Abstract
Here we report the sequence of 569,202 base pairs of Saccharomyces cerevisiae chromosome V. Analysis of the sequence revealed a centromere, two telomeres and 271 open reading frames (ORFs) plus 13 tRNAs and four small nuclear RNAs. There are two Ty1 transposable elements, each of which contains an ORF (included in the count of 271). Of the ORFs, 78 (29%) are new, 81 (30%) have potential homologues in the public databases, and 112 (41%) are previously characterized yeast genes.
As part of an international collaborative effort to sequence the total genome of the yeast Saccharomyces cerevisiae, we have deduced the DNA sequence of 569,202 base pairs of yeast chromosome V. We used an overlapping set of recombinant yeast cosmid and lambda clones that together cover the entire chromosome (except for the extreme ends of the telomeres). A line drawing of chromosome V and the identification of the recombinant DNAs sequenced are shown in Fig. 1. The sequence was broken arbitrarily into 11 slightly overlapping pieces for ease of handling and deposited in Genbank (see Fig. 1 for accession numbers).
Figure 1.
The central line representing S. cerevisiae chromosome V is marked in kilobase pairs, starts at the left at the guanine of the Sau3A site of the leftmost recombinant yeast DNA, and extends to the right to 570 kb. The centromere, CEN, is represented by a solid circle at ~151 kb. Above the line, placed across their map positions, are the individual recombinant yeast DNAs that were sequenced; 16 cosmids (thin lines), 8 lambdas (thick lines), and 1 plasmid (very thick line). Genbank accession numbers are placed below the line, above the bars indicating the corresponding map positions. From left to right, Genbank accession numbers are U18795, U18779, U18530, U18778, U18796, U18813, U18814, U18839, U18916, U18917 and U18922. There is some deliberate overlap between Genbank entries to maintain contiguity.
Sequencing was accomplished in two phases: the ‘shotgun’ phase, using dye-primer chemistry, and the ‘finishing’ phase, using the polymerase chain reaction (PCR) and dye-terminator chemistry. There were no gaps in the sequence at the end of shotgun sequencing and assembly. The assembled, continuous sequence of chromosome V has 569,202 bp, starting from the guanine residue of the Sau3A site on the left vector boundary of the leftmost clone (1160 in Fig. 1). The 569-kilobase sequence is based on the results from 32,631 individual lanes of sequencing gels, or reads. The average depth of coverage was 12.5-fold. The minimum acceptable coverage was three, with at least one read from each strand.
After shotgun sequencing and assembly, problems remained in the sequence at a frequency of (roughly) two per kilobase and were of several types. They included the inability to count unambiguously the number of repeating units, such as poly (dA), and guanine compressions. There were also small regions in which only one of the two strands had been sequenced. These difficulties were resolved during the finishing phase.
After finishing, the 569-kb contig was checked against three external sets of data. First was the use of tetrad segregation data to derive a genetic map for yeast1. The chromosome V gene order based on DNA sequence was in complete agreement with the tetrad segregation data. There were two locations on the genetic map (CENV at 151 kb and PRO3 at 200 kb) where closely spaced loci had been mapped against distant markers and not against each other, resulting in ambiguities of relative locus order1, which were resolved using the DNA sequence. The gene order across the centromere is GLC3 tRNA-Arg GCN4 CENV MNN1. In the region of PRO3, at 200 kb, the gene order is PRO3 GPA2 GCD11 CHO1 GAL83. Second, our sequence was compared to the S. cerevisiae sequences already deposited in Genbank, using both the FASTA and BLAST programs2,3. In the rare cases of sequence difference, we re-examined our trace files. Remaining ambiguities were resolved using the same methodology as finishing. Third, we checked our data against the primary EcoRI/HindIII double-digestion fragment maps of the recombinant yeast DNAs4. Our sequence was examined for EcoRI and HindIII cleavage sites. Of 534 mapped fragments, there were only five discrepancies, which is a tribute to the care taken in preparing the cleavage sites map4. The five apparent discrepancies between the double-digest map4 and our sequence are: the map had doublets where the sequence predicts singlets after bases 272, 193; 280,936; and 441,102; the map has a fragment that was not found in the sequence after base 414,946; and the sequence is missing a cleavage site after base 506,807.
