Abstract
Cultivated groundnut or peanut (Arachis hypogaea L.), an allotetraploid (2n = 4x = 40), is a self pollinated and widely grown crop in the semi-arid regions of the world. Improvement of drought tolerance is an important area of research for groundnut breeding programmes. Therefore, for the identification of candidate QTLs for drought tolerance, a comprehensive and refined genetic map containing 191 SSR loci based on a single mapping population (TAG 24 × ICGV 86031), segregating for drought and surrogate traits was developed. Genotyping data and phenotyping data collected for more than ten drought related traits in 2–3 seasons were analyzed in detail for identification of main effect QTLs (M-QTLs) and epistatic QTLs (E-QTLs) using QTL Cartographer, QTLNetwork and Genotype Matrix Mapping (GMM) programmes. A total of 105 M-QTLs with 3.48–33.36% phenotypic variation explained (PVE) were identified using QTL Cartographer, while only 65 M-QTLs with 1.3–15.01% PVE were identified using QTLNetwork. A total of 53 M-QTLs were such which were identified using both programmes. On the other hand, GMM identified 186 (8.54–44.72% PVE) and 63 (7.11–21.13% PVE), three and two loci interactions, whereas only 8 E-QTL interactions with 1.7–8.34% PVE were identified through QTLNetwork. Interestingly a number of co-localized QTLs controlling 2–9 traits were also identified. The identification of few major, many minor M-QTLs and QTL × QTL interactions during the present study confirmed the complex and quantitative nature of drought tolerance in groundnut. This study suggests deployment of modern approaches like marker-assisted recurrent selection or genomic selection instead of marker-assisted backcrossing approach for breeding for drought tolerance in groundnut.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1517-0) contains supplementary material, which is available to authorized users.
Keywords: Peanut, Drought tolerance, Genetic map, Molecular markers, Main-effect QTLs, Epistatic QTLs, Molecular breeding
Introduction
Cultivated peanut or groundnut (Arachis hypogaea L.) is an allotetraploid (2n = 4x = 40) legume that is widely grown as an oil seed or food crop in more than 100 countries of the world. Over two-third of the groundnut global production occurs in seasonally rainfed regions where drought is the major constraint for crop production (Smartt 1994) and productivity ranges from 0.7 to 1.2 Million mega grams per hectare (Mg ha−1). However, even under a commercial system where groundnut productivity ranges from 2.0 to 4.0 Mg ha−1, water may also be a limiting factor. For both situations, cultivars that are efficient in water utilization are required. Yield loss due to water stress can be partially tackled by developing varieties that are better adapted to water-limited conditions.
Mechanisms of drought adaptation in groundnut are limited to drought escape and drought avoidance (Zhang et al. 2001). In drought escape, plants take advantage of developmental flexibility to match its phenology to the length of the cropping period (early flowering to escape late season drought). The principle of drought avoidance is to either increase water absorption ability (from rooting differences) or decreasing their water loss (shoot/leaf morphological traits or physiological traits). Tolerance to drought is not a simple response, but is mostly conditioned by many genes and has been shown to interact with environment, and thus the networks involved in drought tolerance are quite complex in nature. Therefore, selection based on the phenotype would be difficult for such traits (Collins et al. 2008)
In case of crops like groundnut, which are grown in semi-arid tropics (SAT) environment, which is characterized by short and erratic rainfall and long periods of dry spell, drought avoidance assumes greater importance. Water-use efficiency (WUE) is considered as an important drought avoidance trait, which deals with using soil water more efficiently for biomass production (Blum 2005; Collins et al. 2008). Raising the WUE of both irrigated and rain fed crop production is an urgent imperative (Nigam et al. 2005). Of the world’s allocatable water resource, 80% is currently consumed by irrigated agriculture. Drought tolerance through WUE with transpiration efficiency (TE) as an important component of WUE is one of the target traits for groundnut improvement for water limited environments. Surrogate traits for TE such as carbon discrimination ratio (δ13C), specific leaf area (SLA) and SPAD chlorophyll meter reading (SCMR) have also been used as proxies for TE (Hubick et al. 1986; Nageswara Rao and Wright 1994; Rebetzke et al. 2002), although some recent results are not in complete agreement of this view (Krishnamurthy et al. 2007; Devi et al. 2010).
Conventional breeding for developing drought-tolerant crop varieties is time-consuming and labor intensive due to the quantitative nature of drought tolerance and difficulties in selection for drought tolerance (Ribaut et al. 1997). Recent advances in the area of crop genomics offer tools to assist breeding (Varshney et al. 2005, 2006). The identification of genomic regions associated with drought tolerance would enable breeders to develop improved cultivars with increased drought tolerance using marker-assisted selection (MAS) (Ribaut et al. 1996). To identify the genomic regions suitable for marker-assisted breeding strategies, it is important to establish accurate phenotyping methods, develop highly saturated molecular marker-based genetic linkage maps, and then identify QTLs (quantitative trait loci) associated with traits of interest. Several studies were conducted in the past that reported identification of QTLs for drought tolerance or related traits. For instance, in soybean, 5 QTLs were identified for WUE in an F2 population with 14–20% phenotypic variation explained (PVE) (Mian et al. 1998). In case of wheat, Dashti et al. (2007) identified five QTLs for drought tolerance with 13–34% PVE. In another study, 47 QTLs for different plant stress indicators in rice with 5–59% PVE were identified.
Even though candidate QTLs have been identified in several studies, there have been few attempts to develop strategies to use them in marker assisted selection (MAS) programmes. This can be attributed mainly to following reasons: (a) QTLs for drought tolerance explained only a small proportion of the phenotypic variation, (b) QTLs identified for drought tolerance themselves explained only a portion of the yield variation. Such observations are expected, as drought tolerance is a complex trait involving diverse aspects of phenotype physiology interaction with the environment and presumably, many genes. With these types of complex traits, in addition to simple QTLs, there is a need to identify epistatic QTLs (E-QTLs). Some programmes like QTLNetwork have been used to identify main effect QTLs (M-QTL), epistatic QTLs (E-QTL) and QTL- Environment (QE) effects in several crop species e.g. rice (Xing et al. 2002), wheat (Kulwal et al. 2004; Mohan et al. 2009), cotton (Shen et al. 2006), maize (Yang et al. 2009), etc. Recently, Isobe et al. (2007) developed a new QTL mapping approach that is called genotype matrix mapping (GMM), which searches for QTL interactions in genetic variation.
Based on (TAG 24 × ICGV 86031) RIL mapping population, a framework linkage map was developed earlier for cultivated groundnut that had 135 SSR loci (Varshney et al. 2009a). QTL analysis with genotypic data and phenotypic data obtained from two seasons revealed minor M-QTLs contributing 2.9–17.6% PV. This may be attributed to either the low marker density of the genetic map developed or to the limited range of phenotypic variation existed among the RIL progenies. With this background, the present study was initiated: (a) to saturate the framework map with more marker loci, (b) to phenotype additional drought related traits and attempt to increase the previous range of variation, and (c) to undertake comprehensive QTL analysis to identify M-QTLs, E-QTLs and QE effects for drought and component traits in groundnut.
Materials and methods
Trait phenotyping
A RIL mapping population comprising of 318 F8/F9/F10 lines derived from a cross TAG 24 × ICGV 86031 was phenotyped for transpiration (T, g plant−1), transpiration efficiency (TE, g biomass kg−1 water transpired), SLA (cm2 g−1), SCMR, leaf area (LA, cm2 plant−1), shoot plus pod dry weight (DW, g plant−1), and total dry matter (TDM, g plant−1, which includes root dry weight) and carbon discrimination ratio (δ13C) during post-rainy season in 2004 and 2005. Canopy conductance (ISC) was computed as the ratio of the transpiration to leaf area. The same population was phenotyped for T, TE and shoot plus pod dry weight under well watered and water stress regimes in 2008. The protocols for TE measurements are previously described (Krishnamurthy et al. 2007).
