Abstract
Outbreaks of dengue hemorrhagic fever have coincided with the introduction of the Southeast (SE) Asian genotype of dengue type 2 virus in the Western Hemisphere. This introduced genotype appears to be rapidly displacing the indigenous, American genotype of dengue 2 virus throughout the region. These field observations raise the possibility that the SE Asian genotype of dengue 2 is better adapted for vector transmission than its American counterpart. To evaluate this hypothesis, we compared the ability of viral strains of the SE Asian and American genotypes to infect, replicate, and disseminate within vector mosquitoes (Aedes aegypti). Viral strains of the SE Asian genotype tended to infect and disseminate more efficiently in mosquitoes than did variants of the American genotype. These differences, however, were observed solely in field-derived mosquitoes, whereas viral infection rates were virtually identical in the laboratory-adapted Rockefeller colony of Ae. aegypti. Our findings could provide a physiological basis for the contrasting patterns of dengue virus genotype transmission and spread. Such an understanding of functional differences between viral strains and genotypes may ultimately improve surveillance and intervention strategies.
Keywords: Aedes aegypti, Dengue, Vector competence, Viral genotype
INTRODUCTION
Dengue is among the most common vector-borne diseases worldwide and accounts for an estimated 50–100 million human cases each year (Halstead, 1992). The disease results from infection by one of four antigenically distinct RNA viruses, designated as dengue serotypes 1–4. All four serotypes are transmitted by the yellow fever mosquito (Aedes aegypti) which serves as the main vector species in human populations. Infection may be subclinical or cause illness ranging from a flu-like syndrome with rash, to a life-threatening disease with internal bleeding–dengue hemorrhagic fever (DHF).
Both host-immune factors and viral determinants appear to contribute to DHF pathogenesis. Primary dengue infection confers life-long immunity against that infecting serotype, while increasing the susceptibility to DHF by a second heterologous serotype (Halstead et al. 1970). The pathological mechanisms underlying this phenomenon may be due to antibody-dependent enhancement of viral infection and/or cross reactive T-cell responses (Kliks et al. 1989, Rothman et al. 1996). In addition to the immune status of the host, viral serotypes may differ in their potential to cause severe disease. In Thailand, for example, secondary infection with serotype 2 contributes disproportionately to DHF cases versus other serotypes of dengue virus (Sangkawibha et al. 1984, Vaughn et al., 1997, 2000). Although secondary infection contributes to the pathogenesis of DHF, the sequence of infection by particular serotypes may also affect disease outcome.
Field observations in the Americas suggest that dengue variants within serotypes, or genotypes, differ in their potential to cause DHF. Prior to 1981, dengue outbreaks were not accompanied by DHF in this region, despite simultaneous or sequential transmission of more than one serotype (Pan American Health Organization, 1994). During the 1960s and 1970s dengue outbreaks in Puerto Rico, Colombia, the Dominican Republic, and Jamaica involved dengue serotype 2 and at least one other serotype, yet none were associated with epidemic DHF (Halstead 1980). The virtual absence of DHF in the Americas, however, abruptly ended with the Cuban outbreak of dengue serotype 2 in 1981. Subsequent DHF outbreaks became evident in Brazil, Venezuela, Colombia, and French Guiana and in each of these instances, dengue type 2 was the predominant serotype (Gubler 1997). Phylogenetic analyses indicated that two distinct viral lineages of dengue type 2 are maintained in the Western Hemisphere (Rico-Hesse 1990, Rico-Hesse et al. 1997, Leitmeyer et al. 1999). The American genotype has been detected in the region since the 1950s and, thus far, associated solely with dengue fever. The Southeast (SE) Asian genotype, in contrast, was introduced into the region more recently and is associated with DHF outbreaks (Rico-Hesse et al. 1997, Gubler and Meltzer, 1999). Other evidence comes from epidemiological studies in Peru; despite high rates of secondary infection with the American genotype of dengue 2, no cases of DHF have been detected (Watts et al. 1999).
