Abstract
A hydrophobic aided replica exchange method (HAREM) is introduced to accelerate the simulation of all-atom protein folding in explicit solvent. This method is based on exaggerating the hydrophobic effect of various protein amino acids in water by attenuating the protein-water attractive interactions (mimicking the Chaperon effect) while leaving other interactions among protein atoms and water molecules unchanged. The method is applied to a small representative protein, the α-helix 3K(I), and it is found that the HAREM method successfully folds the protein within 4 ns, while the regular replica exchange method does not fold the same protein within 5 ns, even with many more replicas.
1. Introduction
A detailed understanding of protein folding and misfolding is critical to many problems in computational biology.1 Many believe that “the primary bottleneck to consistent high-resolution protein prediction appears to be conformational sampling”.2 Recent advances in experimental techniques that probe proteins at different stages of the folding process have shed light on the nature of the mechanisms that govern the kinetics and thermodynamics of folding.3-6 However, many of the details of protein folding pathways remain unknown. Computer simulations performed at various levels of complexity can be used to supplement experiment and fill in some of the gaps in our knowledge about protein folding. Despite efforts from many research groups in the past two decades, the atomistic modeling of protein folding still remains a challenging computational task. Difficulties arise not only from inaccuracies in available force fields, but also from the large-scale computations needed because of expensive interaction evaluations as well as rough energy landscapes with many local minima. There is a significant gap between the current routine computer simulation times of nanoseconds for all-atom proteins in explicit solvent and the times on the order of microseconds and longer required for folding. As pointed out by Snow et. al.,7 “performing a molecular simulation for the 10 μs required for the protein BBA5, a 23-residue mini-protein, would require decades for a typical modern CPU.” Simulation times can be shortened by using modern implicit solvent models;8 nevertheless, it is often desirable or necessary to include explicit water molecules to accurately model salt-bridges, bridge waters, and other effects such as dewetting.9 The computation time for BBA5 in explicit solvent will be much longer than that for implicit solvent simulations because the number of interactions to be computed will increase from 106 to 108 for each time step. Some protein folding simulations with explicit solvent were done with supercomputers such as the IBM BlueGene/L or large-scale clusters such as Folding@Home.7,10
It is well-known that hydrophobic interactions between proteins and water play a very important role in protein folding and are in part responsible for the formation of the hydrophobic core of globular proteins. In contrast, hydrophilic residues will distribute themselves wherever possible to be in contact with water. In our recent study of dewetting in protein folding and aggregation,11 we learned that, when the attractive protein-water interactions were turned off (making the hydrophobic residues more hydrophobic), the hydrophobic collapse of the protein was dramatically speeded up. Thirumalai and co-workers have also shown how certain difficult-to-fold lattice sequences can be made to do so either in an optimal hydrophobic environment or by altering the strength of the hydrophobic interactions.12,13 We exploited these observations in our proposed new sampling scheme, the hydrophobic aided replica exchange method (HAREM), in which the hydrophobic interaction between proteins and water is exaggerated by attenuating the attractive interactions between proteins and water (computationally mimicking the “Chaperone effect”). A different prescription for mimicking the Chaperone effect previously used in protein structure refinement was to periodically increase and decrease the partial charges on water between normal and reduced values, thereby modulating the structure of water itself.14 The current scheme, detailed in section 2, when applied to the folding of the α-helix 3K(I) system, is shown to greatly enhance the probability of energy barrier crossing and is capable of finding native-like structures much more efficiently than ordinary replica exchange. The results are shown in section 3, and discussed in section 4.
2. Method
One of the major problems confronting computational biology is the prediction of native states in proteins. These systems have rough energy landscapes. Ordinary sampling methods are very inefficient upon sampling such landscapes because of their large number of energy minima separated by high energy barriers, and are thus usually quasi-ergodic. As a result, there has been a flowering of new sampling methods. One of these, the replica exchange method (REM),15,16 has been attracting more users and is being widely adopted in computer simulations of biological systems.