Field evaluation of populations
In addition to phenotyping conducted in 2004 and 2005, as reported in Krishnamurthy et al. (2007) and used for QTL analysis by Varshney et al. (2009a), the mapping population (318 F10 progenies from the cross TAG 24 (low TE) × ICGV 86031 (high TE) was tested for yield in the field during the post-rainy season (2007–2008), under water stress and well-watered conditions. The trial was planted on December 2007 and stress was imposed on 16 February 2008. A supplemental irrigation of 50 mm was applied to the stress plots on 5 March 2008, whereas the fully irrigated plot received 50 mm of irrigation every 10 days initially and then every 7 days as the season progressed. The trial received 163 mm of unexpected rainfall between 22 and 27 March 2008 (pod maturity stage). Despite this rain, the yield reduction in the water stress treatment was about 20% compared to the fully irrigated control, showing that some of the flowers might have already dried up due to drought at flowering stage and therefore, rain at podding stage could not recover the withered flowers and hence reduction in yield. Harvesting was done in April 2008. Different agronomic traits such as seed weight, pod weight and haulm weight were measured.
SSR loci amplification and genotyping
New set of SSR markers developed by S. J. Knapp at University of Georgia (UGA), USA (unpublished) and Wang et al. (2007a) were used for polymerase chain reaction (PCR) in a 5 μl reaction volume in an ABI 9700 thermal cycler (Applied Biosystems, USA), in 384-well PCR plates (Applied Biosystems, USA). The forward primers for the UGA markers were labeled with one of the fluorescent labels 6-FAM, VIC, NED, PET, HEX or TAMRA and the reverse primers were unlabeled. In case of markers from Wang et al. (2007a), a M13 primer (5′-CGTTGTAAAACGACGGCCAGT-3′) with 6-FAM covalently bound to the 5′-end was used. Two unlabeled primers including a specific SSR-targeting forward primer with the M13 tail and a specific SSR-targeting reverse primer were also used in each reaction. Thus PCRs, in case of Wang et al. (2007a) markers, consisted of three primers in which the M13-labeled primer and reverse primer were in excess of the unlabeled forward primer. This allows the forward M13-tailed primer and reverse primer to initiate the reaction and, when the limited primer is depleted, the labeled primer takes the place of the limited forward primer in the remaining PCR cycles (Schuelke 2000).
In both kinds of markers, the PCR mix consisted of 2 pM of forward (F) and 2 pM of reverse (R) (S. J. Knapp unpublished)/1 pM M13 tailed F: 2 pM R: 2 pM M13 label ratio (Wang et al. 2007a) of primers, 2 mM MgCl2, 2 mM dNTPs, 0.1 U of Taq DNA polymerase (Bioline, USA) and 1X PCR buffer (Bioline, USA). A touch down PCR amplification profile with 3 min of initial denaturation, followed by first 5 cycles of 94°C for 20 s, 60°C for 20 s and 72°C for 30 s, with 1°C decrease in annealing temperature per cycle, then 30 cycles of 94°C for 20 s with constant annealing temperature (59°C) and 72°C for 30 s followed by a final extension for 20 min at 72°C. PCR amplicons generated were first resolved on 1.2% agarose gel to check for the amplification of the PCR products. Subsequently, amplified products for majority of SSR markers were size fractioned and analyzed using capillary electrophoresis on an ABI 3700 automatic DNA sequencer (Applied Biosystems, USA) following the same methods mentioned in Varshney et al. (2009a). Genotyping data on the mapping population were obtained for the polymorphic markers by using the same protocol as mentioned above.
Mapping of polymorphic loci
Genotyping data generated in this study were attempted for integrating the marker loci into the available framework linkage map (Varshney et al. 2009a) using Mapmaker ver. 3.0 (Lander et al. 1987). All polymorphic loci were tried using “build” command with a LOD score of 3 to integrate into the framework map. After integration of new loci, the local marker orders were confirmed by permuting all adjacent triple orders using ‘ripple’ command. Recombination fractions were converted into map distances in centimorgans (cM) based on Kosambi’s mapping function.
Quantitative trait loci (QTLs) detection
For identification of candidate QTL regions for drought component traits, two types of trait mapping were taken up: (a) interval mapping to identify main effect QTLs (M-QTLs) and (b) epistatic interaction analysis (EIA) to identify epistatic interactions between different QTL regions (epistatic QTLs, E-QTLs). The most likely location of QTLs and their genetic effects were initially detected by composite interval mapping (CIM; Zeng 1993, 1994) using the WinQTL Cartographer, version 2.5 (Wang et al. 2007b). CIM was performed using Model 6 after scanning the genetic map and estimating the likelihood of a QTL and its corresponding effects every 1 cM, while using significant marker cofactors to adjust the phenotypic effects associated with other positions in the genetic map. The number of marker cofactors for the background control was set by forward–backward stepwise regression. A window size of 10 cM was used, and therefore cofactors within 10 cM on either side of the QTL test site were not included in the QTL model. When separated by a minimum distance of 20 cM (Ungerer et al. 2002) two peaks on one chromosome were considered as two different QTLs. Otherwise, the higher peak was chosen to more closely approximate the position of the QTL.
EIA was carried out using Genotype Matrix Mapping (GMM) software ver. 2.1 (Isobe et al. 2007, http://www.kajusa.or.jp/GMM) which looks for interactions between different loci. Using GMM, two and three loci interactions were tested. QTLNetwork programme ver. 2.0, based on a mixed linear model (Yang et al. 2005) was also used to identify epistatic QTLs (E-QTL) conditioning drought related traits.
Results
Identification of marker polymorphism and genotyping
In addition to the 1,145 SSRs screened in our earlier study (Varshney et al. 2009a), a set of 2,070 novel SSR markers including 1,947 EST-SSR markers developed at University of Georgia (S. J. Knapp, unpublished) and 123 genomic SSR markers reported by Wang et al. (2007a) were screened on the parental genotypes ICGV 86031 and TAG 24 for polymorphism in this study. Only 51 (2.46%) markers showed polymorphism between these two parental genotypes. In addition, 12 additional polymorphic markers identified in the previous study were included in the dataset of polymorphic markers. All these 63 polymorphic markers were used for genotyping the set of 188 RILs of the mapping population. While genotyping the mapping population, segregation data were scored at two loci for two markers (GM 1971 and GM 1992) and single locus for 61 markers. As a result, segregation data were obtained for 65 loci for 63 polymorphic markers.
In summary, a total of 3,215 SSR markers (1,145 SSRs Varshney et al. 2009a and 2,070 SSRs in this study) were screened on these two parents and segregation data were obtained for 215 marker loci (150 SSR loci Varshney et al. 2009a and 65 SSR loci in this study).
Construction of improved genetic map
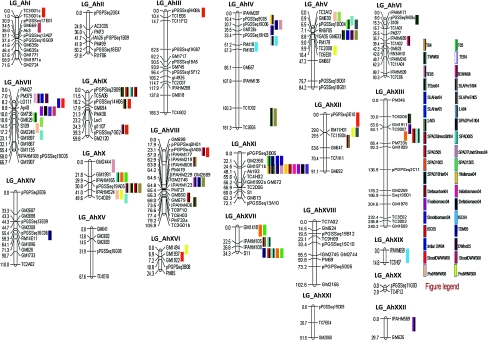
Genotyping data obtained for 65 loci obtained in this study were used to integrate into the framework map comprising of 135 loci (Varshney et al. 2009a). Of the 65 loci tried, 56 loci got integrated into different linkage groups and nine markers remained unlinked. Thus, the present map has a total of 191 loci integrated into 22 linkage groups, covering a length of 1785.4 cM with an average of 9.34 cM between loci along the linkage groups. The 56 new loci got evenly distributed into 17 of the 22 linkage groups. The map locations of the newly mapped markers are shown in Fig. 1. LG_AhII, LG_AhXII, LG_AhXIX, LG_AhXX and LG_AhXXII did not have any new loci integrated. LG_AhXIII has largest number of markers integrated (8). Most of the EST-SSRs used in the present study got mapped into non-centromeric regions which are expected to be gene rich. The present map developed is the most comprehensive cultivated groundnut map available with maximum number of mapped loci on a single RIL population.
Fig. 1.
