Abstract
Avian polyomavirus (APV) causes a fatal, multi-organ disease among several bird species. Using cryogenic electron microscopy and other biochemical techniques, we investigated the structure of APV and compared it to that of mammalian polyomaviruses, particularly JC polyomavirus and simian virus 40. The structure of the pentameric major capsid protein (VP1) is mostly conserved; however, APV VP1 has a unique, truncated C-terminus that eliminates an intercapsomere-connecting β-hairpin observed in other polyomaviruses. We postulate that the terminal β-hairpin locks other polyomavirus capsids in a stable conformation and that absence of the hairpin leads to the observed capsid size variation in APV. Plug-like density features were observed at the base of the VP1 pentamers, consistent with the known location of minor capsid proteins VP2 and VP3. However, the plug density is more prominent in APV and may include VP4, a minor capsid protein unique to bird polyomaviruses.
Keywords: arginine, capsomere connections, cryo-electron microscopy, homology model, human JC polyomavirus, simian virus 40, size-exclusion chromatography, three-dimensional reconstruction, virus assembly, virus structure
Introduction
Polyomaviruses are widespread and infect mammalian and avian hosts. Virions are non-enveloped particles with a diameter of ~50 nm and contain a circular, double-stranded DNA (dsDNA) genome of ~5,000 base pairs. The structures of murine polyomavirus (MPyV) (Finch, 1974; Rayment et al., 1982; Salunke et al., 1986; Stehle and Harrison, 1996; Stehle et al., 1994), simian virus 40 (SV40) (Baker et al., 1988; Belnap et al., 1996; Kawano et al., 2009; Liddington et al., 1991; Stehle et al., 1996), and human BK polyomavirus (BKV) (Nilsson et al., 2005) have been examined by means of X-ray crystallography or electron microscopy.
Mammalian polyomavirus capsids consist of 360 copies of the major capsid protein, VP1, that assemble into T = 7dextro (T, triangulation number) icosahedral shells comprised of sixty hexavalent and twelve pentavalent VP1 pentamers (Liddington et al., 1991; Rayment et al., 1982; Stehle et al., 1996). Each pentamer contains five β-jellyroll folds formed by interdigitating β-strands. Adjacent pentamers are interconnected via invading, C-terminal extensions, or ‘arms’ (Liddington et al., 1991; Stehle et al., 1996; Stehle and Harrison, 1996). The invading arms of five of the six quasi-equivalent VP1 monomers begin with an α-helix, which assemble into two- or three-helix “bundles” at hexavalent-hexavalent and pentavalent-hexavalent boundaries, respectively. In all VP1 subunits, the arm donates a β-strand to an adjacent capsomere. Calcium ions are necessary for capsid stabilization (Liddington et al., 1991; Salunke et al., 1986; Stehle et al., 1996) and presumably support the invading arm (Stehle et al., 1996).
Three of the six arms end in β-hairpins formed by strands K and L (Liddington et al., 1991; Stehle et al., 1996; Stehle and Harrison, 1996). Initially, these hairpins were proposed to plug holes in the capsid (Liddington et al., 1991). More recent studies have shown, however, that partial truncation of the KL residues results in non-uniform particle sizes (Ou et al., 2001; Yokoyama et al., 2007), suggesting an assembly role.
Polyomavirus capsids also contain two minor structural proteins, VP2 and VP3. VP3 and the C-terminal two-thirds of VP2 have identical sequences. Crystallographic studies demonstrated that either VP2 or VP3 (VP2/3) inserts in a prong-like fashion into the axial cavities of the VP1 pentamers (Chen et al., 1998; Griffith et al., 1992). VP2/3 is highly disordered except for a conserved, 29 amino-acid stretch at its C-terminus (Chen et al., 1998). In this region, a conserved α-helix interacts with one of the five VP1 monomers near the base of the capsomere cavity (Chen et al., 1998; Griffith et al., 1992).
Mammalian polyomaviruses usually exhibit host- and tissue-specific tropism and are generally innocuous towards non-immuno-compromised hosts. Conversely, avian polyomavirus (APV) causes acute, sometimes fatal, disease in a variety of species and orders (Arroube et al., 2009; Johne and Müller, 2007; Lafferty et al., 1999) and was detected in nearly all organs of infected birds (Phalen et al., 1993). A phylogenetic analysis of polyomavirus genomes suggested that bird and mammalian polyomaviruses separated early in their evolutionary history and co-evolved with their hosts (Crandall et al., 2006; Pérez-Losada et al., 2006). Differences between mammalian and bird polyomaviruses led to the proposal to separate them into different genera (Johne and Müller, 2007; Stoll et al., 1993). These distinctions include disparate pathology, differences of T-antigen binding to host genome, and the presence of a unique open reading frame in bird viruses (Johne and Müller, 2007). (For a comprehensive review comparing bird and mammalian polyomaviruses, see Johne and Müller (2007).)
The unique open reading frame in bird polyomaviruses encodes the minor capsid protein VP4 (176 amino acids), and its shorter, non-capsid-associated variant, VP4Δ (112 amino acids) (Johne and Müller, 2001). APV VP4 (hereafter, VP4) is a late gene product of APV (Johne and Müller, 2001) and has no sequence similarity with SV40 VP4, which is a non-structural protein that triggers cell lysis and, like VP3, is derived from the VP2 open-reading frame (Daniels et al., 2007). Expression of VP4 induces apoptosis in Sf9 insect cells and in chicken embryonic cells (Johne et al., 2000), though apoptosis is yet to be reported in infected birds. VP4Δ also induces apoptosis, though the protein is not incorporated into capsids (Johne et al., 2000). APV mutants that lack VP4 exhibit reduced virulence, suggesting that VP4 may contribute to acute pathogenesis (Johne et al., 2007).
VP4 likely has at least two roles in particle assembly. First, VP4 interacts with both VP1 and dsDNA, suggesting that VP4 participates in genome packaging (Johne and Müller, 2001); also, VP4 co-localizes with VP1 and VP3 in the nucleus of infected chicken embryonic cells (Johne and Müller, 2004). The VP4 sequence contains a region enriched with basic amino acids near a putative leucine-zipper (i.e. coiled-coil) motif, suggesting dimerization and DNA binding (Johne and Müller, 2001), cf. (Landschulz et al., 1988). Second, deletions in the VP4-encoding region of APV resulted in a high proportion of misassembled particles, suggesting VP4 facilitates capsid formation (Johne et al., 2007). However, the location and arrangement of VP4 within the capsid is unreported.
Here, we used cryogenic electron microscopy (cryo-EM) and other complementary techniques to investigate the three-dimensional (3D) structure of APV and compare it to the structures of SV40, SV40 virus-like particles (VLPs), and human JC polyomavirus (JCV). APV is very similar to the mammalian polyomaviruses except for its size variation, its lack of KL β-hairpin density, and the presence of a larger, electron-dense mass within the axial cavity of the VP1 pentamer, adjacent to the nucleohistone core. Our results suggest that the shorter, nonconserved C-terminus of APV VP1 affects capsid size and conformation and the KL β-hairpin in other polyomaviruses helps lock capsids in the stable conformation. The larger density associated with each APV capsomere may include VP4.
Results
Cryo-EM of polyomaviruses
We recorded cryo-EM images and computed three-dimensional reconstructions of several polyomavirus samples: APV, APV treated with 250 mM L-arginine (APV+R), SV40, SV40 VLPs, JCV, and JCV treated with 250 mM L-arginine (JCV+R) (Table 1).
Table 1.
