Abstract
With the recent advances in high throughput profiling techniques the amount of genetic and phenotypic data available has increased dramatically. Although many genetic diversity studies combine morphological and genetic data, metabolite profiling has yet to be integrated into these studies. For our study we selected 168 accessions representing the different morphotypes and geographic origins of Brassica rapa. Metabolite profiling was performed on all plants of this collection in the youngest expanded leaves, 5 weeks after transplanting and the same material was used for molecular marker profiling. During the same season a year later, 26 morphological characteristics were measured on plants that had been vernalized in the seedling stage. The number of groups and composition following a hierarchical clustering with molecular markers was highly correlated to the groups based on morphological traits (r = 0.420) and metabolic profiles (r = 0.476). To reveal the admixture levels in B. rapa, comparison with the results of the programme STRUCTURE was needed to obtain information on population substructure. To analyze 5546 metabolite (LC–MS) signals the groups identified with STRUCTURE were used for random forests classification. When comparing the random forests and STRUCTURE membership probabilities 86% of the accessions were allocated into the same subgroup. Our findings indicate that if extensive phenotypic data (metabolites) are available, classification based on this type of data is very comparable to genetic classification. These multivariate types of data and methodological approaches are valuable for the selection of accessions to study the genetics of selected traits and for genetic improvement programs, and additionally provide information on the evolution of the different morphotypes in B. rapa.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1516-1) contains supplementary material, which is available to authorized users.
Introduction
In order to assess the levels and patterns of genetic diversity in a core collection of accessions, one of the most important considerations is the choice of informative datasets. The ultimate goal is to examine simultaneously loci or traits to obtain a genome-wide pattern of genetic variation and to be able to classify the accessions. Morphological traits and molecular marker data have been widely used for this purpose and for the selection of lines with maximum variation for plant breeding programmes (Liu et al. 2007; Hartings et al. 2008; Zhang et al. 2008; Smykal et al. 2008). Recently, high throughput metabolomics data have emerged as a valuable resource to measure phenotypic variation in natural populations on a large scale (Keurentjes et al. 2006). Because of its rapid development metabolomics will soon be adopted as an essential component in plant breeding programs (Fernie and Schauer 2008; Verpoorte et al. 2008). Furthermore, because metabolites are the result of the interaction of genes from several pathways, the integration with molecular marker and morphological trait data could increase our understanding of natural variation and can facilitate the identification of valuable genetic resources.
Unravelling the complexity of large datasets presents a challenge in the selection of appropriate multivariate statistical approaches for the identification of subgroups within a core collection. This issue is particularly important in association studies where the relatedness among accessions is complex and the prevalence of a trait of interest in one subpopulation compared to the other subpopulations is known to be an important constraint (Yu et al. 2006; Aranzana et al. 2005).
Brassica rapa is one of the most important members of the Brassica genus and has been cultivated for many centuries across Europe expanding eventually to Central and East Asia (Gomez-Campo 1999; Dixon 2006). Because in B. rapa subgroups representing the different morphotypes have arisen as a result of selection by plant breeders and adaptation to different geographic regions its phenotypic diversity can be related to population structure (Zhao et al. 2007).
In addition to the assessment of the diversity based on morphological characteristics and molecular markers, Brassica morphotypes have been extensively screened for metabolite composition using GC–MS, LC–MS and/or NMR techniques (Abdel-Farid et al. 2007; Chen et al. 2008; Liang et al. 2006; Padilla et al. 2007; Rochfort et al. 2006; Romani et al. 2006). The most frequently studied compounds are glucosinolates, flavonoids, carotenoids, tocopherols and phenylpropanoids. In general, these studies report an overview of compounds found in different organs or under different cultivation methods.
For example, the metabolic characterization of B. rapa leaves using NMR showed the effect of age and type on the resulting NMR spectra of known compounds (Abdel-Farid et al. 2007). In this study NMR spectroscopy was combined with multivariate data analysis to discriminate among cultivars and developmental stages of B. rapa. On the other hand, studies of cultivars within B. rapa types/species showed that no qualitative differences were observed but that differences were mostly quantitative (Rochfort et al. 2006; Romani et al. 2006). The variation of glucosinolates determined in a collection of 113 varieties of turnip greens (B. rapa L.) indicated both variation in total glucosinolate content and glucosinolate profile. Padilla et al. (2007) concluded that qualitative differences observed among aliphatic glucosinolates might be due to allelic variation. Similarly significant genotypic variations in composition and content of glucosinolates were found in five accessions representing different crop types of Chinese Brassica campestris vegetables (Chen et al. 2008).
In Arabidopsis glucosinolate accumulation and profile was also identified in the leaves and seeds of 39 different Arabidopsis ecotypes. Results indicated that like in B. rapa extensive variation in both the composition and total concentration of glucosinolates could be found among different ecotypes (Kliebenstein et al. 2001). In addition, in Arabidopsis the NMR spectra were used to discriminate among nine ecotypes through multivariate analysis and to identify the metabolites responsible for these differences (Ward et al. 2003).
However, in these studies the phytonutrient composition is neither included in a context of correlation to morphological characteristics nor related to molecular genetic diversity and it is restricted to the screening of a limited number of accessions within morphotypes and to known reference compounds.
In the present study a B. rapa core collection of 168 accessions, representing different types and origins, was screened and classified using a genetic approach with two types of molecular markers (AFLP and SSRs), a comprehensive metabolomics approach using LCMS-based untargeted profiling (De Vos et al. 2007), and a phenotypic approach based on 26 different morphological characteristics of vernalized plants.
Population structure was calculated with data of AFLP markers (random, dominant) (Vos et al. 1995) and a set of non-linked multi-allelic SSRs (EST-based and genomic, co-dominant). Accessions were assigned to subgroups in STRUCTURE and the amount of genetic differentiation between and within populations was calculated using F statistics (Wright 1951; Cockerham 1969, 1973). In addition for the analysis of metabolite data we used GeneSrF, a web-based tool and R package to analyze the 5,546 individual LC–MS signals from the 168 accessions that implement individual LCMS signals selection and classification using random forests (Breiman 2001; Diaz-Uriarte and Alvarez de Andres 2006; Diaz-Uriarte 2007; Lunetta et al. 2004).
The large and diverse types of datasets obtained in this study allowed us to (1) use clustering approaches to reveal the diversity of the sample set based on morphological characteristics and metabolite data in correlation to population structure subgroups, (2) reveal the relationship between B. rapa accessions and evolution of B. rapa types and forms based on genetic diversity data and F statistics, and (3) make use of a large data set of LC–MS untargeted metabolite profiling to classify and sub- select a small number of metabolites specific for subgroups from the core collection using random forests.
Materials and methods
Selection of plant material and experimental design
The core collection included a total of 168 accessions representing the different morphotypes and geographic origins of B. rapa (Table S1). The core collection included 132 accessions that were part of the study of Zhao et al. 2005. From the 168 accessions, 137 were obtained from the Dutch Crop Genetic Resources Centre (CGN) in Wageningen, the Chinese Academy of Agricultural Sciences (CAAS)—Institute for Vegetable and Flowers (IVF) and the Oil Crop Research Institute (OCRI)—and the Osborn Lab, while breeding companies provided 31 accessions (hybrid varieties and breeding lines). First, uniformity in growth and appearance was checked in the field. Ten plants per accession were distributed over two blocks in groups of five plants and morphological traits were measured for comparison of plants within accessions (data not shown).
