Abstract
Background
Circulating levels of high-sensitivity C-reactive protein (hsCRP), tumor necrosis factor-alpha (TNF-α) and interleukin-6 (IL-6) have been associated with an increased risk of diabetes.
Methods
To examine the roles of genetic variations in CRP, TNF-α, and IL-6 in the development of diabetes, we conducted a prospective case-control study nested in the Women’s Health Initiative Observational Study. 82,069 postmenopausal women aged 50–79 years with no history of diabetes were followed for incident diabetes during a mean follow-up of 5.5 years. 1,584 cases were identified and matched with 2,198 controls based on age, ethnicity, clinical center, time of blood draw and length of follow-up. A total of 13 tagging single nucleotide polymorphisms (tSNPs) across 2.3 kb of the CRP gene, 16 tSNPs across 2.8kb of the TNF-α gene, and 14 tSNPs across 4.8kb of the IL-6 gene were genotyped. Plasma levels of TNF-α Receptor 2 (TNF-α-R2) and IL-6 were assayed.
Results
After adjusting for matching factors, confounding variables, and multiple comparisons, eight genetic variants of the TNF-α gene were associated with plasma TNF-α-R2 concentrations in whites (q-values<0.05). No association was found with any genetic variants of the IL-6 gene and plasma IL-6 levels after adjusting for multiple comparisons (q-value>0.05). No significant associations were found between any SNPs among the 3 genes (CRP, TNF-α, and IL-6) and diabetes risk (q-values>0.05).
Conclusion
Our data indicate modest associations between TNF-α gene variants and circulating levels of TNF-α-R2. Common variants of the genes coding for CRP, TNF-α and IL-6 were not significantly associated with risk of clinical diabetes in postmenopausal women.
Keywords: C-reactive protein, CRP, Tumor necrosis factor alpha, TNF-α, Interleukin-6, IL-6, Diabetes, Type 2 Diabetes, Women’s Health Initiative, WHI
Introduction
Inflammatory markers such as high-sensitivity C-reactive protein (hsCRP), tumor necrosis factor-alpha (TNF-α), and Interleukin-6 (IL-6) have been implicated as possible etiologic factors in the development of obesity, diabetes, and cardiovascular disease(1–10). We previously reported that among postmenopausal women enrolled in the Women’s Health Initiative Observational Study (WHI-OS), elevated circulating levels of hsCRP, TNF-α, and IL-6 were significantly associated with an increased diabetes risk(12). Recently, common genetic variants in the CRP gene were found to be associated with their corresponding plasma marker concentrations in Europeans(13), European Americans(11, 14), African Americans(7, 11), and Pima Indians(15). To date, relatively few studies have investigated the associations of common variants in the genes coding for TNF-α and IL-6 with their plasma concentrations or the direct associations of the genetic variants of CRP, TNF-α, and IL-6 with diabetes risk, especially in a multiethnic-population.
We conducted a comprehensive assessment of the association of genetic variants of TNF-α and IL-6 with plasma concentrations of these two markers of inflammation in a large case-control study nested in the WHI-OS. We also investigated the association of genetic variations in the CRP, TNF-α, and IL-6 genes with diabetes risk in this same group of women.
Materials and Methods
Participants
Details regarding the case-control study design have been described elsewhere(16, 17). The WHI-OS is a longitudinal study designed to examine the association between clinical, socioeconomic, behavioral, and dietary risk factors with subsequent incidence of health outcomes, including diabetes and cardiovascular disease. In brief, 82,069 of the 93,676 postmenopausal women were enrolled in the WHI-OS had no history of diabetes at baseline. We therefore assumed that baseline plasma marker levels and baseline gene expression were not influenced by the presence of diabetes. Diabetes was self-reported by treatments with diet, oral hypoglycemic agents, or insulin. Incident diabetes cases were identified based on post-baseline self-report of first-time use of oral hypoglycemic agents or insulin, or hospitalization for previously unreported diabetes. Following the principle of risk-set sampling(12), for each new case, controls were selected randomly from women who remained free of diabetes at the time the case was identified during follow-up. 1,584 cases were individually matched with 2,198 controls based on age (±2.5 years), racial/ethnic group, clinical center (geographic location), time of blood draw (±0.10 hours), and length of follow-up. The ethnic groups represented in this study include American whites (n=1,936), African Americans (n=1,098), Hispanic/Latino Americans (n=455), and Asian/Pacific Islanders (n=293). A 1:2 matching ratio was used for minorities to strengthen the power in these smaller case sample sizes(17). Each control was identified and matched with a case only once in the analysis. The study was approved by the Institutional Review Board (IRB) at University of California Los Angeles.
Plasma Marker Measurements
Plasma levels of hsCRP, TNF-α receptor 2 (TNF-α-R2) and IL-6 were measured for each participant. TNF-α-R2 has been shown to be more reliably measured from frozen samples than TNF-α itself and its levels have been shown to correlate well with TNF-α concentrations(18, 19). The details of plasma marker measurements are described elsewhere(12). In brief, the coefficients of variations (CVs) were 1.61% for hsCRP, 3.5% for TNF-α-R2 and 7.6% for IL-6, and the units of measurement were mg/L for hsCRP, pg/mL for TNF-α-R2, and pg/mL for IL-6.
Tagging SNP Selection and genotyping methods
As described previously(20, 21), we implemented a two-stage approach to choose tagging Single Nucleotide Polymorphisms (tSNPs) for genotyping. In the first stage, we comprehensively surveyed common genetic variation using the National Center for Biotechnology Information database SNP (NCBI dbSNP) and HapMap database. In the second stage, we identified tSNPs on the basis of linkage disequilibrium (LD) patterns among 61 individuals from each ethnic population. The rationale and method have been previously reported in detail(21, 22).
For selected SNPs, genotyping was performed using the TaqMan allelic discrimination method. Following polymerase chain reaction amplification, end-point fluorescence was read using the ABI Primer 7900 HT instrument and genotypes were scored using SDS2.2.2 Allelic Differentiation Software (Applied Biosystems, Foster City, CA). We genotyped 5% blind duplicated samples randomly selected to evaluate reproducibility. SNPs with a higher genotyping discordant rate, a higher missing genotype, or with deviations from Hardy-Weinberg equilibrium (HWE) at p<0.001 level were excluded.
