Abstract
The canonical Wnt/β-catenin pathway is an essential component of multiple developmental processes. To investigate the role of this pathway in the ectoderm during facial morphogenesis, we generated conditional β-catenin mouse mutants using a novel ectoderm-specific Cre recombinase transgenic line. Our results demonstrate that ablating or stabilizing β-catenin in the embryonic ectoderm causes dramatic changes in facial morphology. There are accompanying alterations in the expression of Fgf8 and Shh, key molecules that establish a signaling center critical for facial patterning, the frontonasal ectodermal zone (FEZ). These data indicate that Wnt/β-catenin signaling within the ectoderm is critical for facial development and further suggest that this pathway is an important mechanism for generating the diverse facial shapes of vertebrates during evolution.
Keywords: craniofacial, ectoderm, Wnt/β-catenin, FEZ, FGF8
Introduction
The morphogenesis of the vertebrate face, with all its recognizable elements, is highly orchestrated temporally and spatially. This involves the coordinated action of thousands of genes and many signaling pathways, including BMP, SHH, FGF and Wnt/β-catenin (Buchtova et al., 2010; Cai et al., 2005; Chai and Maxson, 2006; Feng et al., 2009a). The face is derived from buds of tissue, termed facial prominences, that undergo both symmetrical and asymmetrical growth so that the appropriate fusion events required to generate the mature facial structure may occur (Rossant and Tam, 2002). These tightly controlled processes take place relatively early and rapidly during embryogenesis. Combined, these particulars make craniofacial development highly susceptible to genetic and environmental perturbation. Indeed, facial dysmorphologies are a component of approximately 75% of birth defects worldwide (CDC, 2010).
Across multiple species, the Wnt/β-catenin signaling pathway components are highly conserved and important for patterning and morphogenesis in a multitude of developmental processes. These include face, organ, and limb development, as well as formation of the skeleton, CNS, skin, and ectodermal appendages such as hair, and teeth (Cadigan and Nusse, 1997; Grigoryan et al., 2008). With regards to craniofacial development, Wnt/β-catenin signaling has been shown to be important for neural crest induction and migration, and for proper fusion of the facial prominences (Brault et al., 2001; Deardorff et al., 2001; Garcia-Castro et al., 2002; LaBonne and Bronner-Fraser, 1998; Lan et al., 2006; Niemann et al., 2004).
Recently, it has been suggested that the canonical Wnt signaling pathway may also have a role in species-specific facial patterning (Brugmann et al., 2007). Indeed, the faces of vertebrate species have undergone extensive evolution and are highly divergent. Studies in chick and mouse have shown that particular regions of Wnt signaling in the facial prominences correlate with outgrowth and fusion processes (Brugmann et al., 2007; Geetha-Loganathan et al., 2009; Hu and Marcucio, 2009a; Mani et al., 2010). Thus, It has been hypothesized that changes in Wnt signaling may underlie evolutionary differences in face shape, such as between the beak and muzzle (Brugmann et al., 2007). Another mechanism controlling mid-facial width, as yet unrelated to canonical Wnt signaling, is the frontonasal ectodermal zone (FEZ) which is defined by the juxtaposition of Fgf8 and Shh expression (Hu et al., 2003). In chick, there is a single FEZ, while the mouse has paired, bilateral FEZ regions separated by the facial midline (Hu and Marcucio, 2009b; Hu et al., 2003). Manipulating the FEZ in chick can indeed alter facial width (Abzhanov et al., 2007), and mutations in SHH signaling are known to cause holoprosencephaly in humans (Nanni et al., 1999). Thus, differential FEZ organization is another mechanism that may contribute to species-specific facial morphology.
In the current study we focused on Ctnnb1, encoding β-catenin, since cytosolic β-catenin is a nodal point for canonical Wnt signaling. In the absence of Wnt ligand, β-catenin is phosphorylated on serine-threonine residues encoded in exon 3 and targeted for proteosome degradation. In the presence of Wnt ligand, β-catenin is translocated to the nucleus, where it acts in concert with transcription factors including LEF/TCF to activate transcription of target genes (Logan and Nusse, 2004). β-catenin is expressed almost ubiquitously in the mouse, and the β-catenin (Ctnnb1) knockout mouse dies early in embryogenesis, around the time of gastrulation, due to defects in the ectodermal cell layer (Haegel et al., 1995). Since the generation of the original knockout mouse, the use of tissue specific Cre recombinase transgenes in conjunction with Ctnnb1 loss-of-function or gain-of-function floxed alleles have been used to study canonical Wnt signaling in development and disease (Grigoryan et al., 2008). However, despite extensive research of this critical pathway, the specific role of Wnt/β-catenin signaling from the ectoderm during the important stages of craniofacial development has not been determined. This is most likely due to the fact that available ectodermal Cre recombinases are expressed after the major growth and fusion events of the facial prominences have occurred, or are not specific to ectoderm (Grigoryan et al., 2008). To address these issues, we have developed a new ectodermal Cre recombinase transgenic mouse line, Crect (Yang et al., in preparation). Cre recombinase activity driven by Crect can be detected in the ectoderm as early as E8.5 as visualized by R26R β-galactosidase function. The Cre transgene is fully expressed in the head by E9.5 and is specific to the ectoderm and its derivatives (Yang et al., in preparation).
Using Crect in conjunction with two different β-catenin alleles, we examined both the loss of, and constitutive activation of, β-catenin in the early embryonic ectoderm of the mouse face. We found growth defects in both models, correlating with a disruption of FEZ formation, suggesting Wnt/β-catenin signaling is upstream of this critical signaling center. Additionally, our data are consistent with the proposed role for Wnt/β-catenin signaling in species-specific facial patterning, and further support the hypothesis that canonical Wnt signaling played a critical role in shaping the diverse facial features present in vertebrate species today.
Materials and Methods
Mouse strains
All animal experiments were performed in accordance with protocols approved by the UCD Animal Care and Usage Committee. Floxed Ctnnb1 mice (B6.129-Ctnnb1tm2Kem/J), used to make β-catenin “loss of function” (LOF) mice, and BATgal mice (B6.Cg-Tg(BAT-lacZ)3Picc/J), were both obtained from The Jackson Laboratory (Bar Harbor, ME). The derivation of mice containing the floxed β-catenin “gain of function” (GOF) allele, β-cateninΔex3/+ (Ctnnb1tm1Mmt) has been described previously (Harada et al., 1999). Crect mice utilize an ectodermal enhancer from Tcfap2a to drive Cre recombinase expression specifically in the ectoderm, and will be described elsewhere (Yang et al., in preparation).
Detection of β-galactosidase (LacZ) Activity
Embryos of the desired time points were dissected in PBS, and stained for LacZ expression as described previously (Reid et al., 2010).
RNA Isolation, Purification, and cDNA Synthesis
Whole embryos were dissected in ice-cold PBS-DEPC and stored in RNA later (Qiagen) at 4°C for the short-term or at −20°C for the long-term (more than 4 weeks). RNA was extracted and purified from embryonic heads using the RNeasy kit from Qiagen (#74104). cDNA was synthesized as previously described (Reid et al., 2010).
Scanning Electron Microscopy
Embryos were dissected in cold PBS and fixed for 48 hours at 4°C in 0.1M Phosphate buffer (PB) pH 7.2 containing 4% EM grade glutaraldehyde (Polysciences, Inc.), washed briefly in 0.1M PB, and then serially dehydrated to 100% ethanol. Specimens were transferred to SEM baskets for critical point drying with CO2, and then mounted onto aluminum stubs with silver paste and coated with gold. Specimens were then viewed and photographed using a JOEL 2005. All comparative images are the same magnification unless otherwise stated.
