Reverse transcriptase
- RP-LC
Reverse phase-liquid chromatography
- TOF
Time of flight
- SA
Shikimic acid
- SDH
Shikimate dehydrogenase
References
- Appeldoorn CCM, Bonnefoy A, Bianca CH, Lutters BCH, Daenens K, van Berkel TJC, Hoylaerts MF, Biessen EAL. Gallic acid antagonizes P-selectin-mediated platelet-leukocyte interactions—Implications for the French paradox. Circulation. 2005;111:106–112. doi: 10.1161/01.CIR.0000151307.10576.02. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. Echerichia coli, plasmids and bacteriophages. In: Asubel FM, Brent R, Kingston RE, Moore D, Seidman J, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York: Wiley and Sons; 1998. pp. 1.8.4–1.8.5. [Google Scholar]
- Balinsky D, Dennis AW, Cleland WE. Kinetic and isotope-exchange studies on shikimate dehydrogenase from Pisum sativum. Biochem. 1971;10:1947–1952. doi: 10.1021/bi00786a032. [DOI] [PubMed] [Google Scholar]
- Chandran SS, Frost JW. Aromatic inhibitors of dehydroquinate synthase: synthesis, evaluation and implications for gallic acid biosynthesis. Bioorg Medicinal Chem Lett. 2001;11:1493–1496. doi: 10.1016/S0960-894X(01)00065-8. [DOI] [PubMed] [Google Scholar]
- Chandran SS, Yi J, Draths KM, von Daeniken R, Weber W, Frost JW. Phosphoenolpyruvate availability and the biosynthesis of shikimic acid. Biotechnol Prog. 2003;19:808–814. doi: 10.1021/bp025769p. [DOI] [PubMed] [Google Scholar]
- Charlton AJ, Baxter NJ, Lilley TH, Haslam E, McDonald CJ, Williamson MP. Tannin interactions with a full-length human salivary proline-rich protein display a stronger affinity than with single proline-rich repeats. FEBS Lett. 1996;382:289–292. doi: 10.1016/0014-5793(96)00186-X. [DOI] [PubMed] [Google Scholar]
- Colich S, Milatovich D, Nikolich D, Zec G. Dehydrogenase isoenzyme polymorphism in selected almond genotypes (Prusus Amygdalus Batsch.) Bulg J Ag Sci. 2009;15:552–556. [Google Scholar]
- Conn EE, Swain T (1961) Biosynthesis of gallic acid in higher plants. Chem Ind: 592–593
- Diaz J, Barcelo AR, De Caceres FM. Changes in shikimate dehydrogenase and the end products of the shikimate pathway, chlorogenic acid and lignins, during the early development of seedlings of Capsicum annuum. New Phytol. 1997;136:183–188. doi: 10.1046/j.1469-8137.1997.00743.x. [DOI] [Google Scholar]
- Diaz J, Bernal A, Pomar F, Merino F. Induction of shikimate dehydrogenase and peroxidase in pepper (Capsicum annuum L.) seedlings in response to copper stress and its relation to lignification. Plant Sci. 2001;161:179–188. doi: 10.1016/S0168-9452(01)00410-1. [DOI] [Google Scholar]
- Escobar MA, Civerolo EL, Summerfelt KR, Dandekar AM. RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc Natl Acad Sci USA. 2001;98:13437–13442. doi: 10.1073/pnas.241276898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross G. Synthesis of β-glucogallin from UDP-glucose and gallic acid by an enzyme preparation from oak leaves. FEBS Lett. 1982;148:67–70. doi: 10.1016/0014-5793(82)81244-1. [DOI] [Google Scholar]
- Grundhoefer P, Niemetz R, Schilling G, Gross GG. Biosynthesis and subcellular distribution of hydrolyzable tannins. Phytochemistry. 2001;57:915–927. doi: 10.1016/S0031-9422(01)00099-1. [DOI] [PubMed] [Google Scholar]
- Hamilton CM. A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene. 1997;200:107–116. doi: 10.1016/S0378-1119(97)00388-0. [DOI] [PubMed] [Google Scholar]
- Haslam ECY. Plant polyphenols (vegetable Tannins)—gallic acid metabolism. Natural Product Rep. 1994;11:41–66. doi: 10.1039/np9941100041. [DOI] [PubMed] [Google Scholar]
- Haslam E. Polyphenols- structure and biosynthesis. In: Haslam E, editor. Practical polyphenolics: from structure to molecular recognition and physiological action. 1. New York: Cambridge University Press; 1998. pp. 51–83. [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Reporter. 1987;5:387–405. doi: 10.1007/BF02667740. [DOI] [Google Scholar]
- Kambourakis S, Draths KM, Frost JW. Synthesis of gallic acid and pyrogallol from glucose: replacing natural product isolation with microbial catalysis. J Am Chem Soc. 2000;122:9042–9043. doi: 10.1021/ja000853r. [DOI] [Google Scholar]
- Khanbabaee K, van Ree T. Tannins: classification and definition. Natural Product Rep. 2001;18:641–649. doi: 10.1039/b101061l. [DOI] [PubMed] [Google Scholar]
- Lee JH, Yang ES, Park J-W. Inactivation of NADP(+)-dependent isocitrate dehydrogenase by peroxynitrite—implications for cytotoxicity and alcohol-induced liver injury. J Biol Chem. 2003;278:51360–51371. doi: 10.1074/jbc.M302332200. [DOI] [PubMed] [Google Scholar]
- Li K, Frost JW. Microbial synthesis of 3-dehydroshikimic acid: a comparative analysis of d-xylose, l-arabinose, and d-glucose carbon sources. Biotechnol Prog. 1999;15:876–883. doi: 10.1021/bp990095c. [DOI] [PubMed] [Google Scholar]