Statistical Analysis
We first assessed the allele frequency and HWE for each SNP among controls in each ethnic group. Next, we tested for heterogeneity in genotype distributions across ethnic groups with a χ2 test (SAS v9.2, SAS Institute, Cary, NC). In multivariable regression models, we adjusted for matching factors (age, clinical center, time of blood draw, and ethnicity) and other potential confounders (body mass index (BMI), cigarette smoking (never, past and current), alcohol intake (never, past and current), family history of diabetes, hormone replacement therapy (HRT) usage (never, past and current), and total metabolic equivalent value (MET, the energy expended by a person at rest; 1 MET=1 kcal·kg−1 body weight·h−1) from recreational physical activity per week at baseline). To investigate the relationship between SNPs and plasma markers, we log-transformed the specific plasma markers with skewed distributions to improve compliance to the normality assumption. We calculated the differences in the mean logarithms of plasma marker concentrations according to each genotyped tSNP by fitting general linear models that treated plasma marker concentrations as dependent variables and tSNPs as independent variables. An additive model was used; the results of this analysis were expressed as an increase or decrease in the difference in mean logarithms of the plasma marker per each additional copy of the reference allele. Likelihood ratio tests were used to test for effects of genotype-ethnicity interaction on inflammatory marker concentrations.
In assessing the relationship between each single SNP and diabetes risk, we employed multivariable logistic regression, conditional on matching, to calculate Odds Ratios (ORs) and 95% Confidence Intervals (CIs). Each SNP was coded as an additive genetic model in the estimation of allelic association with diabetes risk; the likelihood ratio test was used to test the interaction effect between the genotypes and ethnicity on diabetes risk.
To account for potential false positives due to multiple comparisons in this study, we calculated the false discovery rate (FDR) by incorporating all p-values from multiple tests performed for the association of SNPs and plasma markers. The FDR statistics were obtained for each p-value, and the FDR statistics with q < 0.05 were considered significant(23). Proc Mult test procedure in SAS 9.2 was used to obtain the q-values.
Results
Estimation of Allele Frequencies
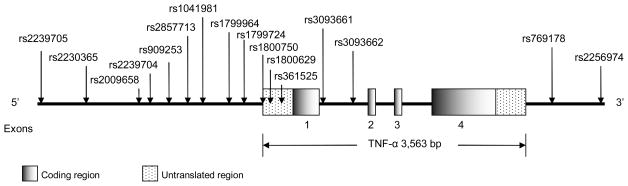
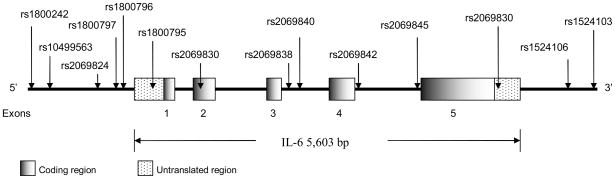
The allele frequencies of 9 SNPs in the TNF-α gene and 13 SNPs in the IL-6 gene differed significantly by ethnicity (Table 1). The 13 SNPs in the CRP gene have been previously published elsewhere(11). Figures 1 and 2 present the locations of the SNPs along the TNF-α and IL-6 genes schematically on the basis of the gene structure in NCBI Entrez Gene. In the TNF-α gene, rs2239704, rs1041981 and rs3093661 in whites were statistically significantly deviated from HWE among the controls. None of the 14 SNPs among the IL-6 gene showed any statistically significant deviation from HWE among the controls in each ethnic group.
Table 1.
Location and allele frequencies of genotyped tagging SNPs in TNF-α and IL-6 genes in controls
| Gene | dbSNP ID a | Location | Alleleb | Allele frequency (%) |
Pc | ||||
|---|---|---|---|---|---|---|---|---|---|
| Whites | Blacks | Hispanics | Asian/Pacific Islanders | Total | |||||
| TNF-α | (n=939) | (n=737) | (n=275) | (n=156) | (n=2,107) | ||||
| rs2239705 | 5′ flanking | C(T) | 18.12 | 13.07 | 25.63 | 20.00 | 17.48 | <0.0001 | |
| rs2230365 | 5′ flanking | C(T) | 15.09 | 6.68 | 17.63 | 25.15 | 13.26 | <0.0001 | |
| rs2009658 | 5′ flanking | C(G) | 16.02 | 13.11 | 18.18 | 17.38 | 15.39 | 0.048 | |
| rs2239704 | 5′ flanking | G(T) | 41.77 | 28.21 | 39.96 | 40.30 | 36.71 | <0.0001 | |
| rs909253 | 5′ flanking | T(C) | 34.22 | 48.59 | 36.05 | 37.35 | 39.73 | <0.0001 | |
| rs2857713 | 5′ flanking | T(C) | 26.70 | 28.36 | 26.71 | 22.26 | 26.93 | 0.28 | |
| rs1041981 | 5′ flanking | C(A) | 33.58 | 48.25 | 35.61 | 38.04 | 39.27 | <0.0001 | |
| rs1799964 | 5′ flanking | T(C) | 21.59 | 18.18 | 21.22 | 20.86 | 20.30 | 0.11 | |
| rs1799724 | 5′ flanking | C(T) | 10.74 | 3.95 | 16.91 | 17.38 | 9.69 | <0.0001 | |
| rs1800750 | 5′ flanking | G(A) | 1.78 | 2.28 | 1.26 | 0 | 1.75 | 0.17 | |
| rs1800629 | 5′ flanking | G(A) | 15.88 | 12.72 | 9.39 | 2.42 | 12.90 | <0.0001 | |
| rs361525 | 5′ flanking | G(A) | 5.67 | 4.62 | 3.06 | 3.44 | 4.80 | 0.22 | |
| rs3093661 | Intron1 | G(A) | 3.98 | 4.10 | 2.36 | 3.46 | 3.77 | 0.50 | |
| rs3093662 | Intron1 | A(G) | 8.15 | 8.67 | 6.27 | 3.61 | 7.73 | 0.045 | |
| rs769178 | 3′ flanking | C(A) | 10.15 | 4.03 | 17.33 | 16.67 | 9.45 | <0.0001 | |
| rs2256974 | 3′ flanking | G(T) | 18.21 | 34.26 | 25.45 | 36.54 | 26.13 | <0.0001 | |
| IL-6 | (n=928) | (n=738) | (n=276) | (n=163) | (n=2,105) | ||||
| rs1880242 | 5′ flanking | G(T) | 53.17 | 79.22 | 56.34 | 26.52 | 60.69 | <0.0001 | |
| rs10499563 | 5′ flanking | C(T) | 77.32 | 82.02 | 80.88 | 82.08 | 79.81 | 0.0069 | |
| rs2069824 | 5′ flanking | C(T) | 91.08 | 85.79 | 91.82 | 99.08 | 89.92 | <0.0001 | |
| rs1800797 | 5′ flanking | A(G) | 62.2 | 90.34 | 78.88 | 96.04 | 76.88 | <0.0001 | |
| rs1800796 | 5′ flanking | C(G) | 93.66 | 90.39 | 78.52 | 30.49 | 85.59 | <0.0001 | |
| rs1800795 | 5′ flanking | C(G) | 61.29 | 90.19 | 78.68 | 97.26 | 76.58 | <0.0001 | |
| rs2069830 | Exon2 | C(T) | 0.05 | 9.14 | 0.36 | 0 | 3.26 | <0.0001 | |
| rs2069838 | Intron3 | C(T) | 0.32 | 6.65 | 1.08 | 0 | 2.62 | <0.0001 | |
| rs2069840 | Intron3 | C(G) | 34.72 | 18.71 | 31.93 | 7.72 | 26.59 | <0.0001 | |
| rs2069842 | Intron4 | A(G) | 99.79 | 92.81 | 99.09 | 100 | 97.26 | <0.0001 | |
| rs2069845 | Intron4 | A(G) | 42.00 | 34.35 | 29.86 | 3.05 | 34.7 | <0.0001 | |
| rs2069861 | 3′ flanking | C(T) | 8.27 | 1.96 | 3.97 | 0.31 | 4.88 | <0.0001 | |
| rs1524106 | 3′ flanking | C(A) | 67.4 | 47.1 | 45.31 | 8.84 | 52.84 | <0.0001 | |
| rs1524103 | 3′ flanking | G(C) | 25.54 | 41.6 | 31.52 | 19.63 | 31.5 | <0.0001 | |
From the National Center for Biotechnology Information (NCBI) dbSNP database.