In ordinary REM, several independent copies of the simulation system, the so-called replicas, are propagated by molecular dynamics (MD) or Monte Carlo (MC) at different temperatures. At specified intervals, an attempt is made to exchange the replicas at neighboring temperatures. The exchange is accepted or rejected based on a well-defined acceptance probability that guarantees detailed balance. Thus, any replica can climb up and down the temperature ladder, and at any given time there will be one replica for each temperature. When a replica is at high temperature, it can overcome large energy barriers separating stable basins on rough energy landscapes. Replica exchange allows the low temperature replicas to sample configurations that would otherwise be reached with very small probability if replica exchange was prohibited. This significantly reduces the often encountered quasi-ergodicity problem in ordinary MC or MD. If one is interested in computing averages at one temperature, one may average over configurations from all the replicas whenever they move on the temperature level of interest. To benefit from the increased sampling rate of high-temperature replicas, one must average over a time sufficient that each replica can sample temperatures from the lowest to the highest temperature for several cycles.
For large proteins solvated in explicit water, many replicas are required because the number of replicas needed scales with f1/2 approximately, where f is the total number of degrees of freedom in the system. For example, 64 replicas have to be used for a β hairpin, a 16-residue polypeptide in explicit solvent, to obtain reasonable acceptance ratios for the neighboring walkers.17
Recently, MD simulations11,18,19 have shown that the strength of the hydrophobic interactions depend critically on the strength of the solute-solvent interactions. During the course of studying the two-domain protein, BphC enzyme, we observed that the collapse of the two domains can speed up by an order of magnitude when the electrostatic water-protein interactions (Eelec) are turned off.11 When both the electrostatic (Eelec) and attractive van der Waals (vdW) (ELJ6) interactions are turned off, the collapse is even faster. The results suggest that the strength of hydrophobic interactions can be tuned by simply changing certain interactions between the protein and water molecules. The exaggeration of the hydrophobic interactions will then help the protein system cross the free energy barrier. This leads us to propose a HAREM for accelerating protein folding in explicit solvent.
In our new scheme, a variant Hamiltonian REM is proposed based on the knowledge gained from previous dewetting studies.11,18,19 Several other groups, including Takada and coworkers20 and Hansmann and co-workers,21 have previously used the Hamiltonian replica exchange (otherwise called model hopping21) for protein simulations. In the Hamiltonian REM, different replicas can have different potential functions, E0(X0), E1(X1), E2(X2), ..., EN(XN), where Xn represents the configurational coordinates of the nth replica system. The potential functions can be tailored to specific problems.20-22 Here, the interaction between the protein and the solvent molecules is scaled to enhance the hydrophobicity (mimicking the Chaperon effect):
| (1) |
with
| (2) |
| (3) |
where rij is the distance between atom i and atom j, qi is the partial charge of atom i, Uother contains all other energy terms in typical force fields, and λ is the scaling parameter for the vdW attractive potential and electrostatic potential between protein and water. It should be noted that separate scaling factors for vdW and electrostatic interactions can be applied (such as λ1, λ2, etc.), and also the scaling can be applied to some subset of amino acids rather than all of them. Thus, different replicas run with different hydrophobic strengths, that is, different protein-water attractive interactions. When the scaling factors are smaller than 1, the hydrophobic interaction will be exaggerated, mimicking the Chaperone effect for protein folding. Thus, the folding event should be accelerated greatly because the hydrophobic effect is one of the main driving forces for protein folding. The replica exchange is applied for the rigorousness, even if only the folded state is of interest here.
It is simple to derive the acceptance probability for the exchange of the nth and mth replicas (see Takada and coworkers20):
| (4) |
where Xm, Em(Xm), and Tm are respectively the configuration, the energy, and the temperature of the mth replica just before an exchange of replicas is attempted (with corresponding expressions for other replicas). The equilibrium probability for this state is
| (5) |
where βm = 1/(kBTm), and Zm is the corresponding configurational partition function. Denoting the transition probability for the exchange i → f, specified in eq 4, by T(i → f), and denoting that for the reverse exchange by T(f → i) and applying the detailed balance condition,
| (6) |
gives the ratio of the transition probabilities
| (7) |
where
| (8) |
| (9) |
Here, the βs, that is, temperatures, are set to be the same. Of course, one can use different temperatures for various replicas as well. If the Metropolis criteria is applied, the acceptance probability can be obtained:
| (10) |
3. Results and Discussion
The representative protein system we study here is the α-helix 3K(I)23 with a sequence of AAAAKAAAAKAAAAKA. The N and C terminals are capped with Ace and Nme groups, respectively. The three lysine residues are all positive charged. This peptide is solvated in a cubic box of 40.3 Å, with 2095 simple point charge (SPC)24 water molecules and three counterions (Cl-). The entire system consists of 6496 atoms. All the MD simulations were carried out with the OPLSAA force field25 with a 13 Å cutoff in nonbonded interactions using GROMACS26 because of its fast speed. We modified GROMACS to selectively turn off or scale certain interactions. The Berendsen thermostat and barostat27 were used to control the temperature and pressure. The internal geometries of the water molecules were constrained using SETTLE,28 and all the bond lengths were fixed by LINCS,29 which allowed the use of a large time step, 2 fs, to propagate the system.