The expanding distribution of DHF may stem, in part, from the rapid spread of the SE Asian genotype of dengue virus type 2. During the years that followed the Cuban outbreak of DHF, the SE Asian genotype has largely supplanted the American genotype in the Western Hemisphere (Rico-Hesse et al. 1997). Refugia of the American variants, however, continue to persist in the northern part of Mexico and in Peru (Watts et al. 1999, Rico-Hesse, unpublished observations). These observations suggest that transmission of the SE Asian genotype is more robust than that of the American genotype. It may be that the SE Asian genotype is better adapted to vector transmission than its American counterpart. To evaluate this hypothesis, we compared the ability of viral strains of either genotype to infect, replicate, and disseminate in laboratory-adapted and field-derived colonies of Ae. aegypti.
MATERIALS AND METHODS
Mosquitoes
Immature stages of Ae. aegypti were collected in McAllen, TX and Iquitos, Peru during the fall of 1999 and winter of 2000, respectively. Each colony was derived from a minimum of 1,000 field-derived mosquitoes. F2 (Iquitos) and F3 (McAllen) generation females were used in this study. In addition, viral infection was tested in the Rockefeller colony of Ae. aegypti for the purposes of comparison. This colony has been maintained in the laboratory for >100 generations.
Mosquitoes were maintained in an insectary at 28°C, 70–80% realtive humidity, and a 12:12 h light–dark cycle. Larvae were reared in pans at a density of 200–400 larvae/L of water and fed a mixture of ground rabbit chow:liver powder:yeast (4:2:1). The resulting pupae were transferred to screened cages, and adults emerged ~2–4 days thereafter. Adults were maintained on a diet of sugar and water, and meals of rabbit blood were provided by means of a water-jacketed membrane feeder. Eggs were collected, kept moist for at least 24 hours, and then air dried for storage. Mosquito colonies were cycled every 3–5 months.
Virus preparation
Six low-passage human isolates representing either the SE Asian or American genotypes of dengue type 2 virus were used for vector infection studies (Table 1). Virus stocks were amplified by passage in C6/36 (Aedes albopictus) cells. Cells were grown to confluency in 75-cm2 flasks, overlaid with each virus diluted in 3 ml of maintenance medium [Minimal Essential Media, 2% fetal bovine serum [FBS], 1× non-essential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin], and incubated for 1 h at 28°C in 5% CO2. Flasks were brought to a final volume of 15 ml with maintenance medium and maintained at 28°C in 5% CO2. The percentage of infected cells was monitored daily by IFA technique (Tesh 1979), and cell supernatants were harvested when >90% of cells were infected, usually 7 or 8 days postinfection. Viral stocks were stored in 20% FBS in individual aliquots at −70°C.
Table 1.
Geographic Origin, Passage History, and Titers of Dengue Strains
| Genotypea | Strain | Passage history | Location (year) | pfu/ml | Viral RNA copies/ml |
|---|---|---|---|---|---|
| SE Asian | Mara3 | C6/36 3 | Maracay, Venezuela (1990) | 4.5 × 106 | 3.4 × 109 |
| 102954 | C6/36 3 | Aragua, Venezuela (1991) | 1.5 × 106 | 2.0 × 109 | |
| K0049 | TS 1, C6/36 3 | Kamphaeng Phet, Thailand (1995) | 9.0 × 105 | 9.9 × 109 | |
| American | Ven2 | AP61 2, C6/36 3 | Maracay, Venezuela (1987) | 1.3 × 106 | 4.0 × 109 |
| 131 | C6/36 3 | Sonora, Mexico (1992) | 2.9 × 106 | 7.0 × 109 | |
| IQT2913 | C6/36 3 | Iquitos, Peru (1996) | 2.1 × 106 | 2.3 × 109 |
Genotype was determined by partial or full genome sequencing (Rico-Hesse et al. 1997, Leitmeyer et al. 1999).
Quantitative reverse transcription (RT)–polymerase chain reaction (PCR)
The concentration of viral RNA was estimated by real-time RT-PCR using the Taqman system (PE Applied Biosystems). Briefly, this technique employs two external primers and an internal probe labeled with both a quencher and reporter dye. When the probe is intact, reporter fluorescence is suppressed by the proximity of the quencher dye linked to the opposite end of the probe. Fluorescence is released when the probe hybridizes during each extension cycle and is cleaved by the exonuclease activity of Amplitaq polymerase. The amount of fluorescence is directly proportional to the starting amount of viral RNA, which is monitored during each extension cycle by an automated fluorometer.