Saturated genetic and QTL map for cultivated groundnut. QTL positions for different drought component traits as colored bars have been shown on right hand side of linkage groups
Trait phenotyping
The two parental genotypes TAG 24 and ICGV 86031 were found to show variation in transpiration (T), transpiration efficiency (TE), specific leaf area (SLA), SPAD chlorophyll meter reading (SCMR), dry weight, total dry matter, biomass, canopy conductance, carbon discrimination ratio, seed weight, pod weight and haulm weight, etc. (Nautiyal et al. 2002; Serraj et al. 2004). Therefore, all 318 RILs were phenotyped for above-mentioned traits for one to three seasons (Table 1). Because a limited range of variation for TE was found among the RILs (2.60–3.60 in 2004 and 2.95–3.40 in 2005), and even between the parents (2.70–3.30 and 2.00–2.20 in 2004 and 2005), detailed studies have been undertaken to assess TE in these parental lines across a range of vapor pressure deficit (VPD) conditions (Vadez et al. unpublished). The underlying hypothesis was that TAG 24 and ICGV 86031 were screened for TE under glasshouse conditions under low VPD, whereas the RILs were screened in outdoor conditions in February and March in India under high VPD. Results indicated a large TE contrast between TAG 24 and ICGV 86031 under glasshouse conditions, but showed decreasing TE differences as VPD increased. Therefore, the population has been re-phenotyped during the rainy season 2008 under well watered and water deficit conditions, therefore, under lower VPD conditions, which has enlarged the range of variation between the RIL progenies.
Table 1.
Trait phenotyping data on ICGV 86031 and TAG 24 and its mapping population
| Trait | Year | ICGV 86031 | TAG 24 | Variation in RILs | Significance | Heritability | Grand Mean | S.E.D. | LSD |
|---|---|---|---|---|---|---|---|---|---|
| Transpiration (T, kg) | 2004 | 1.42 | 1.47 | 1.27–1.51 | <.001 | 0.621 | 1.4169 | 0.08056 | 0.15805 |
| Transpiration (T, kg) | 2005 | 4.47 | 3.65 | 3.18–5.04 | <.001 | 0.782 | 4166.4 | 249.82 | 490.1 |
| Transpiration efficiency | 2004 | 5.56 | 4.36 | 3.98–7.17 | <.001 | 0.717 | 5.169 | 0.4294 | 0.8425 |
| Transpiration efficiency TE, g kg−1 | 2004 | 3.47 | 2.53 | 1.93–4.08 | <.001 | 0.658 | 3.079 | 0.3727 | 0.7311 |
| Transpiration efficiency TE, g kg−1 | 2005 | 2.14 | 1.87 | 1.71–2.56 | <.001 | 0.675 | 2.1183 | 0.16209 | 0.318 |
| Transpiration efficiency under water stress regime | 2008 | 4.57 | 3.42 | 1.95–5.91 | <.001 | 0.738 | 3.748 | 0.5451 | 1.0699 |
| Transpiration efficiency under well watered regime | 2008 | 4.55 | 3.71 | 2.40–5.22 | <.001 | 0.754 | 3.837 | 0.4061 | 0.7971 |
| Transpiration under water stress regime | 2008 | 2.01 | 2.00 | 1.65–2.44 | <.001 | 0.688 | 1993.5 | 149.23 | 292.87 |
| Transpiration under well watered regime | 2008 | 4.15 | 3.61 | 2.92–5.30 | <.001 | 0.765 | 4225 | 333.7 | 655 |
| Specific leaf area (SLA, cm2 g-1) at harvest | 2004 | 122.47 | 151.76 | 102.44–185.80 | <.001 | 0.744 | 147.6 | 10.26 | 20.128 |
| Specific leaf area (SLA, cm2 g-1) at start of stress | 2004 | 147.00 | 153.00 | 137–169 | <.001 | 0.750 | 151.65 | 8.485 | 16.662 |
| Leaf area (LA) | 2004 | 393.40 | 397.94 | 320.30–661.64 | <.001 | 0.701 | 435.8 | 39.86 | 78.2 |
| Specific leaf area (SLA, cm2 g-1) at harvest | 2005 | 143.00 | 174.00 | 112.95–175.71 | <.001 | 0.688 | 144.45 | 10.822 | 21.23 |
| Specific leaf area (SLA, cm2 g-1) at start of stress | 2005 | 165.00 | 192.00 | 161–201 | <.001 | 0.780 | 177.27 | 8.136 | 15.961 |
| Leaf area (LA) | 2005 | 747.00 | 626.00 | 419.05–908.43 | <.001 | 0.698 | 654.4 | 74.62 | 146.38 |
| SCMR after 7 days of treatment | 2004 | 52.80 | 50.30 | 43.1–55.2 | <.001 | 0.707 | 49.82 | 3.461 | 6.79 |
| SCMR at harvest | 2004 | 51.50 | 49.00 | 42.9–55.8 | <.001 | 0.812 | 49.67 | 2.984 | 5.854 |
| SCMR at start of treatment | 2004 | 49.20 | 43.80 | 40.7–50.1 | 0.006 | 0.554 | 45.67 | 2.789 | 5.472 |
| SCMR after 10 days of treatment | 2005 | 45.10 | 41.90 | 39.3–48.0 | <.001 | 0.788 | 44.07 | 1.6617 | 3.26 |
| SCMR after 15 days of treatment | 2005 | 47.60 | 42.20 | 39.9–50.9 | <.001 | 0.800 | 45.795 | 1.6048 | 3.1483 |
| SCMR after 5 days of treatment | 2005 | 43.00 | 36.90 | 34.9–46.0 | <.001 | 0.766 | 40.967 | 2.024 | 3.9708 |
| SCMR at start of treatment | 2005 | 46.90 | 42.30 | 39.1–50.4 | <.001 | 0.758 | 44.262 | 2.23 | 4.3749 |
| Canopy conductance (ISC, g water cm-2) | 2004 | 3.68 | 3.81 | 2.33–4.23 | <.001 | 0.731 | 3.3479 | 0.2505 | 0.49145 |
| Canopy conductance (ISC, g water cm-2) | 2005 | 6.05 | 5.90 | 4.82–8.94 | <.001 | 0.687 | 6.602 | 0.7429 | 1.4574 |
| Carbon isotope discrimination ratio | 2004 | – | – | 17.96–20.32 | – | – | – | – | – |
| Delta biomass | 2004 | 4.86 | 3.73 | 2.44–5.88 | <.001 | 0.658 | 4.39 | 0.6091 | 1.1949 |
| Final biomass | 2004 | 7.78 | 6.41 | 4.51–9.28 | <.001 | 0.771 | 7.304 | 0.6091 | 1.1949 |
| Shoot biomass | 2004 | 5.50 | 4.52 | 3.37–7.07 | <.001 | 0.747 | 5.208 | 0.4897 | 0.9608 |
| Dry weight (DW) increase | 2005 | 9.68 | 6.95 | 6.08–12.60 | <.001 | 0.721 | 8.932 | 0.9723 | 1.9076 |
| Total dry matter (TDM) | 2005 | 13.81 | 10.21 | 9.62–16.89 | <.001 | 0.790 | 12.701 | 0.9723 | 1.9076 |
| Haulm weight under water stress regime | 2008 | 0.38 | 0.48 | 0.25–2.06 | <.001 | 0.925 | 463.4 | 94.18 | 184.93 |
| Haulm weight under well watered regime | 2008 | 0.34 | 0.49 | 0.28–1.66 | <.001 | 0.923 | 404.7 | 83.03 | 163.04 |
| Pod weight under water stress regime | 2008 | 0.90 | 0.96 | 0.68–1.24 | <.001 | 0.692 | 561.2 | 106.44 | 209.02 |
| Pod weight under well watered regime | 2008 | 0.78 | 1.08 | 0.68–1.74 | <.001 | 0.807 | 674.3 | 107.71 | 211.5 |
| Shoot dry weight under water stress regime | 2008 | 12.53 | 11.13 | 7.14–15.75 | <.001 | 0.740 | 10.281 | 1.2712 | 2.4949 |
| Shoot dry weight under well watered regime | 2008 | 20.54 | 16.36 | 12.99–26.77 | <.001 | 0.733 | 18.878 | 2.1067 | 4.1347 |
Main effect QTLs (M-QTLs) identified by QTL Cartographer
For T and TE, good variation and heritability values were observed among RILs across the years (Table 1). QTL analysis using CIM based on QTL Cartographer for T and TE identified two M-QTLs each in 2004 and 2008, three M-QTLs in 2005; in case of TE, two M-QTLs were identified in 2004 and 2008, six M-QTLs were identified in 2008. The phenotypic variation explained (PVE) by the M-QTLs identified for T and that of TE ranged from 5.42–10.86% and 4.47–12.30%, respectively.