The fully extended α-helix is denatured in vacuo at 500 K for 1 ns. The last frame is then minimized for 400 steps. After being solvated in a water box, the peptide is equilibrated for another 1 ns. The last configuration is taken as the initial configuration for the protein folding simulations. Similar to what is done in the temperature REM, short trial runs are performed to determine the suitable set of λs by trial and error required to make the acceptance ratios for replica exchange reasonable (10-30%). Many replicas (64 replicas) would be required in regular REM to sample 3K(I) in explicit water (6496 atoms). However, only seven replicas were required in this new Hamiltonian REM (HAREM) with the λ series (1.0, 0.9, 0.8, 0.7, 0.6, 0.5, 0.4). For this new scheme, the exchange interval was chosen as 6.0 ps, which is longer than the velocity relaxation time of the protein and long enough to allow the system to propagate with the deformed Hamiltonian. The results show that our new scheme (HAREM) can locate the native structure within 4 ns, while the actual folding time for this peptide is on the order of a microsecond.
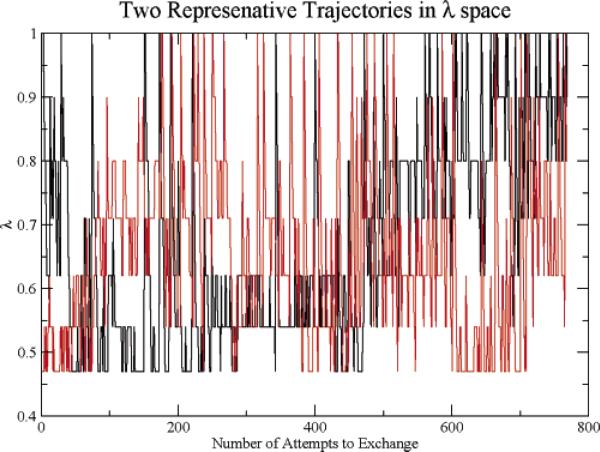
To provide insight into the sampling characteristics of HAREM, we display the trajectories in λ space of two representative replicas, replica 1 and replica 7, in Figure 1. These replicas exhibit random walks spanning the full λ regime accessible, indicating no gaps in the exchange probabilities. Configurations generated in the λ = 1 level, the level of interest, thus has the benefit of the configurations generated in the low λ levels. These results indicate that we have reasonably optimized the spacing of the λ levels.
Figure 1.
The trajectories of λ for two representative replicas.
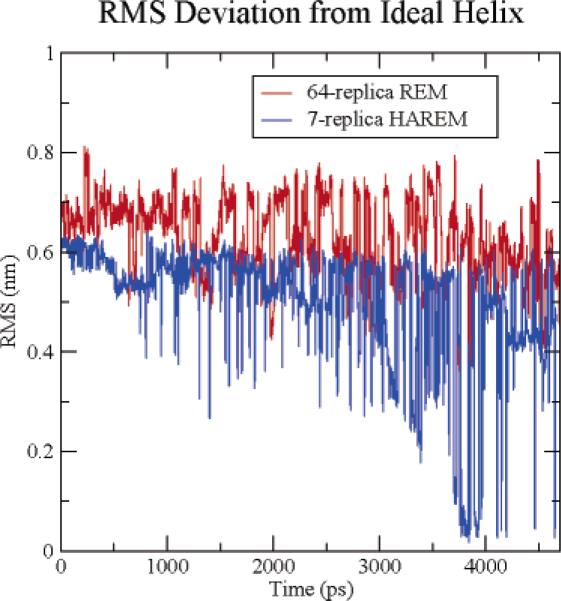
We applied standard REM to the solvated 3K(I) helix system and compared its efficiency to HAREM. A total of 64 replicas were used, with temperatures spanning from 270 to 695 K and a replica exchange acceptance ratio between 20 and 30%. The temperature gaps between neighboring replicas range from 4 to 10 K and give a relatively uniform acceptance ratio.17 All replicas were given the same starting configuration, which was an unfolded conformation. Each replica was run for 5 ns in REM with the same exchange interval of 6.0 ps as in the HAREM. Figure 2 shows the all-heavy-atom root-mean-square deviation (RMSD) from the native structure at 298 K for both REM and HAREM. The RMSD in the REM method is larger than 3.5 Å during the entire 5 ns simulation, indicating that not a single folding event occurred during this run. On the other hand, the RMSD in HAREM decreased to less than 0.5 Å on many occasions, indicating that the helix folded many times in HAREM at 298 K. These results indicate that HAREM not only uses fewer replicas (7 vs 64 here), but also converges faster (4 ns vs >5 ns). On the basis of the relative number of replicas (7 vs 64), we would expect HAREM to be 9 times more efficient than REM. Because HAREM finds the native state in 4 ns while REM does not find it at all in the full run of 5 ns, HAREM appears to be much more than a factor of 9 times more efficient than REM. Much longer runs would be required to determine the relative efficiency.