We examined all six possible reading frames of the 569-kb sequence for ORFs of at least 300 bp that began with a start codon and ended with a stop codon. As a special case, an ORF could be interrupted if there were yeast splice donor/acceptor/branchpoint sequences present at the appropriate intervals. The remaining sequence was examined using FASTA and BLAST for homology to sequences in the public databases. This enabled us to find small ORFs, as well as the centromere, 13 tRNAs, two Ty1 elements (which each contain an ORF), four small nuclear RNAs, many delta and delta-like elements, and the highly conserved X and Y′ sequences characteristic of yeast telomeres (see refs 5, 6) at the far left and right ends.
Initially, 271 ORFs were identified in the 569-kb sequence, although this number has changed as evaluation continued. The 271 ORFs make up roughly 70% of the sequence, with an average of 2.1 kb per ORF. The ‘average’ ORF (1.4 kb) encodes 475 amino acids. Of the ORFs, 112 (41%) have been characterized previously, 81 (30%) have apparent homologues in the public databases, and 78 (29%) are new; six (2%) are spliced. Of the 81 apparent homologues, 55 of these are to other S. cerevisiae sequences.
The fractional G+C content of the 569,202 bp of chromosome V is 0.384. The combined ORF DNAs have a fractional G+C content of 0.401, and the combined ‘non-ORF’ DNA has a G+C content of 0.351.
There are only two places on chromosome V where the quality of the sequence is not high. In the first case, at about 312 kb, there are ~50 bp of unique sequence bounded on both sides by poly (dA): poly (dT). Taq polymerase, and the other DNA polymerases tested, frequently terminated within the homopolymer, and seldom reached the short unique sequence. Therefore, we have only a few reads across the unique sequence. In the second case, at about 450 kb, there is a 5-kb stretch that contains many delta and delta-like sequences interspersed with a small amount of unique sequence. The clone containing this segment was shotgun sequenced to an average of 16-fold redundancy, yet there were relatively few reads in this region. Therefore, for PCR amplification, this 5-kb region was divided into many virtual parts, based on the positions of the unique sequences. Several custom primer pairs, and internal sequencing primers, were designed and synthesized for each part7. These were used in PCR amplification reactions with total yeast genomic DNA as the template. We have sequenced carefully across this region. For most bases, there is sequence from both strands.
There were three special cases that warrant further attention. First, a point mutation had occurred during either cloning or subsequent propagation in Escherichia coli. In an overlap region shared by two recombinant DNAs (lambda 5898 and cosmid 9867; Fig. 1), the sequence should be the same, but in this case there was one reproducible base difference. Lambda 5898 has a guanine residue where cosmid 9867 has an adenine residue. When total yeast genomic DNA was used as a template for PCR amplification, the product of which was used as a template for dye-terminator sequencing, the base at that position was an adenine. The traces showed no indication of a naturally occurring polymorphism. We therefore conclude that the guanine in lambda 5898 was the result of a mutation. In the second special case, we examined the ORFs for any apparently premature, in-frame stop codon, and found two that were puzzling. The first was the TGA stop codon at position (rounded-off) 352 kb. There are three rightward-reading frames, and this stop codon (TGA) is in the first of these. Following the TGA in this frame, there is a lysine codon (AAA) and a methionine codon (ATG) followed by a long ORF. In the second reading frame there are three stop codons: two (TGA, TAA) are next to each other, the third is five triplets further on; this frame is truly stopped. However, there is a long ORF starting with a methionine codon considerably upstream and ending at the double stop codons TGA, TAA. The third reading frame has many stop codons. The two ORFs share one base: A, the first base (ATG) of the first reading frame and the last base (AAA) of the second reading frame. In general, yeast ORFs are separated by several hundred bases. Except for a -1 frame shift in the first reading frame, there would be one ORF rather than two. However, despite sequencing through this position many times, the sequence, the TGA and the five A bases in a row remained invariant. The second apparently premature stop codon occurred in the ORF that corresponds to FLO8 at ~375 kb. The TAG stop codon between YER108c and YER109c appears not just in the recombinant yeast DNA, but also in PCR amplifications from total yeast DNA. Thus it is possible that FLO8 is not functional in yeast strain AB972. In the third special case, in the entire collection of recombinant yeast DNAs, only lambda 3612 (Fig. 1) covers this region. We found lambda 3612 to be highly unstable, giving rise to non-random DNA deletions at extremely high frequency. Starting with 30 individual plaques from a primary stock, only one yielded lambda 3612 DNA without a detectable deletion, as judged by EcoRI/HindIII double-digestion patterns, but even that gave rise to deleted DNAs upon subsequent growth. Therefore, all of the lambda 3612 sequence came (uncharacteristically) from just one preparation of DNA.