SLA measured at the start of drought stress as well as at the time of harvest showed large and significant variation among RILs with good levels of heritability in 2004 and 2005. QTL analysis of SLA at the start of drought stress showed three M-QTLs in 2004 and 2005. For SLA at the time of harvest, two M-QTLs were detected in 2004 and six in 2005. These M-QTLs contributed 4.84–13.94% of PV.
SCMR at the start of stress imposition in both seasons, at 7 and 10 days after imposing the stress in 2004, and at 5, 10 and 15 days after imposing the stress in 2005, showed large and significant variation among RILs (Table 1). Heritability values observed for SCMR were also high. QTL analysis of SCMR measured at different time points showed 13 and 16 M-QTLs in 2004 and 2005, respectively. The PVE of these M-QTLs ranged from 4.0 to 19.53%. The number of QTLs identified with the same data in the previous study (Varshney et al. 2009a) was only eight in each season and the PVE ranged from 2.9 to 11.0%. This can be explained by the reason that the present map is saturated with more number of markers compared to the earlier frame work map.
In case of carbon discrimination ratio (δ13C), QTL analysis identified one M-QTL with 12.15% PVE with QTL Cartographer. Canopy conductance (ISC) showed a limited variation in RILs (2.33–4.23) in 2004, while a higher variation (4.82–8.94) was observed in 2005. QTL analysis of canopy conductance measured in 2004 and 2005 identified seven and five M-QTLs with 3.28–22.24% PVE. Biomass measured at different stages showed good levels of variation and heritability among RILs (Table 1). QTL analysis of biomass measured at different stages has shown seven M-QTLs with 4.25–20.32% PVE.
Dry weight (DW) measured at different stages has shown higher variation among RILs, maximum variation was observed for shoot dry weight measured under well watered and water stress conditions during 2008 (Table 1), indeed the heritability values observed were also good. QTL analysis of initial DW and DW increase measured in 2005 has shown two M-QTLs with 4.69–8.20% PVE. Total dry matter (TDM) measured in 2005 showed three M-QTLs with 4.34–9.85% PVE. Shoot DW measured under well watered and water stress conditions in 2008 showed four and six M-QTLs, respectively, with 5.03–8.50% of PVE. Using QTLNetwork, only two and four M-QTLs for DW increase and TDM in 2005 were identified and the identified QTLs were same as those identified by QTL Cartographer (ESM 1).
Pod weight and haulm weight showed moderate levels of variability among RILs with high heritability values, the heritability value observed for haulm weight was highest (0.923–0.925) (Table 1) among all the traits evaluated. QTL analysis of pod weight and haulm weight under well-watered conditions revealed three M-QTLs each for pod weight, seed weight and four QTLs for haulm weight in 2008. PVE by these M-QTLs varied from 3.78–33.36%. Under water stress conditions two M-QTLs each for seed weight and haulm weight and four QTLs for pod weight were identified and PVE varied from 4.18 to 8.78%.
M-QTLs identified by QTLNetwork
In order to compare the M-QTLs identified by QTL Cartographer, QTL analysis was also carried out with another programme, QTLNetwork. As a result, a total of 65 M-QTLs were identified of which 53 M-QTLs were also identified by QTL Cartographer. Apart from these, eight new M-QTLs for traits SLAHar05_XIb, LA05_VII, SPAD505_XIII, SPAD1505_IX, SPAD1505_XIII, ISC04_VI, TDM05_VIIb and SeedWtWS08_Vb were identified by QTLNetwork. A summary and comparison of number of M-QTLs identified by QTL Cartographer and QTLNetwork is given in Table 2. The phenotypic variation explained by the M-QTLs identified by QTLNetwork for different drought component traits was comparatively lower than that of the QTL Cartographer (ESM1). In summary, a total of 117 M-QTLs were identified by QTL Cartographer and QTLNetwork together.
Table 2.
Main effect QTLs (M-QTLs) for drought tolerance component traits identified by QTL Cartographer and QTLNetwork
| Trait | QTL Cartographer | QTLNetwork | ||
|---|---|---|---|---|
| No. of QTLs | Range of PVE (R 2 %) | No. of QTLs | Range of PVE (R 2 %) | |
| Transpiration (T) | 7 | 4.36–10.86 | 2 | 3.48–6.27 |
| Transpiration efficiency (TE) | 7 | 4.47–12.30 | 5 | 3.13–6.33 |
| Specific leaf area (SLA) | 13 | 3.48–13.29 | 7 | 1.3–10.97 |
| Leaf area | 3 | 7.24–11.51 | 3 | 2.93–7.53 |
| SPAD chlorophyll meter readings (SCMR) | 29 | 4.00–19.53 | 19 | 3.72–7.71 |
| Carbon isotope discrimination ratio | 1 | 12.15 | 1 | 7.45 |
| Biomass | 7 | 4.25–20.32 | 5 | 4.97–9.65 |
| Canopy conductance | 7 | 3.28–22.24 | 5 | 3.05–15.01 |
| Total dry matter (TDM) | 3 | 4.34–9.85 | 4 | 2.48–8.72 |
| Dry weight (DW) | 10 | 4.69–9.18 | 5 | 3.38–9.00 |
| Pod weight | 7 | 4.17–7.23 | 5 | 4.55–9.64 |
| Seed weight | 5 | 4.18–8.22 | 4 | 5.09–7.93 |
| Haulm weight | 6 | 4.19–33.36 | – | – |
PVE Phenotypic variation explained
In the present study, M-QTLs identified for drought component traits were distributed on 17 of the 22 linkage groups. While five linkage groups (LG_AhII, LG_AhXV, LG_AhXVIII, LG_AhXX and LG_AhXXI) did not show any M-QTL, 16 M-QTLs were identified on LG_AhXVI.
Since drought is a complex polygenic trait and involves complex interactions with several other parameters, digenic epistatic interaction analysis were undertaken to identify epistatic QTLs (E-QTLs) using genotype matrix mapping (GMM) software (Isobe et al. 2007) and QTLNetwork ver. 2.0 software (Yang et al. 2005).
Epistatic QTLs (E-QTLs) identified by Genotype Matrix Mapping (GMM)
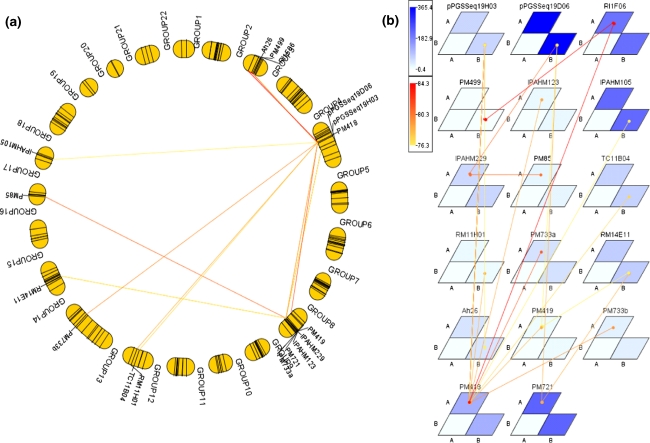
For epistatic interaction analysis (EIA), the locus combinations representing interacting QTLs for two and three loci were considered. A number of significant loci combinations for drought and component traits were identified by GMM (ESM2). For 2 loci combinations, 5 E-QTLs were detected for T, 13 E-QTLs for TE, 1 E-QTL for SLA, 3 E-QTLs for LA, 6 E-QTLs for DW, 14 E-QTLs for biomass, 2 E-QTLs for TDM, 5 E-QTLs for canopy conductance, 7 E-QTLs for pod weight, 6 E-QTLs for seed weight and 1 E-QTL for haulm weight (Table 3). For three loci combinations, 16 E-QTLs were detected for T, 27 E-QTLs for TE, 13 E-QTLs for SLA, 42 E-QTLs for DW, 18 E-QTLs for biomass, 5 E-QTLs for TDM, 27 E-QTLs for canopy conductance and 19 E-QTLs for pod weight, 14 E-QTLs for seed weight and 5 E-QTLs for haulm weight (Table 3). Graphical representation of combinations of interacting triple loci and their position on groundnut linkage map are shown in Fig. 2. Locations of interacting loci are interlinked by lines on the linkage groups. PVE by two QTL combinations ranged from 5.92 to 20.20% for T, 7.11–15.44% for TE, 16.2% for SLA, 10.37–11.18% for LA, 10.10–18.97% for DW, 5.57–11.18% for biomass, 12.29–13.15% for TDM, 18.28–19.02% for canopy conductance, 11.47–13.74% for pod weight, 10.47–12.58% for seed weight and 21.13% for haulm weight. The PVE for three QTL combinations were 10.23–11.29% for T, 9.16–22.06% for TE, 15.68–25.03% for SLA, 10.10–44.72% for DW, 8.54–16.28% for biomass, 12.84–13.67% for TDM, 18.99–22.50% for canopy conductance, 12.79–21.83% for pod weight, 13.01–16.96% for seed weight and 25.35–31.43% for haulm weight.