The reference alleles were indicated in parenthesis.
P-values were estimated by a chi-square test (df=3) for genotype distribution across the four ethnic groups.
Figure 1.
Human TNF-α gene (Chromosome 6p21.3) and SNP locations
Figure 2.
Human IL-6 gene (Chromosome 7p21) and SNP locations
Associations of genetic variants with plasma biomarkers and diabetes risk
Half of the 16 SNPs in TNF-α gene were associated with plasma TNF-α-R2 concentrations in whites (Table 2). For four SNPs, carriers of each additional copy of the reference allele had lower TNF-α-R2 concentrations (decrease in mean logarithm per allele [standard error], range: −0.03 [0.01] to −0.04 [0.02]; all adjusted q-values<0.05 after FDR). In contrast, carriers of the reference alleles of the four other SNPs (rs909253, rs1041981, rs1800629, and rs2256974) had higher TNF-α-R2 concentrations (increase in mean logarithm per allele [standard error], range: 0.04 [0.01] to 0.05 [0.01]; all adjusted q-values<0.05 after FDR). We found no significant association between any of the IL-6 genetic variants and IL-6 concentrations after adjusting for multiple testing.
Table 2.
Geometric mean differences in plasma concentrations according to corresponding SNPs stratified by ethnicity
| Gene | SNP ID | White | Black | Hispanics | Asian/Pacific Islanders | All | P-value for ethnic interactionb |
|---|---|---|---|---|---|---|---|
| Difference in mean logarithms (SE)a | Difference in mean logarithms (SE) | Difference in mean logarithms (SE) | Difference in mean logarithms (SE) | Difference in mean logarithms (SE) | |||
| TNF-α | (n=1,592) | (n=912) | (n=328) | (n=209) | (n=3,041) | ||
| rs2239705 | −0.02 (0.01) | −0.03 (0.02) | 0.002 (0.03) | 0.04 (0.04) | 0.003 (0.01) | 0.03 | |
| rs2230365 | −0.04 (0.01)c | −0.04 (0.03) | −0.02 (0.03) | −0.06 (0.03) | −0.02 (0.01) | 0.86 | |
| rs2009658 | −0.03 (0.01) | −0.04 (0.02) | 0.01 (0.03) | −0.05 (0.04) | −0.02 (0.01) | 0.73 | |
| rs2239704 | −0.02 (0.01) | −0.002 (0.02) | −0.005 (0.02) | −0.01 (0.03) | 0.001 (0.01) | 0.25 | |
| rs909253 | 0.04 (0.01)d | 0.01 (0.02) | 0.02 (0.02) | 0.06 (0.03) | 0.02 (0.01) | 0.70 | |
| rs2857713 | −0.03 (0.01)e | −0.02 (0.02) | 0.001 (0.03) | −0.07 (0.04) | −0.03 (0.01)k | 0.38 | |
| rs1041981 | 0.05 (0.01)f | 0.003 (0.02) | 0.01 (0.02) | 0.06 (0.03) | 0.02 (0.01) | 0.51 | |
| rs1799964 | −0.03 (0.01)g | −0.02 (0.02) | 0.01 (0.03) | −0.07 (0.04) | −0.02 (0.01) | 0.65 | |
| rs1799724 | −0.03 (0.02) | −0.08 (0.04) | −0.01 (0.03) | 0.05 (0.04) | −0.002 (0.01) | 0.01 | |
| rs1800750 | 0.03 (0.04) | 0.04 (0.05) | 0.08 (0.08) | −0.35 (0.31) | 0.03 (0.03) | 0.93 | |
| rs1800629 | 0.04 (0.01)h | −0.01 (0.02) | 0.03 (0.04) | 0.01 (0.08) | 0.02 (0.01) | 0.07 | |
| rs361525 | −0.03 (0.02) | 0.02 (0.04) | −0.02 (0.06) | −0.09 (0.08) | −0.02 (0.02) | 0.58 | |
| rs3093661 | −0.04 (0.02) | −0.03 (0.04) | −0.08 (0.08) | −0.08 (0.08) | −0.05 (0.02) | 0.44 | |
| rs3093662 | −0.04 (0.02) | 0.01 (0.03) | −0.05 (0.05) | −0.10 (0.08) | −0.03 (0.01) | 0.44 | |
| rs769178 | −0.03 (0.02) | −0.07 (0.04) | −0.01 (0.03) | 0.04 (0.04) | −0.001 (0.01) | 0.02 | |
| rs2256974 | 0.04 (0.01)i | 0.01 (0.02) | 0.02 (0.03) | 0.05 (0.03) | 0.01 (0.01) | 0.54 | |
| IL-6 | (n=1,608) | (n=918) | (n=327) | (n=207) | (n=3,060) | ||
| rs1880242 | −0.01 (0.01) | −0.03 (0.03) | −0.06 (0.04) | −0.01 (0.05) | −0.01 (0.01) | 0.64 | |
| rs10499563 | −0.003 (0.01) | −0.003 (0.02) | 0.003 (0.03) | 0.02 (0.05) | −0.002 (0.01) | 0.69 | |
| rs2069824 | −0.01 (0.02) | −0.01 (0.03) | −0.02 (0.05) | −0.08 (0.09) | −0.01 (0.01) | 0.28 | |
| rs1800797 | −0.05 (0.03) | −0.05 (0.07) | −0.10 (0.08) | −0.28 (0.26) | −0.04 (0.02) | 0.02 | |
| rs1800796 | 0.03 (0.05) | 0.06 (0.07) | −0.10 (0.07) | 0.02 (0.10) | 0.04 (0.03) | 0.07 | |
| rs1800795 | −0.04 (0.03) | −0.05 (0.07) | −0.10 (0.08) | −0.23 (0.27) | −0.03 (0.02) | 0.01 | |
| rs2069830 | −0.32 (0.50) | 0.05 (0.07) | −0.06 (0.53) | ---j | 0.09 (0.06) | 0.62 | |