Microscopy and image reconstruction data.
| Sample | Voltage (kV)* | Number of images | Reconstruction resolution‡ (Å) | Particlediameters (nm) | Plug length (Å) | Plug volume (kÅ3) | EM Data Bank ID |
|---|---|---|---|---|---|---|---|
| APV ‘tiny’ (pH 7.4)¥ | 250, 300 | 168** | - | < 46 | - | - | - |
| APV ‘small’ (pH 7.4) | 250, 300 | 434** | 31 | 46-51 | n/a§ | n/a§ | - |
| APV ‘medium’ (pH 7.4) | 250, 300 | 570** | 35 | 51-57 | n/a§ | n/a§ | - |
| APV ‘large’ (pH 7.4)¥ | 250, 300 | 56** | - | > 57 | - | - | - |
| APV+R (0.25M arginine, pH 10.7) | 200 | 5,338 | 11¶ | 47-53 | 43±2 | 33±3 | 5180 |
| APVempty+R (empty particles) | 200 | 671 | 18 | 47-53 | 39±3 | 23±3 | 5181 |
| SV40 (pH 7.4) | 80† | 1,155 | 23 | 47-53 | 29±1 | 13±2 | 5187 |
| SV40 VLPs (pH 7.4) | 200 | 13,857 | 14 | 47-53 | 31±2 | 18±4 | 5182 |
| JCV (pH 7.4) | 200 | 1,139 | 19 | - | 32±1 | 17±2 | 5183 |
| JCV+R (0.25M arginine, pH 10.7) | 200 | 1,044 | 18 | - | n/a§ | n/a§ | 5184 |
Except where noted, micrographs were recorded on a FEI Tecnai F30 microscope at a nominal magnification of 39,000×
The relatively low number of particles used in the APV reconstructions (without arginine) was a result of difficulties in extracting individual particle images from particle aggregates (particles must be freestanding). Reconstructions of APV ‘small’ and ‘medium’ are not shown.
Datasets failed to produce consistent reconstructions.
SV40 data set recorded on a Philips EM420 electron microscope at a nominal magnification of 36,000×.
Resolution was estimated by using the Fourier shell correlation criterion (van Heel, 1987) cutoff of 0.5.
By means of a quasi-”local-resolution” test (G. Cardone, in preparation), we determined that the core, β-jellyroll fold was rendered at ~8.5–10-Å resolution (data not shown). Similar resolution was achieved in the αC helices that stabilize intercapsomere interactions. Other more flexible regions, such as those corresponding to exterior loops, were less well resolved (~10–14-Å). In principle, the more rigid a structure is, the higher the resolution that can be attained in crystal structure and cryo-EM studies.
Plugs were not observed in the JCV+R reconstruction, but plugs were observed in the non-arginine treated APV ‘small’ and ‘medium’ reconstructions. The poor resolution and size uncertainties made measurement of the ‘small’ and ‘medium’ APV plugs unfeasible.
Cryo-EM of APV reveals variably sized capsids
Cryo-EM analysis of APV revealed extensive particle aggregation and variable particle sizes (Fig. 1A). With murine polyomavirus (MPyV, ~51 nm diameter) (Stehle and Harrison, 1996) serving as a control, we determined that relative size factors (RSFs) of APV particle images ranged between 0.75 – 1.27 (diameter range 38–65 nm). Unaggregated APV particles with circular profiles were divided into four size classes, designated ‘tiny’, ‘small’, ‘medium’, and ‘large’ (Table 1, TFig. 1C). Conversely, SV40 particles exhibited a narrower size distribution (Fig. 1E). Each particle class appeared to have filled cores (Fig. 1D), suggesting that each contained DNA. We attempted to compute a 3D reconstruction from the four classes of APV particle images, but only the ‘small’ and ‘medium’ classes yielded consistent results—similar = 7, icosahedral structures at 31- and 35-Å resolution, respectively (Table 1).
Fig. 1.

Effect of L-arginine on APV. (A) Cryo-EM image of APV shows particle aggregation and size variation (particles classified according to size). (B) Cryo-EM image of APV after arginine treatment shows significantly less particle aggregation and size variation than those without treatment (A). Scale bars, 100 nm (for A and B). (C) Image averages of four classes of variably sized APV particles observed in preparations like that shown in (A). Particle images were aligned, summed, and normalized via a procedure described previously (Belnap et al., 1993). Scale bar (for C), 20 nm. (D) Radial density profiles of averaged images from C and from images of arginine-treated APV. (E) Relative size factor (RSF) distributions as assessed by PFT2 of three different cryo-EM data sets, with 1.00 set as the standard based on the MPyV reference model. Legend shows sample type and the mean ± standard deviation for each data set (bin size = 0.01).
Cryo-EM of arginine-treated APV (APV+R)
We attempted to improve the resolution in our APV reconstructions by minimizing aggregation of APV particles. Aggregation persisted over a large pH range (4 – 11), even in the presence of mild detergents. Higher resolution reconstructions were not obtained if the ‘small’ and ‘medium’ classes of particle images were pooled into a single dataset or if a narrower range of particle sizes (0.99 – 1.01 RSF) was used (data not shown).
Treatment of APV with 250 mM L-arginine (APV+R) markedly reduced particle aggregation and substantially narrowed the distribution of APV particle sizes to a range that is comparable to that observed for SV40 particles under mild buffer conditions (Fig. 1B,E). In addition, the average size of the APV+R particles lies between the ‘small’ and ‘medium’ classes on non-arginine treated APV (Fig. 1D). The addition of arginine led to a cryo-reconstruction of APV+R at ~1-nm resolution (Table 1, Fig. 2). Addition of L-arginine raised the pH of the APV sample from 7.4 to 10.7. Readjustment of the pH to 7.4 led to re-aggregation of the particles (Fig. S1). Furthermore, aggregation was not reduced by raising the pH of the APV solution to 10.7 in the absence of arginine (Fig. S1). Although a change from pH 7.4 to 10.7 might cause significant conformational changes in some proteins, a reconstruction of JCV treated with 250 mM L-arginine (JCV+R) showed a similar VP1 structure as untreated JCV (Fig. 2). Arginine treatment does appear to lower the density inside the particle (Figs. 1D, S2).
Fig. 2.

Views of pentavalent and hexavalent capsomeres based on cryo-EM reconstructions of APV+R, APV+R empty capsids, SV40, SV40 VLPs, JCV, and JCV+R all rendered at 25-Å resolution to allow for direct comparison. Left column, surface rendering views of fivefold symmetric pentavalent (top, filled pentagon) and non-symmetric hexavalent (bottom, empty hexagon) capsomeres and inter-capsomere connections rendered at 1σ density level. Top, VP1 monomers labeled according to Liddington et al. (1991). Right column, cross-sectional density slices of pentavalent (left) and hexavalent (right) capsomeres. Plug densities were observed at the VP1 pentamer-dsDNA interface, but not when JCV was treated with arginine (black arrowheads). Black (APV+R) or white lines overlaid against plug densities show the measured length of the plug-like density. Lines at the bottom right corner of each panel compare the average pentavalent and hexavalent plug length against the APV+R plug length, with percentage given. Scale bar (striped), 5 nm, for right column only.
The APV+R reconstruction closely resembled cryo-reconstructions of SV40 and JCV (Fig. 2, 3C). The APV+R capsid has the same quaternary structure as mammalian polyomaviruses: pentameric, barrel-shaped capsomeres arranged with T=7 icosahedral symmetry. Inter-capsomere densities are high where αC helices from two (β-β’) or three (α, α’, and α’’) VP1s are known to interact and where twofold-related hexavalent capsomeres lie close together (Liddington et al., 1991; Stehle et al., 1996; Stehle and Harrison, 1996).
Fig. 3.
The structure of APV VP1 compared to other polyomaviruses. Where colored, VP1 subunits are α (gray), α’ (dark blue), α” (green), β (red), β’ (turquoise), and γ (yellow).
(A) C-terminal alignment of 12 variants of polyomavirus VP1. Secondary structure, based on MPyV coordinates (Stehle and Harrison, 1996), is shown above alignment. Black boxes show identical residues, where at least 50% of the aligned amino acids are conserved. Gray boxes show amino acids with conserved properties. Dots show putative Ca2+ binding residues. The APV sequence is not conserved beyond the J′ strand (green arrow) and terminates at the loop between strands K and L. Virus name or host is given on left.
(B) Ribbon diagrams of the homology model of APV VP1 γ subunit (yellow) superimposed over the crystal coordinates of the MPyV VP1 γ subunit (blue). Conserved structures are colored green. β-strands and loops labeled in non-italicized and italicized fonts, respectively. The BC, DE, EF, and HI loops lie on the exterior of the capsid. As shown in (A), the APV sequence terminates after the K β-strand. The C-terminus of APV VP1 (open arrowhead) does not end in a freestanding β-strand and likely does not follow the K β-strand of MPyV.