RNA was extracted from viral stocks, in duplicate, using the Viral RNA kit (Qiagen). RNA was eluted in a total volume of 140 μl of water, and 5 μl of this preparation was added to a 50-μl reaction using the TaqMan Gold RT-PCR kit (PE Applied Biosystems). Each 50-μl reaction contained a final concentration of 1× TaqMan Buffer, 5.5 mM MgCl2, 300 μM ATP, 300 μM CTP, 300 μM GTP, 300 μM UTP, 100 nM PA-1 (5′-CAGATCTCTGATGAATAACCAACG-3′), 100 nM PA-4 (5′-CATTCCAAGTGAGAATCTCTTTGTCA-3′), 100 nM PRB (TET-ATGCTGAAACGCGAGAGAAACCGCTAMARA), 1.25 U of Amplitaq Gold DNA polymerase, 12.5 U of Multiscribe reverse transcriptase, and 20 U of RNase inhibitor. PCR primers and probes were designed to target regions of the capsid gene that are strictly conserved among variants of the SE Asian and American genotypes (Table 2). Amplification was performed using a ABI Prism 770 Sequence Detection Instrument (PE Biosystems) as follows: 50°C for 2 min (1 cycle), 95°C for 10 min (1 cycle), followed by 40 cycles of 95°C for 15 s, and 64°C for 1 min. RNA copy number was estimated from a standard curve generated by in vitro transcribed RNA standards. Viral RNA standards were prepared by amplifying a 670-bp fragment of the dengue type 2 genome (NGC strain) with primers D2/11V and D2/662 (Leitmeyer et al. 1999). The resulting PCR product was ligated into PCR 2.1 using the TA cloning kit (Invitrogen). Plasmid DNA was linearized with HindIII and RNA transcripts were generating using the T7 promotor Megascript Kit (Ambion) according to the manufacturer’s specifications. The concentration of transcribed RNA was estimated by UV spectrophotometry.
Table 2.
Nucleotide Alignment of Dengue Type 2 Virus Sequences Corresponding to PCR Primer–Probe Target Sites
| PA-1 | PRB | PA-4 | |
|---|---|---|---|
| Map no. | 87 | 139 | 182 |
| NGC | CAGAUCUCUGAUGAAUAACCAACG | AUGCUGAAACGCGAGAGAAACCGC | UGACAAAGAGAUUCUCACUUGGAAUG |
| Ven2 | ------------------------ | ------------------------ | -------------------------- |
| 131 | ------------------------ | ------------------------ | -------------------------- |
| Iqt2913 | ------------------------ | ------------------------ | -------------------------- |
| 16681 | ------------------------ | ------------------------ | -------------------------- |
| Mara4 | ------------------------ | ------------------------ | -------------------------- |
| Co390 | ------------------------ | ------------------------ | -------------------------- |
| 1409 | ------------------------ | ------------------------ | -------------------------- |
Map numbers correspond to the sequence of the NGC strain of dengue virus. Dashed lines indicate identical sequence.
Plaque assay
Plaque titrations were performed in six-well plates of confluent LLC-MK2 cells as previously described (Huang et al. 2000). Serial dilutions of each virus isolate were absorbed onto cell monolayers for 1.5 h at 37°C, 5% CO2. Cell monolayers were then overlaid with 4 ml of 1% SeaKem LE agarose (FMC Bioproducts) in nutrient medium (0.165% lactalbumin hydrolysate, 0.033% yeast extract, Earle’s balanced salt solution, 25 μg/ml gentamicin sulfate, 1.0 μg/ml amphotericin B, and 2% FBS). Monolayers were incubated at 37°C 5% CO2 for 7 days and then received a second 2-ml overlay containing the addition of 80 μg/ml neutral red stain (Gibco BRL). Cells were incubated for an additional 2–6 days before plaques were counted.
Vector infection
Ae. aegypti females were transferred to mesh-covered cages 7–10 days after emergence and then deprived access to sugar 24 hours prior to blood feeding. Each dengue strain was diluted to a final concentration of 5 × 108 viral RNA copies/ml in EDTA-treated rabbit blood and presented to mosquitoes via a water-jacketed membrane feeder. During each experimental trial, one group of mosquitoes was fed a virus of the SE Asian genotype, while a second group was simultaneously fed an American genotype virus. Engorged mosquitoes were selected and maintained at 30°C on a diet of 4% sucrose. Mosquitoes were harvested at various time intervals after infection and then tested for the presence of dengue antigen in the head by IFA as previously described (Kuberski and Rosen 1977). In addition, some of the mosquitoes were analyzed by quantitative PCR to compare the efficiency of viral replication. The heads of these mosquitoes were reserved for IFA, while the abdomen and thorax was stored in 0.5 ml of Trizol (Life Technologies). Total RNA was extracted according to the manufacturer’s specifications except that 10 μg of glycogen was added as a carrier during the precipitation step; 5 μl of this preparation was added to a 50 μl RT-PCR mixture using the same primers and conditions described above.