For the metabolite profiling two plants per accession were sown in the greenhouse September 2006 and September 2007 under the following conditions: 16 h light and temperature between 18 and 21°C. The plants were distributed over two tables in a randomized design with one plant per accession on each table. In the 5th week after transplanting, the leaf material (youngest expanded leaves) was harvested from one plant per accession and directly frozen in liquid nitrogen, ground and stored at −70°C.
DNA was extracted from the ground and frozen material, from the same plant as selected for metabolite profiling, and isolated with the DNAeasy kit (Qiagen, USA).
Morphological data collection
For the assessment of morphological traits, ten seeds per accession were sown on filter paper on a Petri dish. After germination, the plates with seedlings were transferred to a dark cold room (5°C) for 4 weeks. After vernalization treatment, four germinated seeds were transferred to pots in the greenhouse and distributed over four tables, with one plant per table in a completely randomized design.
Twenty-six morphological characteristics were measured at the time when the first flower opened, these included leaf, flower and plant architecture traits (Table 2).
Table 2.
Descriptors of morphological traits
| PC 1 | PC 2 | PC 3 | |||
|---|---|---|---|---|---|
| A. Leaf traits | |||||
| Leaf length | LL | Length from base of petiole to tip of lamina (cm) | 0.88 | 0.31 | 0.22 |
| Lamina blade length | Lbl | Distance from the tip lamina to the fist lobe (cm) | 0.39 | 0.65 | 0.36 |
| Lamina width | LW | Lamina width at the widest point (cm) | 0.43 | 0.74 | −0.05 |
| Leaf index | LI | Ratio of Lbl/LW | −0.04 | −0.06 | 0.71 |
| Leaf area | LA | The whole surface of full leaf including lobes (cm2) | 0.58 | 0.68 | 0.05 |
| Leaf perimeter | LP | The edge of full leaf (cm) | 0.9 | 0.25 | 0.14 |
| Petiole length | LPL | Distance from the base of the petiole to bottom of lamina (cm) | 0.95 | 0.07 | 0.1 |
| Leaf lobes | LB | Number of lobes on the leaf | 0.82 | −0.13 | 0.11 |
| Leaf color | LC | Visual score (1 = dark, 2 = high green, 3 = medium green, 4 = light green, 5 = green–yellow, 6 = yellow) | −0.42 | 0.53 | 0.25 |
| Leaf edge shape | LES | Score (1 = entire, 2 = slightly serrated, 3 = intermediate serrated, 4 = very serrated) | 0.46 | 0.09 | 0.22 |
| Presence of petiole | LPP | Score (0 = absent, 1 = present) | 0.41 | −0.46 | 0.013 |
| SPAD | SPAD | Chlorophyll content | 0.42 | −0.27 | −0.19 |
| B. Flower traits | |||||
| Corolla length | CL | Symmetric length between petals (mm) | −0.11 | 0.76 | 0.09 |
| Corolla width | CW | Symmetric width between petals (mm) | −0.11 | 0.76 | −0.2 |
| Petal length | pL | Distance from base to the top of the petal (mm) | −0.02 | 0.48 | 0.47 |
| Petal width | pW | Petal width at the widest point (mm) | −0.07 | 0.85 | −0.37 |
| Petal index | pI | Ratio of pL/pW | 0.06 | −0.47 | 0.74 |
| Petal area | pA | The whole surface of petal (mm2) | −0.08 | 0.87 | −0.14 |
| Petal perimeter | pP | The edge of petal (mm) | −0.06 | 0.74 | 0.08 |
| Petal shape | pS | Scored (1 = round, 2 = oval, 3 = elongate) | 0.06 | −0.46 | 0.58 |
| Petal color | pC | Visual screening of petal color (1 = orange, 2 = high yellow, 3 = Yellow, 4 = medium yellow, 5 = light yellow) | −0.76 | −0.1 | 0.05 |
| Flowering in time | DTF | Number of days from transplant till the appearance of the first open flower (days) | 0.86 | 0.04 | −0.07 |
| C. Plant architecture trait | |||||
| Leaf number | LN | Number of the leaves when the first flower opens | −0.47 | 0.22 | 0.53 |
| Plant branch | PB | Number of the branches at flowering time | −0.66 | 0.07 | 0.18 |
| Plant height | PH | Distance from the cotyledons to the top of the plant at pre-mature stage (cm) | −0.8 | 0.33 | 0.15 |
| Plant final height | PfH | Distance from the cotyledons to the top of the plant at mature stage (cm) | −0.66 | 0.44 | 0.11 |
| Variance | 31.22 | 23.87 | 9.58 | ||
Variance and Loading values of the first three Principal Components using morphological data
Photos of the different plant organs, one flower and one fully developed leaf (third leaf) from four plants were analyzed using ImageJ software. (http://rsb.info.nih.gov/ij/).
The different morphological data values (continuous and categorical variables) were averaged over the four plants per accession and auto-scaled within each variable using the formula [z = x − mean/SD] (x variable to be standardized and SD standard deviation). Data analysis of the auto-scaled data, correlations between morphological variables and hierarchical clustering using the unweighted pair group method with arithmetic averages (UPGMA) of the accessions were performed using Genemaths XT (Applied Maths, Belgium). The dissimilarity matrix was calculated based on Euclidean distances between the morphological variables.
LC–MS metabolic profiling
Brassica leaf samples were analyzed for variation in semi-polar metabolite composition using LC-QTOF MS, essentially as described in De Vos et al. (2007). In short, 0.5 g FW of frozen leaf powder, from one individual plant per accession, was weighed in 10 ml glass tubes and extracted with 1.5 ml of methanol containing 0.1% formic acid. Samples were sonicated and then filtered (Captiva 0.45 μm PTFE filter plate, Ansys Technologies) into 96-well plates with 700 μl glass inserts (Waters) using a TECAN Genesis Workstation equipped with a 4-channel pipetting robot and a TeVacS 96-wells filtration unit. Samples were injected (5 μl) using an Alliance 2795 HT instrument (Waters), separated on a Phenomenex Luna C18 (2) column (2.0 × 150 mm, 3 mm particle size) using a 5–35% acetonitrile gradient in water (acidified with 0.1% formic acid) and then detected on-line first by a Waters photodiode array detector [(wavelength 220–600 nm (Waters)] and secondly by a Water-Micromass QTOF Ultima MS with negative electrospray ionization (m/z 80–1,500).
Metalign software (http://www.metalign.nl) was used to automatically extract and align all relevant mass signals (signal to local noise ratio >3) from the raw data files. The total of 46,788 signals was filtered for signals present in at least 10 samples and having amplitudes of at least 200 (about 8 times the noise value) in at least one of the samples. Then, all signals eluting within 3 min of retention time (i.e. the injection peak, mostly consisting of signals from non-retained highly polar compounds) were removed from the dataset.
A total of 5,546 LC–MS peaks defined by mass and scan number from 168 accessions were included in the subsequent data analysis. Hierarchical cluster analysis of the accessions was done in GeneMaths XT (Applied Maths, Belgium). The similarity matrix was calculated on the log2 transformed 5,546 LC–MS data with Pearson’s correlation and UPGMA clustering in the program DARwin (Perrier and Jacquemoud-Collet 2006) and drawn with TreeDyn (Chevenet et al. 2006).