| rs2069838 | 0.05 (0.27) | 0.005 (0.08) | −0.36 (0.38) | ---j | 0.04 (0.07) | 0.57 | |
| rs2069840 | −0.01 (0.03) | 0.08 (0.05) | −0.17 (0.06) | −0.06 (0.17) | −0.01 (0.02) | 0.66 | |
| rs2069842 | −0.16 (0.27) | −0.05 (0.08) | 0.46 (0.26) | ---j | −0.08 (0.06) | 0.17 | |
| rs2069845 | 0.04 (0.03) | −0.001 (0.04) | 0.12 (0.07) | 0.10 (0.40) | 0.04 (0.02) | 0.01 | |
| rs2069861 | 0.05 (0.05) | 0.13 (0.15) | 0.21 (0.14) | ---j | 0.07 (0.04) | 0.04 | |
| rs1524106 | 0.03 (0.03) | 0.05 (0.04) | −0.01 (0.06) | −0.02 (0.15) | 0.03 (0.02) | 0.04 | |
| rs1524103 | −0.03 (0.03) | −0.03 (0.04) | −0.05 (0.07) | 0.002 (0.12) | −0.01 (0.02) | 0.50 | |
Difference in mean logarithms and standard error per additional reference allele of each SNP was calculated using general linear regression models with adjustment for matching factors (age, clinical center, and time of blood draw), incidence of diabetes, and other confounders including BMI, hormone replacement therapy, alcohol consumption, cigarette smoking, family history of diabetes, and physical activity.
P-value was estimated based on a log-likelihood ratio test for interaction between each genotype and ethnicity on plasma concentrations.
P-value was 0.003 with q-value = 0.01 after FDR.
P-value was 0.000041 with q-value = 0.0003 after FDR.
P-value was 0.005 with q-value = 0.02 after FDR.
P-value was 0.000006 with q-value < 0.0001 after FDR.
P-value was 0.007 with q-value = 0.02 after FDR.
P-value was 0.006 with q-value = 0.02 after FDR.
P-value was 0.004 with q-value = 0.01 after FDR.
Result is difficult to interpret because of small sample size within strata.
P-value was 0.0009 with q-value = 0.01 after FDR.
After adjusting for matching factors, other potential confounders, and multiple comparisons, we found no evidence of any significant associations between any of the SNPs among the three genes (CRP, TNF-α, and IL-6) and diabetes risk (all q-values >0.05) (Table 3). Our findings were confirmed when we analyzed four additional models using various covariates, in particular looking at the effect of controlling for family history of diabetes and BMI, to investigate the potential independent associations of the inflammatory variants of interest with diabetes risk (data not shown).
Table 3.
The multivariable-adjusted odds ratio (95% CI) for diabetes risk associated with genetic variants using the additive effect model
| Gene | SNP ID | Adjusted OR (95% CI)a |
P-value for ethnic interactionc | ||||
|---|---|---|---|---|---|---|---|
| Whites | Blacks | Hispanics | Asian/Pacific Islanders | All | |||
| CRP | (870/865)b | (303/638) | (115/242) | (67/154) | (1355/1899) | ||
| rs4275453 | 0.87(0.71–1.07) | 0.85(0.67–1.09) | 0.93(0.57–1.52) | 1.04(0.51–2.15) | 0.85(0.74–0.98)i | 0.92 | |
| rs2808634 | 0.82(0.66–1.03) | 0.74(0.51–1.07) | 0.84(0.48–1.49) | 1.93(0.73–5.11) | 0.81(0.68–0.97)j | 0.34 | |
| rs3093059 | 0.86(0.59–1.25) | 1.17(0.88–1.56) | 0.62(0.30–1.25) | 0.76(0.32–1.80) | 1.03(0.84–1.26) | 0.38 | |
| rs2794521 | 0.82(0.65–1.02) | 0.78(0.54–1.14) | 0.76(0.41–1.38) | 1.66(0.66–4.19) | 0.81(0.68–0.97)k | 0.43 | |
| rs1417938 | 1.03(0.84–1.28) | 1.36(0.93–2.00) | 1.07(0.65–1.76) | 0.75(0.23–2.47) | 1.09(0.92–1.28) | 0.91 | |
| rs1800947 | 1.25(0.85–1.84) | 0.52(0.15–1.81) | 1.46(0.40–5.31) | 1.00(0.33–2.96) | 1.18(0.86–1.63) | 0.55 | |
| rs1130864 | 1.09(0.88–1.35) | 1.18(0.86–1.63) | 0.91(0.56–1.48) | 1.15(0.40–3.31) | 1.09(0.93–1.28) | 0.76 | |
| rs1205 | 1.10(0.89–1.37) | 0.80(0.59–1.10) | 1.19(0.74–1.92) | 0.82(0.43–1.56) | 1.01(0.86–1.17) | 0.61 | |
| rs3093075 | 0.90(0.61–1.32) | 1.17(0.88–1.55) | 0.57(0.28–1.16) | 1.09(0.47–2.51) | 1.06(0.87–1.31) | 0.76 | |
| rs3093068 | 0.81(0.59–1.13) | 1.30(0.97–1.73) | 0.84(0.42–1.66) | 1.11(0.47–2.61) | 1.05(0.86–1.28) | 0.62 | |
| rs2808629 | 1.15(0.93–1.43) | 0.73(0.54–0.99)d | 1.30(0.81–2.10) | 0.72(0.39–1.35) | 1.00(0.86–1.16) | 0.34 | |
| rs2369146 | 0.79(0.62–1.00) | 0.96(0.74–1.24) | 1.23(0.76–2.00) | 2.19(0.80–5.97) | 0.92(0.78–1.08) | 0.02l | |
