Abstract
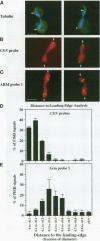
Although general features of chromosome movement during the cell cycle are conserved among all eukaryotic cells, particular aspects vary between organisms. Understanding the basis for these variations should provide significant insight into the mechanism of chromosome movement. In this context, establishing the types of chromosome movement in the budding yeast Saccharomyces cerevisiae is important since the complexes that mediate chromosome movement (microtubule organizing centers, spindles, and kinetochores) appear much simpler in this organism than in many other eukaryotic cells. We have used fluorescence in situ hybridization to begin an analysis of chromosome movement in budding yeast. Our results demonstrate that the position of yeast centromeres changes as a function of the cell cycle in a manner similar to other eukaryotes. Centromeres are skewed to the side of the nucleus containing the spindle pole in G1; away from the poles in mid-M and clustered near the poles in anaphase and telophase. The change in position of the centromeres relative to the spindle poles supports the existence of anaphase A in budding yeast. In addition, an anaphase A-like activity independent of anaphase B was demonstrated by following the change in centromere position in telophase-arrested cells upon depolymerization and subsequent repolymerization of microtubules. The roles of anaphase A activity and G1 centromere positioning in the segregation of budding yeast chromosomes are discussed. The fluorescence in situ hybridization methodology and experimental strategies described in this study provide powerful new tools to analyze mutants defective in specific kinesin-like molecules, spindle components, and centromere factors, thereby elucidating the mechanism of chromosome movement.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Afshar K., Barton N. R., Hawley R. S., Goldstein L. S. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell. 1995 Apr 7;81(1):129–138. doi: 10.1016/0092-8674(95)90377-1. [DOI] [PubMed] [Google Scholar]
- Aist J. R., Williams P. H. Ultrastructure and time course of mitosis in the fungus Fusarium oxysporum. J Cell Biol. 1972 Nov;55(2):368–389. doi: 10.1083/jcb.55.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley B. R., Ouspenski I., Zinkowski R. P. Structure and molecular organization of the centromere-kinetochore complex. Trends Cell Biol. 1992 Jan;2(1):15–21. doi: 10.1016/0962-8924(92)90139-e. [DOI] [PubMed] [Google Scholar]
- Brown M. T. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995 Jul 4;160(1):111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975 Oct;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C., Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996 Jul 26;86(2):275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D., Urrestarazu L. A., Vissers S., Jauniaux J. C., van Vliet-Reedijk J. C., Planta R. J., Gibbons I. R. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M., Ward D. C. Cell cycle dependent chromosomal movement in pre-mitotic human T-lymphocyte nuclei. Chromosoma. 1992 Aug;101(9):557–565. doi: 10.1007/BF00660315. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Funabiki H., Hagan I., Uzawa S., Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993 Jun;121(5):961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P. Y., Kilmartin J. V. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993 May;121(3):503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G. J. Kinetochores, microtubules and the metaphase checkpoint. Trends Cell Biol. 1995 Apr;5(4):143–148. doi: 10.1016/s0962-8924(00)88968-0. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994 May;125(3):517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Yamamoto A., Strunnikov A., Kingsbury J., Hogan E., Meluh P., Koshland D. Structure and function of chromosomes in mitosis of budding yeast. Cold Spring Harb Symp Quant Biol. 1993;58:677–685. doi: 10.1101/sqb.1993.058.01.075. [DOI] [PubMed] [Google Scholar]
- Heath I. B. Behavior of kinetochores during mitosis in the fungus Saprolegnia ferax. J Cell Biol. 1980 Mar;84(3):531–546. doi: 10.1083/jcb.84.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., He L., Loo K. K., Saunders W. S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992 Jul;118(1):109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A. A., Middleton K., Centola M., Mitchison T. J., Carbon J. Microtubule-motor activity of a yeast centromere-binding protein complex. Nature. 1992 Oct 8;359(6395):533–536. doi: 10.1038/359533a0. [DOI] [PubMed] [Google Scholar]
- Inoué S., Salmon E. D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995 Dec;6(12):1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. R., Michaud W. A., Sikorski R. S., Hieter P. A. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 1994 Sep 15;13(18):4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991 Feb 22;64(4):717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Li Y. Y., Yeh E., Hays T., Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995 Jul;6(7):793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Salmon E. D. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J Cell Biol. 1992 Nov;119(3):569–582. doi: 10.1083/jcb.119.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D., Bagget M., Tu Y. H., Fink G. R., Tu H. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J Cell Biol. 1995 Sep;130(6):1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. B., Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976 Nov;22(2):219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Pluta A. F., Mackay A. M., Ainsztein A. M., Goldberg I. G., Earnshaw W. C. The centromere: hub of chromosomal activities. Science. 1995 Dec 8;270(5242):1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Salmon E. D. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol. 1994 Feb;124(3):223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof D. M., Meluh P. B., Rose M. D. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992 Jul;118(1):95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K. C., Curnick K. E., Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995 Mar 1;9(5):573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Tugendreich S., Tomkiel J., Earnshaw W., Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995 Apr 21;81(2):261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Vernos I., Raats J., Hirano T., Heasman J., Karsenti E., Wylie C. Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995 Apr 7;81(1):117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- Vourc'h C., Taruscio D., Boyle A. L., Ward D. C. Cell cycle-dependent distribution of telomeres, centromeres, and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp Cell Res. 1993 Mar;205(1):142–151. doi: 10.1006/excr.1993.1068. [DOI] [PubMed] [Google Scholar]
- Wan J., Xu H., Grunstein M. CDC14 of Saccharomyces cerevisiae. Cloning, sequence analysis, and transcription during the cell cycle. J Biol Chem. 1992 Jun 5;267(16):11274–11280. [PubMed] [Google Scholar]
- Wang S. Z., Adler R. Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J Cell Biol. 1995 Mar;128(5):761–768. doi: 10.1083/jcb.128.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Mamay C. L., O'Toole E. T., Mastronarde D. N., Giddings T. H., Jr, McDonald K. L., McIntosh J. R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995 Jun;129(6):1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996 Apr;133(1):85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Lambie E. J., Hardin J., Craft J., Snyder M. Higher order structure is present in the yeast nucleus: autoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma. 1989 Aug;98(2):123–128. doi: 10.1007/BF00291048. [DOI] [PubMed] [Google Scholar]