(C) APV homology model (wires) fit into 3D reconstruction (gray mesh) of APV+R. Representative α, α’, α”, β, β’, and γ subunits are labeled. A fivefold symmetric pentavalent capsomere is labeled with a filled pentagon. Non-symmetric hexavalent capsomeres (unfilled hexagon) contain multi-colored subunits and are numbered. Three- and twofold symmetry axes are labeled with triangle or oval, respectively. Boxes D and E and numbered capsomeres (1–5) refer to panels below. APV reconstruction was rendered at 1σ. C-terminal residues corresponding to the K β-strand in MPyV and SV40 (α’, β’, and γ subunits, e.g. open arrowhead) protrude out of the APV+R reconstruction density, showing that the homology model in this region is incorrect.
(D,E) Inter-capsomere interactions of KL β-hairpins adjacent to twofold (D) and threefold (E) axes rendered at 1.2σ density level. MPyV panels show only the crystal structure (Stehle and Harrison, 1996). JCV and JCV+R panels show MPyV structure fitted into cryo-EM density maps and have density corresponding to the KL β-hairpin. APV+R panels show absence of density where the KL β-hairpin is located. Identified hydrogen bonds are orange with the KL residue labeled (see Table 2). Individual protein subunits are identified by the pentamer where the subunit originates (Arabic number) and by the subunit within the pentamer (Greek letter). Subunits with the same Greek letter are given a different shade of the same color.
(F) Inter-capsomere interactions seen in the SV40 crystal structure (Stehle et al., 1996) shown as in (D), left, and (E), right. Some interactions are similar to those in MPyV but others differ (cf. Table 2).
(G) The KL β-hairpin is analogous to a mechanical cotter or retaining pin, which is a small pin that inserts into one or both ends of a larger and stronger clevis pin used to connect two objects. A cotter pin prevents removal of the clevis pin. In polyomaviruses, the KL β-hairpin functions like a cotter pin in the sense that it more securely fastens the stronger, remaining parts of the C-terminal arm (e.g. the J strand, αC, and Ca2+ chelation) that form intercapsomere connections. Therefore, interactions of the KL β-hairpin may serve as a final “lock” that secures pentamers in the stable, mature conformation.
Sequence analysis and homology modeling of APV VP1
We investigated the basis of APV size variation by comparing the VP1 sequences of several polyomaviruses. With the exception of the more distantly related WU and KI polyomaviruses, alignment of eighteen polyomavirus VP1 sequences revealed 41–62% amino acid identity between bird and mammalian sequences (Fig. S3). High sequence conservation occurs in the β-jellyroll fold, the disulfide bond-forming residues, and most motifs that tether adjacent capsomeres (Fig. S3).
The C-terminal, 12-residue sequence following the J’-strand (amino acids 332–343) is not conserved in APV VP1 and is truncated (Figs. 3A and S3). Other polyomavirus VP1 sequences are significantly longer and are generally conserved in this region. The shorter C-terminus of APV VP1 lacks residues that would normally form a terminal β-hairpin loop composed of strands K and L (KL β-hairpin). The non-conserved sequence is present among all currently sequenced APV strains, including BFDV1 strains 1, 4, and 5, and APV1–7 (data not shown, see Supplementary Data for Genbank accession numbers). Thus, the “premature” truncation is not limited to a single isolate. If the APV stop codon was ignored or various frame shifts were introduced, subsequent codons did not code for amino-acid residues that were similar to those in the β-hairpin of other polyomavirus VP1s.
Other significant differences between the bird and mammalian VP1 sequences occur at the N-terminus and in four external loops (BC, DE, EF, and HI) (Fig. S3). The N-termini in mammalian polyomaviruses contain the nuclear localization signal and the DNA-binding domains, but these are not conserved in APV (Johne and Müller, 2004). Regions corresponding to external loops BC and HI contain residues that mediate receptor binding in SV40 (Neu et al., 2008) and MPyV (Stehle and Harrison, 1997).
High conservation of the core jellyroll fold and inter-capsomere connections provided a rationale for using the MPyV structure (Stehle and Harrison, 1996) as a template to create a homology model for residues 21–343 of APV VP1 (Fig. 3B). The APV homology model fit nicely into the electron density map of the APV+R reconstruction, confirming structural conservation of the major jellyroll fold and T = 7d, pentameric quaternary structure (Fig. 3C). Inter-capsomere stabilization motifs, mediated by the C-terminal arm, are also structurally homologous. Differences between the APV and MPyV structures were seen in the external loops and KL β-hairpin (Fig. 3B). C-terminal residues following the J’-strand protrude out of the APV+R reconstruction, demonstrating that the homology model is incorrect in this region (Fig. 3C).
KL-hairpin interactions in mammalian polyomaviruses
Densities attributable to the KL β-hairpin interactions, as observed in the crystal structures of SV40 (Liddington et al., 1991; Stehle et al., 1996) and MPyV (Stehle and Harrison, 1996), were observed in all the moderate-resolution, 3D reconstructions of JCV and SV40 samples, but were absent in the higher-resolution APV+R 3D map (Figs. 2; 3D,E). The lack of hairpin density was likely not an effect of arginine treatment because the densities remained in the JCV+R reconstruction (Fig. 3D,E). Although the KL β-hairpin structures were observed in α’, β’, and γ subunits of the crystal structures of SV40 (Liddington et al., 1991; Stehle et al., 1996) and MPyV (Stehle and Harrison, 1996), some details of their structures have not been described. The KL-hairpins in MPyV provide additional stability to virions through several inter-capsomere interactions, primarily involving hydrogen bonds (Table 2; Fig. 3D,E). The KL-hairpins of SV40 involve similar inter-capsomere interactions (Table 2, Fig. 3F)—for instance, a conserved arginine in MPyV (R368) and SV40 (R348) hydrogen-bonds with a conserved lysine residue. The SV40 hairpins also participate in non-conserved interactions, such as the hydrophobic interface formed between I350 of a α’ hairpin and F353 of a γ hairpin (Table 2, Fig. 3F). The stabilizing function of the KL-hairpin is non-existent in APV.
Table 2.
Inter-subunit, inter-pentamer interactions that include a residue in K β-strand, hairpin turn, or L β-strand.*, ¶
| Virus | KL β-hairpin | Interacting Entity | ||||
|---|---|---|---|---|---|---|
| Subunit‡ | Residue | Group or Atom | Subunit‡ | Residue(s) | Group or Atom | |
| SV40** | 3γ | P h e 3 5 3 | hydrophobic§ | 1α’ | Ile350 | hydrophobic§ |
| 3γ | Arg348 | main-chain O | 1γ | Lys29 | main-chain NH | |
| 3γ | Thr358 | main-chain O | 1α’ | Tyr299 | side-chain OH | |
| 1α’ | Gln355 | side-chain NH | 2α” | Leu26 | main-chain O | |
| 1α’ | Arg348 | main-chain O | 2α” | Lys29 | main-chain NH | |
| 1α’ | Arg348 | side-chain NH | 2α” | Leu26 | main-chain O | |
| 1β’ | Arg348 | main-chain O | 4β | Lys29 | main-chain NH | |
| 1β’ | Gln355 | side-chain NH | 4β | Val27 | main-chain O | |
| 1β’ | Thr357 | main-chain NH | 5β | Asp101 | side-chain O | |
| MPyV*** | 3γ | Arg372 | main-chain O | 1α’ | Arg372 | main-chain NH |
| 3γ | Lys377 | side-chain NH | 1α’ | Asn317, Pro318 | main-chain O | |
| 3γ | Arg368 | main-chain O | 1γ | Lys31 | main-chain NH | |
| 3γ | Arg368 | side-chain NH | 1γ | Leu28 | main-chain O | |
| 1α’ | Tyr369 | side-chain OH | 2α” | Glu35 | side-chain O | |
| 1α’ | Lys375 | side-chain NH | 2α” | Leu28 | main-chain O | |
| 1α’ | Arg368 | main-chain O | 2α” | Lys31 | main-chain NH | |
| 1α’ | Lys377 | side-chain NH | 2β | Asn317 | main-chain O | |
| 1β’ | Arg368 | main-chain O | 4β | Lys31 | main-chain NH | |
| 1β’ | Met366¶ | main-chain O | 4β | Lys31 | side-chain NH | |
No intra-subunit interactions are listed. All interactions are hydrogen bonds unless stated otherwise. Hydrogen-bonding interactions were identified using UCSF Chimera (Pettersen et al., 2004) with relaxed constraints of 0.4 Å and 20º, based on geometric criteria previously reported (Mills and Dean, 1996).