| rs1470515 | 1.21(0.99–1.48) | 0.70(0.52–0.94)e | 0.97(0.62–1.53) | 0.95(0.50–1.83) | 1.01(0.87–1.17) | 0.29 | |
| TNF-α | (867/882) | (306/638) | (115/244) | (68/155) | (1356/1919) | ||
| rs2239705 | 1.02(0.79–1.31) | 0.74(0.50–1.09) | 0.96(0.61–1.53) | 0.75(0.31–1.79) | 0.92(0.76–1.10) | 0.39 | |
| rs2230365 | 1.06(0.81–1.40) | 0.91(0.53–1.54) | 0.77(0.42–1.44) | 0.86(0.47–1.55) | 0.96(0.78–1.17) | 0.79 | |
| rs2009658 | 0.96(0.73–1.27) | 0.90(0.60–1.33) | 0.75(0.42–1.33) | 0.92(0.44–1.94) | 0.92(0.76–1.11) | 0.75 | |
| rs2239704 | 1.09(0.90–1.31) | 0.79(0.59–1.07) | 1.33(0.92–1.92) | 0.66(0.35–1.23) | 1.00(0.88–1.15) | 0.36 | |
| rs909253 | 0.99(0.81–1.21) | 1.40(1.06–1.85) | 0.83(0.54–1.28) | 1.77(0.92–3.38) | 1.09(0.95–1.25) | 0.52 | |
| rs2857713 | 0.84(0.67–1.05) | 0.94(0.70–1.27) | 0.90(0.55–1.46) | 0.97(0.50–1.91) | 0.90(0.77–1.05) | 0.27 | |
| rs1041981 | 1.02(0.83–1.24) | 1.39(1.06–1.83) | 0.84(0.55–1.28) | 1.79(0.93–3.43) | 1.11(0.97–1.28) | 0.60 | |
| rs1799964 | 0.89(0.70–1.13) | 0.86(0.61–1.20) | 1.01(0.60–1.72) | 1.01(0.50–2.03) | 0.91(0.76–1.07) | 0.28 | |
| rs1799724 | 1.04(0.76–1.43) | 0.62(0.30–1.28) | 1.14(0.67–1.94) | 0.80(0.34–1.92) | 0.97(0.77–1.23) | 0.57 | |
| rs1800750 | 0.51(0.24–1.09) | 1.22(0.55–2.69) | 2.58(0.55–12.2) | ---g | 0.90(0.54–1.50) | 0.01m | |
| rs1800629 | 0.91(0.70–1.18) | 1.24(0.86–1.78) | 1.19(0.56–2.52) | 6.29(1.14–34.8)h | 1.04(0.85–1.27) | 0.02n | |
| rs361525 | 0.74(0.49–1.11) | 1.04(0.57–1.87) | 2.23(0.77–6.46) | 1.56(0.39–6.17) | 0.93(0.69–1.26) | 0.09 | |
| rs3093661 | 0.79(0.50–1.26) | 0.66(0.33–1.32) | 1.56(0.44–5.50) | 0.48(0.08–2.92) | 0.79(0.56–1.12) | 0.95 | |
| rs3093662 | 0.87(0.62–1.23) | 0.70(0.43–1.12) | 1.09(0.47–2.52) | 1.24(0.33–4.63) | 0.84(0.66–1.08) | 0.61 | |
| rs769178 | 1.06(0.77–1.47) | 0.58(0.28–1.20) | 1.09(0.64–1.86) | 0.94(0.39–2.27) | 0.98(0.77–1.24) | 0.70 | |
| rs2256974 | 1.10(0.85–1.42) | 1.26(0.96–1.67) | 0.80(0.49–1.30) | 1.49(0.76–2.93) | 1.14(0.97–1.34) | 0.71 | |
| IL-6 | (876/888) | (304/641) | (115/244) | (67/154) | (1362/1927) | ||
| rs1880242 | 1.01 (0.90–1.13) | 1.04 (0.89–1.22) | 1.14 (0.74–1.76) | 1.14 (0.81–1.60) | 1.03 (0.95–1.12) | 0.36 | |
| rs10499563 | 0.97 (0.88–1.08) | 0.96 (0.83–1.11) | 0.97 (0.76–1.23) | 0.80 (0.58–1.11) | 0.95 (0.89–1.03) | 0.36 | |
| rs2069824 | 0.93 (0.81–1.08) | 1.06 (0.89–1.27) | 0.66 (0.45–0.98)f | ---g | 0.98 (0.89–1.09) | 0.57 | |
| rs1800797 | 1.10 (0.90–1.34) | 0.84 (0.57–1.26) | 1.12 (0.62–2.04) | 0.66 (0.09–4.77) | 1.04 (0.89–1.23) | 0.82 | |
| rs1800796 | 1.18 (0.79–1.76) | 1.34 (0.88–2.04) | 1.02 (0.60–1.73) | 0.90 (0.52–1.55) | 1.10 (0.89–1.37) | 0.66 | |
| rs1800795 | 1.06 (0.87–1.29) | 0.93 (0.61–1.41) | 1.35 (0.76–2.41) | 0.31 (0.01–6.92) | 1.05 (0.89–1.23) | 0.77 | |
| rs2069830 | 1.81 (0.09–38.7) | 0.99 (0.62–1.56) | ---g | ---g | 1.00 (0.64–1.57) | 0.66 | |
| rs2069838 | 0.76 (0.15–3.88) | 1.22 (0.76–1.97) | ---g | ---g | 1.08 (0.69–1.69) | 0.76 | |
| rs2069840 | 1.16 (0.93–1.43) | 0.89 (0.65–1.23) | 0.94 (0.61–1.44) | 0.64 (0.24–1.69) | 1.06 (0.91–1.23) | 0.28 | |
| rs2069842 | 1.07 (0.11–10.4) | 1.43 (0.87–2.37) | 1.07 (0.14–8.55) | ---g | 1.26 (0.79–2.02) | 0.98 | |
| rs2069845 | 0.91 (0.75–1.11) | 1.13 (0.87–1.48) | 0.82 (0.51–1.32) | ---g | 0.95 (0.82–1.09) | 0.49 | |
| rs2069861 | 0.88 (0.62–1.25) | 1.12 (0.43–2.91) | 1.16 (0.43–3.16) | ---g | 0.98 (0.73–1.33) | 0.92 | |
| rs1524106 | 0.86 (0.70–1.06) | 1.13 (0.88–1.44) | 0.87 (0.58–1.31) | 0.97 (0.40–2.34) | 0.95 (0.83–1.10) | 0.53 | |
| rs1524103 | 1.17 (0.93–1.47) | 0.94 (0.73–1.20) | 1.16 (0.73–1.85) | 0.75 (0.38–1.47) | 1.02 (0.88–1.18) | 0.33 | |
Odds Ratio (OR) per additional reference allele of each SNP was calculated for additive genetic effect model; ORs were estimated using conditional logistic regression models adjusted for matching factors (age, clinical center, time of blood draw, and ethnicity), BMI, cigarette smoking, alcohol intake, hormone replacement therapy, family history of diabetes and physical activity.