Figure 2.
Comparison of the RMSD of the helix during the folding trajectory at 298 K for the 7-replica HAREM and the 64-replica normal REM.
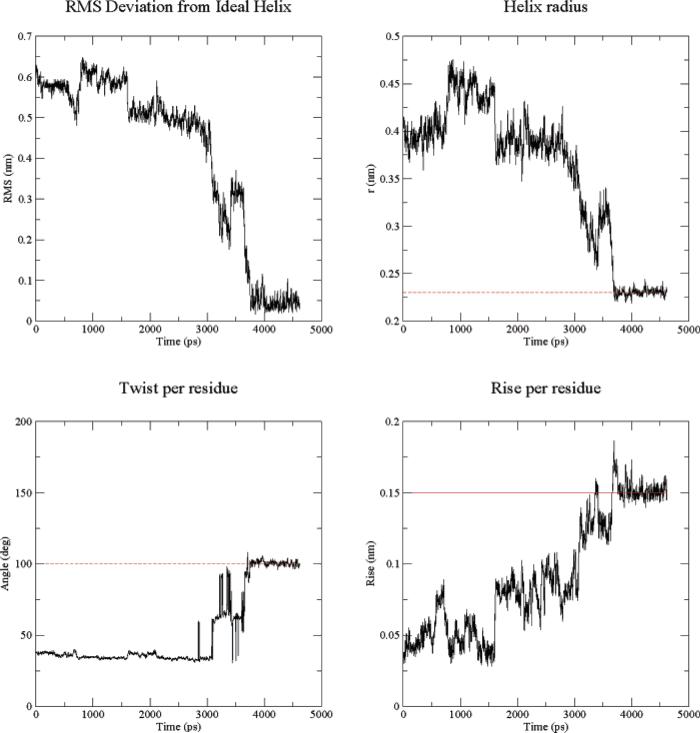
In the following, we investigate how various properties evolve for representative replicas in HAREM (or REM) as they traverse different λ (or temperature) levels. The RMSD from the ideal helix conformation as a function of simulation time is shown in Figure 3a. The initial RMSD is about 6.4 Å, which is large for such a small polypeptide as 3K(I), containing only 16 residues. During the first 800 ps, the RMSD gradually decreases. At around 900 ps, the RMSD increases abruptly, showing that there is a finite probability for accepting high energy configurations. Similarly, there is another bounce in the RMSD curve at about 3.4 ns. Thereafter, the RMSD monotonically decreases. Especially between 3.5 ns and 3.8 ns, the RMSD drops from 3.5 to 0.5 Å. This peptide folds into the ideal helix structure after this transition.
Figure 3.
(a) RMS deviation from an ideal helix, (b) helix radius, (c) twist per residue, and (d) rise per residue.