Table 3.
Epistatic QTLs (E-QTLs) for drought tolerance component traits at three and two loci identified with Genotype Matrix Mapping (GMM)
| Trait | Three loci interactions | Two loci interactions | ||
|---|---|---|---|---|
| No. of QTLs | Range of PVE (R 2 %) | No. of QTLs | Range of PVE (R 2 %) | |
| Transpiration (T) | 16 | 10.23–11.29 | 5 | 5.92–20.20 |
| Transpiration efficiency (TE) | 27 | 9.16–22.06 | 13 | 7.11–15.44 |
| Specific leaf area (SLA) | 13 | 15.68–25.03 | 1 | 16.2 |
| Leaf area (LA) | – | – | 3 | 10.37–11.18 |
| SPAD Chlorophyll meter readings (SCMR) | – | – | – | – |
| Carbon discrimination ratio | – | – | – | – |
| Biomass | 18 | 8.54–16.28 | 14 | 5.57–11.18 |
| Canopy conductance (ISC) | 27 | 18.99–22.50 | 5 | 18.28–19.02 |
| Total dry matter (TDM) | 5 | 12.84–13.67 | 2 | 12.29–13.15 |
| Dry weight (DW) | 42 | 10.10–44.72 | 6 | 10.1–18.97 |
| Pod weight | 19 | 12.79–21.83 | 7 | 11.47–13.74 |
| Seed weight | 14 | 13.01–16.96 | 6 | 10.47–12.58 |
| Haulm weight | 5 | 25.35–31.43 | 1 | 21.13 |
PVE Phenotyping variation explained
Fig. 2.
Graphical representation of combinations of interacting marker loci on genetic map of groundnut detected by Genotype Matrix Mapping (GMM). a Graphical presentation of interacting triple loci and their positions on the genetic linkage map for the trait ISC04 identified by GMM. Linkage groups are arranged tandemly as a circle. Triangles in the circle indicate triple loci combination. b Graphical presentation of interacting loci and allele type by genotype matrices (GMs) and a genotype matrix network (GMN). Significant locus/allele combinations of three interacting loci are shown by GMs and GMN. Matrices and connecting lines indicate GMs and GMNs, respectively
E-QTLs identified by QTLNetwork
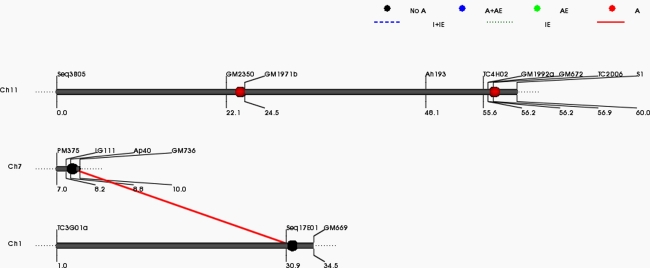
In contrast to large number of interacting QTL identified by GMM, QTLNetwork could detect only 15 E-QTLs (2 for SCMR, 4 for SLA, 3 for ISC, 2 for pod weight, 2 for seed weight and 2 for shoot DW) involved in 8 epistatic interactions (ESM3). Interestingly, QTLNetwork identified E-QTLs for SLA (Fig. 3) and SCMR for which GMM could not identify any E-QTL. Nevertheless the PVE by the QTLs is very low as indicated by the heritability estimate of epistatic allele, 8.34% for SCMR, 2.85–4.84% for SLA, 1.44–1.58% for ISC, 1.7% for pod weight, 5.51% for haulm weight and 3.11% for shoot dry weight.
Fig. 3.
A representative figure showing epistatic interaction identified by QTLNetwork. The figure shows epistatic QTLs for the trait SLA Har05. The black ball represents epistatic QTLs without individual effect while interacting loci are shown by red colored bar
Co-localized QTLs identified through QTL Cartographer and QTLNetwork
QTL analysis during the present study for all the 13 traits detected 105 M-QTLs in 54 genomic regions (using QTL Cartographer) and 65 M-QTLs in 40 genomic regions (using QTLNetwork) on 17 linkage groups (except LG-AhII, LG-AhXV, LG-AhXVIII, LG-AhXXI and LG-AhXX) (ESM1). Among the 54 genomic regions identified by QTL Cartographer, 22 genomic regions harboured a total of 73 QTLs for different traits, with QTLs for 2–9 traits in an individual genomic region (ESM 4). The remaining 32 genomic regions each contained a QTL for a solitary trait. Similarly, among 40 genomic regions identified using QTLNetwork, 12 genomic regions were such which harboured a total of 37 QTLs for different traits, with QTLs for 2–8 traits in an individual genomic region (ESM 4). The remaining 28 genomic regions each contained a QTL for a solitary trait. Interestingly all the genomic regions (containing QTLs for different traits) identified by QTLNetwork were also identified by software QTL Cartographer. In this analysis, the QTL for different traits that were available in a particular genomic region (QTL clusters) with same/overlapping marker intervals were considered co-localized QTL.
Discussion
Marker polymorphism and genetic map
Molecular markers and genetic linkage maps are pre-requisites for crop improvement through molecular breeding in any crop species. Recent advances in the area of crop genomics have offered tools to assist breeding by speeding up the process of introgression of beneficial traits into preferred varieties, especially for complex traits, such as drought (Varshney et al. 2005, 2006). However, for groundnut, although several hundreds of microsatellite markers have been developed (see Varshney et al. 2007; Liang et al. 2009), a comprehensive genetic map based on a cultivated × cultivated cross was not available until 2009 (Varshney et al. 2009a; Hong et al. 2010). In this scenario, the current study was aimed at saturating the existing genetic map for cultivated groundnut for identification of M-QTLs and E-QTLs related to several drought component traits.
After screening a new set of 2,070 SSR markers coming from UGA and Wang et al. (2007a), 51 (2.46%) markers showed polymorphism between parental genotypes. The very low level of polymorphism observed in the present study is because the majority of SSRs (1,947) were from ESTs that represent conserved genic regions. The low levels of polymorphism observed in case of EST derived SSRs has been reported in several other studies (Gupta et al. 2004; Varshney et al. 2005). Low level of polymorphism observed can also be attributed to the origin of groundnut from a recent and single polyploidization event from one or a few individuals of each diploid parental species, and self-pollination (Halward et al. 1991). Two EST-derived primers GM 1971 and GM 1992 amplified more than one segregating fragment. Amplification of more than one fragment has been reported in several other studies (Kottapalli et al. 2007; Varshney et al. 2009a, b). Amplification of more than one fragment in our study can be attributed to amplification of duplicated locus or a different locus due to the allotetraploid nature of cultivated groundnut genome.