Residues 347-358 form the KL β-hairpin, reference (Stehle et al., 1996), Protein Data Bank entry 1SVA.
Residues 367-378 form the KL β-hairpin for the β’ subunit, 368-377 for the α’ and γ subunits, reference (Stehle and Harrison, 1996) Protein Data Bank entry 1SIE.
Residue Met366 in MPyV is on the N-terminal side of the K β-strand.
Arabic numbers show the hexavalent pentamer from which the designated subunit originates, as labeled in Fig. 3C.
Hydrophobic interface.
Axial density at base of VP1 pentamer is larger in APV+R than in other polyomaviruses
The axial cavity of each VP1 pentamer in the APV+R reconstruction is plugged by an electron-dense mass at the inner capsid surface (Fig. 2, right). Previous studies demonstrated that VP2/3 insert into the cavity in a prong-like fashion (Chen et al., 1998; Griffith et al., 1992). Sodium dodecyl sulfate – polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analyses of the APV+R sample used in our cryo-EM studies confirmed the presence of VP2 and VP3 in the virion (Fig. 4A).
Fig. 4.
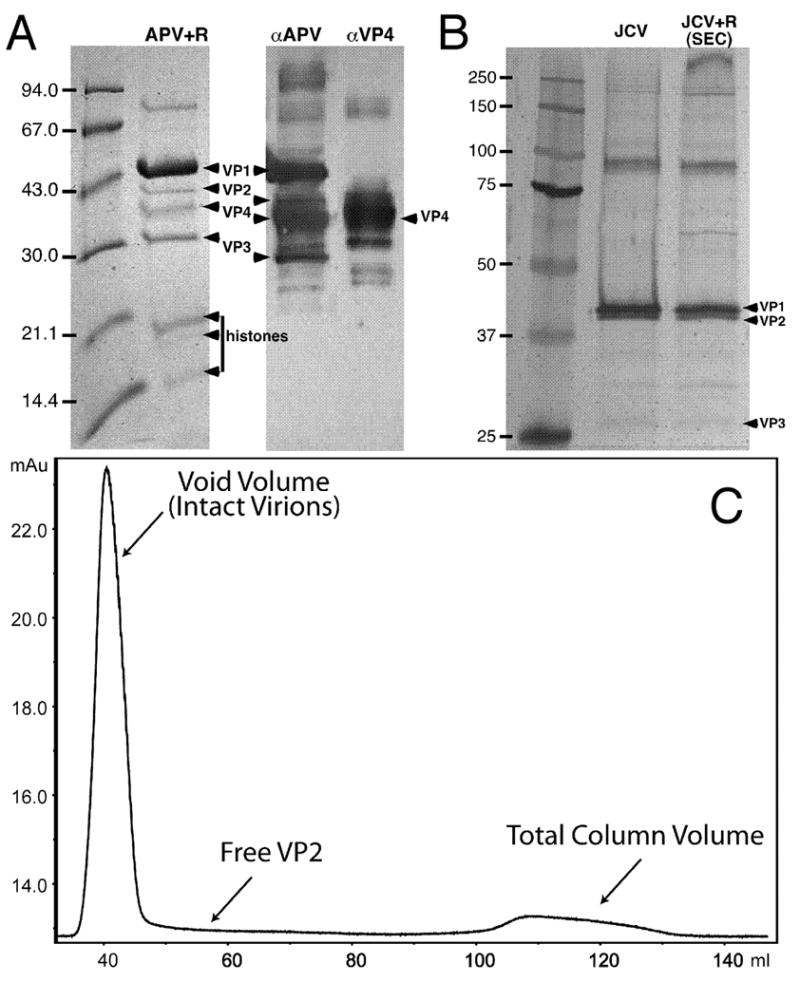
Minor capsid proteins after treatment with L-arginine: APV+R (A) and JCV and JCV+R (B,C). (A) Coomassie-stained SDS-PAGE (left) and immunoblot (right) analysis confirms the presence of VP4 in the sample used to image APV. In the immunoblot, antibodies to all APV structural proteins (left, αAPV) or VP4-specific antibodies (right, αVP4) were used. Note that our APV samples retained protein components, including histones, suggesting that the KL β-hairpin is not a plug as previously suggested (Liddington et al., 1991). (B) Silver-stained SDS-PAGE gel confirms the presence of VP2 and VP3 in JCV virions without and with L-arg treatment. Second lane, JCV without arginine (VP2:VP1 ratio, 0.47). Third lane, JCV+R purified from S100 column (VP2:VP1 ratio, 0.48). High molecular weight bands are seen in both preparations and are likely cross-linked VP1 that did not dissociate during treatment with denaturing buffer. (C) Chromatography of JCV+R purified by size exclusion chromatography (SEC) on a Sephacryl S100 HR column. Plot shows absorbance (milli-absorbance units, mAu, at 280 nm) vs. accumulated fraction volume (ml). Intact virions are eluted in the void volume, whereas any free VP2 should have eluted later (arrow). Although we cannot exclude the fact that some VP2 could be lost on the column, the ratio of VP1-to-VP2 in the control and SEC lanes are nearly identical (see B), indicating that most VP2 remained with the virion after arginine treatment.
The 3D reconstructions of SV40, SV40 VLPs, and JCV revealed similar, yet smaller, ‘plugs’ at the same location as in APV+R (Fig. 2, right). SDS-PAGE analysis also confirmed the presence of VP2 and VP3 in the JCV sample (Fig. 4B). The plug densities in all reconstructions spanned between the axial cavity of the VP1 pentamer and the nucleohistone core of the virion and were observed in both hexavalent and pentavalent capsomeres. The magnitude of these densities was similar to that seen in the VP1 portion of the capsomeres, indicating that most, if not all, of the 72 capsomeres in the capsid are plugged.
For each reconstruction, the approximate volume and length of the plugs in the hexavalent and pentavalent capsomeres were measured from density slices (Table 1; Fig. 2, right). The APV+R plugs were larger compared to those in the mammalian polyomavirus reconstructions, even though the polypeptide-chain lengths of VP2 and VP3 are nearly identical and the VP1-binding helix is conserved in all polyomaviruses (Fig. S4) (Chen et al., 1998). We computed a reconstruction of empty capsids of APV+R (APVempty+R), and these also had larger plugs compared to SV40, SV40 VLPs, and JCV (Table 1, Fig. 2, right). On average, the plug occupied a volume of 28 (± 4) · 103 Å3 in the APV+R and APVempty+R reconstructions. An unsymmetrized model of APV+R was also computed (Sanz-Garcia et al., 2010), and this confirmed the presence of plugs in the same locations (data not shown). The plugs in SV40, SV40 VLPs, and JCV occupied significantly less volume (16 (± 4) · 103 Å3). Length measurements showed that APV+R and APVempty+R plugs were 9 (± 4) Å longer than those in SV40 and JCV (Table 1, Fig. 2, right).