A comparison of the DNA base sequence of chromosome V to that of the other S. cerevisiae chromosomes shows that there are two stretches that have similar genes in the same order on two other yeast chromosomes. A portion of the left arm of chromosome V, containing CYC7 and RAD, shows the same relative gene order as chromosome X, but in the opposite orientation, as noted previously8. In addition, a 60-kb region of the left arm of chromosome IX contains nine genes or ORFs for which each has an apparent homologue within a 60-kb region of the right arm of chromosome V. The nine putative protein pairs and their calculated similarity (identicality)9 are: (1) YIL045w/YER054c, 63 (44) %; (2) YIL050w/YER059w, 71 (52) %; (3) YIL051c/YER057c, 87 (70) %; (4) YIL053w/YER062c, 97 (92) %; (5) YIL056w/YER064c, 63 (47) %; (6) tRNA-ser, 100%; (7) YIL057c/YER067w, 85 (67) %; (8) RNR3/YER070w, 90 (82) %; and (9) YIL074c/YER081w, 95 (91) %. On chromosome V itself, the FCY2 protein product10 and the putative YER060w translation product are 87 (75) % related.
When considering the accuracy of our 569,202-bp sequence of S. cerevisiae chromosome V, we must emphasize that (essentially) all of the sequence was determined from recombinant DNA propagated in E. coli. Even if our sequence of the recombinant DNAs were 100% accurate, there may be sequence differences between the recombinant DNAs and the yeast genome. We identified one apparent point mutation solely because it occurred within a region common to two recombinant DNAs. Other point mutations, occurring during cloning or propagation in E. coli, would probably not be detected.
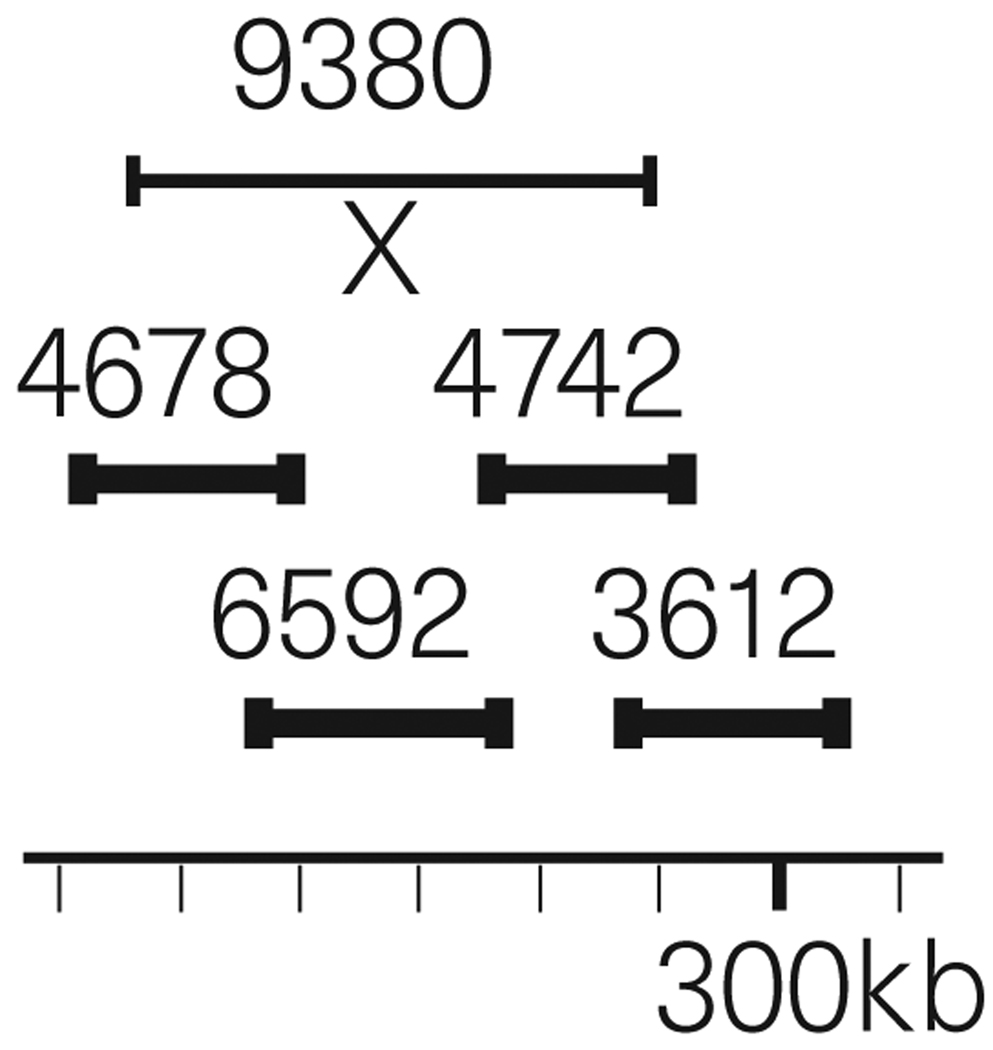
There is a much more dramatic example of a discrepancy between the sequence of a recombinant DNA and the yeast genome. We deposited our chromosome V sequence in Genbank and SacchDB (http://genome-www.stanford.edu/) in December 1994. On 18 April 1996, we received an e-mail from J.-L. Souciet, J. de Montigny and S. Potier (CNRS, Strasbourg) politely telling us that ~2 kb were missing from our sequence. They had found that, in addition to FCY2 (YER056c; encoding a purine-cytosine permease10) at ~267 kb and a closely related ORF (YER060w) at ~275 kb, there is also a third closely related ORF in this region (Genbank accession no. X97346) (Fig 2.). Our sequence across the apparent deletion came from