The present study integrated 56 new markers into the linkage map which now has a total of 191 markers covering 1,785.4 cM of total map distance. Most of the newly integrated markers derived from ESTs mapped into the non-centromeric regions; this can be explained by these regions being gene rich. Interestingly, some of the new EST-SSR markers and genomic SSR markers were mapped into the gaps on linkage groups LG_AhVI, LG_AhVIII and LG_AhXV. For instance, IPAHM 509 was mapped into 28.4 cM gap between TC11A04 and TC7C06 on LG_AhXV and pPGPSeq3A06 was mapped into the 36.4 cM gap between pPGPSeq8H01 and IPAHM 177 on LG_AhVIII. Similarly, GM 2602 and GM 2603 were mapped into the 34.8 cM gap between GM 641 and pPGPSeq16G08 on LG_AhXV.
To the best of our knowledge, this is the most comprehensive genetic map of cultivated groundnut based only on a single mapping population from cultivated tetraploid genotypes. Although Hong et al. (2010) reported a composite genetic map for tetraploid groundnut, it is based on three mapping populations and have a total of 175 loci as compared to the 191 mapped loci based on single population in the present study. As SSR markers are the markers of choice for plant geneticists and breeders (Gupta and Varshney 2000) and a larger number of SSR markers are available for groundnut (see Varshney et al. 2007), it is anticipated that future groundnut genetic maps will involve mapped SSR markers. Therefore, the present SSR genetic map of cultivated groundnut should be very useful to the groundnut community to compare the future genetic maps of groundnut with the map developed during the present study.
M-QTLs for drought component traits
In the present study, a large number of QTLs for several drought component traits have been identified by CIM analysis. In case of CIM using QTL Cartographer, 105 M-QTLs were identified for various drought component traits. As the QTL identification is a statistical approach, the possibility of identifying false positive and false negative QTL for the thresholds and mapping approaches used exists (McElroy et al. 2006; Mackay and Powell 2007). However, reliability of QTLs identified may be enhanced by identification of QTL using more than one software. Therefore, two software namely QTL Cartographer and QTLNetwork were employed to identify M-QTLs. Interestingly, M-QTLs identified for T on LG_AhVII, TE on LG_AhVII, LG_AhX and LG_AhXI, canopy conductance on LG_AhIII, LG_AhIV, LG_AhIX, SLA on LG_AhIV, LG_AhXI, LG_AhXIII and LG_AhXXII, SCMR on LG_AhIV, LG_AhVIII, LG_AhIX, LG_AhX, LG_AhXI, LG_AhXVII, DW on LG_AhV, LG_AhVII LG_AhXVI, δ13C on LG_AhXI, dry matter on LG_AhVII, biomass on LGAh_VII and LG_AhXI, pod weight on LG_AhV, LG_AhVII, LG_AhX, LG_AhXVI, seed weight on LG_AhV and LG_AhVII were identified by both programmes. Moreover, the genomic locations of these M-QTLs were similar based on analysis with these programme, and therefore these M-QTLs may be considered as reliable QTLs. On the other hand, in the case of haulm weight, QTLNetwork identified none of the six M-QTLs identified by QTL Cartographer. Therefore, these M-QTLs may be false positives and there is a need for their validation by other approaches. Alleles with moderate additive effects were identified for most of the evaluated traits. These alleles, which should confer more tolerance to drought, were derived from both the tolerant (positive additive effect, ICGV 86031) and the susceptible (negative effect, TAG 24) parents (ESM1). Alleles that improve the trait being derived from parents agronomically inferior have been identified for several plant species (Xiao et al. 1998; Frary et al. 2004; Wang et al. 2004; Yoon et al. 2006).
Even though several M-QTLs were identified for all traits in different seasons, the majority of the identified M-QTLs did not reveal a high phenotypic variance. However, given the highly polygenic nature of the traits analyzed (Krishnamurthy et al. 2007) and the relatively high number of progenies, QTLs with lower phenotypic variation is expected. Based on QTL mapping studies in other species, it can be generalized that higher phenotypic variation for the given trait in the mapping population and high/reasonable marker density genotyping data are the pre-requisites to identify the major QTL explaining higher phenotypic variation. However, in the present study, the range of variations for the targeted traits was not very high in RILs. For instance, the range of TE value was only between 2.53 and 3.47 g kg-1 water transpired in 2004, between 1.87 and 2.14 g kg-1 water transpired in 2005, between 3.61 and 4.15 under well watered regime in 2008 and between 2.00 and 2.01 under water stress regime (Table 1). In addition, the targeted trait, TE, is a ratio (biomass accumulation divided by transpired water), with several physiological mechanisms leading to TE differences, and TE itself. For instance, a low stomatal conductance would reduce transpiration, but a low stomatal conductance would also reduce biomass accumulation. Therefore, depending on the magnitude of the conductance changes, and the interaction with other factors impacting biomass accumulation and transpiration (photosynthesis, leaf area, etc.), one can see a number of factors that can have either a positive or a negative relationship with TE, thereby precluding the chance to identify a major M-QTL for TE, but rather a number of small M-QTLs that have high interactions. Even though the marker density on present genetic map is reasonable, consistent QTLs with higher phenotypic variance were not identified. This could be explained by the complex trait of drought tolerance being governed by several small effect QTLs/genes present on different chromosomal regions.
E-QTLs for drought component traits
To date, most findings have suggested that quantitative variation is determined by a few QTLs with a relatively large effect and a large number of genes having progressively smaller effects. Jannink (2007) recently identified QTLs by analyzing genetic background interactions in association studies, and was able to detect loci that have no main effect but which influence a trait only through their interactions with other loci. Multiple QTL interactions might be buried under the smaller effect of single QTL (Isobe et al. 2007). In the present study EIA undertaken with GMM and QTLNetwork revealed several epistatic QTLs. In the case of GMM, two (63) and three loci (186) interactions were identified for different drought component traits. The largest number of interactions were identified for DW (42) followed by TE and ISC (27) measured at different time points, in contrast epistatic interactions were not observed for SCMR and δ13C. Interestingly the number of E-QTLs identified by GMM was more than the main effect QTLs and also the PVE by these QTL combinations was comparatively higher than that identified by QTL Cartographer. In an earlier study, Klimenko et al. (2010) identified hub regions harboring QTL interactions in a rye mapping population segregating for plant persistency. In the present study even though a large number of interacting QTLs were identified the loci that are interacting for different drought component traits were found to be diverse and hub regions associated for drought component traits were not identified. This may be attributed to the variability of various traits across parents and seasons. In the case of EIA using QTLNetwork a much lower number of epistatic loci were identified, two each for SCMR, pod weight, haulm weight, dry weight and four for SLA and three for ISC. This can be explained by the fact that GMM searches for QTL interactions and interaction–interaction relationships which is not the case for the other programmes. In GMM, each marker is given a matrix in which each of the total number of alleles for the marker in the tested population is represented by intersecting lines and rows. QTL interactions are estimated and compared through virtual networks generated among the locus matrices. When a particular network indicates a significant relationship to the phenotype, the marker-allele combinations assigned on the genotype matrices are considered a QTL interaction combination. The algorithm used in GMM which is capable of comparing multiple QTL interactions at the same time makes it more advantageous in identifying epistatic interactions.
In addition, in the case of EIA, even though a large number of QTLs were identified for drought and component traits, considerable amount of phenotypic variation was not explained for different drought component traits. These results suggested that drought tolerance in groundnut is governed by a large number of M-QTLs and E-QTLs each with a small phenotypic variation.
Pyramiding of all these minor QTLs for the improvement of drought tolerance in groundnut is not possible through marker-assisted backcrossing (MABC), since MABC involved the transfer of limited number of QTLs from one genetic background to another (Ribaut et al. 2010). Therefore, alternative and more efficient approaches like MARS and GWS, which allows selection for several QTL with small effects (Ribaut and Ragot 2007; Bernardo 2008; Varshney and Dubey 2009) will be useful for the improvement of drought tolerance in groundnut.
Co-localized M-QTLs
The detection of co-localized QTLs during the present study for drought tolerance suggested that either a single pleiotropic QTL controlled a number of traits or more than one tightly linked QTL for different traits are present together in the same region. The issue of pleiotropy versus tight linkage of QTL may be resolved in future through fine mapping of the target genomic regions. The results also suggested that QTL for drought related traits are not evenly dispersed throughout the genome but are rather clustered in numerous specified genomic regions. Therefore, these co-localized QTLs could be very useful for the simultaneous improvement of more than one trait, if the desirable alleles at these QTLs are contributed by a single parent.