Effect of L-arginine on interactions between major and minor capsid proteins
Although the JCV and JCV+R capsid structures are nearly identical, our JCV+R reconstruction surprisingly showed no detectable densities in the axial cavities of the VP1 pentamers (Fig. 2, right). To see whether arginine treatment caused VP2/3 to be discharged from the JCV capsid, we used size-exclusion chromatography on a JCV+R sample, which should separate virions from any VP2 free in solution. No protein absorbance peaks (λ = 280nm) were detected in fractions following virion elution (Fig. 4C). SDS-PAGE analysis of the virion fraction demonstrated that VP2 and VP3 remained with the particles in a similar ratio as untreated JCV (Fig. 4B). The chromatography and cryo-EM results suggest that, after arginine treatment, JCV VP2 and VP3 have tenuous connections to VP1. The increased pH caused by arginine treatment may be the basis for dissociation of VP2 and VP3; indeed, treatment of SV40 particles with DTT at pH 9.8 dislodged most VP2 and VP3 from VP1 (Christiansen et al., 1977).
Disruption of VP1 pentamer by EDTA, DTT, and L-arginine
For our SV40 VLP and APV samples, we investigated if minor capsid proteins remained connected to the VP1 pentamer after arginine treatment. Virions and VLPs were dissociated into VP1 pentamers using the required EDTA and DTT (Brady et al., 1977) with and without arginine. Solutions without arginine clearly showed co-sedimentation of VP1 and minor capsid proteins (Fig. S5). Surprisingly, solutions with arginine showed co-sedimentation of capsid proteins at a much slower rate, indicating that the VP1 pentamer had been disrupted by treatment with EDTA, DTT, and L-arginine (Fig. S5A,B). Negative-stain electron microscopy of the dissociated APV capsids showed pentamers in the EDTA-DTT solution, but pentamers were not observed in the EDTA-DTT-arginine solution (Fig. S5C,D). Therefore, we were unable to assess the composition of minor capsid proteins that remain attached to VP1 pentamers after arginine treatment.
Discussion
Non-conserved C-terminus of VP1 may cause size variation in APV capsids
Our cryo-EM analysis of APV showed significant size variation among particles (38–65 nm) (Fig. 1). Size variation has been previously reported for APV isolated from infected birds or cell cultures (42–56 nm) (Bernier et al., 1981; Davis et al., 1981; Dykstra and Bozeman, 1982; Randall et al., 1987; Rodgers et al., 1994) and for VLPs made from recombinant VP1 (52–58 nm) (Rodgers et al., 1994). Attempts to correct for relative size differences did not yield improved reconstructions, suggesting that size variation was accompanied by conformational variation.
The crucial role of the C-terminus in forming polyomavirus inter-capsomere connections suggests that the unusual size variation in APV is caused by the unique, truncated C-terminus of VP1 and not by any other intra- or inter-pentamer interaction, all of which are conserved (Figs. 3, S3). The C-terminal extension in APV VP1 is at least nine amino-acid residues shorter than the extension in any other polyomavirus—including those that infect other birds (Figs. 3A, S3). Compared to other polyomaviruses analyzed by cryo-EM by us (Fig. 1E) and others (e.g. Baker et al., 1988; Baker et al., 1989; Belnap et al., 1993; Belnap et al., 1996), the size variation of wild-type particles seems unique to APV. The biological role, if any, of APV size and conformational variation is unknown, but pliable capsids might allow more flexible and advantageous interactions with cellular receptors or may facilitate uncoating after cell entry. The variability may also mean that only certain particle classes are infectious.
Stabilizing function of the KL β-hairpin
Our results suggest that the KL β-hairpin acts to stabilize the polyomavirus capsid and is not a plug as previously suggested (Liddington et al., 1991). The β-hairpins interact with two to four other VP1 subunits and connect two or three hexavalent pentamers (Table 2, Fig. 3D–F). Although the hairpin interactions are relatively few and weak, the C-termini of the α’, β’, and γ subunits do not simply insert completely into the target pentamer (Wolf et al., 2010) but also link additional pentamers. Mammalian polyomaviruses show density consistent with the interacting KL β-hairpins, but APV does not (Fig. 2; Fig. 3D,E). Observation of KL β-hairpin density at moderate resolutions suggests that these hairpins are stable entities and add rigidity to the overall virion structure. Indeed, genomic deletions of the KL β-hairpin in SV40 (residues 345–361) (Yokoyama et al., 2007) and JCV (residues 339–354) (Ou et al., 2001) resulted in particles that exhibited significant size variation.
We liken the relatively weak KL β-hairpin interactions and the other, stronger C-terminal interactions to mechanical cotter and clevis pins, respectively (Fig. 3G). As a cotter pin stabilizes the clevis pin, the KL β-hairpin locks the stronger inter-capsomere interactions in place. Some T=3 viruses have components analogous to the β-hairpin of polyomavirus. For example, cowpea chlorotic mottle virus (CCMV) contains a β-hexamer motif made from six strands from six different subunits. Truncation of the CCMV β-hexamer did not prevent particle assembly, but the β motif does provide increased stability (Willits et al., 2003). Residues in a three-stranded, triangular-shaped β-annulus in Sesbania mosaic virus are not needed for particle assembly (Pappachan et al., 2008), but presumably stabilize the capsid. In polyomaviruses, CCMV, and Sesbania mosaic virus, respectively, the β-hairpin, β-hexamer, and β-annulus each seems to lock the capsid in a stable conformation.
Possible location of minor capsid protein VP4
At the base of the axial cavities of the VP1 capsomeres, we observed larger plug-like densities in APV+R than in SV40 or JCV, and the plug-like density disappeared for JCV+R (Fig. 2). Minor capsid proteins were retained in APV and JCV capsids after arginine treatment (Fig. 4). At least three possibilities explain these phenomena: 1) VP4 is bound in the axial channel adjacent to VP2/3 and shields these minor proteins from the dissociating effects of arginine; 2) VP1, VP2/3, or both are more ordered and stable in APV+R than in mammalian polyomaviruses and VP4 is disordered in the icosahedrally averaged structure; and 3) as in JCV, APV VP2/3 dissociates from the chamber in the presence of arginine, leaving VP4 to form the complete plug.
The sequences of VP2 and VP3 are comparable between bird and mammalian polyomaviruses, with conserved and non-conserved residues in the same regions (Fig. S4). Chen et al. (1998) observed that the C-terminal portion of VP2/3 forms an α-helix that interacts specifically with VP1 and postulated that this interaction is conserved among all polyomaviruses. Therefore, if the only ordered portion of VP2/3 is the conserved VP1-binding region, then the extra density in APV+R would be something other than VP2/3, or the entire plug-like density would be VP4.
Placement of VP4 between VP1 and the genome is consistent with the demonstrated binding of VP4 to both VP1 and dsDNA (Johne and Müller, 2001). A VP1-VP4 interaction was also shown by their co-localization in the nucleus (Johne and Müller, 2004). VP4 is required for proper APV assembly, as deletion of VP4 resulted in a higher percentage of mis-assembled particles (Johne et al., 2007). Recombinant baculovirus expression of only VP1–3 was insufficient to form APV VLPs (An et al., 1999b), whereas expression of mammalian polyomavirus VP1–3 does form VLPs (An et al., 1999a). Although no proof of the VP4 location is possible from our data, a position at the base of the axial cavity, in proximity to VP2/3, would be consistent with the proposed function of VP4 in APV genome packaging and virion assembly (Johne and Müller, 2001; Johne et al., 2007).
Materials and Methods
Cultivation and purification of APV
APV (strain, BFDV-5, GenBank accession number AF241170) was cultivated as previously described (Müller and Nitschke, 1986). It was purified according to the protocol used by Johne and Müller (2001); briefly, infected chicken embryonic cells were sonicated on ice in the presence of Freon-TF. After removing cellular debris by low-speed centrifugation, the supernatant was loaded onto a two-step, CsCl density gradient (ρ = 1.3 and 1.4 g/ml). Two well-separated viral bands were observed. The top band (lowest density) had a higher percentage of empty particles than the bottom band (data not shown). Particles from the bottom band were used in all subsequent analyses. Viral bands were collected after spinning at 53,000 · g for 3 hours, and CsCl was removed by dialysis (D-tube Mini Dialyzers, MWCO (molecular weight cutoff) 12-14 kDa, Novagen, Gibbstown, New Jersey, USA) and the sample was retained in GP buffer (200 mM NaCl, 20 mM Tris-HCl, 1 mM CaCl2 buffer, pH 7.4).