Real-Time PCR
RNA for real-time PCR was isolated and purified from embryonic heads as described above. The primers and TaqMan® probe set used to amplify and detect Fgf8 was adapted from Grieshammer, U. et al., 2005 (Grieshammer et al., 2005), and spans exons 2 and 3. All reactions were done as a one-step amplification on an Applied Biosystems 7300/7500 Fast Real-Time PCR Thermal Cycler. A small region (66bp) spanning exons 2 and 3 of Fgf8 was Topo cloned and used in known amounts for a standard curve. All samples were normalized to the mouse house-keeping gene TBP (Valente et al., 2009).
RNA for SABioscience arrays was isolated as described above, except cDNA was generated using the RT2 First Strand Kit (C-03, SABioscience). Real-Time PCR was performed as described in the SABiosciences user manual, using 1ug of total RNA for the 96-well plate format. The following arrays were used: Mouse Signal Transduction PathwayFinder (PAMM-014C), Mouse WNT Signaling Pathway (PAMM-043C), Mouse MAP Kinase Signaling Pathway (PAMM-061C), and two custom arrays.
For ABI custom plates, RNA was isolated as described above and cDNA was synthesized using the ABI High-Capacity cDNA Reverse Transcription Kit (cat no. 4368813). Real-time PCR was performed as described in the manufacturer’s protocol on an ABI 7900HT using the following cycling parameters: 50°C for 2 mins and 95°C for 10 mins, followed by 40 cycles of 95°C for 15 secs and 60°C for 1 minute.
Whole-mount in situ hybridization
Embryos of the desired time points were dissected in DEPC-PBS, and WMISH was performed as described previously (Feng et al., 2009b).
Whole-mount TUNEL detection
Embryos were dissected in sterile PBS and fixed for 2 hours in 4% paraformaldehyde (PFA) in PBS at 4°C. After a brief washing in PBT (PBS plus 0.1% Tween-20) embryos were serially dehydrated to 100% methanol and stored at −20°C. For TUNEL, embryos were bleached in methanol containing 6% H2O2 for one hour, washed in methanol, and serially rehydrated to PBT. Samples were treated with 5µg/ml proteinase K in PBT for varying times (6.5 minutes for an E10.5 embryo), washed twice with PBT, and re-fixed in 0.2% glutaraldehyde in 4% PFA at room temperature for 20 minutes, followed by 3 washes in PBT. Embryos were then treated with freshly prepared 0.1% sodium borohydride in PBT for 20 minutes at room temperature, washed 3 times in PBT, and then placed in 1ml of 1X TdT buffer (30mM Tris-HCl, 140mM Sodium Cacodylate pH 7.2, and 1mM CoCl2) for 10 minutes. Buffer was replaced with 200µl of reaction mix (1X TdT buffer (5X stock from Invitrogen), 20µM dig-dUTP (Roche), 20µM dTTP (Roche), 0.3U/µl TdT (Invitrogen) in sterile water), and embryos were rocked at 37°C for 2–3 hours. Embryos were then washed twice in TBST (1X TBS, 0.1% Tween-20, 2mM Levamisole) and heated at 70°C for 20 minutes in TBST, and blocked in 0.5% BMB blocking reagent (Roche) in TBST at room temperature for 1 hour. Anti-dig-AP antibody was prepared as for WMISH (Feng et al., 2009b), diluted 1:5000 in blocking solution, and placed on embryos overnight at 4°C. Afterwards embryos were rinsed three times in TBST, washed four times for 1 hour each in TBST, followed by two 20 minute washes in NTMT (100mM NaCl, 100mM Tris pH 9.5, 50mM MgCl2, 0.1% Tween-20, 2mM Levamisole). The reaction was developed with BM purple reagent (Roche) to a desired color, washed, photographed, and stored in PBT protected from light.
Analysis of cell proliferation
For whole-mount immunoperoxidase staining, embryos of the desired time points were dissected in PBS, and cell proliferation was detected using the p-Histone H3 (Ser 10)-R (sc-8656-R) antibody from Santa Cruz Biotechnology, Inc. The procedure was performed as described previously (Reid et al., 2010). Alternatively, 7µm frozen sections of E9.5 and E10.5 embryos were stained using a pHistone H3 antisera (Cell Signaling, #9701) and an Alexa Flour© 488 goat anti-rabbit secondary antisera (Invitrogen A31627).
β-catenin protein detection on frozen sections
Embryos were fixed and set for cryosectioning as previously described (Reid et al., 2010), and cut at 7µm. For detection of β-catenin, room temperature slides were washed for 10 minutes in PBS, followed by two 10 minute washes in PBST (PBS plus 0.1% Triton X), and one 15 minute wash in PBST (PBS plus 0.25% Triton X). Sections were blocked in blocking solution (0.1M PB, 0.15M NaCl, 0.3% Triton X-100, 30mg Bovine serum albumin) with 5% normal goat serum for 2 hours at room temperature and then incubated in primary antibody (Sigma C2206) diluted 1:2000 in blocking solution overnight at 4°C. Sections were washed 3 times for 10 minutes each in PBST, and incubated in goat anti-rabbit Alexa 488 (Molecular Probes) diluted 1:1000 in blocking solution for 2 hours at room temperature, counterstained with HOECHST, and washed in PBS. Sections were then coverslipped and photographed.
Skeletal stains
Bone and cartilage was detected as described previously (Reid et al., 2010).
Results and Discussion
Severe facial defects are caused by either loss or constitutive stabilization of β-catenin
To investigate the role of canonical Wnt signaling in the facial ectoderm of developing mouse embryos, we utilized two different β-catenin alleles in combination with our novel Crect transgenic line. To generate β-catenin loss-of-function mutants, females homozygous for the β-catenin floxed allele were bred to males positive for Crect and heterozygous for floxed β-catenin. One quarter of resultant offspring were β-cateninflox/flox;Crect, hereafter referred to as LOF (Supplemental Fig. 1A). Compared with wild-type littermates, the LOF mice displayed hypoplasia of all facial prominences (Fig. 1A, B).
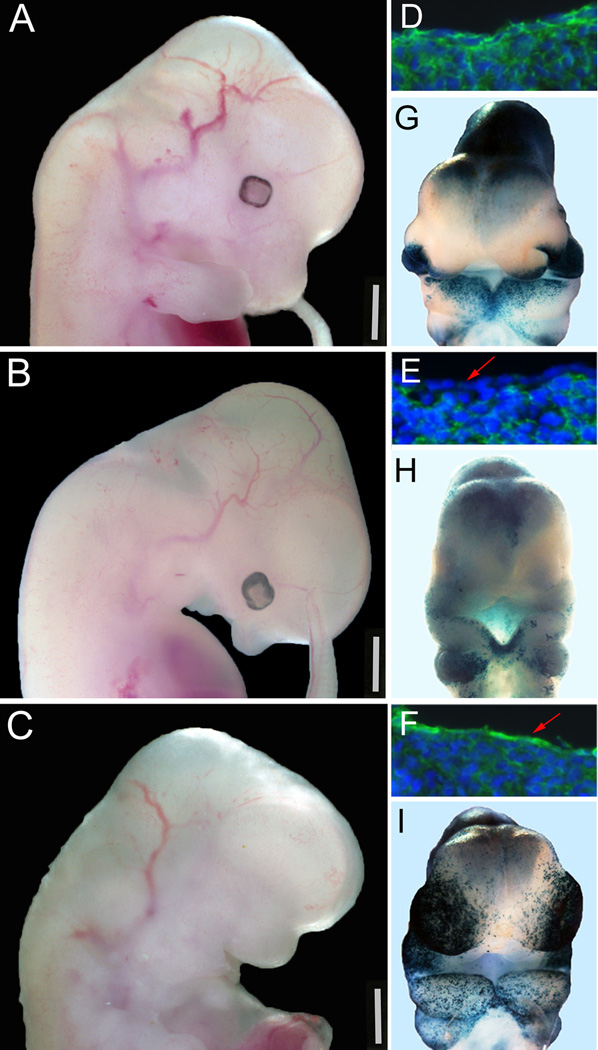
Figure 1. Loss or constitutive activation of ectodermal β-catenin via Crect results in severe developmental defects.