Sample size for each ethnic group was shown for each plasma marker in the format of (cases/controls).
P-value was estimated based on a log-likelihood ratio test for interaction between each genotype and ethnicity on diabetes risk.
P-value was 0.04 with q-value = 0.28 after FDR.
P-value was 0.02 with q-value = 0.25 after FDR.
P-value was 0.04 with q-value = 0.56 after FDR.
Result was difficult to interpret because of small sample size within strata.
P-value was 0.04 with q-value = 0.46 after FDR.
P-value was 0.03 with q-value = 0.12 after FDR.
P-value was 0.02 with q-value = 0.12 after FDR.
P-value was 0.02 with q-value = 0.12 after FDR.
The P-value became 0.24 after FDR.
The P-value became 0.15 after FDR.
The P-value became 0.15 after FDR.
Discussion
In this large multiethnic cohort of postmenopausal women, eight common genetic variants of the gene coding for TNF-α were associated with plasma concentrations of TNF-α-R2 in whites, whereas there was no association between common genetic variants of the IL-6 gene and its plasma concentration. No genetic variants of CRP, TNF-α, or IL-6 were statistically associated with elevated diabetes risk after correcting for multiple comparison.
One of the 8 SNPs (rs1800629) in the TNF-α gene that we found to be associated with plasma TNF-α concentrations in whites has been associated with TNF-α concentrations in the same ethnic group in a prior study(24). The null findings between the common variants of the same gene with diabetes risk were consistent with previous studies in European and Chinese populations(25–28). The presence of the A allele of SNP rs1800629 in the TNF-α gene among Brazilian individuals over the age of 48 years was associated with elevated levels of hsCRP levels(29). Although our results were not statistically significant, the direction of this association between this genetic variant of TNF-α and hsCRP concentration was the same in our samples. Furthermore, as shown in our prior study, elevated levels of hsCRP were associated with an increased diabetes risk(12). Therefore, if this genetic variant of TNF-α is in fact associated with elevated hsCRP levels, then it may play an indirect role in the pathogenesis of type 2 diabetes. A meta-analysis indicated that individuals who carried this same TNF-α variant are at higher risk of developing obesity compared with controls, suggesting the TNF-α gene is involved in the pathogenesis of the metabolic syndrome(30). Obesity is a well-known risk factor for the development of type 2 diabetes. On the other hand, another study indicated that the same TNF-α variant was not associated with insulin resistance in young Asian Indians(31). Taken together, the TNF-α gene may not directly lead to the development of diabetes, but may play an interactive role with other factors, such as CRP, in the pathogenesis of diabetes.
We did not observe any significant associations between genetic variants of IL-6 gene with plasma concentrations of IL-6 in our samples. One study showed the genetic variant rs10499563 was significantly associated with increased IL-6 concentrations in an acute inflammatory state30. The presence of acute inflammation may affect the association between this genetic variant with plasma IL-6 levels. In general, we would presume the women in our study would not have acute inflammation at the time of their blood draw which may account for this discrepancy. A study of 1953 Korean men and women reported that the rs1800796 G/G genotype was associated with increased serum concentrations of IL-6(32). This is consistent with our analysis which showed that carriers of each additional copy of the G allele in this SNP were associated with increased IL-6 concentrations in the Asian population, although our result was not statistically significant. Inconsistent findings regarding the association between this gene and diabetes risk have been reported previously in several case-control, prospective population-based studies and meta-analyses(33–37). A joint analysis of individual participants’ data from 21 studies observed that the C allele of the rs1800795 SNP in the IL-6 gene was associated with reduced risk of diabetes(34), while a meta-analysis indicated a null association between the same SNP in the IL-6 gene with diabetes risk(36). In general, the literature lacks studies that examine the associations of common variants in the TNF-α and IL-6 genomic regions with their plasma marker concentrations and diabetes risk, particularly in a multiethnic cohort.
Assuming an additive model, we did not observe any significant associations between the genetic variants in the CRP gene with risk of clinical diabetes, which is consistent with prior findings(38). Although these genetic variants were found to have substantial and independent associations with plasma hsCRP concentrations(11), our prospective data do not support a direct heritable role of CRP in the development of diabetes.
The lack of significant genetic associations in the current study may be due to insufficient statistical power, especially among Hispanic and Asian women. Nevertheless, our study was well-powered to detect effects for alleles that were shared across all ethnic groups. In fact, we had >80% power to detect a relative risk of ≥1.25 for a risk alleles with frequencies ranging from 10% to 70%. Additionally, our study only included postmenopausal women and therefore our results may not be generalized to men or younger women.
In conclusion, eight common genetic variants of the TNF-α gene were associated with plasma TNF-α-R2 concentration among whites in this large multiethnic case-control study of postmenopausal women, although these common genetic variants of TNF-α were not associated with risk of clinical diabetes. Common genetic variants of IL-6 were not associated with IL-6 concentrations nor diabetes risk. Neither were common genetic variants of CRP associated with risk of clinical diabetes. Our data indicate modest associations between TNF-α gene variants and circulating levels of TNF-α-R2. Common variants of the genes coding for CRP, TNF-α and IL-6 were not significantly associated with risk of clinical diabetes in postmenopausal women.
Supplementary Material
Acknowledgments
The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. A list of WHI investigators is available in Supplementary Data online.