L-arginine has been used to reduce protein aggregation (Shiraki et al., 2002), and we treated purified APV with GP buffer plus 250 mM L-arginine (APV+R) by dialysis (D-tube Mini Dialyzers, MWCO 12-14 kDa, Novagen) to stabilize and de-aggregate particles. The pH of the solution was 10.7. The sample was subjected to centrifugal filtration (YM-100 centrifugal devices, Millipore, Billerica, Massachusetts, USA) for a final washing and concentrating step. Particle deaggregation also was attempted by treating samples with mild detergents, 0.1-0.5% Triton X-100 or Igepal (Sigma-Aldrich, St. Louis, Missouri, USA), and adjusting the pH between 4.0 and 10.7.
Purified APV was subjected to SDS-PAGE and immunoblotting as previously described (Johne and Müller, 2001; Stoll et al., 1993). Rabbit polyclonal antisera αAPV (specific for APV structural proteins VP1, VP2, VP3 and VP4) and αVP4 (specific for VP4) were used for immunodetection of the proteins (Johne and Müller, 2001).
Cultivation and purification of SV40 and SV40 VLPs
SV40 virions were cultivated and purified as described previously (Baker et al., 1988). SV40 VLPs were produced and isolated from Sf-9 cells using a recombinant baculovirus system as reported (Kawano et al., 2009). Briefly, wild-type VP1, and recombinant VP2 and VP3 fused with enhanced green fluorescent protein were cloned individually into pFastBac-1 plasmid vectors (Invitrogen, Carlsbad, California, USA) and introduced into BacMid vectors (Invitrogen). The BacMid vectors were transfected into Sf-9 cells to produce recombinant baculovirus. Sf-9 cells were then infected with plaque-purified, recombinant baculoviruses to generate recombinant SV40 VLPs. The insect cells were harvested three days after infection and cell lysates were prepared via sonication. The resulting VLPs were purified on a 20-50% CsCl density gradient and the virus-containing band was further concentrated with a 37% CsCl density gradient. Finally, the fractions containing VLPs were collected and dialyzed against a buffer containing 20 mM Tris (pH = 7.9) and 50 mM NaCl.
Cultivation and purification of JCV
The Mad-1SVEΔ strain of JCV was used in this study (Vacante et al., 1989). JCV was propagated in SVG-A cells as previously described (Major et al., 1985). JCV was purified using a modified version of previously published results (Jiang et al., 2009; Pelkmans et al., 2001; Pho et al., 2000). At 16-19 days post infection, cells were scraped from the flasks in the presence of growth media, and then pelleted by centrifugation. The virus-containing pellet was resuspended in 10 mL of Buffer A (10mM Tris, 50mM NaCl, 0.1 mM CaCl2, pH 7.4) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, Missouri, USA). The pellet was frozen and thawed three times, after which the lysate was treated with 0.5M HEPES buffer (pH 5.4) to lower the pH to 6.0. Next, five units of type V neuraminidase (Sigma-Aldrich) were added to the lysate for 1 hr at 37°C, then the pH was raised back to 7.5 with 0.5M HEPES (pH 8.0). CaCl2 was added to a final concentration of 0.5mM and cell lysates were treated with 1000 units of DNase for 1 hr at 37°C (New England Biolabs, Ipswitch Massachusetts, USA), after which the lysate was sonicated. The suspension was then pelleted at 4° C, resuspended in 5 mL of Buffer A, sonicated, and centrifuged again as above. The supernatants were pooled following a final round of resuspension and sonication. The virus-containing supernatant was pelleted through a 20% sucrose cushion in at ~155,000 · g for 2.5 hrs (Beckman-Coulter, Brea, California, USA). The resulting pellets were pooled together and resuspended in Buffer A containing CsCl to a final density of 1.34 g/ml; this mixture was centrifuged at 152,000 · g for 16 hr. The virus-containing band was extracted and then dialyzed against either Buffer A alone or Buffer A with 250 mM L-arginine (JCV+R, pH 10.7) (Slide-A-Lyzer, 10 kDa MWCO, Thermo Fisher Scientific, Rockford, Illinois, USA).
Analysis of minor capsid protein retention in polyomaviruses following arginine treatment
To assess whether VP2 remained associated with the virion after arginine treatment, JCV+R was purified by size-exclusion chromatography (SEC) on a Sephacryl S100 column (GE Healthcare, Piscataway, New Jersey, USA) and absorbance (λ = 280 nm) of each fraction was measured. Fractions corresponding to intact virions were pooled and loaded onto 10% SDS-PAGE gels. Gels were color silver stained (Pierce), and imaged on a Chemidoc XRS gel scanner (Bio-Rad, Hercules, California, USA). The presence of VP2 was compared to a non-SEC-purified control to determine whether VP2 was dissociated from virions after L-Arg treatment.
APV, APV+R, SV40 VLPs, and SV40 VLPs+R were dissociated into their pentameric subunits (Brady et al., 1977) by using overnight dialysis in a buffer containing 200 mM NaCl, 5% glycerol (v/v), 10 mM dithiothreitol, 10 mM EDTA, and Tris-HCl, at pH 7.4 and 4 ºC (“+R” buffers also contained 250 mM L-arginine). The samples were then placed on top of a 5-40 %, 5-30 %, or 5-20 % sucrose gradient made by gently layering sucrose solutions in equal amounts and allowing the layered solution to equilibrate overnight at 4 ºC. The samples were centrifuged at ~285,000 · g for 3 hours at 4 ºC and equal volume fractions were extracted from the top of the gradient. Fractions were analyzed via SDS-PAGE.
Cryo-EM and image reconstruction
SV40 virions were imaged as described previously (Baker et al., 1988; Belnap et al., 1996). For all other samples, 3.5 μl of purified virus solution was placed on a glow-discharged, holey-carbon-coated copper grid, blotted, and plunge frozen in liquid ethane with an FEI Vitrobot (Hillsboro, Oregon, USA). Specimens were transferred to a Gatan 626 cryoholder (Pleasanton, California, USA) cooled with liquid nitrogen. Cryo-EM images were recorded at either 200, 250, or 300keV and at 39,000 · magnification via low-dose methods at objective lens underfocus levels ranging from 0.5–4.0 μm on Kodak SO-163 film in an FEI Tecnai F30 transmission electron microscope (Hillsboro, Oregon, USA) (Table 1).
All electron micrographs were digitized on a Nikon Super Coolscan 9000 ED scanner. Particle images were extracted from electron micrographs using X3DPREPROCESS (Conway and Steven, 1999) (Table 1). Using Bsoft (bshow and bctf functions) (Heymann and Belnap, 2007), we determined contrast transfer function signal and decay parameters and applied them to correct the images. Origins and orientations of the extracted particles were determined using the model-based technique of PFT2 (Baker and Cheng, 1996), which was adapted to use phase and amplitude information in orientation selection (Sanz-Garcia et al., 2010). For APV and SV40, a reference model for PFT2 analysis was generated by the random-model method with imposed icosahedral symmetry (Yan et al., 2007). For JCV and SV40 VLPs, reference models were the same as the APV starting model. The final 3D reconstructions were calculated using EM3DR2 (Crowther et al., 1970; Fuller et al., 1996). Bsoft (bresolve and blocres functions) (Heymann and Belnap, 2007) was used to estimate the resolution of reconstructions (Table 1). Two reconstructions were computed, each from alternating particle images in the data set, and used to assess resolution. Quasi-local-resolution was determined by comparing 323 voxel (1.42 · 105 Å3/pixel3) sub-volumes (from the two reconstructions) centered at each voxel in the capsid region of the APV+R structure via a routine implemented in Bsoft (G. Cardone, unpublished) (Gao et al., 2004; Heymann and Belnap, 2007; Mancini et al., 2000). Contour levels are given in terms of σ, which was calculated as the number of standard deviations relative to the average map density.