Data analysis
We conducted two separate analyses to compare the proportion of mosquitoes becoming infected by viruses of each dengue genotype. Initially, we pooled disseminated infection frequencies for the strains making up each genotype and compared genotype frequencies using Fisher’s exact tests for each time point. Because we observed significant variation among individual strains within a genotype, we reanalyzed the data by analysis of covariance (ANCOVA) to account for this extra source of variation. Infection frequencies for each strain were transformed by the arcsine transformation for proportions as previously described (Freeman and Tukey 1950) and were weighted for unequal sample sizes. Regression lines were fitted over time for viral genotypes and were tested for differences in slope and elevation by ANCOVA.
RESULTS
In preliminary experiments, we evaluated the sensitivity and reproducibility of our quantitative RT-PCR assay. Tenfold dilutions of viral RNA standard were subjected to RT-PCR amplification in triplicate. The assay consistently detected 1 × 108–1 × 102 copies of RNA standard but failed to detect 10 copies in all three RT-PCR mixtures. Frozen stocks of dengue virus (Ven2) were subsequently extracted in triplicate using Trizol reagent (Life Technologies) or the viral RNA kit (Qiagen). RNA preparations were tested by quantitative RT-PCR to estimate the extent of variation between replicate samples. The coefficient of variance for triplicate extractions using the Qiagen kit was 13%, whereas that of the Trizol method was 27%. Based on these results, we employed the Qiagen kit for estimating RNA concentration of our viral stocks. Trizol reagent was used for preparing individual mosquitoes because of cost considerations.
Viral stocks were titrated by means of plaque assay and quantitative RT-PCR (Table 1). Viral RNA loads did not consistently correlate with the number of infectious plaques for each viral strain. The number of plaque-forming units (pfu)/ml and RNA copies/ml deviated greatest for strain K0049, which yielded the highest concentration of viral RNA and the lowest concentration of infectious plaques among the 6 strains tested. This finding is not surprising given that low-passage strains of dengue virus may vary in their ability to form infectious plaques (Mangada et al., 1998). Because plaque assay measures a restricted phenotype, the ability to form plaques in LLC-MK2 cells, virus loads were standardized on the basis of viral RNA concentration.
To determine whether dengue viruses of a particular genotype infect and disseminate in a greater proportion of mosquitoes, we allowed mosquitoes to engorge on virus–blood meals and tested them for the presence of dengue antigen in head squashes. Mosquitoes of the Rockefeller colony appeared to be about equally susceptible to infection by members of the SE Asian and American genotypes (Table 3). In contrast, field-derived strains of Ae. aegypti tended to be more susceptible to infection by variants of the SE Asian genotype. Mosquitoes of the McAllen colony appeared to be more susceptible to infection by members of the SE Asian genotype but these differences were not statistically significant except at day 10 (Fisher’s exact test, p < 0.03). The proportion of Iquitos mosquitoes infected by SE Asian genotype viruses was significantly greater at days 6, 8, 10, and 14 (Fisher’s exact test, p < 0.005 at days 6, 8, and 14 and p < 0.02 at day 10). Dengue strains Mara3, 102954, IQT2913, and 131 were responsible for the observed difference among genotypes, whereas virus strains K0049 (SE Asian) and Ven2 (American) did not conform to this pattern of differential infection. We did not expect to find so much variation among virus strains within a particular genotype. Accordingly, we reanalyzed the data presented in Table 3 by ANCOVA to account for variation among individual viral strains and to adjust for different time intervals. Vector infection rates were regressed over time to compare slopes and y-intercepts for each genotype. In the Rockefeller colony, ANCOVA revealed no significant differences in slope or elevation among the SE Asian and American genotype regression lines (ANCOVA, slope p = 0.94, y-intercept p = 0.50). In the Iquitos colony, significant differences in mosquito infection rates were detected between the two viral genotypes (ANCOVA, slope p < 0.03, y-intercept p < 0.0001). Likewise, mosquitoes of the McAllen colony tended to be more susceptible to infection by members of the SE Asian genotype and these differences were also significant (ANCOVA, slope p < 0.02, y-intercept p < 0.0001).