Similar behavior was found for other properties calculated by GROMACS's g_helix analysis tools, which computes various features of helices. First, the peptide is checked to find the longest helical part. This is determined by criteria involving hydrogen bonds and φ/ψ angles. Then the helical part of the peptide is fit to an ideal helix around the Z-axis and centered around at the origin. The following properties are then computed: (1) Helix radius. This is merely the RMSD in two (X,Y) dimensions for all Cα atoms; it is calculated as , where N is the number of backbone atoms. For an ideal helix, the radius is about 2.3 Å. (2) Twist. This calculates the average helical angle per residue. For the α-helix, it is ~100°; for 310-helices it will be smaller, and for π-helices it will be larger. (3) Rise per residue. The helical rise per residue is plotted as the difference in the Z-coordinate between Cα atoms. For an ideal helix, this is 1.5 Å. Figure 3b shows the helix radius of 3K(I) as a function of simulation time. All the abrupt transitions seen in the above RMSD curve have counterparts in this helix radius curve, indicating that the results are consistent. Additionally, the twist per residue and the rise per residue are shown in Figure 3c,d, which characterizes the types of helices. The value for the twist per residue is found to be 100°, and the rise per residue is approximately 1.5 Å, which are very close to the ideal values of the α-helix, respectively. Thus, both indicators show that the α-helix, but not the 310- or π-helices, were formed in the simulation around 3.8 ns. We also found that the average helicity of the protein increases slightly when the parameter λ is decreased from 1.0 to about 0.7, but, when λ is decreased further, the helicity starts to decrease because the increased hydrophobicity favors a more globular rather than helical structure.
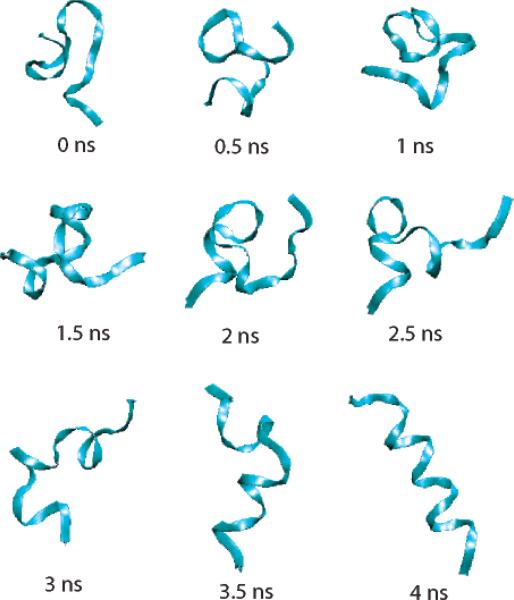
Figure 4 shows how the helix structure evolves during a 4 ns simulation in HAREM for a representative trajectory. The peptide forms a half to one turn of the helix in the early stages of folding, but these small helical seeds are not very stable: they arise and regress. During this, a large turn forms a sort of hairpin structure. After 3-3.5 ns, the helix starts to develop firmly at one end, the N terminal. After 4 ns, the entire helix has formed with a less than 0.5 Å RMSD from the ideal helical structure, indicating that the protein has folded to its most stable structure.
Figure 4.
Snapshots of the peptide in one replica during the folding process.
4. Conclusion
In this paper, we introduce HAREM, a new variant of Hamiltonian replica exchange,20-22 for the efficient sampling of complex biological systems such as protein in explicit solvent. This scheme is inspired by the observation that the hydrophobic driving force in folding can be enhanced simply by scaling the protein-water interaction, thus greatly accelerating hydrophobically generated folding events. In this new algorithm, we partition the system into protein and solvent. By scaling the interaction between the solvent and protein, we mimic the Chaperone effect for protein folding. Since only part of the Hamiltonian is involved in the exchange potential, the acceptance probability for the swap is much larger, making the number of replicas required greatly reduced. This also leads to the benefit of a much better scaling with system size as compared to the regular REM. In this respect, HAREM resembles the replica exchange with solute tempering (REST) method we recently introduced.22
We have applied HAREM to an α-helix, 3K(I), in explicit solvent. In the current HAREM implementation for this peptide, only seven replicas were needed compared to the 64 replicas required in ordinary replica exchange (REM). Even with such a small number of replicas, HAREM is much more efficient in finding the folded conformations than REM starting from the same initial configurations. The HAREM method located the native structure of 3K(I) within 4 ns, whereas the 64-replica regular REM did not find a single folding event within a 5 ns simulation time and nevertheless cost 9 times more, despite its lack of success.