Because it is known that many metabolites can be influenced by environmental conditions, we tested the repeatability of the metabolic profiles. For this purpose we selected homogeneous accessions, which correspond to the ones obtained from Dutch seed companies. The 17 accessions were grown in the greenhouse during the same season of two consecutive years, each year at two separate locations in the greenhouse (blocks). We thus obtained four biological repetitions from two consecutive years. For each replicate, leaf material from two plants was collected at the same plant age (5 weeks after sowing). Upon harvest, the leaves were immediately frozen in liquid nitrogen, ground into a fine powder in liquid nitrogen, and stored at −80°C until use. The 4 replicates of the 17 accessions were simultaneously extracted and analyzed for variation in metabolic composition, using the untargeted LC–MS profiling approach described above (De Vos et al. 2007). We subsequently analyzed by multiple linear regressions the correlation (R 2) between metabolite signals within each year (by comparing the different blocks) and between years. In addition, we also selected nine pak choi accessions, from which leaf material of plants of each of the two blocks was sampled in 2006. The samples were simultaneously extracted and analyzed for variation in metabolic composition, using the untargeted LC–MS profiling approach. These data provide information on the variation in metabolite composition due to environment (different blocks) and due to heterogeneity of the gene bank accessions.
Genotypic datasets
The AFLP procedure was performed as described by Vos et al. (1995). Total genomic DNA (200 ng) was digested with two restriction enzymes PstI and MseI and ligated to adaptors. Pre-amplifications were performed in 20 μl volume of 1× PCR buffer, 0.2 mM dNTPs, 30 ng of adaptor primer, 0.4 Taq polymerase and 5 μl of a 10× diluted restriction ligation mix, using 24 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 60 s. Pre-amplifications products were used as template for selective amplification with three primers combinations (P23M48, P23M50 and P21M47).
The NBS-profiling protocol (van der Linden et al. 2004) was extended to motif directed profiling to target Myb motifs. For The MYB targeted profiling total genomic DNA was digested using the following enzymes per reaction: HaeIII, RsaI, AluI and MseI and ligated to an adaptor. Pre-amplifications with one primer directed to a common myb motif (Gerard van der Linden, unpublished results) and one adaptor primer were performed in 25 μl of 1× PCR buffer (with 15 Mm MgCl2), 0.2 mM dNTPs, 0.8 pMol Gene specific primer, 0.8 pMol Adapter primer, U Hotstar Taq polymerase (Qiagen) and 5 μl of a 10× diluted restriction ligation mix. Amplification products were used as template for selective amplification.
For microsatellite (SSR) screening twenty-eight primers were selected for amplification in the core collection accessions. From the primers 10 were genomic and 18 were new est-SSRs (Dr MaRongcai, Dr Tang Jifeng personal communication). The primers were selected because of their map position in different maps of B. rapa and distribution over all the linkage groups (A1–A10) (linkage groups listed in Table 1).
Table 1.
Number of SSR alleles found in the core collection and per population as defined by STRUCTURE
| Locus# | Name | LG | Population 1 | Population 2 | Population 3 | Population 4 | Mean | SD | Number | H e | Forward primer | Reverse primer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Br46 | R1 | 3 | 2 | 2 | 2 | 2.25 | 0.5 | 3 | 0.4145 | AGGTTTCGAGGTTTGTGGCTTCT | CTAAACTCATCGCTTCCGTAAACA |
| 2 | br333 | R1 | 5 | 4 | 3 | 3 | 3.75 | 0.957 | 6 | 0.5833 | AGTTGGCCCCATTTCATTGTTAT | CATCTTGACGGCCTCCATCTCCA |
| 3 | KS50420 | R10 | 17 | 15 | 8 | 11 | 12.75 | 4.031 | 21 | 0.9112 | TTCACACAAGGTTTGTGCC | CGTAAAGGCATCAAGGAAAA |
| 4 | Br27 | R2 | 5 | 4 | 4 | 4 | 4.25 | 0.5 | 5 | 0.526 | AAGTACATGGTCATCCAAGG | AAGGATCCATCACATGGTAA |
| 5 | Br48 | R2 | 4 | 5 | 4 | 3 | 4 | 0.816 | 5 | 0.6257 | GGTGGTGGGCTGGGGAGTA 3′ | CGTCGATCGATTCATAACCGTAGA |
| 6 | br323 | R2 | 8 | 4 | 4 | 6 | 5.5 | 1.915 | 8 | 0.7442 | GTGGTGAACGTGCTTAAGAT | ACGAGCTGGTTGAAAGTTTA |
| 7 | F3H-SSR2 | R3 | 4 | 4 | 3 | 4 | 3.75 | 0.5 | 4 | 0.7432 | GTCATCTCCAGGTAAATCCA | TCTTGAACAACCTCTCCCTA |
| 8 | fito63 | R3 | 6 | 5 | 4 | 6 | 5.25 | 0.957 | 9 | 0.6985 | GTTCAGTTCCCAGATTCCTAA | TTTCCTCTTCCTTCTCTCTTC |
| 9 | br356 | R3 | 3 | 3 | 3 | 2 | 2.75 | 0.5 | 4 | 0.4978 | GCATCTCAGCCTTACAACTT | AGCAAGAACCCAGAAACATA |
| 10 | br377 | R4 | 4 | 2 | 2 | 2 | 2.5 | 1 | 4 | 0.352 | GAAATGAGCGACAGTGTGAT | ACAAACGACCAGTTCATAGG |
| 11 | Br65 | R4 | 3 | 4 | 2 | 3 | 3 | 0.816 | 4 | 0.3567 | TTCCGTCCCTTCCCTAAACAA | TGAACACTACTGCCCAGAGAACAC |
| 12 | Na10D09 | R4 | 8 | 7 | 5 | 4 | 6 | 1.826 | 8 | 0.7452 | Lowe et al. (2003) | |
| 13 | br384 | R4 | 6 | 6 | 5 | 4 | 5.25 | 0.957 | 6 | 0.7669 | TTCAATCACTTCTTCGTTTG | GAAGTAGCAGAAACAGCACC |
| 14 | brms34 | R5 | 8 | 8 | 6 | 7 | 7.25 | 0.957 | 9 | 0.7947 | GATCAAATAACGAACGGAGAGA | GAGCCAAGAAAGGACCTAAGAT |
| 15 | br378 | R5 | 8 | 5 | 5 | 3 | 5.25 | 2.062 | 8 | 0.6027 | TTCATCCATCCATCTTTCTC | ATGATTCCTCCATGTTCATC |
| 16 | Br51 | R6 | 3 | 3 | 2 | 3 | 2.75 | 0.5 | 3 | 0.4109 | CCGAGGAAGAAAGCTGTTGAGTTG | ATCGCTTCCGTAGACACCTTCGTT |
| 17 | Na12h07 | R6 | 6 | 5 | 4 | 3 | 4.5 | 1.291 | 6 | 0.5812 | Lowe et al. (2003) | |
| 18 | Br89 | R6 | 6 | 6 | 4 | 6 | 5.5 | 1 | 9 | 0.6583 | CGTCCGTAGCGCTATTTTTCAGA3 | ACGTTGTCGATCGCCCAGTTC |
| 19 | BR372-WU | R7 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 0.4491 | AACGTAGTCACCAACGAAAC | TCTGAGAAAAGAAGGAGCTG |
| 20 | brms36 | R7 | 6 | 5 | 4 | 4 | 4.75 | 0.957 | 7 | 0.7416 | Suwabe et al. (2002) | |
| 21 | Ra2A01 | R7 | 8 | 16 | 7 | 10 | 10.25 | 4.031 | 17 | 0.8576 | Lowe et al. (2003) | |
| 22 | br319 | R8 | 4 | 3 | 3 | 3 | 3.25 | 0.5 | 4 | 0.671 | TCTATGATCATGGCTTCCTC | TCTCCGGTGTAGAGTTTGTT |
| 23 | br321 | R9 | 3 | 3 | 2 | 2 | 2.5 | 0.577 | 3 | 0.3669 | CCTTATCCCATCTCTCCTCT | GAGATCAAAGTCGTAGTGGC |
| 24 | Br360a | R3 | CATCGTCGTCTCCAATACTA | GAGTTGAGATCGTTCCTCTG | ||||||||
| 25 | Br63a | R3 | TTCCGTCCCTTCCCTAAACA | GAACACTACTGCCCAGAGAACAC | ||||||||
| 26 | brms43a | R3 | Suwabe et al. (2002) | |||||||||
| 27 | ol11b05a | R3 | Lowe et al. (2003) | |||||||||
| 28 | brms50a | R3 | Suwabe et al. (2002) | |||||||||
| Mean | 5.652 | 5.261 | 3.826 | 4.217 | 4.739 | 0.859 | 6.739 | 14.0992 | ||||
| SD | 3.142 | 3.583 | 1.642 | 2.449 | 2.704 | 0.848 | 4.366 | 0.166630462 |
He gene diversity corrected for sample size (Nei 1978); LG chromosome location; Br prefix for an EST SSR
aNot considered for Fst statistics analysis
AFLP and MYB profiling images were analyzed using Quantar PRO software; marker data were scored as present (1) or absent (0) and treated as dominant markers. Microsatellites scores were converted to binary data per observed allele (fragment of defined size) as present (1) or absent (0) and were also treated as dominant markers.