two libraries made from lambda 6592. The sequence is 12-fold deep, and there are no traces that diverge from our Genbank sequence. The Genbank sequence is therefore an accurate sequence of this particular recombinant DNA: lambda 6592. We had intended to sequence cosmid 9380. Repeated attempts to prepare 9380 yielded only minute amounts of DNA. We abandoned cosmid 9380, and instead sequenced three lambda DNAs: 4678, 6592 and 4742. From the tiny amount of cosmid 9380 DNA, we constructed (and sequenced) a Sau3A library. The data (traces) from the library of Sau3A-cleaved 9380 DNA were used within the assemblies of the three lambda DNAs. There were some data from the 9380 library that were left over, mostly vector without insert, low-quality traces, etc., which we put aside. We searched our ‘left-over’ 9380 traces for homology to unique sequences in Genbank X97346 and found one excellent (97.3% identity) match for 294 bp (bounded by Sau3A sites). We conclude that lambda 6592 has a deletion relative to the yeast genome and that one additional ORF should be added, bringing the ORF total to 272.
Figure 2.
The bottom line is a schematic map of yeast chromosome V from 240 kb to 310 kb. Individual, sequenced recombinant DNAs are placed above the line and across the appropriate map positions. Cosmid 9380 covers map positions ~240 kb to 300 kb and overlaps lambda 3612. The latter is the only recombinant DNA in the entire Olson collection4,14 that covers map positions 300 kb to 310 kb. Because 9380 could only be produced in minute amounts, three lambda DNAs (thick lines) were substituted: 4678, 6592 and 4742. The X marks the position of the ~2 kb deletion in 6592 DNA.
To complete the sequence of chromosome V, an insertion of 2,011 bp (Genbank accession number X97346) should be made at base 275,951 of our sequence as has been done in SacchDB. In addition, H. Wedler and R. Wambutt have sequenced the left (Genbank accession no. U73806) and right (Genbank accession no. U34775) telomeres of yeast chromosome V. Within SacchDB, 2,477 bp have been placed to the left of the leftmost base (the G of the leftmost Sau3A site) of our sequence. That G is no longer base 1 but base 2,478. Within the left telomere, there is an ORF, YEL077c, which brings the current ORF total to 273. Concomitantly in SacchDB, 3,181 bp have been placed to the right of our sequence.