We calibrated the image pixel size by comparing radially averaged density plots (Belnap et al., 1993) with Bsoft (bradial function) (Heymann and Belnap, 2007) (Fig. S2). Density computed from the atomic coordinates of SV40 (PDB ID, 1SVA, (Stehle et al., 1996)) was our standard. Relative size factors were determined via a routine within PFT2 that changes the size of the reference projection and determines the best correlation with respect to each particle image. Following size calibration, Bsoft (bshow function) (Heymann and Belnap, 2007) was used to estimate the lengths and volumes of densities found inside capsomeres.
Sequence and homology model analysis
Polyomavirus VP1 and VP2 sequences, obtained from GenBank (see Supplemental Data for accession numbers), were aligned using ClustalW2 (Larkin et al., 2007). We created APV VP1 homology models of each quasi-equivalent subunit (α, α’, α’’, β, β’, and γ ) using the SWISS-MODEL server (http://swissmodel.expasy.org) under the program’s alignment mode (Arnold et al., 2006). Bond energies were minimized using UCSF Chimera (Pettersen et al., 2004), after which PROCHECK was used to check structural soundness by a Ramachandran plot (Laskowski et al., 1993).
The APV homology model was fit as a rigid body into the electron density map (Pettersen et al., 2004; Wu et al., 2003). Molecular images were produced using the UCSF Chimera package (Pettersen et al., 2004). Full capsid coordinates of murine polyomavirus (Stehle and Harrison, 1996), SV40 (Stehle et al., 1996), and the APV homology model were assembled by applying icosahedral symmetry using UCSF Chimera (Pettersen et al., 2004) or the ViperDB oligomer generator (Carrillo-Tripp et al., 2009). Hydrogen-bonding sites were determined using UCSF Chimera (Pettersen et al., 2004) with relaxed constraints of 0.4 Å and 20º, based on geometric criteria previously reported (Mills and Dean, 1996).
Supplementary Material
Acknowledgments
This research was supported by a Brigham Young University Mentoring Environment Grant and other BYU institutional funds to DMB; BYU Cancer Research Center fellowships, a BYU Graduate Research Fellowship, a Roland K. Robins Graduate Research Fellowship, and a Jennie R. Swensen Graduate Research Fellowship to PSS; a grant from the Deutsche Forschungsgesellschaft (project JO 369/3–2) to DE; Ruth L. Kirschstein National Research Service Award (F32) 1F32NS070687 to CDSN; National Institutes of Health grant R37-GM033050 to TSB; University of California Discovery Grant 05-10505, NIH grant AI066287-01A1, and National Cancer Institute Pilot Grant (CA093373-06S1) to RHC; and NIH grant R01NS043097 to WJA. The BYU Fulton Supercomputing Lab provided computer resources. We thank Matthew Masner, Jenny Shen, Daniel Shen, and David Eng for help with image processing; Kristin Pande for laboratory assistance; Rebecca Page for help with size-exclusion experiments; Irene Larsen for early work on this project; Jeffrey Farrer and Michael Standing for microscopy support; J. Bernard Heymann for software support; J. Zachary Porterfield for suggesting arginine treatment to deaggregate APV; Hiroshi Handa for SV40 VLP expression vectors; Adam Zlotnick for helpful discussions; our peer reviewers for helpful suggestions on the manuscript; Eduardo Sanz-Garcia for insightful discussions and help with computer programs; and Hermann Müller for ongoing encouragement of this project. The UCSF Chimera package (http://www.cgl.ucsf.edu/chimera) is supported by NIH P41 RR-01081. The APV homology model is available as entry 3IYS in the Protein Data Bank.
Footnotes
budgerigar fledgling disease virus, APV was originally isolated from budgerigars.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An K, Gillock ET, Sweat JA, Reeves WM, Consigli RA. Use of the baculovirus system to assemble polyomavirus capsid-like particles with different polyomavirus structural proteins: analysis of the recombinant assembled capsid-like particles. J Gen Virol. 1999a;80:1009–1016. doi: 10.1099/0022-1317-80-4-1009. [DOI] [PubMed] [Google Scholar]
- An K, Smiley SA, Gillock ET, Reeves WM, Consigli RA. Avian polyomavirus major capsid protein VP1 interacts with the minor capsid proteins and is transported into the cell nucleus but does not assemble into capsid-like particles when expressed in the baculovirus system. Virus Res. 1999b;64:173–185. doi: 10.1016/s0168-1702(99)00093-3. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Arroube AS, Halami MY, Johne R, Dorrestein GM. Mortality due to polyomavirus infection in two nightjars (Caprimulgus europaeus) J Avian Med Surg. 2009;23:136–140. doi: 10.1647/2008-007.1. [DOI] [PubMed] [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryo-electron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. Reconstruction of the Three-Dimensional Structure of Simian Virus 40 and Visualization of the Chromatin Core. Proc Natl Acad Sci USA. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. The Capsid of Small Papova Viruses Contains 72 Pentameric Capsomeres: Direct Evidence from Cryo-Electron-Microscopy of Simian Virus 40. Biophys J. 1989;55:243–253. doi: 10.1016/S0006-3495(89)82799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap DM, Grochulski WD, Olson NH, Baker TS. Use of radial density plots to calibrate image magnification for frozen-hydrated specimens. Ultramicroscopy. 1993;48:347–358. doi: 10.1016/0304-3991(93)90110-j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belnap DM, Olson NH, Cladel NM, Newcomb WW, Brown JC, Kreider JW, Christensen ND, Baker TS. Conserved Features in Papillomavirus and Polyomavirus Capsids. J Mol Biol. 1996;259:249–263. doi: 10.1006/jmbi.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier G, Morin M, Marsolais G. A generalized inclusion body disease in the budgerigar (Melopsittacus undulatus) caused by a papova-like agent. Avian Dis. 1981;25:1083–1092. [PubMed] [Google Scholar]
- Brady JN, Winston VD, Consigli RA. Dissociation of polyoma virus by the chelation of calcium ions found associated with purified virions. J Virol. 1977;23:717–724. doi: 10.1128/jvi.23.3.717-724.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks ICL, Reddy VS. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nuc Acid Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Stehle T, Harrison SC. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 1998;17:3233–3240. doi: 10.1093/emboj/17.12.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G, Landers T, Griffith J, Berg P. Characterization of components released by alkali disruption of simian virus 40. J Virol. 1977;21:1079–1084. doi: 10.1128/jvi.21.3.1079-1084.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Steven AC. Methods for reconstructing density maps of “single” particles from cryoelectron micrographs to subnanometer resolution. J Struct Biol. 1999;128:106–118. doi: 10.1006/jsbi.1999.4168. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Pérez-Losada M, Christensen RG, McClellan DA, Viscidi RP. Phylogenomics and molecular evolution of polyomaviruses. Adv Exp Med Biol. 2006;577:46–59. doi: 10.1007/0-387-32957-9_3. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Amos LA, Finch JT, DeRosier DJ, Klug A. Three Dimensional Reconstructions of Spherical Viruses by Fourier Synthesis from Electron Micrographs. Nature. 1970;226:421–425. doi: 10.1038/226421a0. [DOI] [PubMed] [Google Scholar]
- Daniels R, Sadowicz D, Hebert DN. A very late viral protein triggers the lytic release of SV40. PLoS Pathogens. 2007;3:e98. doi: 10.1371/journal.ppat.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RB, Bozeman LH, Gaudry D, Fletcher OJ, Lukert PD, Dykstra MJ. A viral disease of fledgling budgerigars. Avian Dis. 1981;25:179–183. [PubMed] [Google Scholar]
- Dykstra MJ, Bozeman LH. A light and electron microscopic examination of budgerigar fledgling disease virus in tissue and in cell culture. Avian Pathol. 1982;11:11–28. doi: 10.1080/03079458208436078. [DOI] [PubMed] [Google Scholar]
- Finch JT. The Surface Structure of Polyoma Virus. J Gen Virol. 1974;24:359–364. doi: 10.1099/0022-1317-24-2-359. [DOI] [PubMed] [Google Scholar]
- Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles--the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- Gao H, Valle M, Ehrenberg M, Frank J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J Struct Biol. 2004;147:283–290. doi: 10.1016/j.jsb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Griffith JP, Griffith DL, Rayment I, Murakami WT, Caspar DLD. Inside polyomavirus at 25-Å resolution. Nature. 1992;355:652–654. doi: 10.1038/355652a0. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Tsai B, Imperiale MJ. Early events during BK virus entry and disassembly. J Virol. 2009;83:1350–1358. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R, Jungmann A, Müller H. Agnoprotein 1a and agnoprotein 1b of avian polyomavirus are apoptotic inducers. J Gen Virol. 2000;81:1183–1190. doi: 10.1099/0022-1317-81-5-1183. [DOI] [PubMed] [Google Scholar]
- Johne R, Müller H. Avian polyomavirus agnoprotein 1a is incorporated into the virus particle as a fourth structural protein, VP4. J Gen Virol. 2001;82:909–918. doi: 10.1099/0022-1317-82-4-909. [DOI] [PubMed] [Google Scholar]
- Johne R, Müller H. Nuclear localization of avian polyomavirus structural protein VP1 is a prerequisite for the formation of virus-like particles. J Virol. 2004;78:930–937. doi: 10.1128/JVI.78.2.930-937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R, Müller H. Polyomaviruses of Birds: Etiologic Agents of Inflammatory Diseases in a Tumor Virus Family. J Virol. 2007;81:11554–11559. doi: 10.1128/JVI.01178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R, Paul G, Enderlein D, Stahl T, Grund C, Müller H. Avian polyomavirus mutants with deletions in the VP4-encoding region show deficiencies in capsid assembly and virus release, and have reduced infectivity in chicken. J Gen Virol. 2007;88:823–830. doi: 10.1099/vir.0.82506-0. [DOI] [PubMed] [Google Scholar]
- Kawano M-a, Xing L, Tsukamoto H, Inoue T, Handa H, Cheng RH. Calcium bridge triggers capsid disassembly in the cell entry process of simian virus 40. J Biol Chem. 2009;284:34703–34712. doi: 10.1074/jbc.M109.015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty SL, Fudge AM, Schmidt RE, Wilson VG, Phalen DN. Avian polyomavirus infection and disease in a green aracaris (Pteroglossus viridus) Avian Dis. 1999;43:577–585. [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK- a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. Structure of simian virus 40 at 3.8-Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci USA. 1985;82:1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Molecular Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- Mills JEJ, Dean PM. Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J Comput-Aided Mol Des. 1996;10:607. doi: 10.1007/BF00134183. [DOI] [PubMed] [Google Scholar]
- Müller H, Nitschke R. A polyoma-like virus associated with an acute disease of fledgling budgerigars (Melopsittacus undulatus) Med Microbiol Immunol. 1986;175:1–13. doi: 10.1007/BF02123124. [DOI] [PubMed] [Google Scholar]
- Neu U, Woellner K, Gauglitz G, Stehle T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc Natl Acad Sci USA. 2008;105:5219–5224. doi: 10.1073/pnas.0710301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Miyazaki N, Xing L, Wu B, Hammar L, Li TC, Takeda N, Miyamura T, Cheng RH. Structure and Assembly of a T = 1 Virus-Like Particle in BK Polyomavirus. J Virol. 2005;79:5337–5345. doi: 10.1128/JVI.79.9.5337-5345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou WC, Chen LH, Wang M, Hseu TH, Chang D. Analysis of minimal sequences on JC virus VP1 required for capsid assembly. J Neurovirol. 2001;7:298–301. doi: 10.1080/13550280152537139. [DOI] [PubMed] [Google Scholar]
- Pappachan A, Subashchandrabose C, Satheshkumar PS, Savithri HS, Murthy MRN. Structure of recombinant capsids formed by the β-annulus deletion mutant — rCP (Δ48–59) of Sesbania mosaic virus. Virology. 2008;375:190–196. doi: 10.1016/j.virol.2008.01.023. [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Kartenbeck J, Helenius A. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat Cell Biol. 2001;3:473–483. doi: 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- Pérez-Losada M, Christensen RG, McClellan DA, Adams BJ, Viscidi RP, Demma JC, Crandall KA. Comparing phylogenetic codivergence between polyomaviruses and their hosts. J Virol. 2006;80:5663–5669. doi: 10.1128/JVI.00056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phalen DN, Wilson VG, Graham DL. Organ distribution of avian polyomavirus DNA and virus-neutralizing antibody titers in healthy adult budgerigars. Am J Vet Res. 1993;54:2040–2047. [PubMed] [Google Scholar]
- Pho MT, Ashok A, Atwood WJ. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J Virol. 2000;74:2288–2292. doi: 10.1128/jvi.74.5.2288-2292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall CJ, Lees S, Inglis DM. Papovavirus-like infection in budgerigars (Melopsittacus undulatus) Avian Pathol. 1987;16:623–633. doi: 10.1080/03079458708436411. [DOI] [PubMed] [Google Scholar]
- Rayment I, Baker TS, Caspar DLD, Murakami WT. Polyoma Virus Capsid Structure at 22.5 Å Resolution. Nature. 1982;295:110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers RED, Chang D, Cai X, Consigli RA. Purification of Recombinant Budgerigar Fledgling Disease Virus VP1 Capsid Protein and Its Ability for In Vitro Capsid Assembly. J Virol. 1994;68:3386–3390. doi: 10.1128/jvi.68.5.3386-3390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salunke DM, Caspar DLD, Garcea RL. Self-Assembly of Purified Polyomavirus Capsid Protein VP1. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- Sanz-Garcia E, Stewart AB, Belnap DM. The random-model method enables ab initio three-dimensional reconstruction of asymmetric particles and determination of particle symmetry. J Struct Biol. 2010 doi: 10.1016/j.jsb.2010.03.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki K, Kudou M, Fujiwara S, Imanaka T, Takagi M. Biophysical Effect of Amino Acids on the Prevention of Protein Aggregation. J Biochem. 2002;132:591–595. doi: 10.1093/oxfordjournals.jbchem.a003261. [DOI] [PubMed] [Google Scholar]
- Stehle T, Gamblin SJ, Yan Y, Harrison SC. The structure of simian virus 40 refined at 3.1 Å resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- Stehle T, Harrison SC. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- Stehle T, Harrison SC. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 1997;16:5139–5148. doi: 10.1093/emboj/16.16.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T, Yan Y, Benjamin TL, Harrison SC. Structure of Murine Polyomavirus Complexed with an Oligosaccharide Receptor Fragment. Nature. 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- Stoll R, Luo D, Kouwenhoven B, Hobom G, Müller H. Molecular and biological characteristics of avian polyomaviruses: isolates from different species of birds indicate that avian polyomaviruses form a distinct subgenus within the polyomavirus genus. J Gen Virol. 1993;74:229–237. doi: 10.1099/0022-1317-74-2-229. [DOI] [PubMed] [Google Scholar]
- Vacante DA, Traub R, Major EO. Extension of JC virus host range to monkey cells by insertion of a simian virus 40 enhancer into the JC virus regulatory region. Virology. 1989;170:353–361. doi: 10.1016/0042-6822(89)90425-x. [DOI] [PubMed] [Google Scholar]
- van Heel M. Similarity measures between images. Ultramicroscopy. 1987;21:95–99. [Google Scholar]
- Willits D, Zhao X, Olson N, Baker TS, Zlotnick A, Johnson JE, Douglas T, Young MJ. Effects of the Cowpea chlorotic mottle bromovirus β-hexamer structure on virion assembly. Virology. 2003;306:280–288. doi: 10.1016/s0042-6822(02)00054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci USA. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Milne JLS, Borgnia MJ, Rostapshov AV, Subramaniam S, Brooks BR. A core-weighted fitting method for docking atomic structures into low-resolution maps: application to cryo-electron microscopy. J Struct Biol. 2003;141:63–76. doi: 10.1016/s1047-8477(02)00570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N, Kawano M-a, Tsukamoto H, Enomoto T, Inoue T, Takahashi R-u, Nakanishi A, Imai T, Wada T, Handa H. Mutational analysis of the carboxyl-terminal region of the SV40 major capsid protein VP1. J Biochem. 2007;141:279–286. doi: 10.1093/jb/mvm038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