The genetic distance values were calculated using Jaccard’s coefficient for 412 polymorphic AFLP and SSR fragments. Marker data were used for cluster analysis using the UPGMA clustering in MEGA 4.0 (Kumar et al. 2008).
Assessment of genetic diversity
The genetic diversity was assessed in 166 accessions (excluding accession numbers 164 and 165 because of large number of missing values) with 23 SSR primer pairs, which represented loci from different linkage groups. These 23 SSRs were subselected from 28 SSRs scored over the core collection not to over represent any of the linkage groups in the analysis (Table 1).
The SPAGeDi 1.2 g program was used to calculate Dst, the average gene diversity between subpopulations, the allele frequency per locus and the genetic diversity corrected for sample size (H e) (Hardy and Vekemans 2002; Nei 1978). A hierarchical analysis of molecular variance (AMOVA) was performed on the SSR data with Arlequin 3.11 software (Excoffier and Laval 2005). Data conversion was done in SPAGeDi and Genepop web application (http://genepop.curtin.edu.au/). Fst values were computed according to Weir and Cockerham 1984, to quantify the extent of between–within population differentiation (FCT between populations, FSC within population).
Fst values equal 0 when subpopulations are identical in allele frequencies and 1 when they have different alleles. Populations with little divergence have Fst values less than 0.05, moderately differentiated populations have values between 0.05 and 0.15, greatly differentiated populations have values between 0.15 and 0.25 and very greatly differentiated populations have values greater than 0.25 (Hartl and Andrew 1997; Mohammadi and Prasanna 2003).
Assessment of population structure
Marker data (AFLP and SSR) were used to identify the different subgroups and admixture within the accessions of the core collection through a model of Bayesian clustering for inferring population structure.
To be included in this analysis the SSR alleles were scored as dominant data making a total of 412 markers for the analysis. The number of subpopulations was determined using the software STRUCTURE 2.2 (http://pritch.bsd.uchicago.edu/software), assuming a model for B. rapa of K between 1 and 10 subpopulations, with a total of 300,000 iterations for MCMC repetitions and burn in of 100,000.
Principal components analysis
The auto-scaled data from the 26 morphological traits were used for principal components analysis (PCA). PCA is the most commonly used visualization technique in multivariate statistics, which also identifies eigenvectors and amounts of variance and cumulative explained variances per component. The PCA analysis was conducted using the “FactoMineR” package in R-software (Husson et al. 2008).
Mantel test
To test the correlation between the clusters calculated with the phenotypic and genotypic marker data, a Mantel test was applied in R packages ape4 and ecodist (Goslee and Urban 2007; Mantel 1967).
Random forests classification of LC–MS data
We used GeneSrF, http://genesrf.bioinfo.cnio.es (Diaz-Uriarte 2007), a web-based tool originally implemented for microarray data to select very small sets of genes that preserve classification accuracy. The output includes bootstrapped estimates of prediction error rate and assessment of the prediction error. Based on the allelic frequency classification, STRUCTURE result assigns the 168 accessions to subgroups. In the GeneSrF web-based application this subgroup classification was inserted as the class file and the 5,546 LC–MS mass scan data were input as the equivalent of the expression data file. We considered the mean class membership probabilities obtained from the random forests output for comparison with the probabilities of inferred ancestry of individuals (membership probabilities) from AFLP and SSR markers obtained with STRUCTURE.
Results
Population structure
The genetic structure of 168 accessions was inferred using 412 markers (AFLP and SSR polymorphic bands). The Bayesian clustering implemented in the STRUCTURE software revealed four subpopulations. The selection of the subgroups (K = 4) was done since the average likelihood value of runs for a given K value increased gradually until K = 4. The selection of a K > 4 would result in groups with no relationship to morphotype and/or origin of the accessions.
Population 1, includes mostly turnip accessions from European origin and broccoletto accessions (VT + FT); population 2, includes several types: pak choi, winter oil, mizuna, mibuna, komatsuna, turnip green, oil rape and Asian turnip (PC + T); population 3, includes annual oil type accessions (SO, YS and RC) and population 4 includes, mainly accessions of Chinese cabbage (CC) (Table S1).
Of the 168 accessions, 112 were assigned to a group with a probability value of p > 0.70. Fifty-six accessions have p < 0.7 probability values with different levels of admixture between subgroups (Table S1). The morphotypes with highest levels of admixture were the winter oil accessions from Pakistan with six out of seven accessions having probability values of p < 0.7 The highest membership probability values of the six winter oil accessions corresponded to population 1 (VT + FT) and population 2 (PC + T).
Multivariate analyses
Molecular marker data
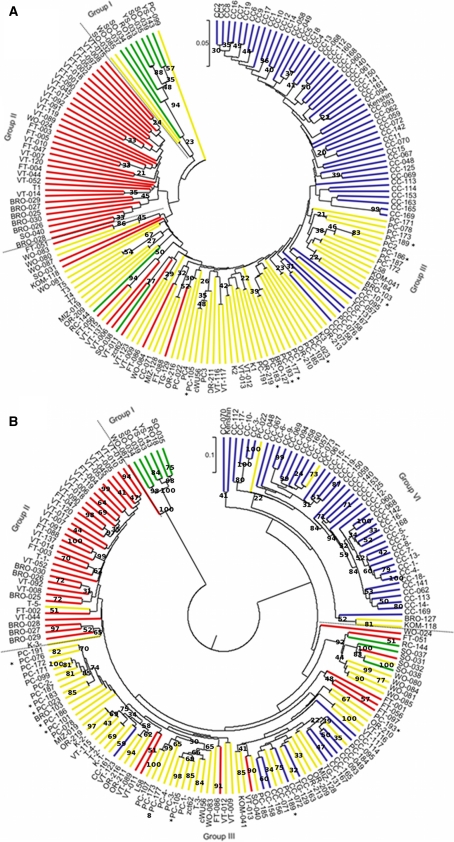
The clustering based on 412 AFLP and SSR markers gave as a result the separation of the collection into three groups. The three groups were composed as follows: I, a small group of oil types; II, a group of European turnips (vegetable and fodder turnip) and broccoletto and III, a group of morphotypes of Asian origin (CC, Pak choi and few Asian turnips).