- Li K, Mikola MR, Draths KM, Worden RM, Frost JW. Fed-batch fermentor synthesis of 3-dehydroshikimic acid using recombinant Escherichia coli. Biotechnol Bioeng. 1999;64:61–73. doi: 10.1002/(SICI)1097-0290(19990705)64:1<61::AID-BIT7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Michel G, Roszak AW, Sauve V, Maclean J, Matte A, Coggins JR, Cygler M, Lapthorn AJ. Structures of shikimate dehydrogenase AroE and its paralog YdiB—a common structural framework for different activities. J. Biol.Chem. 2003;278:19463–19472. doi: 10.1074/jbc.M300794200. [DOI] [PubMed] [Google Scholar]
- Ossipov V, Bonner C, Ossipova S, Jensen R. Broad-specificity quinate (shikimate) dehydrogenase from Pinus taeda needles. Plant Physiol Biochem. 2000;38:923–928. doi: 10.1016/S0981-9428(00)01203-1. [DOI] [Google Scholar]
- Ossipov V, Salminen J-P, Ossipova S, Haukioja E, Pihlaja K. Gallic acid and hydrolysable tannins are formed in birch leaves from an intermediate compound of the shikimate pathway. Biochem Sys Ecol. 2003;31:3–16. doi: 10.1016/S0305-1978(02)00081-9. [DOI] [Google Scholar]
- Padyana AK, Burley SK. Crystal structure of shikimate 5-dehydrogenase (SDH) bound to NADP: insights into function and evolution. Structure. 2003;11:1005–1013. doi: 10.1016/S0969-2126(03)00159-X. [DOI] [PubMed] [Google Scholar]
- Salminen JP. Effects of sample drying and storage, and choice of extraction solvent and analysis method on the yield of birch leaf hydrolyzable tannins. J Chem Ecol. 2003;29:1289–1305. doi: 10.1023/A:1024249016741. [DOI] [PubMed] [Google Scholar]
- Sharma SB, Dawra RK. A spectrophotometric method for assay of tannase using rhodanine. Analyt Biochem. 2000;279:85–89. doi: 10.1006/abio.1999.4405. [DOI] [PubMed] [Google Scholar]
- Singh SA, Christendat D. Structure of Arabidopsis dehydroquinate dehydratase-shikimate dehydrogenase and implications for metabolic channeling in the shikimate pathway. Biochem. 2006;45:7787–7996. doi: 10.1021/bi060366+. [DOI] [PubMed] [Google Scholar]
- Smeekens S, Binsbergen JV, Weisbeek P. The plant ferredoxin precursor—Nucleotide-sequence of a full length cDNA clone. Nucleic Acids Res. 1985;13:3179–3194. doi: 10.1093/nar/13.9.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan E. Shikimate dehydrogenase structure reveals novel fold. Structure. 2003;11:902–903. doi: 10.1016/S0969-2126(03)00165-5. [DOI] [PubMed] [Google Scholar]
- Walker JM. Nondenaturing polyacrylamide gel electrophoresis of proteins. In: Walker JM, editor. Basic protein and peptide protocols. 1. Totowa, NJ: Humana Press Inc; 1994. pp. 17–22. [DOI] [PubMed] [Google Scholar]
- Werner I, Bacher A, Eisenreich W. Retrobiosynthetic NMR studies with C-13-labeled glucose—formation of gallic acid in plants and fungi. J Biol Chem. 1997;272:25474–25482. doi: 10.1074/jbc.272.41.25474. [DOI] [PubMed] [Google Scholar]
- Werner RA, Bacher A, Eisenreich W. Analysis of gallic acid biosynthesis via quantitative prediction of isotope labeling patterns. In: Gross GG, Hemingway RW, Yoshida T, editors. Plant polyphenols 2: chemistry, biology, pharmacology, ecology. 1. NY: Kluwer Academic/Plenum Publishers; 1999. pp. 43–61. [Google Scholar]
- Werner RA, Rossmann A, Schwarz C, Bacher A, Schmidt H-L, Eisenreich W. Biosynthesis of gallic acid in Rhus typhina: discrimination between alternative pathways from natural oxygen isotope abundance. Phytochemistry. 2004;65:2809–2813. doi: 10.1016/j.phytochem.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Ye S, von Delft F, Brooun A, Knuth MW, Swanson RV, McRee DE. The crystal structure of shikimate dehydrogenase (AroE) reveals a unique NADPH binding mode. J Bacteriol. 2003;185:4144–4151. doi: 10.1128/JB.185.14.4144-4151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Tolstikov V. Probing genetic algorithms for feature selection in comprehensive metabolic profiling approach. Rapid Commun Mass Spectro. 2008;22:1312–1324. doi: 10.1002/rcm.3507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Schematic of binary vectors pDU04.2601 and pPTP. CaMV35S 5’, 35S promoter from cauliflower mosaic virus; TP, transit peptide from silene ferrodoxin; SDH cDNA, shikimate dehydrogenase cDNA from J. regia cv. Tulare; 3’ocs, terminator sequence from octopine synthase gene; ubi3-5’, 5’ regulatory sequence from the potato ubi3 gene; nos3’, 3’ regulatory sequence from the nopaline synthase genes; uidA, β-glucuronidase, GUS scorable marker gene; mas5’/mas3’, 5’ and 3’ regulatory sequences from the mannopine synthase gene, nptII, neomycin phosphotransferase gene encoding resistance to kanamycin, a selectable marker gene. (DOC 61 kb)