Table 3.
Infection and Dissemination of Dengue 2 Viruses of the SE Asian and American Genotypes in Rockefeller, McAllen, and Iquitos Strains of Ae. aegypti
| Mosquito strain | Days post- exposure | SE Asian genotype |
American genotype |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Viral strains |
Total | % | Viral strains |
Total | % | ||||||
| Mara3 | 102954 | K0049 | Ven2 | IQT2913 | 131 | ||||||
| Rockefeller | 3 | 0/15 | 0/15 | 0/15 | 0/45 | 0 | 0/15 | 0/15 | 0/15 | 0/45 | 0 |
| 6 | 1/15 | 7/15 | 1/24 | 9/54 | 17 | 3/15 | 5/15 | 2/28 | 10/58 | 17 | |
| 9 | 11/15 | 12/15 | 6/24 | 29/54 | 54 | 12/16 | 5/11 | 12/24 | 29/51 | 57 | |
| 12 | 11/12 | 14/16 | 8/23 | 33/51 | 65 | 10/14 | 8/15 | 11/27 | 29/56 | 52 | |
| 15 | 13/15 | 15/23 | 9/24 | 37/62 | 60 | 14/16 | 11/16 | 14/26 | 39/58 | 67 | |
| McAllen | 2 | 0/15 | 0/15 | 0/15 | 0/45 | 0 | 0/15 | 0/15 | 0/15 | 0/45 | 0 |
| 4 | 0/15 | 0/15 | 0/15 | 0/45 | 0 | 0/15 | 0/15 | 0/15 | 0/45 | 0 | |
| 6 | 5/21 | 9/22 | 0/17 | 14/60 | 23 | 3/21 | 1/20 | 2/20 | 6/61 | 10 | |
| 8 | 7/20 | 14/21 | 5/19 | 26/60 | 43 | 9/20 | 5/20 | 7/20 | 21/60 | 35 | |
| 10 | 14/20 | 14/19 | 10/16 | 38/55 | 69 | 11/20 | 6/20 | 11/18 | 28/58 | 48 | |
| 14 | 22/26 | 28/30 | 11/19 | 61/75 | 81 | 21/29 | 10/17 | 15/23 | 46/69 | 67 | |
| Iquitos | 2 | 0/20 | 0/20 | 0/10 | 0/50 | 0 | 0/20 | 0/20 | 0/10 | 0/50 | 0 |
| 4 | 1/20 | 0/20 | 0/10 | 1/50 | 2 | 0/20 | 0/20 | 0/10 | 0/50 | 0 | |
| 6 | 9/20 | 4/20 | 0/19 | 13/59 | 22 | 1/20 | 0/20 | 1/18 | 2/58 | 3 | |
| 8 | 16/30 | 12/30 | 5/20 | 33/80 | 41 | 9/28 | 2/25 | 2/20 | 13/73 | 18 | |
| 10 | 18/21 | 15/17 | 11/33 | 44/71 | 62 | 14/20 | 12/27 | 1/19 | 27/66 | 41 | |
| 14 | 24/27 | 22/28 | 9/25 | 55/80 | 69 | 21/30 | 13/39 | 10/38 | 44/107 | 41 | |
Data are no. of mosquitoes positive/no. tested. Mosquitoes were scored positive if dengue antigen was detected in head squashes by IFA. Each time point represents the proportion of mosquitoes positive by IFA.