The results obtained with the hierarchical clustering showed that most of the accessions were included into subgroups that had non-significant bootstrap values. However, the hierarchical clustering results are mostly supported by the results obtained with STRUCTURE and represent the classification based on morphotype and the geographical origin.
Comparison of the three groups identified with hierarchical clustering with the 4 subpopulations found in STRUCTURE indicated the subdivision of cluster/group III with the morphotypes of Asian origin into two different groups with also mainly accessions of Asian origin (II and IV) in STRUCTURE (Fig. 2a). When the four groups found in STRUCTURE are compared with the oil group I, the European turnips group II and the pak choi and CC group III found with the hierarchical clustering, 19 accessions are differently classified in both methods.
Fig. 2.
Hierarchical cluster UPGMA obtained with a molecular markers and b 5,546 LC–MS mass scan signal. The colors indicate STRUCTURE subgroups red population 1, yellow population 2, green population 3 and blue population 4. Bootstrap values higher than 20 are indicated. Accessions indicated with an asterisk were selected for the study of metabolic variation within morphotypes (color figure online)
Morphological traits
The hierarchical cluster analysis of 26 morphological traits using the UPGMA method showed three distinct groups and few small groups of accessions (Fig. 1). The first group (I) consisted of 28 accessions mainly European vegetable and fodder turnips; a second group (II) consisted of 62 accessions, including 51 CC accessions, landraces/cultivars and modern breeding lines from companies and 11 accessions corresponding to different types; the third (III) group consisted of 50 accessions, composed by a combination of pak choi, broccoletto, Asian turnips and few annual oil accessions. The separation into these subgroups is not supported by high bootstrap values. Instead, the variation between subgroups is less significant in comparison with the within group variation The comparison with the molecular marker data classification based on STRUCTURE results showed that the turnip cluster (I) is formed mostly by accessions of European origin in correspondence to the STRUCTURE subgroup (population 1) without the broccoletto’s. The CC cluster (II) is very similar to the STRUCTURE subgroup (population 4) and independent of cultivated form (landrace or modern breeding lines from companies) plus several accessions from STRUCTURE population 2. The admixed cluster (III) group accessions are independent of cultivated form, similar to the STRUCTURE population 2 with several Asian types (pak choi, Chinese Cabbage, Winter oils, Asian turnips, etc.) plus the European broccoletto’s from STRUCTURE population 1.
Fig. 1.
Hierarchical cluster UPGMA obtained with 26 morphological traits. Colors in the first column indicate STRUCTURE subgroups; red population 1, yellow population 2, green population 3 and blue population 4. Colors in the second column indicate geographic origin; red company line, yellow Asia, blue Europe, green America. Bootstrap values higher than 20 are indicated (color figure online)
The results of the PCA indicated that more than 50% of the total variance was explained by the first two PCs (31.22% by PC1 and 23.87% by PC2). The most relevant loadings for the PC1 were mostly leaf characteristics and flowering time and for PC2 the most important loadings were a combination of leaf and flower characteristics (Table 2).
Correlation analysis of the 26 morphological traits showed significant and high correlations within leaf, flower and plant architecture traits, high values were also found between leaf (area and width) and flower traits (area and width) in correspondence to a possible modularity and genetic co-regulation of these developmental traits (data not shown).
Metabolite LC–MS data
The clustering based on the log2 transformed LC–MS data from the core collection (5,546 mass scan numbers) showed 4 main groups and few small mixed groups (Fig. 2b).
Similar to the results obtained with the morphological traits and molecular data the subgroup separation was not supported by high bootstrap values; higher values were obtained for the within group variation. However, the composition of each group showed a high correspondence with the four subpopulations found with molecular marker information in STRUCTURE, especially in the case of groups I, II and IV and to a lesser extent for group III.
Group I consists of 7 accessions, including spring oil (YS and SO) and 1 winter oil accession; group II includes 33 accessions of the following types: broccoletto, European vegetable and fodder turnips; group III, the most admixed group in terms of morphotypes and its relationship to the STRUCTURE subgroups, is composed by 81 accessions of the following types: pak choi, Asian turnips, turnip rape, winter oil, mizuna and few Chinese cabbages; and group IV consists of 47 accessions mainly CC accessions and modern company CC accessions.
In the hierarchical clustering based on LC–MS data, 31 accessions were differently classified with respect to the subpopulations found with STRUCTURE. From these 31 accessions, 12 were also differently classified when molecular marker data were used for the hierarchical clustering (Fig. 2a, b). These accessions have an admixed genetic nature, which allows for flexibility in terms of group assignment (Table S1). In the case of the remainder 19 accessions the genetic admixture is not significant, but still they are assigned to a different group based on this high number of metabolite mass scan signals (5,546) compared to their assignment to STRUCTURE subpopulations.
Diversity of LC–MS profiles within accessions
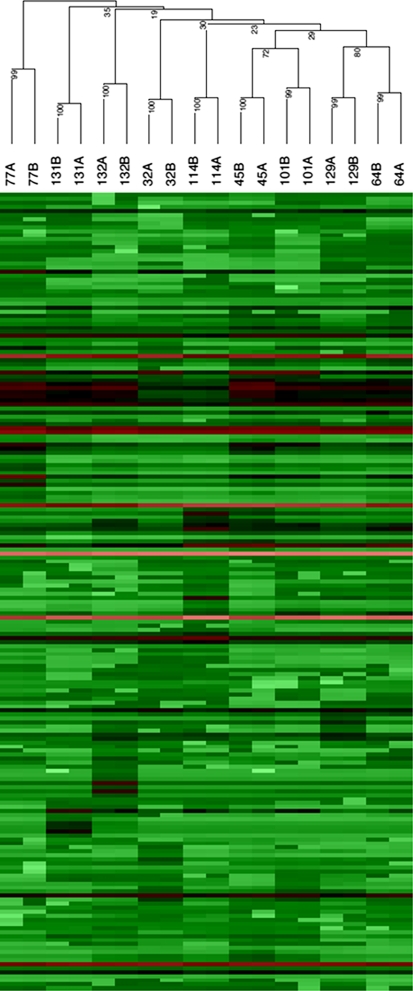
Data of 3,564 mass scan signals from the 4 biological repetitions of the 17 homogenous company lines were evaluated through a linear regression on each individual mass scan signal. Comparison of blocks from the same year, by means of linear correlation regression analysis, resulted in average r 2 values of 0.84 for year 1 and 0.86 for year 2. Comparison of results between years gave an average r 2 value of 0.79. These results indicate that the LC–MS profiles of mass scan signals obtained from each accession are highly repeatable within and between years. Based on this biological repetition experiment on 17 homogeneous lines, we can assume that the metabolite profiles observed within the B. rapa core collection are consistent for most metabolites over different years and biological repeats. Furthermore, the LC–MS profiling was repeated with leaf samples from a group of nine accessions of the pak choi morphotype, which are genetically heterogeneous. The samples were taken from two plants grown in two blocks in the same year (2006). Clustering based on information from 198 centrotypes indicated that the variation found within accessions (bootstrap values 99 or 100) is lower than the variation between accessions as observed by clustering (Fig. 3).