A–C) Lateral views of E12.5 wild-type (A), LOF (B), and GOF (C) embryos. D–F) Sagittal head sections of E10.5 wild-type (D), LOF (E), and GOF (F) stained for β-catenin protein (green) and counterstained with Hoechst (blue) to show nuclei. G–J) E10.5 ventral views of wild-type (G), LOF (H), and GOF (I) stained with X-gal to visualize BATgal reporter activity. Red arrows in E & F point to lack or presence of β-catenin in the ectodermal cell layer, respectively. Scale bars in A–C represent 1mm.
To generate β-catenin gain-of-function mutants, females homozygous for the β-catenin activating allele (Harada et al., 1999) were bred to males positive for Crect. One half of resultant offspring were β-cateninΔex3/+;Crect, hereafter referred to as GOF (Supplemental Fig. 1B). Most GOF mutants did not exhibit any recognizable facial features except for a widened oral cavity (Fig. 1C). GOF mutants died between E12.5 and E16.5, most likely due to vasculature defects.
To determine the efficacy of Crect to delete or stabilize β-catenin, we examined β-catenin localization using immunohistochemistry on sagittal sections of E10.5 embryonic heads (Fig. 1D–F). In the ectoderm of wild-type embryos, β-catenin protein is mainly observed in the cytoplasm, with lower levels detected in the nucleus (Fig. 1D). In LOF mutants, β-catenin was absent from the facial ectoderm (Fig. 1E, red arrow), and in GOF mutants β-catenin staining in the ectoderm was more intense in both the cell cytoplasm and nucleus (Fig. 1F, red arrow). These data demonstrate tissue-specific loss or stabilization of β-catenin in the facial ectoderm of LOF and GOF embryos, respectively.
Embryonic mouse facial ectoderm is competent to respond to stabilized β- catenin
In mouse and chick, the components of canonical Wnt/β-catenin signaling, including β-catenin itself, are nearly ubiquitously expressed (Geetha-Loganathan et al., 2009). However, differential expression of molecules that support or antagonize β-catenin mediated signaling impart specificity and regulation (Huang and He, 2008). To visualize the facial regions that were affected in our studies, we crossed the LOF and GOF lines to BATgal reporter mice and examined transcription activity via LacZ expression (Fig. 1G–I) (Maretto et al., 2003). At E9.5, β-gal activity driven by BATgal was detected around the optic placode and in the distal mandibular prominence (MnP) (Supplemental Fig. 2A, C), also in the brain and otic vesicle (Supplemental Fig. 2A, C). At E10.5, BATgal was expressed around the rim of the nasal pit, and in the distal regions of the MnPs and maxillary prominences (MxP) (Fig. 1G). Consistent with the immunohistochemistry results in LOF mutants (Fig. 1E), we did not observe LacZ reporter activity in facial ectoderm (Fig. 1H & data not shown). Staining was still present in the underlying mesenchyme of LOF embryos, but was decreased compared to wild-type, consistent with the overall reduction in size of the facial prominences in the mutants (Compare Fig. 1G and 1H). In E9.5 GOF mutants we observed an increase in BATgal expression in the optic and olfactory placodes, ectopic activity in the MnP and MxP, and punctate expression was observed in lateral facial regions (Supplemental Fig. 2B, D). At E10.5, GOF mutants exhibited high BATgal activity in the MxP and MnP, and expression was particularly strong in the nasal region (Fig. 1I). These data demonstrate that the embryonic facial ectoderm is sensitive to changes in the stabilization of β-catenin, and gene transcription via LEF/TCF is affected in both mutant models.
Canonical Wnt signaling from the ectoderm controls facial growth
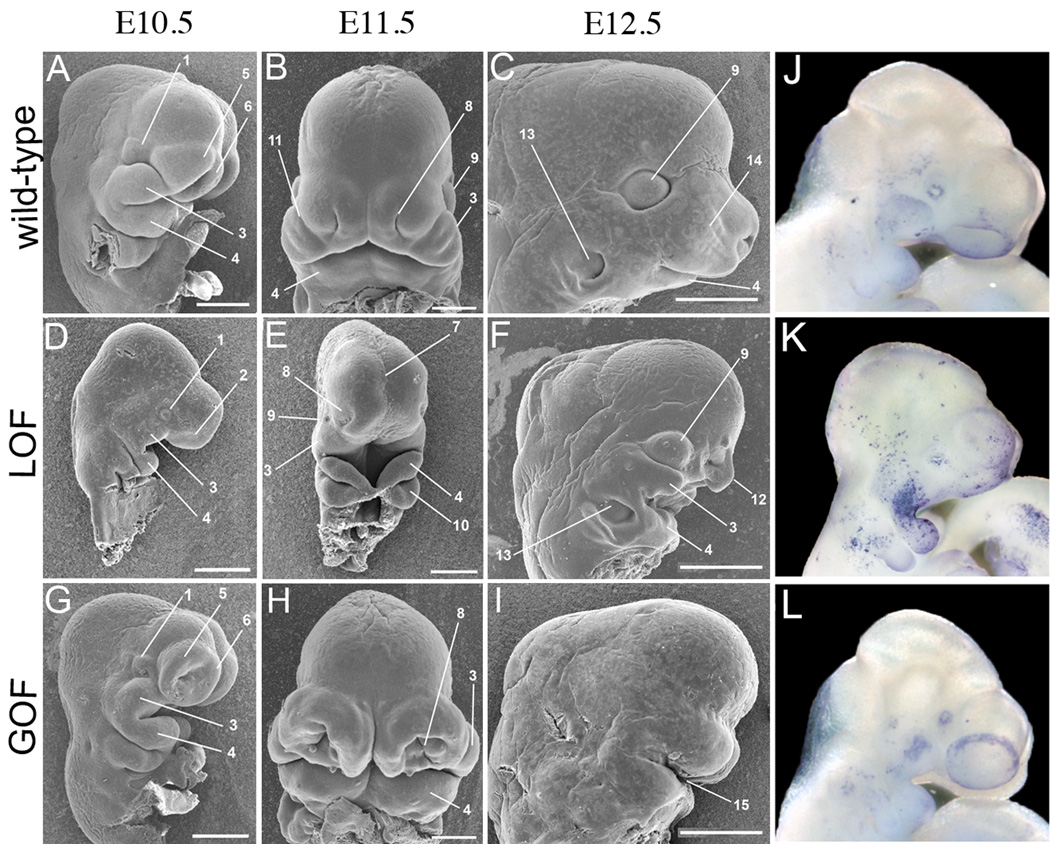
To examine the pathology of LOF and GOF mutants, and how the facial defects manifested during development, we examined external morphology from E9.5 through E12.5 using scanning electron microscopy (SEM) (Fig. 2A–I & data not shown). In wild-type mice, the time period between E9.5 to E12.5 is critical for facial development, as growth and fusion processes occur among the facial prominences (Fig. 2A–C). In LOF mutants, the facial prominences were comparable to wild-type at E9.0 (data not shown). Shortly thereafter, the prominences failed to grow, such that by E10.5 LOF mutants exhibited a hypoplastic facial morphology, and in general had features more characteristic of an E9.5 embryo (Fig. 2D, & data not shown). The optic eminence and the corneal ectoderm were reduced (Fig. 2D), and the eyelids, which form from the corneal ectoderm, did not form (see Fig. 3B). Preliminary data also suggest defects in the formation of the lens (data not shown); similar to previous reports examining β-catenin function during eye development (Smith et al., 2005). The olfactory placode of LOF mutants was underdeveloped, consistent with the hypoplasia of the medial and lateral nasal processes (Fig. 2D, E). At E11.5, the nasal slit observed in wild-type embryos has been replaced by a shallow nasal depression in the LOF (Fig. 2E). Highlighting the lack of facial growth, the groove overlying the lamina terminalis of forebrain, which normally disappears shortly after E10.5, was still present at E11.5 (Fig. 2E). While the MnPs fuse at the midline of LOF mutants, they too were severely underdeveloped at both E11.5 and E12.5 (Fig. 2E, F). Most notably, by E12.5 the frontonasal process of LOF mutants was narrowed, producing a protuberance with a superficial avian beak-like appearance rather than the normal muzzle of a mouse (compare Fig. 2C & F).