Basically, there are two types of errors: random and systematic. If there is a random error in an individual sequence read, we will find and correct that error because we sequenced both strands to high redundancy (average of 12.5-fold). It is much more difficult to identify a systematic error that is inherent in, for example, the dye-primer chemistry, polyacry-lamide gel electrophoresis, Taq polymerase, or base-calling software, that systematically misreads or deletes a base(s) within a particular sequence. Taq polymerase seemed to have systematic difficulties synthesizing across short repeating units; not only is the number of repeats often ambiguous, the sequence traces following a repeat are often diminished in signal quality. We believe that this observation reflects an inherent characteristic of Taq polymerase. Of the several DNA polymerases tested in an attempt to solve this problem, Amplitaq FS polymerase (Perkin-Elmer 402079) yielded the best number of good sequence calls, but did not solve the problem completely. However, the problem in counting the short repeating units unambiguously may not be a sequencing problem but in some cases may reflect true biological heterogeneity. A second possible systematic error arises from the well-known guanine compressions. Guanine compressions are usually identified when the base-calling software identifies fewer guanines on one strand than cytosines on the opposing strand. However, if two (or more) guanine compressions are positioned symmetrically on opposing strands, the compression on one strand is compensated by an analogous compression on the other strand. There are no ‘extra’ cytosines, and the existence of the compressions could be missed.
One reason for sequencing all of the S. cerevisiae DNA is that yeast is important as a model organism. A second reason is to test the approaches to, and develop technologies for, large-scale DNA sequencing in preparation for the sequencing of the human genome. In this regard, we would like to describe some important lessons learned during the sequencing of yeast chromosome V. First, 800 kb were shotgun sequenced to achieve 569,202 bp of contiguous sequence, an inefficiency of 40%. Considerable time and money would have been saved if the ends of the recombinant yeast DNAs had been mapped relative to each other (a ‘sequence-ready’ contig of cosmid DNAs). Second, a large amount of freezer space was used in archiving recombinant M13 DNAs, a small percentage of which were later used as templates for finishing. An important reason in the delay of finishing was the cost of oligonucleotide primers for PCR. Finishing has been made economical by the availability of low-cost oligonucleotides7, so long-term storage of M13 DNAs is no longer necessary. Third, when the Yeast Genome Project was started, the conventional wisdom had that it was necessary to sequence a set of overlapping cosmids. However, we now know that the sequence of DNA as large as bacterial genomes can be assembled using a shotgun approach11,12. If we started again, we would purify S. cerevisiae chromosome V directly by pulse-field gel electrophoresis, hydrodynamically shear the DNA to an average size of 1 kb (ref 13) and shotgun clone the sheared DNA directly into the M13 sequencing vector. The yeast genome could probably have been sequenced by the direct shotgun cloning of total genomic DNA to generate one M13 sequencing library. Individual cosmid and lambda clones could have been used to fill holes and resolve ambiguities.
Methods
All of the S. cerevisiae recombinant DNAs sequenced in this study were constructed in the laboratory of M. Olson4,14. With the exception of plasmid 7990, which was derived from two yeast strains15, all of the recombinant DNAs were derived from yeast strain AB972. Those recombinant DNAs with number designations less than 8000 are lambdas (except for plasmid 7990), those with numbers over 8000 are cosmids. We sequenced 16 cosmids (8198, 8199, 8229, 8334, 9115, 9132, 9163, 9379, 9537, 9581, 9669, 9747, 9781, 9867, 9871 and 9981), eight lambdas (1160, 3612, 4678, 4742, 5898, 6134, 6592 and 6693), and one plasmid (7990) (Fig. 1). We also obtained some sequence from another five cosmids (8063, 9268, 9380, 9495 which contained a large deletion and 9675) and two lambdas (3955 and 6052). These S. cerevisiae recombinant DNAs (except 9495) are available from the American Type Culture Collection.
The ‘shotgun’ sequencing strategy was to reduce randomly the size of the yeast recombinant DNAs (‘inserts’) to approximately 1 kb. The inserts were ligated to the M13 sequencing vector by using a ‘linker–adaptor’ system, which minimizes the formation of chimaeric DNAs. The recombinant M13 ‘sequencing library’ was electroporated into E. coli and plated. Individual M13 plaques were picked and grown, and recombinant M13 DNAs were purified. (Our detailed laboratory protocols are freely available on the World-Wide Web at http://sequence-www.stanford.edu.)