Conclusions
This study reports a comprehensive genetic linkage map for cultivated groundnut which will be a valuable genomic resource for groundnut community to align future genetic maps. Based on extensive phenotyping data and updated genotyping data, a large number of QTLs have been identified using CIM and EIA approaches. Phenotypic variation explained by all these QTLs was found to be small; this suggested that drought tolerance in groundnut is governed by several M-QTLs and E-QTLs each with a small phenotypic variation. Therefore, genome wide marker approaches, such as MARS and GS should be more effective approaches as compared to marker-assisted backcrossing (MABC) in case of groundnut to develop the varieties with enhanced drought tolerance through molecular breeding (Charmet et al. 1999; Bernardo and Charcosset 2006; Bernardo and Yu 2007; Mayor and Bernardo 2009).
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1. Comparision of M-QTLs identified by QTL Cartographer and QTLNetwork for droughttolerance traits in groundnut (XLS 52 kb)
ESM 2. Two and three loci epistatic interactions identified by genotype matrix mapping (GMM) (XLS 74 kb)
ESM 3: Epistatic QTLs identified for drought component traits using QTLNetwork (XLS 27 kb)
ESM4: Co-localized QTLs identified using QTL Cartographer and QTLNetwork (XLS 24 kb)
Acknowledgments
Thanks are due to Mr T. Jayakumar, Ms B. Gautami and Mr B. J. Moss for conducting lab experiments and collection of data and to Mr V. Sujay for constructive criticisms. Financial support from National Fund for Basic and Strategic Research (NFBSRA), of Indian Council of Agricultural Research (ICAR), New Delhi, India and Generation Challenge Programme (http://www.generationcp.org) of Consultative Group on International Agricultural Research (CGIAR) is gratefully acknowledged. Authors are also thankful to Dr A. Bandyopadhyay, NFBSRA, New Delhi, for his valuable suggestions and support throughout this study.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bernardo R, Charcosset A. Usefulness of gene information in marker-assisted recurrent selection: a simulation appraisal. Crop Sci. 2006;46:614–621. doi: 10.2135/cropsci2005.05-0088. [DOI] [Google Scholar]
- Bernardo R, Yu J. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 2007;47:1082–1090. doi: 10.2135/cropsci2006.11.0690. [DOI] [Google Scholar]
- Bernardo R. Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci. 2008;48:1649–1664. doi: 10.2135/cropsci2008.03.0131. [DOI] [Google Scholar]
- Blum A. Drought resistance, water-use efficiency, and yield potential: are they compatible, dissonant, or mutually exclusive? Aust J Agric Res. 2005;56:1159–1168. doi: 10.1071/AR05069. [DOI] [Google Scholar]
- Collins NC, Tardieu F, Tuberosa R. Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiol. 2008;147:469–486. doi: 10.1104/pp.108.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmet G, Robert N, Perretant MR, Gay G, Sourdille P, Groos C, Bernard S, Bernard M. Marker-assisted recurrent selection for cumulating additive and interactive QTLs in recombinant inbred lines. Theor Appl Genet. 1999;99:1143–1148. doi: 10.1007/s001220051318. [DOI] [Google Scholar]
- Dashti H, Yazdi-Samadi B, Ghannada M, Naghavi MR, Quarri S. QTL analysis for drought resistance in wheat using doubled haploid lines. Int J Agric Biol. 2007;9:98–101. [Google Scholar]
- Devi MJ, Sinclair TR, Vadez V. Genotypic variation in peanut for transpiration response to vapor pressure deficit. Crop Sci. 2010;50:191–196. doi: 10.2135/cropsci2009.04.0220. [DOI] [Google Scholar]
- Frary A, Fulton TM, Zamir D, Tanksley SD. Advanced backcross QTL analysis of a Lycopersicon esculentum × L. pennellii cross and identification of possible orthologs in the Solanaceae. Theor Appl Genet. 2004;108:485–496. doi: 10.1007/s00122-003-1422-x. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Varshney RK. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113:163–185. doi: 10.1023/A:1003910819967. [DOI] [Google Scholar]
- Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS. Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genom. 2004;270:315–323. doi: 10.1007/s00438-003-0921-4. [DOI] [PubMed] [Google Scholar]
- Halward TM, Stalker HT, Larue EA, Kochert G. Genetic variation detectable with molecular markers among unadapted germplasm resources of cultivated peanut and related wild species. Genome. 1991;34:1013–1020. [Google Scholar]
- Hong Y, Chen X, Liang X, Liu H, Zhou G, Li S, Wen S, Holbrook CC, Guo B. A SSR-based composite genetic linkage map for the cultivated peanut (Arachis hypogaea L.) genome. BMC Plant Biol. 2010;10:17. doi: 10.1186/1471-2229-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubick KT, Farquhar GD, Shorter R. Correlation between water-use efficiency and carbon isotope discrimination in diverse peanut (Arachis) germplasm. Aust J Plant Physiol. 1986;13:803–816. doi: 10.1071/PP9860803. [DOI] [Google Scholar]
- Isobe S, Nakaya A, Tabata S. Genotype matrix mapping: searching for quantitative trait loci interactions in genetic variation in complex traits. DNA Res. 2007;14:217–225. doi: 10.1093/dnares/dsm020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannink J. Identifying quantitative trait locus by genetic background interactions in association studies. Genetics. 2007;176:553–561. doi: 10.1534/genetics.106.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimenko I, Razgulayeva N, Gau M, Okumura K, Nakaya A, Tabata S, Kozlov NN, Isobe S. Mapping candidate QTLs related to plant persistency in red clover. Theor Appl Genet. 2010;120:1253–1263. doi: 10.1007/s00122-009-1253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottapalli KR, Burow MD, Burow G, Burke J, Puppala N. Molecular characterization of the U.S. peanut mini core collection using microsatellite markers. Crop Sci. 2007;47:1718–1727. doi: 10.2135/cropsci2006.06.0407. [DOI] [Google Scholar]
- Krishnamurthy L, Vadez V, Jyotsna Devi M, Serraj R, Nigam SN, Sheshshayee MS, Chandra S, Aruna R. Variation in transpiration efficiency and its related traits in a groundnut (Arachis hypogaea L.) mapping population. Field Crops Res. 2007;103:189–197. doi: 10.1016/j.fcr.2007.06.009. [DOI] [Google Scholar]
- Kulwal PL, Singh R, Balyan HS, Gupta PK. Genetic basis of pre-harvest sprouting tolerance using single-locus and two-locus QTL analyses in bread wheat. Funct Integr Genom. 2004;4:94–101. doi: 10.1007/s10142-004-0105-2. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Liang X, Chen X, Hong Y, Liu H, Zhou G, Li S, Guo B. Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biol. 2009;9:35. doi: 10.1186/1471-2229-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I, Powell W. Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 2007;12:57–63. doi: 10.1016/j.tplants.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Mayor PJ, Bernardo R. Genomewide selection and marker-assisted recurrent selection in doubled haploid versus F2 populations. Crop Sci. 2009;47:1719–1725. doi: 10.2135/cropsci2008.10.0587. [DOI] [Google Scholar]
- McElroy JP, Zhang W, Koehler KJ, Lamont SJ, Dekkers JC. Comparison of methods for analysis of selective genotyping survival data. Genet Sel Evol. 2006;38:637–655. doi: 10.1186/1297-9686-38-6-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian MAR, Ashley DA, Boerma HR. An additional QTL for water use efficiency in soybean. Crop Sci. 1998;38:390–393. doi: 10.2135/cropsci1998.0011183X003800020020x. [DOI] [Google Scholar]