Figure 2. Conditional mutants exhibit complementary growth defects during craniofacial development.
Scanning electron microscopy images of wild-type (A–C), LOF (D–F) and GOF (G–I) embryos shown at E10.5 (A, D, G), E11.5 (B, E, H), and E12.5 (C, F, I). Images in B, E, & H are shown in a ventral view, all others are lateral views. J–L) Lateral views of E10.5 wild-type (J), LOF (K), and GOF (L) stained for cell death using TUNEL. e – eye, 1 – optic eminence, 2 – olfactory placode, 3 – maxillary prominence (upper jaw), 4 – mandibular prominence (lower jaw), 5 – lateral nasal process, 6 – medial nasal process, 7 – groove overlying the lamina terminalis of forebrain, 8 – nasal pit, 9 – eye, 10 – branchial arch two, 11 – lacro nasal groove, 12 – frontal nasal process, 13 – ear, 14 – vibrissae, 15 – oral cavity. Scale bars in C, F, & I represent 1mm, all others represent 500µm.
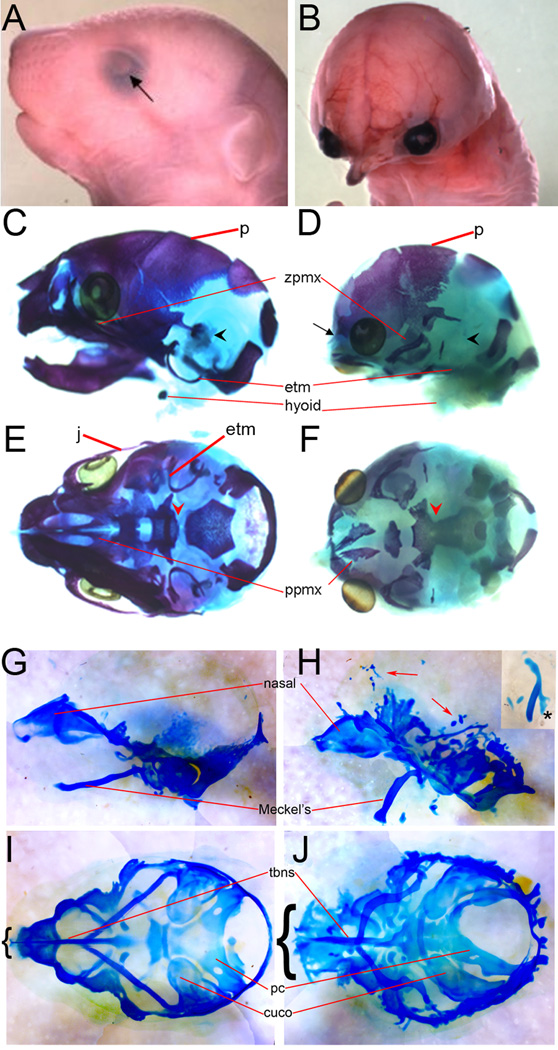
Figure 3. Cranial skeletal defects in LOF and GOF mutants.
A, B) E18.5 wild-type (A) and LOF (B) embryos. C–F) Alizarin red and alcian blue skeletal stains of embryos shown in A & B, respectively, shown in lateral (C, D) and norma verticalis (E, F) views. G–J) Alcian blue cartilage stains of E14.5 wild-type (G, I) and GOF (H, J) embryos shown in lateral (G, H) and norma verticalis (I, J) views. cuco – cupola cochlearis, etm – ectotympanic, j – jugal, p – parietal bone, pc – parachordal, ppmx – palatal process of premaxilla, tbns – trabecular plate-nasal septum, zpmx – zygomatic process of maxilla. Arrow in A points out normal eye formation, compared to the LOF (B). Arrowheads in C & D point to presence and absence of middle ear bones, respectively. Black arrow in D points to residual nasal bone. Red arrowheads in E & F indicate region between the basisphenoid and the basioccipital in wild-type and LOF skulls. Red arrows in H point to ectopic cartilages in GOF mutants. Inset in H shows isolated Meckel’s cartilage in same proximal distal orientation as main panel, but rotated 90 degrees to show duplication on internal aspect of Meckel’s cartilage (asterisk). Brackets in I & J compare the width of the nasal cartilages in wild-type and GOF embryos.
Similar to LOF mutants, facial development in GOF mutants appeared normal at E9.0 (data not shown). However, shortly thereafter the prominences began to enlarge at a faster rate than wild-type (Fig. 2G, H). The nasal process was severely affected, and did not exhibit the controlled, directional growth that gives rise to a properly shaped nasal pit (Fig. 2H). The MxP and MnP also exhibited an increase in overall size, and therefore the upper and lower jaws did not form or fuse properly, leading to a widened oral cavity (Fig. 2I). By E12.5, most GOF mutants did not possess any features characteristic of a mouse face, such as vibrissae, or external eyes or ears – although rudimentary eyes were found internalized (Fig. 2I & data not shown). However, we note that a small percentage of mutants exhibited recognizable facial features, but the defects were still severe, suggesting some variability in the GOF craniofacial phenotype (Supplemental Fig. 3).
We next studied cell proliferation and cell death to ascertain whether the changes in facial size and shape were due to differences in these processes. We did not detect major differences in cell proliferation in LOF and GOF mutants compared to wild-type embryos at either E9.5 or E10.5 as measured using phospho-histone H3 (Ser10) immunohistochemistry (data not shown). In contrast, using whole-mount TUNEL analysis at E10.5, we detected a clear increase in cell death in the LOF mutants (Fig. 2K), compared to lower levels in both wild-type and GOF embryos (Fig. 2 J and L). Thus, the facial hypoplasia characteristic of the LOF mutants appears to be caused at least in part by an increase in cell death. In contrast, the altered growth of the GOF facial prominences does not seem to correlate with either a widespread increase in cell proliferation or changes in cell death. At this juncture we cannot exclude subtle differences in the rate of cell growth as the mechanism underlying the altered morphology of the GOF facial prominences. However, one alternative possibility is that stabilization of β–catenin in the facial ectoderm may cause hyperplasia by promoting increased neural crest cell migration into the facial prominences. Although further studies will be necessary to test these hypotheses, our data nevertheless clearly demonstrate that ectodermal β-catenin regulates outgrowth of the facial prominences.
Aberrant Wnt signaling in the ectoderm causes defects in the NC derived head skeleton
The proper growth of the facial prominences is essential for cartilage formation. During development, the mesenchymal cells beneath the facial ectoderm must receive proliferation signals for the appropriate amount of time to establish a critical number of progenitor cells (Chai and Maxson, 2006). These progenitor cell populations then respond to localized and intrinsic patterning signals, as well as inhibitory signals from the ectoderm, to form a cartilage of the correct shape and size (Chai and Maxson, 2006). Both LOF and GOF mutants displayed dramatic changes in the overall shape of the face, which would be expected to alter the underlying cranial skeleton. LOF mice survive until birth, when they display a superficial beak-like facial morphology compared with wild-type (Fig. 3A, B). We therefore examined cartilage and bone in LOF mutants using alcian blue and alizarin red staining (Fig. 3D, F). However, since GOF mutants die prior to ossification (which is normally first seen at ~E15.5), only cartilage stains were performed on GOF mutants (Fig. H, J).