Fig. 3.
Hierarchical cluster analysis of 198 centrotypes identified in nine pak choi accessions. The numbers indicate the accession order as indicated in Table S1, A and B correspond to leaf samples from two block repeats
Comparison of morphological, metabolic and molecular group classification
Although the bootstrap values obtained with hierarchical clustering were low the distance values were used to test the correlation between results with the different datasets. The Mantel test between morphological and molecular distances revealed a strong and significant correlation value of r = 0.420 (p < 0.01). In the comparison between metabolite and molecular distances the result is also strong and significant with a value of r = 0.476 (p < 0.01). The congruence between metabolite and morphological data resulted in a weak but significant correlation with a value of r = 0.174 (p < 0.01).
Population differentiation with microsatellite marker data
The molecular marker data set of 23 SSRs and 166 genotypes was used for calculation of between and within population differentiation. In the core collection the allele number ranged from 2 to 21 amplified fragments (alleles) per SSR. The mean allele number per SSR over all loci in the four sub-populations is 4.7. The highest mean value for alleles was found in population 1 (VT + FT) (European turnip and broccoletto group) and population 2 (PC + T) with a value of 5.6 and 5.3, respectively (Table 1).
The genetic diversity corrected for sample size (H e) calculated over the four populations ranged from 0.91 for KS50420 (A10) to 0.352 for br377 (A04). Pairwise Ds (Nei’s 1978 standard distance) between populations, as defined in STRUCTURE, indicated a high genetic distance value between population 1 (VT + FT) (European turnip and broccoletto group) and population 3 (SO, YS and RC) (oil group) of 0.4267 and a low value between population 1 (VT + FT) (European turnip and broccoletto group) and population 2 (PC + T) (pak choi and turnip group) of 0.1539 (Table 3). Global F statistics results indicate the presence of moderately differentiated populations (Fst = 0.1534), which points towards the occurrence of population structure. The Fst values of differentiation between populations ranged from 0.09 (population 1 (VT + FT) − population 2 (PC + T)) to 0.23 (population 3 (SO, YS and RC) − population 4 (CC)) (Table 4). AMOVA results indicate that the highest percentage of variation is found within populations (84.66%) compared to among populations (15.34%). (Table 5).
Table 3.
Pairwise Ds (Nei’s 1978 standard distance) for all 23 loci profiled with SSR markers in the populations as defined by STRUCTURE
| Population 1 | Population 2 | Population 3 | Population 4 | |
|---|---|---|---|---|
| Population 1 | 0.1539 | 0.4267 | 0.3151 | |
| Population 2 | 0.1539 | 0.2637 | 0.1797 | |
| Population 3 | 0.4267 | 0.2637 | 0.3245 | |
| Population 4 | 0.3151 | 0.1797 | 0.3245 |
Table 4.
Fst values of differentiation between populations as defined in STRUCTURE
| Population 1 | Population 2 | Population 3 | Population 4 | |
|---|---|---|---|---|
| Population 1 | ||||
| Population 2 | 0.08961 | |||
| Population 3 | 0.19046 | 0.14982 | ||
| Population 4 | 0.19843 | 0.14113 | 0.23374 |
Table 5.
AMOVA result of global Fst statistics
| Source of variationa | df | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|
| Among populations | 3 | 251.524 | 1.0058 | 15.34 |
| Within populations | 328 | 1,820.609 | 5.55064 | 84.66 |
| Total | 331 | 2,072.133 | 6.55643 | |
| Fixation index FST: 0.15341 |
aSource of variation within and among populations as defined in STRUCTURE
Random forest classification and identification of variables
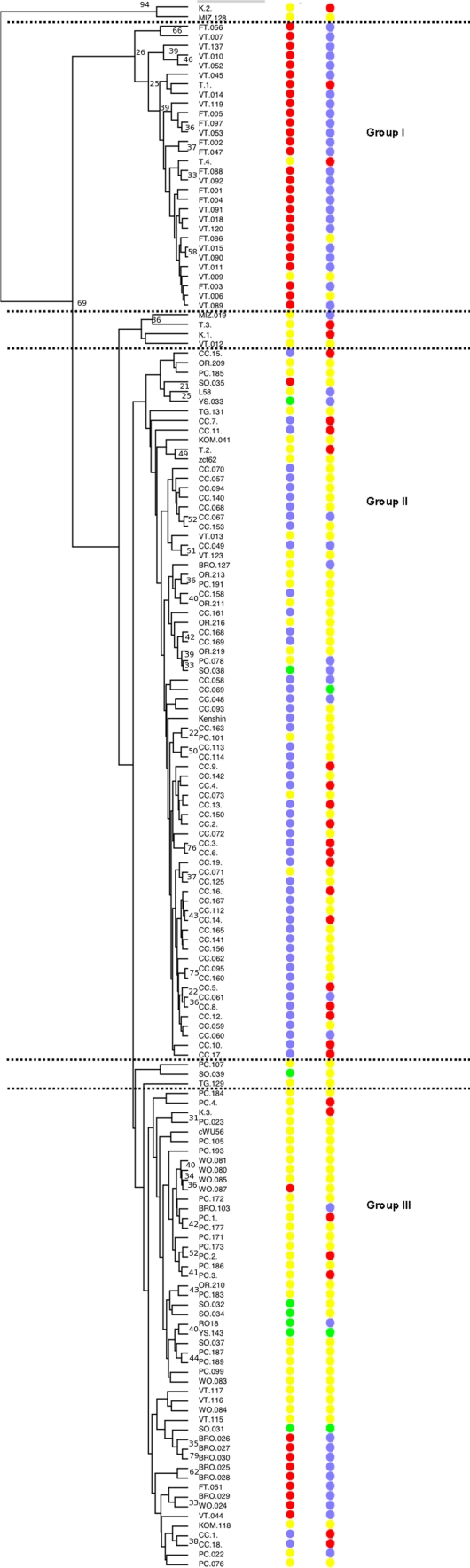
Genotype information of the STRUCTURE subgroup classification was used in the random forest web-based application to account for the estimated relatedness between accessions. The number of variables (metabolite: mass scan number) from the original dataset that can summarize, in a small subset, the differences between the classes (4 population subgroups) was estimated. The mean class membership probabilities based on metabolite data obtained for each accession were plotted in a similar way to the STRUCTURE membership probabilities (Figure S1). Based on the random forest probabilities using metabolite data, 145 out of the 168 accessions were allocated into the same class compared to the one assigned using the molecular markers with a Bayesian clustering algorithm in STRUCTURE (Table S1).
In the variable selection using all data a number of 100 variables were found to be the set with a good predictive accuracy for the classes (4 population subgroups). For the variable selection the bootstrap estimate of prediction error was 13.3% (using 200 bootstrap iterations).
Correlation analysis between these 100 individual mass signals indicated that these might represent 35 different metabolites (Dr. Ric de Vos unpublished results). Between the metabolites identified are isopropyl glucosinolate, methylpropyl glucosinolate, hexyl glucosinolate 2, caffeoylquinic acid, chlorogenic acid, coumaroylquinic acid, quercetin 3-(2-feruloylsophoroside) 7-diglucoside and kaempferol coffeoyl tetraglucoside. The mass signals representing these 35 compounds were further used to construct an UPGMA dendrogram. Two biological replicates were included in these cluster analyses, an oil type RO18 and a caixin type L58, which showed consistency in the clustering. In the hierarchical clustering three very well defined groups are separated in accordance with the STRUCTURE groups: Group I (VT + FT: population 1), Group 2 (CC: population 4) and group 3 (PC + T: population 2) and few admixed subgroups (Figure S2). The oil group (SO, YS and RC: population 3) was found to be subdivided into two groups, the summer and spring oil accessions, with STRUCTURE membership probability 0.79–0.98, were grouped based on the identified metabolites in the sub-cluster oil I, while the other summer and spring oil accessions, with STRUCTURE membership probabilities 0.38–0.43, were grouped in the sub-cluster oil II.