Abbreviations
- CRP
C-reactive protein
- TNF-α
Tumor necrosis factor alpha
- IL-6
Interleukin-6
- tSNPs
haplotype tagging SNPs
- hsCRP
high-sensitivity C-reactive protein
- IRB
Institutional Review Board
- WHI-OS
Women’s Health Initiative Observational Study
- SNP
single nucleotide polymorphism
- NCBI dbSNP
National Center for Biotechnology Information database SNP
- ABI
Applied Biosystems
- LD
linkage disequilibrium
- HWE
Hardy-Weinberg Equilibrium
- BMI
body mass index
- HRT
hormone replacement therapy
- FDR
false discovery rate
- NHLBI
National Heart, Lung, and Blood Institute
- NIH
National Institutes of Health
- US HHS
US Department of Health and Human Services
- NIDDK
National Institute of Diabetes and Digestive and Kidney Diseases
Footnotes
Human Genes Discussed in this Manuscript: CRP with HUGO approved names “C-reactive protein” and “pentraxin-related” (HGNC: 2367), alias: PTX1; TNF with HUGO approved names “tumor necrosis factor” (HGNC: 11892), alias: TNFSF2, DIF, TNF-alpha; IL-6 with HUGO approved names “interleukin 6 (interferon, beta 2)” (HGNC: 6018), alias: IL-6, BSF2, HGF, HSF.
References
- 1.Pickup JC, Crook MA. Is type II diabetes mellitus a disease of the innate immune system? Diabetologia. 1998;41:1241–8. doi: 10.1007/s001250051058. [DOI] [PubMed] [Google Scholar]
- 2.Duncan BB, Schmidt MI, Pankow JS, Ballantyne CM, Couper D, Vigo A, et al. Low-grade systemic inflammation and the development of type 2 diabetes: the atherosclerosis risk in communities study. Diabetes. 2003;52:1799–805. doi: 10.2337/diabetes.52.7.1799. [DOI] [PubMed] [Google Scholar]
- 3.Han TS, Sattar N, Williams K, Gonzalez-Villalpando C, Lean ME, Haffner SM. Prospective study of C-reactive protein in relation to the development of diabetes and metabolic syndrome in the Mexico City Diabetes Study. Diabetes Care. 2002;25:2016–21. doi: 10.2337/diacare.25.11.2016. [DOI] [PubMed] [Google Scholar]
- 4.McMillan DE. Increased levels of acute-phase serum proteins in diabetes. Metabolism. 1989;38:1042–6. doi: 10.1016/0026-0495(89)90038-3. [DOI] [PubMed] [Google Scholar]
- 5.Freeman DJ, Norrie J, Caslake MJ, Gaw A, Ford I, Lowe GD, et al. C-reactive protein is an independent predictor of risk for the development of diabetes in the West of Scotland Coronary Prevention Study. Diabetes. 2002;51:1596–600. doi: 10.2337/diabetes.51.5.1596. [DOI] [PubMed] [Google Scholar]
- 6.Hu FB, Meigs JB, Li TY, Rifai N, Manson JE. Inflammatory markers and risk of developing type 2 diabetes in women. Diabetes. 2004;53:693–700. doi: 10.2337/diabetes.53.3.693. [DOI] [PubMed] [Google Scholar]
- 7.Carlson CS, Aldred SF, Lee PK, Tracy RP, Schwartz SM, Rieder M, et al. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet. 2005;77:64–77. doi: 10.1086/431366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakanishi S, Yamane K, Kamei N, Okubo M, Kohno N. Elevated C-reactive protein is a risk factor for the development of type 2 diabetes in Japanese Americans. Diabetes Care. 2003;26:2754–7. doi: 10.2337/diacare.26.10.2754. [DOI] [PubMed] [Google Scholar]
- 9.Hu G, Jousilahti P, Tuomilehto J, Antikainen R, Sundvall J, Salomaa V. Association of serum C-reactive protein level with sex-specific type 2 diabetes risk: a prospective finnish study. J Clin Endocrinol Metab. 2009;94:2099–105. doi: 10.1210/jc.2008-2260. [DOI] [PubMed] [Google Scholar]
- 10.Onat A, Can G, Hergenc G. Serum C-reactive protein is an independent risk factor predicting cardiometabolic risk. Metabolism. 2008;57:207–14. doi: 10.1016/j.metabol.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Lee CC, You NC, Song Y, Hsu YH, Manson J, Nathan L, et al. Relation of genetic variation in the gene coding for C-reactive protein with its plasma protein concentrations: findings from the Women’s Health Initiative Observational Cohort. Clin Chem. 2009;55:351–60. doi: 10.1373/clinchem.2008.117176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Tinker L, Song Y, Rifai N, Bonds DE, Cook NR, et al. A prospective study of inflammatory cytokines and diabetes mellitus in a multiethnic cohort of postmenopausal women. Arch Intern Med. 2007;167:1676–85. doi: 10.1001/archinte.167.15.1676. [DOI] [PubMed] [Google Scholar]
- 13.Dehghan A, Kardys I, de Maat MP, Uitterlinden AG, Sijbrands EJ, Bootsma AH, et al. Genetic variation, C-reactive protein levels, and incidence of diabetes. Diabetes. 2007;56:872–8. doi: 10.2337/db06-0922. [DOI] [PubMed] [Google Scholar]
- 14.Suk HJ, Ridker PM, Cook NR, Zee RY. Relation of polymorphism within the C-reactive protein gene and plasma CRP levels. Atherosclerosis. 2005;178:139–45. doi: 10.1016/j.atherosclerosis.2004.07.033. [DOI] [PubMed] [Google Scholar]
- 15.Wolford JK, Gruber JD, Ossowski VM, Vozarova B, Antonio Tataranni P, Bogardus C, Hanson RL. A C-reactive protein promoter polymorphism is associated with type 2 diabetes mellitus in Pima Indians. Mol Genet Metab. 2003;78:136–44. doi: 10.1016/s1096-7192(02)00230-5. [DOI] [PubMed] [Google Scholar]
- 16.Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 17.Anderson GL, Manson J, Wallace R, Lund B, Hall D, Davis S, et al. Implementation of the Women’s Health Initiative study design. Ann Epidemiol. 2003;13:S5–17. doi: 10.1016/s1047-2797(03)00043-7. [DOI] [PubMed] [Google Scholar]
- 18.Aderka D. The potential biological and clinical significance of the soluble tumor necrosis factor receptors. Cytokine Growth Factor Rev. 1996;7:231–40. doi: 10.1016/s1359-6101(96)00026-3. [DOI] [PubMed] [Google Scholar]