Wild-type skeletons are shown at E18.5 (Fig. 3C, E) and E14.5 (Fig. 3G, I) to highlight the craniofacial skeletal defects observed in both LOF (Fig. 3D, F) and GOF (Fig. 3H, J) mutants. In LOF mutants, most of the bones of the face failed to develop. Underlying the superficial beak-shaped structure, a trace of nasal bone was present together with some cartilage (Fig. 3D, arrow). Additionally, part of the palatal processes of the maxilla (ppmx) and a fragment of the zygotic process of the maxilla were also present (Fig. 3, compare E & F). In contrast, the middle ear ossicles, most of the ectotympanic, the jugal, the mandible, and the ossifying center of the hyoid bone were all absent (Fig. 3D, F). The bones forming the neurocranium were all present, although with minor defects. In addition, the basisphenoid and basioccipital, which are normally separated by cartilage, were connected by calcified tissues to form a trabecular basal plate in the LOF mutant embryo (Fig. 3E & F red arrowhead).
GOF mutants exhibited extensive ectopic cartilages throughout the head region resulting in cartilage fusions and malformations (Fig. 3H, red arrows & J). Thus, identifying specific skeletal elements was difficult. Cartilages of the head that were identifiable, such as the parachordal plate (pc) and those of the ear (cuco) were grossly malformed (Fig. 3J). Meckel’s cartilage, which forms the lower jaw, was turned downwards and in some mutants, appeared to be duplicated on the interior aspect (Fig. 3H, asterisk in cut out). The nasal cartilages were wider and misshapen (compare brackets in Fig. 3I, J). Taken together, these data demonstrate that ectodermal β-catenin signaling plays a critical role in the formation and patterning of the underlying cranial skeleton.
Gene expression profiling in β-catenin conditional mutants
To investigate changes in gene regulation that may presage the growth and skeletal defects observed in mutants we initially utilized a combination of commercially available PCR arrays and real-time RT-PCR to focus on the expression of a limited number of genes involved in signal transduction. For these analyses we isolated RNA from E10.5 wild-type, LOF, and GOF heads, and comparison of their gene expression profiles indicated changes in the expression of several key molecules (Table 1). Consistent with modification of β-catenin expression and function, we detected associated changes in genes involved in canonical Wnt signaling. When compared to wild-type mice, GOF mutants showed increased expression of transcripts for the secreted WNT inhibitors Wif1, Dkk1 and Dkk4 (Table 1). These results indicate that stabilizing β-catenin in the facial ectoderm resulted in the induction of genes involved in negative feedback regulation of Wnt signaling. In support of this conjecture, LOF mutants showed a corresponding down-regulation of Dkk4 transcripts (Table 1). In GOF mutants, we observed increased expression of the genes encoding the FGF ligands Fgf4, Fgf8, Fgf9, and Fgf20 (Table 1). The most dramatic change was in Fgf4 expression, which was increased >100 fold compared with controls and is a known target of LEF1 (Kratochwil et al., 2002). However, the activation of Fgf expression was limited to a subset of the gene family, as we did not detect major differences in the expression of Fgf10, Fgf17, or Fgf18 using RT-PCR analysis (data not shown). We also detected significant changes in the expression of genes associated with other signaling events in GOF mice. Cdkn1a expression was increased >4 fold in the GOF embryos, suggesting alterations in cell cycle regulation. There was also an ~5 fold rise in Bmp2 expression, whereas Bmp4 expression only showed a modest increase in GOF mice. None of these three genes exhibited significant expression changes in LOF mutants (data not shown).
Table 1. Representative gene expression changes in E10.5 heads from LOF and GOF mutants compared with wild-type controls.
All values are the mean + standard error for at least n=3 samples using the ABI platform, and all have P<0.05. Positive or negative value indicates the fold-increase or decrease, respectively, compared to wild-type control samples. The dash indicates no significant change. All genes in the Table also showed similar positive or negative gene expression changes on the SAB platform.
| Gene | LOF | GOF |
|---|---|---|
| Dkk4 | − 26 ± 6.7 | + 45.5 ± 5.2 |
| Dkk1 | − | + 8.8 ± 1.0 |
| Wif1 | − | + 7.8 ± 0.9 |
| Fgf4 | − | + 163 ± 6.4 |
| Fgf20 | − | + 17.9 ± 4.3 |
| Fgf9 | − | + 3.3 ± 0.18 |
| Fgf8 | − | + 3.2 ± 0.25 |
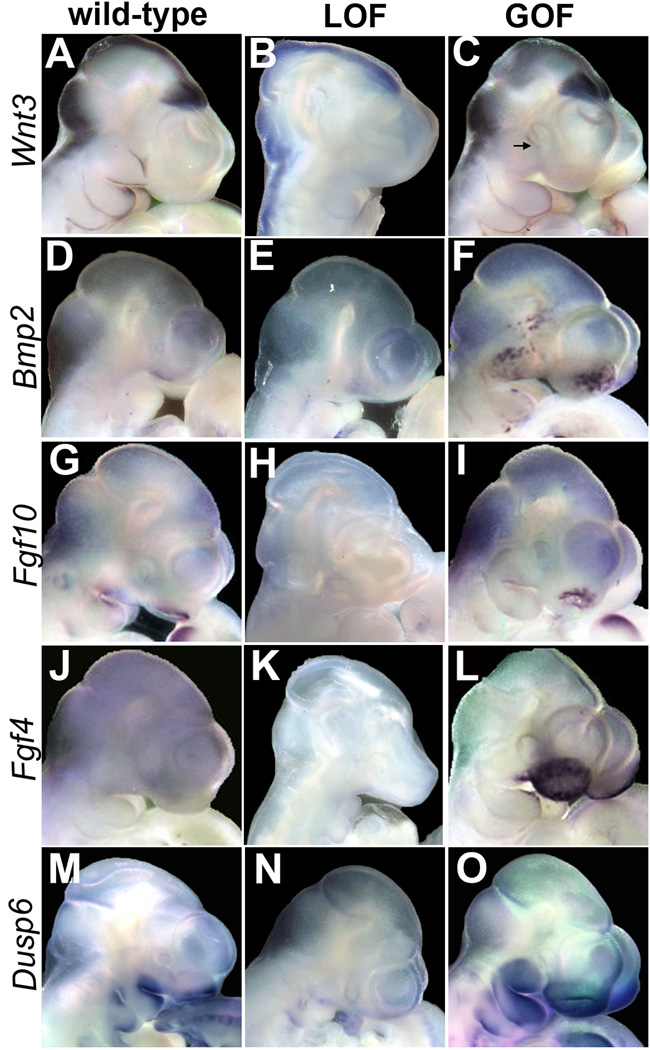
We next employed whole-mount in situ hybridization (WMISH) to verify and extend the analysis of gene expression changes in the context of the developing face (Fig. 4). We first examined Wnt3, a gene implicated in human facial clefting, which is normally expressed at the margins of the fusing facial prominences (Fig. 4A). We determined that Wnt3 expression was drastically reduced in LOF facial prominences (Fig. 4B), but appeared relatively normal in the branchial arches of GOF mutants, although slightly down-regulated in the maxilla (Fig. 4C, black arrow). In contrast, Bmp2 expression was comparable in wild-type and LOF embryos, occurring in the mesenchyme of the facial prominences (Fig 4D, E). In GOF embryos, though, Bmp2 was expressed in a patchy and ectopic manner in the lateral facial prominences and nasal pit, in agreement with the increased expression evident from our real-time data (Fig 4F and data not shown). BMP proteins, particularly BMP2 and 4, are known to affect facial shape and orofacial clefting in avian and mammalian systems (Chai and Maxson, 2006; Foppiano et al., 2007). Moreover, excess BMP signaling is associated with altered beak shape in avians and can cause facial dysmorphology in humans (SYNS1) (van den Ende et al., 2005). Therefore, the increased levels of Bmp2 expression may underlie some of the aberrant facial development in the GOF mice. We next examined Fgf10, which is normally expressed in the MnP and MxP as well as around the nasal pit (Fig. 4G), and is also associated with craniofacial dysmorphology (Hajihosseini et al., 2009; Veistinen et al., 2009). Fgf10 transcripts were greatly decreased in the facial prominences of LOF embryos in comparison to wild-type controls (Fig 4G, H). However, similar to Bmp2, Fgf10 displayed a disorganized expression pattern in GOF embryonic facial prominences, but its level of expression was comparable with wild-type controls (Fig 4I, & data not shown). We next examined Fgf4. Fgf4 is not normally expressed in the mouse face at E10.5 (Fig 4J), but is a known target of canonical Wnt signaling in other developmental processes (Kratochwil et al., 2002), and was found to be >100 fold increased in our GOF mutant Real-time PCR array. GOF embryos showed considerable ectopic Fgf4 expression associated with the expanded nasal placodes, and expression was also apparent in the MxP and MnP (compare Fig. 4J & L). We next studied Dusp6 (also known as Mkp3) since this is known target gene of FGF signaling (Kawakami et al., 2003). Compared to wild-type controls (Fig 4M), we found a reduction of Dusp6 expression in LOF embryos (Fig. 4N), and an increase in Dusp6 expression in the underlying mesenchyme of GOF embryos (Fig 4O). These findings are consistent with altered FGF ligand expression leading to changes in downstream effectors of FGF signaling responses in the facial mesenchyme.