In general, the HAREM method should be useful for simulating biological processes where the hydrophobic effect is the main driving force. However, if some other interactions dominate, such as hydrogen bonds or strong electrostatic interactions among charges, this method might not be as efficient or might even lead to a misfolded state. In other cases, a similar strategy might prove useful when applied to other parts of the protein-water force field. For example, it might prove useful to modify the interaction between hydrophilic residues and water molecules. By exaggerating both the hydrophobic interactions and hydrophilic interactions, the folding event might be further accelerated.
Acknowledgment
This work was supported by a grant awarded to B.J.B. from the NSF (CHE-03-16896). This work was partially supported by the National Center for Supercomputing Applications under CHE050081N.
Footnotes
Part of the special issue “Robert J. Silbey Festschrift”.
References and Notes
- 1.Fersht AR. Structure and Mechanism in Protein Science. W. H. Freeman and Company; New York: 1999. [Google Scholar]
- 2.Bradley P, Misura KM, S l, Baker D. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 3.Blanco FJ, Rivas G, Serrano L. Nat. Struct. Biol. 1994;1:584–590. doi: 10.1038/nsb0994-584. [DOI] [PubMed] [Google Scholar]
- 4.Blanco FJ, Serrano L. Eur. J. Biochem. 1995;230:634–649. doi: 10.1111/j.1432-1033.1995.tb20605.x. [DOI] [PubMed] [Google Scholar]
- 5.Munoz V, Thompson PA, Hofrichter J, Eaton WA. Nature. 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 6.Munoz V, Henry ER, Hofrichter J, Eaton WA. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5872–5879. doi: 10.1073/pnas.95.11.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snow C, Nguyen H, Pande V, Gruebele M. Nature. 2002;420:102–106. doi: 10.1038/nature01160. [DOI] [PubMed] [Google Scholar]
- 8.Gallicchio E, Zhang LY, Levy RM. Proteins. 2004;56:310–321. doi: 10.1002/prot.20104. [DOI] [PubMed] [Google Scholar]
- 9.Zhou R, Berne B. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12777–12782. doi: 10.1073/pnas.142430099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan Y, Kollman PA. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 11.Zhou R, Huang X, Margulius C, Berne B. Science. 2004;305:1605–1609. doi: 10.1126/science.1101176. [DOI] [PubMed] [Google Scholar]
- 12.Betancourt MR, Thirumalai D. J. Mol. Biol. 1999;287:627–644. doi: 10.1006/jmbi.1999.2591. [DOI] [PubMed] [Google Scholar]
- 13.Cheung MS, Thirumalai D. J. Mol. Biol. 2006;357:632–643. doi: 10.1016/j.jmb.2005.12.048. [DOI] [PubMed] [Google Scholar]
- 14.Fan H, Mark AE. Prot. Sci. 2004;13:992–999. doi: 10.1110/ps.03449904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hukushima K, Nemoto K. J. Phys. Soc. Jpn. 1996;65:1604–1608. [Google Scholar]
- 16.Marinari E, Parisi G, Ruiz-Lorenzo JJ. World Sci., Singapore. 1998:59–98. [Google Scholar]
- 17.Zhou R, Berne B, Germain R. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14931–14936. doi: 10.1073/pnas.201543998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, Margulis C, Berne B. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11953–11958. doi: 10.1073/pnas.1934837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu P, Huang X, Zhou R, Berne BJ. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 20.Fukunishi H, Watanabe O, Takada S. J. Chem. Phys. 2002;116:9058–9067. [Google Scholar]
- 21.Kwak W, Hansmann UH. Phys. Rev. Lett. 2005;95:138102–138104. doi: 10.1103/PhysRevLett.95.138102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu P, Kim B, Friesner RA, Berne BJ. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13749–13754. doi: 10.1073/pnas.0506346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smythe M, Nakie C, Marshall G. J. Am. Chem. Soc. 1995;117:10555–10562. [Google Scholar]
- 24.Berendsen H, Postma J, van Gunsteren W, Hermans J. In: Intermolecular Forces. Pullman B, editor. Reidel; Dordrecht, Holland: 1981. p. 331. [Google Scholar]
- 25.Jorgensen W, Maxwell D, Tirado-Rives J. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 26.Lindahl E, van der Spoel D. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 27.Berendsen H, Postma J, DiNola A, Haak J. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 28.Miyamoto S, Kollman P. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 29.Hess B, Bekker H, Berendsen H, Fraaije J. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]