Discussion
With the recent advances in high throughput profiling techniques the amount of genetic and phenotypic data available has increased dramatically. Although many studies combine morphological and genetic data, metabolite profiling has yet to be integrated into diversity studies. More importantly, to establish the existence of relationships between accessions with such a multivariate approach could lead us to a better understanding of the processes that shape natural variation.
For our research we composed a B. rapa core collection with accessions widely diverse in geographical origin, morphotype and phenotypic characteristics, including genebank accessions and modern cultivars/breeding lines provided by five breeding companies. B. rapa is mainly an out-crosser and self incompatible, except for some annual oil types; as a result landraces are heterogeneous. In contrast, modern cultivars and breeding lines from seed companies are homogeneous hybrids with homozygous inbred lines.
We consider B. rapa as suitable for genetic diversity studies because of the selection and adaptation this crop has undergone during centuries of cultivation. Hence, the effect of these processes can be measured on the evolution of morphotypes and in the interaction of metabolite composition and developmental traits.
In our study the hierarchical cluster analysis based on molecular markers identified three groups, very similar to the groups found in previous studies in B. rapa (Zhao et al. 2005, 2007, 2010). However, like in the study of Zhao et al. (2005), the bootstrap values were low, and support the hypothesis that most polymorphisms do not contribute to the phenotypic variation. Because the same result is obtained when the morphological and metabolic data are included in the cluster analysis we can also conclude that only a few genes are involved in the emergence of crop types with different morphology and metabolic profiles. In addition, using hierarchical clustering the group number and composition identified with molecular markers is highly correlated to the groups based on morphological traits of vernalized accessions (r = 0.420) and based on metabolic profiling of leaves (r = 0.476).
With STRUCTURE the number of groups identified is four because group III (Asian crop types) defined using hierarchical clustering is represented by two structured groups (II and IV, comprising Chinese cabbages, and Asian turnips and pak choi, respectively). Furthermore, the four subpopulations show correspondence to the B. rapa morphotypes (turnip, oil, leafy types) and their geographic origin (Asia and Europe).
Because the output obtained with STRUCTURE reflects the value of allele exchange (admixture) and relatedness among individuals we decided to use the structured sub-groups as a priori reference to define populations/classes for the random forests and genetic diversity estimation with microsatellites.
A constraint in the use of AFLP profiling in a core collection is that the chromosomal map position of the AFLP markers is mostly unknown and that the markers are scored dominantly. In our study we made use of SSRs with known map position, which allowed us to overcome this issue and to confirm with AMOVA the presence of moderately to strong population differentiation (Fst = 0.15341). The distribution of variation in the group of accessions was found to be larger at the within population level (84.66%) compared to the variation between populations (15.34%). This suggests that subpopulations harbor enough genetic variation and could be enlarged to study morphotype-specific characteristics in association studies avoiding in this way the effect of population confounding.
Although we collected data from leaf material at one developmental stage only, metabolite data proved to be very valuable in the classification of morphotypes, comparable to the genetic profiling and reflected the level of admixture found with STRUCTURE (Figure S1). Random forests provided a valuable tool to select a small group of variables from the unidentified LC–MS signals that represent/define these sub-populations. For future research high throughput metabolite profiling in combination with random forests and clustering analysis can be worthy for the identification of lines that carry interesting metabolites for crop improvement and/or for the selection of parental lines to create populations for metabolic QTL studies or to define subgroups with wide variation for association studies.
Besides the classification purposes of this study, the value of multivariate data analysis to reveal morphotypes ancestry and evolutionary processes should be acknowledged. It has been suggested that in B. rapa similar morphotypes have an independent origin and/or a long and separate domestication and breeding history in Asia and Europe (Zhao et al. 2005). In the study of genetic relationships among cultivated types of B. rapa with AFLP markers it is considered that turnip was the primitive type of cultivated B. rapa which originated in Central Asia or in Europe and spread both to East Asia and to Europe and India (Takuno et al. 2007).
Our results indicate that genetically the winter oil accessions from Pakistan show higher levels of population admixture, which indicate the presence of genetic background shared both with European turnips and Asian pak choi types (STRUCTURE and F statistics). This ancestry of the winter oil type is reflected, for example in morphological characteristics like the weedy-type look, the hairy, lobulated and rosette arrangement of the leaves, which resemble the European turnip type. If indeed winter oils are the ancient crop types that further developed into turnips and broccoletto in Europe and into turnips and pak choi in Asia then both turnip and pak choi types are a consequence of adaptation and selection on each continent. Further studies that will include sequencing of genes both affected and not affected by selection, will clarify the evolution of the B. rapa forms in different geographical regions.
In this study we have estimated the genetic diversity in a group of 168 accessions with different types of data. Our statistical approaches have been successful to unravel the complexity of the data and to establish relationships between accessions using genetic markers, morphological data and metabolite profiles.
The phenotypic diversity, represented by the evaluated traits (metabolite and morphology) from a large group of accessions, in this B. rapa core collection showed generally a high correlation with the genetic classification. However, hierarchical clustering of molecular data was not sufficient to reveal the level of admixture in B. rapa and comparison of clustering results with STRUCTURE is needed, especially when association studies in B. rapa are the goal, since it gives additional information on the level of population substructure.