Figure 4. Gene expression profiling in conditional mutants.
WMISH of E10.5 wild-type (A, D, G, J, M), LOF (B, E, H, K, N), and GOF (C, F, I, L, O) shown in lateral views. Gene RNA probes for each row are indicated on the left. Arrow in C indicates weak expression associated with boundary of maxilla.
FEZ morphology is disrupted in LOF and GOF mutants
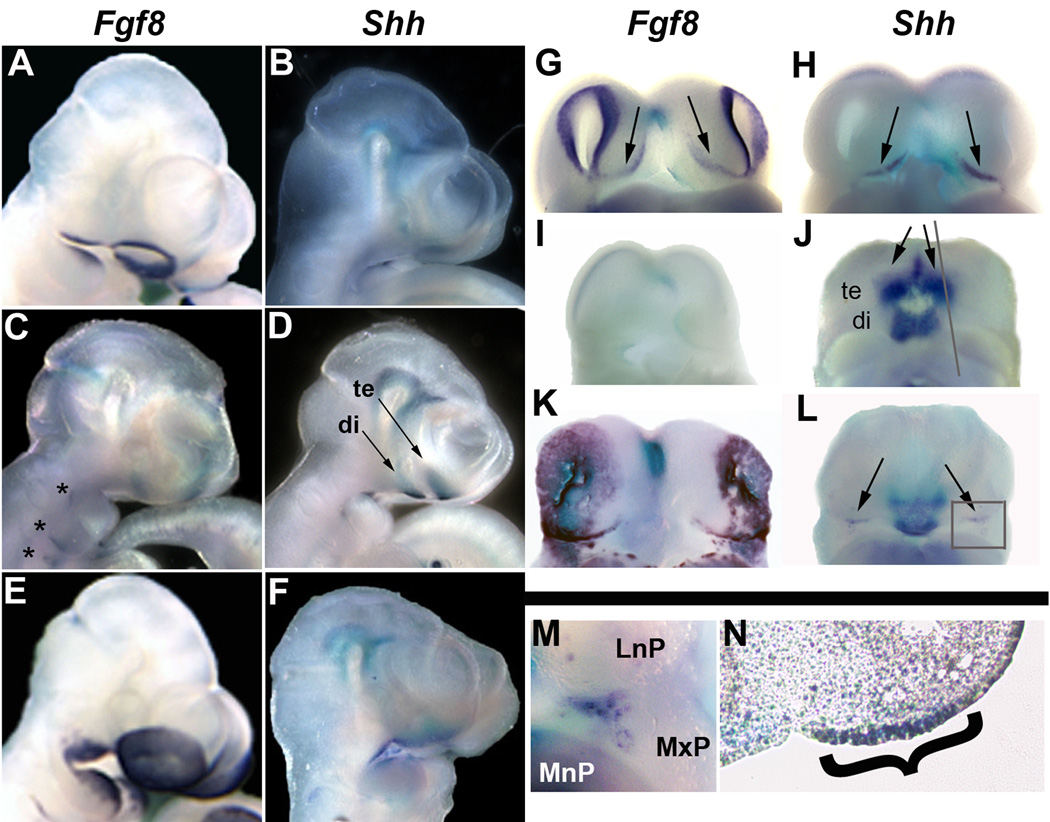
Changes in facial width and morphology prompted us to study Fgf8 and Shh, since these are the morphogens responsible for FEZ function (Hu et al., 2003). At E10.5, Fgf8 is normally expressed in the rim of olfactory epithelium surrounding the invaginating nasal pit, the MxP, and in the proximal region of the MnP (Fig. 5A), in addition to being expressed in the FEZ (Fig. 5G arrows). In LOF mutants, Fgf8 expression was absent from all facial ectoderm at E10.5 (Fig. 5C, I), though it was still present in facial endoderm (Fig. 5C, asterisks). In E10.5 GOF mutants, Fgf8 expression was expanded and up-regulated - consistent with our Real-time PCR data - and sectional analysis showed that this expansion was confined to the ectoderm (Fig. 5E, K & data not shown). By E11.5 we observed ectopic Fgf8 expression in a punctate pattern throughout the head region (Supplemental Fig. 4A, B). Interestingly, Fgf8 expression was unaltered in E9.5 LOF and GOF mutants (Supplemental Fig. 2F, G), though changes in BATgal reporter activity were visible at this time (Supplemental Fig. 2B, D). Thus, while it is clear there is an epistatic relationship between Wnt/β-catenin signaling and Fgf8 expression, these data suggest that either Wnt/β-catenin does not begin to regulate Fgf8 expression until E10.5, or that the regulation is indirect, occurring through one or more intermediates. The altered expression of Fgf8 in LOF and GOF embryonic facial ectoderm correlates with the changes observed in the expression of its downstream target, Dusp6, in the underlying mesenchyme (Fig 4N, O).
Figure 5. Fgf8 and Shh expression in conditional mutants.
A–M) Whole-mount in situ hybridization of Fgf8 (A, C, E, G, I, & K) and Shh (B, D, F, H, J, L, M) in wild-type (A, B, G, H), LOF (C, D, I, J) and GOF (E, F, K, L, M) shown in lateral views (A–F) and ventral (G–M). M is magnification of boxed region of Shh expression in L. N) Sagittal section of embryo shown in J, along plane of section shown by gray line, bracket indicates Shh expression in the ectoderm of the nasal process. Asterisks in C indicate expression of Fgf8 in pouch endoderm. Arrows in D point to Shh expression in the telencephalon (te) and diencephalon (di). Arrows in G, H, J, & L point to FEZ.