- Mohan A, Kulwal P, Singh R, Kumar V, Mir RR, Kumar J, Prasad M, Balyan HS, Gupta PK. Genome-wide QTL analysis for pre-harvest sprouting tolerance in bread wheat. Euphytica. 2009;168:319–328. doi: 10.1007/s10681-009-9935-2. [DOI] [Google Scholar]
- Nageswara Rao RC, Wright GC. Stability of the relationship between specific leaf area and carbon isotope discrimination across environments in peanut. Crop Sci. 1994;34:98–103. doi: 10.2135/cropsci1994.0011183X003400010017x. [DOI] [Google Scholar]
- Nautiyal PC, Rachaputi NR, Joshi YC. Moisture-deficit induced changes in leaf-water content, leaf carbon exchange rate and biomass production in groundnut cultivars differing in specific leaf area. Field Crops Res. 2002;74:67–79. doi: 10.1016/S0378-4290(01)00199-X. [DOI] [Google Scholar]
- Nigam SN, Chandra S, Rupa Sridevi K, Manohar B, Reddy AGS, Nageswara Rao RC, Wright GC, Reddy PV, Deshmukh MP, Mathur RK, Basu MS, Vasundhara S, Vindhiya Varman P, Nagda AK. Efficiency of physiological trait-based and empirical selection approaches for drought tolerance in groundnut. Ann Appl Biol. 2005;146:433–439. doi: 10.1111/j.1744-7348.2005.040076.x. [DOI] [Google Scholar]
- Rebetzke GJ, Condon AG, Richards RA, Farquhar GD. Selection for reduced carbon isotope discrimination increases aerial biomass and grain yield of rain fed bread wheat. Crop Sci. 2002;42:739–745. doi: 10.2135/cropsci2002.0739. [DOI] [Google Scholar]
- Ribaut JM, Hoisington DA, Deutsch JA. Identification of quantitative trait loci under drought conditions in tropical maize. 1. Flowering parameters and the anthesis-silking interval. Theor Appl Genet. 1996;92:905–914. doi: 10.1007/BF00221905. [DOI] [PubMed] [Google Scholar]
- Ribaut JM, Jiang C, Gonzalez-de-Leon D, Edmeades GO, Hoisington DA. Identification of quantitative trait loci under drought conditions in tropical maize. 2. Yield components and marker-assisted selection strategies. Theor Appl Genet. 1997;94:887–896. doi: 10.1007/s001220050492. [DOI] [Google Scholar]
- Ribaut JM, Ragot M. Marker-assisted selection to improve drought adaptation in maize: the backcross approach, perspectives, limitations, and alternatives. J Exp Bot. 2007;58:351–360. doi: 10.1093/jxb/erl214. [DOI] [PubMed] [Google Scholar]
- Ribaut JM, de Vicente MC, Delannay X. Molecular breeding in developing countries: challenges and perspectives. Curr Opin Plant Biol. 2010;13:213–218. doi: 10.1016/j.pbi.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Schuelke M. An economic method for the fluorescent labeling of PCR fragments. A poor man’s approach to genotyping for research and high-throughput diagnostics. Nature. 2000;18:233–234. doi: 10.1038/72708. [DOI] [PubMed] [Google Scholar]
- Serraj R, Krishnamurthy L, Jyotsna Devi M, Reddy MJV, Nigam SN. Variation in transpiration efficiency and related traits in a groundnut mapping population. Int Arachis Newslett. 2004;24:42–45. [Google Scholar]
- Shen X, Zhang T, Guo W, Zhu X, Zhang X. Mapping fiber and yield QTLs with main, epistatic and QTL × environment interaction effects in recombinant inbred lines of upland cotton. Crop Sci. 2006;46:61–66. doi: 10.2135/cropsci2005.0056. [DOI] [Google Scholar]
- Smartt J. The groundnut crop—a scientific basis for improvement. London: Chapman & Hall; 1994. [Google Scholar]
- Ungerer MC, Halldorsdottir SS, Modliszewski JL, Mackay TFC, Purugganan MD. Quantitative trait loci for inflorescence development in Arabidopsis thaliana. Genetics. 2002;160:1133–1151. doi: 10.1093/genetics/160.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Graner A, Sorrells ME. Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005;23:48–55. doi: 10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Hoisington DA, Tyagi AK. Advances in cereal genomics and applications in crop breeding. Trends Biotechnol. 2006;24:490–499. doi: 10.1016/j.tibtech.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Hoisington DA, Upadhyaya HD, Gaur PM, Nigam SN, Saxena K, Vadez V, Sethy NK, Bhatia S, Aruna R, Gowda MVC, Singh NK. Molecular genetics and breeding of grain legume crops for the semi-arid tropics. In: Varshney RK, Tuberosa R, editors. Genomic assisted crop improvement: genomics approaches and platforms. The Netherlands: Springer; 2007. pp. 207–242. [Google Scholar]
- Varshney RK, Dubey A. Novel genomic tools and modern genetic and breeding approaches for crop improvement. J Plant Biochem Biotechnol. 2009;18:127–138. [Google Scholar]
- Varshney RK, Bertioli DJ, Moretzsohn MC, Vadez V, Krishnamurty L, Aruna R, Nigam SN, Ravi K, He G, Knapp SJ, Hoisington DA. The first SSR based genetic linkage map for cultivated groundnut (Arachis hypogaea L.) Theor Appl Genet. 2009;118:729–739. doi: 10.1007/s00122-008-0933-x. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Mahendar T, Aruna R, Nigam SN, Neelima K, Vadez V, Hoisington DA. High level of natural variation in a groundnut (Arachis hypogaea L.) germplasm collection assayed by selected informative SSR markers. Plant Breed. 2009;128:486–494. doi: 10.1111/j.1439-0523.2009.01638.x. [DOI] [Google Scholar]
- Wang CT, Yang XD, Chen DX, Yu SL, Liu GZ, Tang YY, Xu JZ. Isolation of simple sequence repeats from groundnut. Electron J Biotechnol. 2007;10:473–480. doi: 10.2225/vol10-issue3-fulltext-10. [DOI] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB (2007b) Windows QTL cartographer 2.5. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
- Wang D, Graef GL, Procopiuk AM, Diers BW. Identification of putative QTL that underlie yield in interspecific soybean backcross populations. Theor Appl Genet. 2004;108:458–467. doi: 10.1007/s00122-003-1449-z. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Tanksley SD, McCouch SR. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics. 1998;150:899–909. doi: 10.1093/genetics/150.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing YZ, Tan YF, Hua JP, Sun XL, Xu CG, Zhang QF. Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet. 2002;105:248–257. doi: 10.1007/s00122-002-0952-y. [DOI] [PubMed] [Google Scholar]
- Yang J, Hu CC, Ye XZ, Zhu J (2005) QTLNetwork 2.0. Available at http://ibi.zju.edu.cn/software/qtlnetwork. Institute of Bioinformatics, Zhejiang University, Hangzhou, China
- Yang X, Guo Y, Yan J, Zhang J, Song T, Rocheford T, Li JS. Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor Appl Genet. 2009;120:665–678. doi: 10.1007/s00122-009-1184-1. [DOI] [PubMed] [Google Scholar]
- Yoon DB, Kang KH, Kim HJ, Ju HG, Kwon SJ, Suh JP, Jeong OY, Ahn SN. Mapping quantitative trait loci for yield components and morphological traits in an advanced backcross population between Oryza grandiglumis and the O. sativa japonica cultivar Hwaseongbyeo. Theor Appl Genet. 2006;112:1052–1062. doi: 10.1007/s00122-006-0207-4. [DOI] [PubMed] [Google Scholar]
- Zeng ZB. Theoretical basis of separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zheng HG, Aarti A, Pantuwan G, Nguyen TT, Tripathy JN, Sarial AK, Robin S, Babu RC, Nguyen BD, Sarkarung S, Blum A, Nguyen HT. Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theor Appl Genet. 2001;103:19–29. doi: 10.1007/s001220000534. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ESM 1. Comparision of M-QTLs identified by QTL Cartographer and QTLNetwork for droughttolerance traits in groundnut (XLS 52 kb)
ESM 2. Two and three loci epistatic interactions identified by genotype matrix mapping (GMM) (XLS 74 kb)
ESM 3: Epistatic QTLs identified for drought component traits using QTLNetwork (XLS 27 kb)
ESM4: Co-localized QTLs identified using QTL Cartographer and QTLNetwork (XLS 24 kb)