In addition, this type of multivariate type of data and methodological approach is valuable for the selection of accessions to study the genetics of selected traits and for genetic improvement programs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. Membership probabilities output for each accession represented as a barplot: (A) Random forests, (B) STRUCTURE. (TIFF 1,307 kb)
Figure S2 Hierarchical cluster analysis of 35 centrotypes selected from the total 5546 LC–MS mass scan signal. The signals were selected variables of the random forests output. Bootstrap values higher than 20 are indicated. (TIFF 1,830 kb)
Table S1 Membership probabilities and group assignment based on metabolic and genetic data: random forests (RF) and STRUCTURE. Values p < 0.70 are indicated with an asterisk and differences between RF and STRUCTURE are indicated in bold and italics in the group column. (XLS 73 kb)
Acknowledgments
We thank the assistance of Cristina Requena Robles and Johan Bucher (Wageningen University) in the greenhouse work and data collection. This project was (co)financed by the Centre for BioSystems Genomics (CBSG) which is part of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abdel-Farid IB, Hye KK, Young HC, Verpoorte R. Metabolic characterization of Brassica rapa leaves by NMR spectroscopy. J Agric Food Chem. 2007;55:7936–7943. doi: 10.1021/jf071294b. [DOI] [PubMed] [Google Scholar]
- Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, Toomajian C, Traw B, Zheng H, Bergelson J, Dean C, Marjoram P, Nordborg M. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 2005;1:e60. doi: 10.1371/journal.pgen.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L (2001) Random Forests. Mach Learn 45(1):5–32
- Chen X, Zhu J, Gerendas J, Zimmermann N. Glucosinolates in Chinese Brassica campestris vegetables: Chinese cabbage, purple cai-tai, choysum, pakchoi, and turnip. HortScience. 2008;43:571–574. [Google Scholar]
- Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees BMC. Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham CC. Variance of gene frequencies. Evolution. 1969;23:72–84. doi: 10.2307/2406485. [DOI] [PubMed] [Google Scholar]
- Cockerham CC. Analysis of gene frequencies. Genetics. 1973;74:679–700. doi: 10.1093/genetics/74.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos RCH, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD. Untargeted large- scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protoc. 2007;2:778–791. doi: 10.1038/nprot.2007.95. [DOI] [PubMed] [Google Scholar]
- Diaz-Uriarte R. GeneSrF and varSelRF: a web-based tool and R package for gene selection and classification using random forest. BMC Bioinform. 2007;8:328. doi: 10.1186/1471-2105-8-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon GR. Vegetable Brassicas and related crucifers. UK: CABI; 2006. [Google Scholar]
- Excoffier LG, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Schauer N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 2008;25:39–48. doi: 10.1016/j.tig.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Gomez-Campo C. Developments in plant genetics and breeding, vol 4. Biology of Brassica coenospecies. Amsterdam: Elsevier; 1999. [Google Scholar]
- Goslee SC, Urban DL. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw. 2007;22:1–19. [Google Scholar]
- Hardy OJ, Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. doi: 10.1046/j.1471-8286.2002.00305.x. [DOI] [Google Scholar]
- Hartings H, Berardo N, Mazzinelli GF, Valoti P, Verderio A, et al. Assessment of genetic diversity and relationships among maize (Zea mays L.) Italian landraces by morphological traits and AFLP profiling. Theor Appl Genet. 2008;117:831–842. doi: 10.1007/s00122-008-0823-2. [DOI] [PubMed] [Google Scholar]
- Hartl DLC, Andrew G. Principles of population genetics. Sunderland: Sinauer Associates; 1997. [Google Scholar]
- Husson F, Josse J, Le S, Mazet J (2008) Factor analysis and data mining with R. R package version 1.10. http://factominer.free.fr; http://www.agrocampus-rennes.fr/math/
- Keurentjes JB, Fu J, de Vos CHR, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M. The genetics of plant metabolism. Nat Genet. 2006;38(7):842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 2001;126:811–825. doi: 10.1104/pp.126.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dudley J, Nei M, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YS, Kim HK, Lefeber AWM, Erkelens C, Choi YH, et al. Identification of phenylpropanoids in methyl jasmonate treated Brassica rapa leaves using two-dimensional nuclear magnetic resonance spectroscopy. J Chromatogr. 2006;1112:148–155. doi: 10.1016/j.chroma.2005.11.114. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu L, Hou N, Zhang A, Liu C. Genetic diversity of wheat gene pool of recurrent selection assessed by microsatellite markers and morphological traits. Euphytica. 2007;155:249–258. doi: 10.1007/s10681-006-9326-x. [DOI] [Google Scholar]
- Lowe AJ, Moule C, Trick M, Edwards KJ (2003) Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor Appl Genet 108:1103–1112 [DOI] [PubMed]
- Lunetta KL, Hayward LB, Segal J, van Eerdewegh P. Screening large-scale association study data: Exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Mohammadi SA, Prasanna BM. Analysis of genetic diversity in crop plants—salient statistical tools and considerations. Crop Sci. 2003;43:1235–1248. doi: 10.2135/cropsci2003.1235. [DOI] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla G, Cartea ME, Velasco P, de Haro A, Ordas A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry. 2007;68:536–545. doi: 10.1016/j.phytochem.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin
- Rochfort SJ, Imsic M, Jones R, Trenerry VC, Tomkins B. Characterization of flavonol conjugates in immature leaves of pak choi [Brassica rapa L. Ssp. chinensis L. (Hanelt.)] by HPLC-DAD and LC-MS/MS. J Agric Food Chem. 2006;54:4855–4860. doi: 10.1021/jf060154j. [DOI] [PubMed] [Google Scholar]
- Romani A, Vignolini P, Isolani L, Ieri F, Heimler D. HPLC-DAD/MS characterization of flavonoids and hydroxycinnamic derivatives in turnip tops (Brassica rapa L. Subsp. sylvestris L.) J Agric Food Chem. 2006;54:1342–1346. doi: 10.1021/jf052629x. [DOI] [PubMed] [Google Scholar]
- Smykal P, Hybl M, Corander J, Jarkovsky J, Flavell AJ, et al. Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theor Appl Genet. 2008;117:413–424. doi: 10.1007/s00122-008-0785-4. [DOI] [PubMed] [Google Scholar]
- Suwabe K, Iketani H, Nunome T, Kage T, Hirai M (2002) Isolation and characterization of microsatellites in Brassica rapa L. Theor Appl Genet 104:1092–1098 [DOI] [PubMed]
- Takuno S, Kawahara T, Ohnishi O. Phylogenetic relationships among cultivated types of Brassica rapa L. em. Metzg. as revealed by AFLP analysis. Genet Resour Crop Evol. 2007;54:279–285. doi: 10.1007/s10722-005-4260-7. [DOI] [Google Scholar]
- van der Linden CG, Wouters DCAE, Mihalka V, Kochieva EZ, Smulders MJM, Vosman B (2004) Efficient targeting of plant disease resistance loci using NBS profiling. Theor Appl Genet 109:384–393 [DOI] [PubMed]
- Verpoorte R, Choi YH, Mustafa NR, Kim HK. Metabolomics: back to basics. Phytochem Rev. 2008;7:525–537. doi: 10.1007/s11101-008-9091-7. [DOI] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Rijan M, van der Lee T, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acid Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JL, Harris C, Lewis J, Beale MH. Assessment of 1H NMR spectroscopy and multivariate analysis as a technique for metabolite fingerprinting of Arabidopsis thaliana. Phytochemistry. 2003;62:949–957. doi: 10.1016/S0031-9422(02)00705-7. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F statistics for the analysis of population structure. Evol Bioinform Online. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zhang X, Blair MW, Wang S. Genetic diversity of Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeat markers. Theor Appl Genet. 2008;117:629–640. doi: 10.1007/s00122-008-0807-2. [DOI] [PubMed] [Google Scholar]
- Zhao J, Wang X, Deng B, Lou P, Wu J, et al. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor Appl Genet. 2005;110:1301–1314. doi: 10.1007/s00122-005-1967-y. [DOI] [PubMed] [Google Scholar]
- Zhao J, Paulo MJ, Jamar D, Lou P, Van Eeuwijk F, et al. Association mapping of leaf traits, flowering time, and phytate content in Brassica rapa. Genome. 2007;50:963–973. doi: 10.1139/G07-078. [DOI] [PubMed] [Google Scholar]
- Zhao J, Artemyeva A, Pino Del Carpio D, Basnet R.K, Zhang N, Gao J, Bucher J, Wang X, Visser RGF, Bonnema G (2010) Design of a Brassica rapa core collection for association mapping studies. Genome 53:884–898 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Membership probabilities output for each accession represented as a barplot: (A) Random forests, (B) STRUCTURE. (TIFF 1,307 kb)
Figure S2 Hierarchical cluster analysis of 35 centrotypes selected from the total 5546 LC–MS mass scan signal. The signals were selected variables of the random forests output. Bootstrap values higher than 20 are indicated. (TIFF 1,830 kb)
Table S1 Membership probabilities and group assignment based on metabolic and genetic data: random forests (RF) and STRUCTURE. Values p < 0.70 are indicated with an asterisk and differences between RF and STRUCTURE are indicated in bold and italics in the group column. (XLS 73 kb)