In wild-type E10.5 embryos, Shh is expressed in the oral ectoderm of the distal mandibular prominence, in the basal forebrain, and in the FEZ (Fig. 5B, H, arrows & data not shown). In LOF embryos, Shh was still expressed in these domains (Fig. 5D, J), however, the bilateral FEZ domains of Shh appeared to have fused at the midline, and this medio-lateral expression had expanded along the dorso-ventral axis (Fig. 5J arrows, N bracket). This difference may be due to the lack of Fgf8 negative regulation, and failure of lateral facial growth. In GOF mutants, Shh expression was only observed in the most lateral regions of the FEZ, and was disorganized and punctate (Fig. 5L, M). We also observed ectopic punctate expression in the MnP, and in the oral cavity (data not shown). At E12.5, Shh expression was observed in a punctate pattern throughout the head ectoderm (Supplemental Fig. 4C, D), similar to what was observed with Fgf8 expression. FGF8 and SHH have been shown to act synergistically during chondrogenesis (Abzhanov and Tabin, 2004), and have significant roles in the patterning of the MnP (Hu and Helms, 1999; Trumpp et al., 1999; Tucker et al., 1999). Therefore, the misexpression of these two morphogens during facial development may be responsible for the ectopic cartilages seen in the GOF mutants throughout the head region and the misshapen Meckel’s cartilage (Fig. 3 H, J). We also note that the altered expression of other signaling molecules, especially BMP2, might also influence chondrogenesis.
Conclusions
Implications for facial evolution
It has been hypothesized that changes in canonical Wnt signaling have been a contributing factor to generating the divergent facial shapes present in vertebrate species (Brugmann et al., 2007). Our data strongly support this conjecture, since ectodermal ablation or stabilization of β-catenin during early mouse facial development leads to drastic differences in craniofacial morphology. However, we note that the alternative experimental approaches used by Brugmann et al and by ourselves produce subtly different conclusions concerning the mechanism of Wnt/β-catenin action in facial development. The initial theory that canonical Wnt signaling was critical for the evolution of facial shape was based in part on a comparison of Wnt responsive reporter gene expression between chick and mouse. The midline was found to be the major site of Wnt-responsiveness in chick and this was postulated to drive outgrowth of the beak from the frontonasal mass (Brugmann et al., 2007). Note, however, that recent studies in chick have argued that Wnt responsiveness is broadly distributed throughout the facial prominences (Geetha-Loganathan et al., 2009; Hu and Marcucio, 2009a). Analogous studies in mouse demonstrated that the lateral nasal prominences were the primary region of Wnt/β-catenin signaling, leading to a broader muzzle shaped face with a narrower midline contribution (Brugmann et al., 2007). Furthermore, it was shown that inhibition of canonical Wnt signaling throughout the mouse embryo by targeting the transcriptional effectors Lef1 and Tcf4 resulted in hypoplastic development of the lateral facial prominences and an expanded midface (Brugmann et al., 2007).
Using LacZ reporter transgenic mice, we show that the lateral facial regions are highly responsive to changes in β-catenin activity (Fig 1G–I). These findings are in agreement with previous conclusions regarding the regions of Wnt responsiveness in the developing mouse face (Brugmann et al., 2007; Mani et al., 2010). Moreover, in mice, the activity of canonical Wnt signaling in the lateral facial prominences correlates with their growth and morphogenesis. Specifically, the absence of ectodermal β-catenin leads to hypoplasia of these prominences, whereas stabilization of ectodermal β-catenin causes expansion of the lateral facial regions. In addition, as the midline of LOF mutants is relatively unaffected, our data further suggest that active Wnt signaling is not necessary for midfacial outgrowth in mouse. In this regard a narrow, elongated frontonasal shape – superficially resembling an avian beak in external appearance - is essentially a default face that develops when little or no active β-catenin signaling is present in the ectoderm of the lateral facial regions. This mechanism stands in contrast to that proposed for the avian embryo, in which active Wnt/β-catenin signaling was postulated to cause midfacial outgrowth (Brugmann et al., 2007). We also note that the removal of β-catenin from the ectoderm of the mouse face as reported here produced a narrow midfacial region, whereas the global loss of the canonical Wnt transcriptional activators Lef1 and Tcf4 resulted in a minor widening of the midface (Brugmann et al., 2007). Both approaches lead to hypoplasia of the more lateral facial regions and we suspect that the observed differences in facial shape reflect the various tissues and genes targeted in the two experimental approaches. The results of these two independent studies are also consistent with the possibility that the addition of increased Wnt/β-catenin signaling or responsiveness in the lateral facial prominences led to a more rounded, muzzle-shaped face during the process of mammalian evolution.
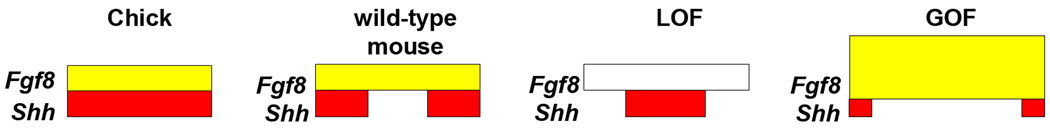
Our data show that disturbing ectodermal β-catenin signaling led to altered expression of multiple signaling molecules, notably Fgf4, Fgf8, Bmp2, and Shh. Alterations of any one of these, with the exception of Fgf4, has been shown to influence facial development and patterning (Chai and Maxson, 2006). Nevertheless, the most dramatic changes we observed were in FGF ligand expression. This latter finding provides a plausible connection between Wnt signaling, FEZ organization, and ultimately gross facial morphology. Recent data places the FEZ as an important regulator of mid-facial width in chick and mouse (see model, Figure 6). Facial width was altered in both LOF and GOF models (Fig. 6). To summarize, in LOF embryos the FEZ is characterized by a loss of Fgf8 expression and the consolidation of Shh expression to a single midline domain. This arrangement may mimic the operations of a single FEZ as observed in chick facial development (Hu et al., 2003), giving rise to the narrow, pointed, upper face of LOF mutants. In contrast, GOF mutants exhibited an increased distance between Shh domains and an expansion of Fgf8 expression. Previous data obtained in the chick model (Abzhanov et al., 2007), predict that this would cause the wider facial profile observed in GOF mutants.
Figure 6. FEZ organization in mutants.
Model depicting the expression domains of Fgf8 (yellow) and Shh (red) in chick, wild-type mouse, LOF, and GOF FEZ (chick and mouse model adapted from (Hu and Marcucio, 2009b)).
In conclusion, our data support a model in which ectodermal Wnt/β-catenin signaling lies upstream of molecules known to regulate facial shape, including the FEZ associated molecules Shh and Fgf8. In the future it will be important to understand how these various signaling pathways interact to regulate mouse facial development.
Research Highlights
Stabilization of β-catenin in ectoderm alters facial morphology
Loss of β-catenin in facial ectoderm causes hypoplasia of maxilla and mandible
Stabilization of β-catenin in facial ectoderm activates Fgf4 and Fgf8
Supplementary Material
A, B) Schematic showing genetic crosses of mice to obtain LOF (A) and GOF (B) mutants.
A–D) E9.5 wild-type (A, C) and GOF (B, D) embryos shown in lateral (A, B) and ventral (C, D) views, stained with X-gal to visualize LacZ expression. E–G) Whole-mount in situ hybridization of E9.5 Fgf8 expression in wild-type (E), LOF (F), and GOF (G). MnP – mandibular prominence, op – optic placode, ot – otic placode.
Ventral view of an E13.5 GOF mutant embryo showing facial dysplasia and orofacial clefting.
A, B) Fgf8 expression in E11.5 wild-type (A) and GOF (B) embryos. C, D) Shh expression in E12.5 wild-type (C) and GOF (D) embryos.
Acknowledments
We thank Dr. Ralph Marcucio for discussions, Dr. Anne Hansen for her assistance with the SEM images, and Dr. Brian Parr and Dr. Lee Niswander for in situ probes. We are grateful to Uma Pugazhenthi, University of Colorado Cancer Center PCR Core Facility, for assistance with microarray analysis using real-time PCR. We also thank Dr. Chengji Zhou for sharing unpublished data concerning related studies using a different Cre recombinase transgene. This research was supported by NIH grant 1 R01 DE019843-01 to T.W.