The shotgun sequencing used dye-primer chemistry in cycle sequencing reactions, followed by fluorescence detection using an ABI 373A automated sequencer. Most of the sequence data from individual lanes (‘traces’ or ‘reads’) were edited automatically using custom software, with borderline cases being edited manually using the TED software16. Individual sequence reads were assembled using the XBAP program16. The final sequence was determined by editing manually the assembled reads.
Where there seemed to be overlapping ORFs, in either the same or in the opposite direction, the conventional assumption was made that yeast seldom uses both overlapping frames. Three criteria were used to determine which was the most likely ORF. First, using both FASTA and BLAST programs, each of the overlapping reading frames was examined for homology to known genes in the public databases; an ORF with homology was chosen over one without. Second, each organism has its own distinctive preference for certain codons over others. This preference can be expressed in arithmetic terms, as it is within the GeneFinder program (L. Hiller and P. Green, 1990–1993; documentation, software and yeast codon usage data files. Genome Sequencing Center, Washington University School of Medicine, St Louis, MO 63108, USA). GeneFinder was used to compare the codon usage for each of the overlapping ORFs. The ORF that more closely matched yeast’s codon usage was chosen; in almost all cases, this distinction was unequivocal. Third, the longer ORF was selected. The nomenclature for yeast ORFs is composed of five letter/number combinations: Y (for yeast), E (the fifth letter of the alphabet for V), L or R (for the left or right arm, as defined genetically), a number (counted sequentially from the centromere in both directions), and w or c (for the transcribed strand); for example, the URA3 gene, encoding orotidine-5′-phosphate decarboxylase, is YEL021w.
Acknowledgements
We thank the members of the informal, international group who sequenced the entire yeast genome for their generosity and cooperation that enabled the Yeast Genome Project to be completed a year earlier than scheduled and under budget. In particular, we thank L. Riles and M. Olson for sending us their set of recombinant yeast DNAs and unpublished mapping data; E. Gilbertson for assistance in preparing this manuscript; and N. Schroff and A. Wynant for technical assistance. This work was supported by grants from the NIH (U. S. Public Health Service).
Footnotes
The sequence of yeast chromosome V has been deposited in Genbank.
References
- 1.Mortimer RK, Contopoulou CR, King JS. Yeast. 1992;8:817–902. doi: 10.1002/yea.320081002. [DOI] [PubMed] [Google Scholar]
- 2.Pearson WR, Lipman DJ. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Riles L, et al. Genetics. 1993;134:81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis EJ, Haber JE. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis EJ, Naumova ES, Lee A, Naumov G, Haber JE. Genetics. 1994;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lashkari DA, Hunicke–Smith SP, Norgren RM, Davis RW, Brennan T. Proc. Natl Acad. Sci. 1995;92:7912–7915. doi: 10.1073/pnas.92.17.7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiles JI, Friedman LR, Helms C, Consaul S, Sherman F. J. Mol. Biol. 1981;148:331–346. doi: 10.1016/0022-2836(81)90179-0. [DOI] [PubMed] [Google Scholar]
- 9.Smith T, Waterman M, Burks C. Nucleic Acids Res. 1985;13:645–656. doi: 10.1093/nar/13.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber E, Rodriguez C, Chevallier MR, Jund R. Mol. Microbiol. 1990;4:585–596. doi: 10.1111/j.1365-2958.1990.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 11.Fleischmann RD, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Fraser CM, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 13.Oefner PS, et al. Nucleic Acids Res. 1996;24:3879–3886. doi: 10.1093/nar/24.20.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson MV, et al. Proc. Natl. Acad. Sci. USA. 1986;83:7826–7830. doi: 10.1073/pnas.83.20.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleer R, Nicolet CM, Pure GA, Friedberg EC. Mol. Cell. Biol. 1987;7:1180–1192. doi: 10.1128/mcb.7.3.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gleeson TJ, Staden R. Comp. Appl. Biosci. 1991;7:398. doi: 10.1093/bioinformatics/7.3.398. [DOI] [PubMed] [Google Scholar]