- 19.Aderka D, Engelmann H, Maor Y, Brakebusch C, Wallach D. Stabilization of the bioactivity of tumor necrosis factor by its soluble receptors. J Exp Med. 1992;175:323–9. doi: 10.1084/jem.175.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao K, Liu S, Niu T. A sparse marker extension tree algorithm for selecting the best set of haplotype tagging single nucleotide polymorphisms. Genet Epidemiol. 2005;29:336–52. doi: 10.1002/gepi.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu YH, Niu T, Song Y, Tinker L, Kuller LH, Liu S. Genetic variants in the UCP2-UCP3 gene cluster and risk of diabetes in the Women’s Health Initiative Observational Study. Diabetes. 2008;57:1101–7. doi: 10.2337/db07-1269. [DOI] [PubMed] [Google Scholar]
- 22.Song Y, You NC, Hsu YH, Sul J, Wang L, Tinker L, et al. Common genetic variation in calpain-10 gene (CAPN10) and diabetes risk in a multi-ethnic cohort of American postmenopausal women. Hum Mol Genet. 2007;16:2960–71. doi: 10.1093/hmg/ddm256. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y, YH Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 24.Spinarova L, Spinar J, Vasku A, Pavkova-Goldbergova M, Ludka O, Tomandl J, Vitovec J. Genetics of humoral and cytokine activation in heart failure and its importance for risk stratification of patients. Exp Mol Pathol. 2008;84:251–5. doi: 10.1016/j.yexmp.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Shiau MY, Wu CY, Huang CN, Hu SW, Lin SJ, Chang YH. TNF-alpha polymorphisms and type 2 diabetes mellitus in Taiwanese patients. Tissue Antigens. 2003;61:393–7. doi: 10.1034/j.1399-0039.2003.00059.x. [DOI] [PubMed] [Google Scholar]
- 26.Zeggini E, Groves CJ, Parkinson JR, Halford S, Owen KR, Frayling TM, et al. Large-scale studies of the association between variation at the TNF/LTA locus and susceptibility to type 2 diabetes. Diabetologia. 2005;48:2013–7. doi: 10.1007/s00125-005-1902-4. [DOI] [PubMed] [Google Scholar]
- 27.Koch M, Rett K, Volk A, Maerker E, Haist K, Weisser M, et al. The tumour necrosis factor alpha -238 G --> A and -308 G --> A promoter polymorphisms are not associated with insulin sensitivity and insulin secretion in young healthy relatives of Type II diabetic patients. Diabetologia. 2000;43:181–4. doi: 10.1007/s001250050027. [DOI] [PubMed] [Google Scholar]
- 28.Rasmussen SK, Urhammer SA, Jensen JN, Hansen T, Borch-Johnsen K, Pedersen O. The -238 and -308 G-->A polymorphisms of the tumor necrosis factor alpha gene promoter are not associated with features of the insulin resistance syndrome or altered birth weight in Danish Caucasians. J Clin Endocrinol Metab. 2000;85:1731–4. doi: 10.1210/jcem.85.4.6563. [DOI] [PubMed] [Google Scholar]
- 29.Araujo F, Pereira AC, Mota GF, Latorre Mdo R, Krieger JE, Mansur AJ. The influence of tumor necrosis factor -308 and C-reactive protein G1059C gene variants on serum concentration of C-reactive protein: evidence for an age-dependent association. Clin Chim Acta. 2004;349:129–34. doi: 10.1016/j.cccn.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Sookoian SC, Gonzalez C, Pirola CJ. Meta-analysis on the G-308A tumor necrosis factor alpha gene variant and phenotypes associated with the metabolic syndrome. Obes Res. 2005;13:2122–31. doi: 10.1038/oby.2005.263. [DOI] [PubMed] [Google Scholar]
- 31.Ranjith N, Pegoraro RJ, Naidoo DP, Shanmugam R, Rom L. Genetic variants associated with insulin resistance and metabolic syndrome in young Asian Indians with myocardial infarction. Metab Syndr Relat Disord. 2008;6:209–14. doi: 10.1089/met.2008.0023. [DOI] [PubMed] [Google Scholar]
- 32.Koh SJ, Jang Y, Hyun YJ, Park JY, Song YD, Shin KK, et al. Interleukin-6 (IL-6) -572C-->G promoter polymorphism is associated with type 2 diabetes risk in Koreans. Clin Endocrinol (Oxf) 2009;70:238–44. doi: 10.1111/j.1365-2265.2008.03315.x. [DOI] [PubMed] [Google Scholar]
- 33.Herbert A, Liu C, Karamohamed S, Schiller J, Liu J, Yang Q, et al. The -174 IL-6 GG genotype is associated with a reduced risk of type 2 diabetes mellitus in a family sample from the National Heart, Lung and Blood Institute’s Framingham Heart Study. Diabetologia. 2005;48:1492–5. doi: 10.1007/s00125-005-1830-3. [DOI] [PubMed] [Google Scholar]
- 34.Huth C, Heid IM, Vollmert C, Gieger C, Grallert H, Wolford JK, et al. IL6 gene promoter polymorphisms and type 2 diabetes: joint analysis of individual participants’ data from 21 studies. Diabetes. 2006;55:2915–21. doi: 10.2337/db06-0600. [DOI] [PubMed] [Google Scholar]
- 35.Illig T, Bongardt F, Schopfer-Wendels A, Huth C, Heid I, Rathmann W, et al. Genetics of type 2 diabetes: impact of interleukin-6 gene variants. Gesundheitswesen. 2005;67 (Suppl 1):S122–6. doi: 10.1055/s-2005-858396. [DOI] [PubMed] [Google Scholar]
- 36.Qi L, van Dam RM, Meigs JB, Manson JE, Hunter D, Hu FB. Genetic variation in IL6 gene and type 2 diabetes: tagging-SNP haplotype analysis in large-scale case-control study and meta-analysis. Hum Mol Genet. 2006;15:1914–20. doi: 10.1093/hmg/ddl113. [DOI] [PubMed] [Google Scholar]
- 37.Vozarova B, Fernandez-Real JM, Knowler WC, Gallart L, Hanson RL, Gruber JD, et al. The interleukin-6 (-174) G/C promoter polymorphism is associated with type-2 diabetes mellitus in Native Americans and Caucasians. Hum Genet. 2003;112:409–13. doi: 10.1007/s00439-003-0912-x. [DOI] [PubMed] [Google Scholar]
- 38.Zee RY, Germer S, Thomas A, Raji A, Rhees B, Ridker PM, et al. C-reactive protein gene variation and type 2 diabetes mellitus: a case-control study. Atherosclerosis. 2008;197:931–6. doi: 10.1016/j.atherosclerosis.2007.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.