Grant sponsor: NIDCR
Grant number: DE019843-01
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abzhanov A, Cordero DR, Sen J, Tabin CJ, Helms JA. Cross-regulatory interactions between Fgf8 and Shh in the avian frontonasal prominence. Congenit Anom (Kyoto) 2007;47:136–148. doi: 10.1111/j.1741-4520.2007.00162.x. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Tabin CJ. Shh and Fgf8 act synergistically to drive cartilage outgrowth during cranial development. Dev Biol. 2004;273:134–148. doi: 10.1016/j.ydbio.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development - Supplement. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, Clevers H, Nusse R, Helms JA. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3283–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- Buchtova M, Kuo WP, Nimmagadda S, Benson SL, Geetha-Loganathan P, Logan C, Au-Yeung T, Chiang E, Fu K, Richman JM. Whole genome microarray analysis of chicken embryo facial prominences. Dev Dyn. 2010;239:574–591. doi: 10.1002/dvdy.22135. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Cai J, Ash D, Kotch LE, Jabs EW, Attie-Bitach T, Auge J, Mattei G, Etchevers H, Vekemans M, Korshunova Y, Tidwell R, Messina DN, Winston JB, Lovett M. Gene expression in pharyngeal arch 1 during human embryonic development. Hum Mol Genet. 2005;14:903–912. doi: 10.1093/hmg/ddi083. [DOI] [PubMed] [Google Scholar]
- CDC. 2010 http://www.cdc.gov/ncbddd/bd/facts.htm.
- Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Tan C, Saint-Jeannet JP, Klein PS. A role for frizzled 3 in neural crest development. Development. 2001;128:3655–3663. doi: 10.1242/dev.128.19.3655. [DOI] [PubMed] [Google Scholar]
- Feng W, Leach SM, Tipney H, Phang T, Geraci M, Spritz RA, Hunter LE, Williams T. Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS One. 2009a;4:e8066. doi: 10.1371/journal.pone.0008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Simoes-de-Souza F, Finger TE, Restrepo D, Williams T. Disorganized olfactory bulb lamination in mice deficient for transcription factor AP-2epsilon. Mol Cell Neurosci. 2009b;42:161–171. doi: 10.1016/j.mcn.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foppiano S, Hu D, Marcucio RS. Signaling by bone morphogenetic proteins directs formation of an ectodermal signaling center that regulates craniofacial development. Dev Biol. 2007;312:103–114. doi: 10.1016/j.ydbio.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Geetha-Loganathan P, Nimmagadda S, Antoni L, Fu K, Whiting CJ, Francis-West P, Richman JM. Expression of WNT signaling pathway genes during chicken craniofacial development. Dev Dyn. 2009;238:1150–1165. doi: 10.1002/dvdy.21934. [DOI] [PubMed] [Google Scholar]
- Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, Martin GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- Hajihosseini MK, Duarte R, Pegrum J, Donjacour A, Lana-Elola E, Rice DP, Sharpe J, Dickson C. Evidence that Fgf10 contributes to the skeletal and visceral defects of an Apert syndrome mouse model. Dev Dyn. 2009;238:376–385. doi: 10.1002/dvdy.21648. [DOI] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development - Supplement. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development. 2009a;136:107–116. doi: 10.1242/dev.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS. Unique organization of the frontonasal ctodermal zone in birds and mammals. Dev Biol. 2009b;325:200–210. doi: 10.1016/j.ydbio.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130:1749–1758. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- Huang H, He X. Wnt/beta-catenin signaling: new (and old) players and new insights. Curr Opin Cell Biol. 2008;20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez-Leon J, Koth CM, Buscher D, Itoh T, Raya A, Ng JK, Esteban CR, Takahashi S, Henrique D, Schwarz MF, Asahara H, Izpisua Belmonte JC. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat Cell Biol. 2003;5:513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- Kratochwil K, Galceran J, Tontsch S, Roth W, Grosschedl R. FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(−/−) mice. Genes Dev. 2002;16:3173–3185. doi: 10.1101/gad.1035602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, Lidral AC, Jiang R. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn. 2006;235:1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Mani P, Jarrell A, Myers J, Atit R. Visualizing canonical Wnt signaling during mouse craniofacial development. Dev Dyn. 2010;239:354–363. doi: 10.1002/dvdy.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Bocian M, Steinhaus K, Bianchi DW, Die-Smulders C, Giannotti A, Imaizumi K, Jones KL, Campo MD, Martin RA, Meinecke P, Pierpont ME, Robin NH, Young ID, Roessler E, Muenke M. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet. 1999;8:2479–2488. doi: 10.1093/hmg/8.13.2479. [DOI] [PubMed] [Google Scholar]
- Niemann S, Zhao C, Pascu F, Stahl U, Aulepp U, Niswander L, Weber JL, Muller U. Homozygous WNT3 mutation causes tetra-amelia in a large consanguineous family. Am J Hum Genet. 2004;74:558–563. doi: 10.1086/382196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BS, Sargent TD, Williams T. Generation and characterization of a novel neural crest marker allele, Inka1-LacZ, reveals a role for Inka1 in mouse neural tube closure. Dev Dyn. 2010;239:1188–1196. doi: 10.1002/dvdy.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Tam PPL. Mouse development:patterning, morphogenesis, and organogenesis. San Diego, Calif: Academic; 2002. [Google Scholar]
- Smith AN, Miller LA, Song N, Taketo MM, Lang RA. The duality of beta-catenin function: a requirement in lens morphogenesis and signaling suppression of lens fate in periocular ectoderm. Dev Biol. 2005;285:477–489. doi: 10.1016/j.ydbio.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Yamada G, Grigoriou M, Pachnis V, Sharpe PT. Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development. 1999;126:51–61. doi: 10.1242/dev.126.1.51. [DOI] [PubMed] [Google Scholar]
- Valente V, Teixeira SA, Neder L, Okamoto OK, Oba-Shinjo SM, Marie SK, Scrideli CA, Paco-Larson ML, Carlotti CG., Jr Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol Biol. 2009;10:17. doi: 10.1186/1471-2199-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ende JJ, Mattelaer P, Declau F, Vanhoenacker F, Claes J, Van Hul E, Baten E. The facio-audio-symphalangism syndrome in a four generation family with a nonsense mutation in the NOG-gene. Clin Dysmorphol. 2005;14:73–80. [PubMed] [Google Scholar]
- Veistinen L, Aberg T, Rice DP. Convergent signalling through Fgfr2 regulates divergent craniofacial morphogenesis. J Exp Zool B Mol Dev Evol. 2009;312B:351–360. doi: 10.1002/jez.b.21276. [DOI] [PubMed] [Google Scholar]
- Yang H, Melvin V, Zhang J, Williams T. Tracking the fate of epithelial seams during facial prominence fusion using a new ectodermal Cre recombinase derived from Tcfap2a. in preparation. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, B) Schematic showing genetic crosses of mice to obtain LOF (A) and GOF (B) mutants.
A–D) E9.5 wild-type (A, C) and GOF (B, D) embryos shown in lateral (A, B) and ventral (C, D) views, stained with X-gal to visualize LacZ expression. E–G) Whole-mount in situ hybridization of E9.5 Fgf8 expression in wild-type (E), LOF (F), and GOF (G). MnP – mandibular prominence, op – optic placode, ot – otic placode.
Ventral view of an E13.5 GOF mutant embryo showing facial dysplasia and orofacial clefting.
A, B) Fgf8 expression in E11.5 wild-type (A) and GOF (B) embryos. C, D) Shh expression in E12.5 wild-type (C) and GOF (D) embryos.