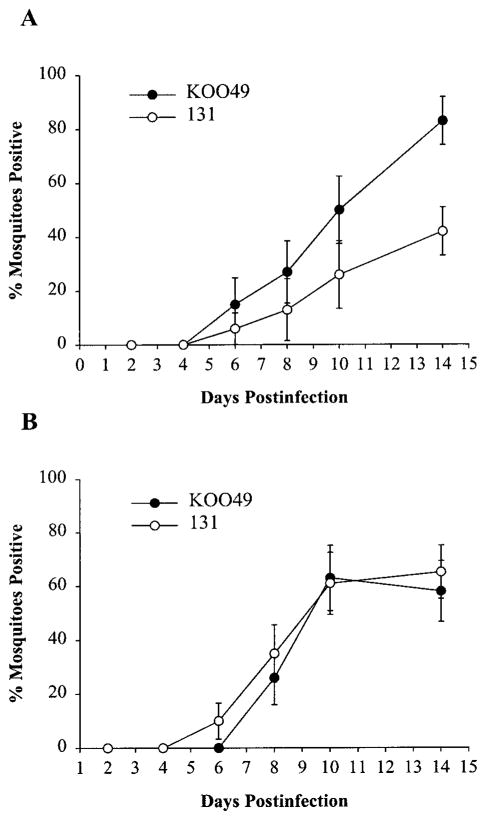
Mosquitoes were subsequently infected with dengue viruses using an equivalent concentration of pfu rather than viral RNA copies. Dengue strain K0049 was selected for this analysis because of the observed difference between RNA copy number and pfu concentration for this particular strain. When mosquitoes were infected with an equivalent concentration of viral genome copies, strains K0049 and 131 were equally infectious to mosquitoes of the McAllen colony (Fig. 1). When virus concentrations were standardized on the basis of pfu/ml, strain K0049 infected a greater proportion of mosquitoes than strain 131. These differences, however, were not significant except at day 14 (Fisher’s exact test, p < 0.02).
FIG. 1. Susceptibility of McAllen strain of Ae. aegypti to infection by dengue type 2 strains K0049 and 131.
A: Mosquitoes fed virus at a concentration of 1 × 105 pfu/ml. B: Mosquitoes fed virus at a concentration of 5 × 108 viral RNA copies/ml. Error bars represent SD of means.
To determine whether viruses of a particular dengue genotype replicate faster within the vector, we quantified viral RNA in the thorax and abdomen of orally infected mosquitoes by quantitative RT-PCR. Six to 10 mosquitoes were analyzed at each time point for a total of 18 virus–vector pairs (three different mosquito colonies versus six different virus strains). In all of the mosquito colonies, virus strains exhibited typical growth dynamics with viral RNA loads peaking at about day 6 (Table 4). Replication rates for viruses of the American and SE Asian genotypes appeared to be virtually identical, and no statistical difference was observed when comparing standard error values.
Table 4.
Dengue Virus Titers (Log10 Viral RNA Copies per Mosquito) of Orally Infected Ae. aegypti Estimated by Quantitative RT-PCR
| Mosquito strain | Days post- exposure | SE Asian genotype |
American genotype |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Viral strains |
Mean | SE | Viral strains |
Mean | SE | ||||||
| Mara3 | 102954 | K0049 | Ven2 | IQT2913 | 131 | ||||||
| Rockefeller | 0 | 6.1 | 5.8 | 5.8 | 5.9 | 5.3 | 6.4 | 6.4 | 6.3 | 6.4 | 5.5 |
| 3 | 6.5 | 6.3 | 6.9 | 6.5 | 6.0 | 6.6 | 6.2 | 6.9 | 6.6 | 6.0 | |
| 6 | 7.8 | 7.7 | 7.9 | 7.8 | 7.2 | 7.8 | 7.9 | 8.0 | 7.9 | 7.3 | |
| 9 | 8.0 | 7.9 | 8.2 | 8.0 | 7.2 | 8.0 | 7.7 | 8.1 | 7.9 | 7.2 | |
| 12 | 8.0 | 8.0 | 8.8 | 8.4 | 7.9 | 8.0 | 7.8 | 8.5 | 8.2 | 7.7 | |
| 15 | 7.6 | 7.8 | 8.6 | 8.1 | 7.6 | 7.6 | 7.9 | 8.5 | 8.1 | 7.6 | |
| McAllen | 2 | 5.2 | 5.0 | 5.0 | 5.1 | 4.6 | 5.1 | 4.8 | 5.5 | 5.3 | 5.1 |
| 4 | 7.1 | 7.2 | 7.1 | 7.1 | 6.6 | 7.6 | 6.2 | 6.9 | 7.2 | 7.1 | |
| 6 | 6.9 | 7.7 | 7.8 | 7.6 | 7.0 | 7.8 | 6.4 | 8.1 | 7.9 | 7.4 | |
| 8 | 7.5 | 7.4 | 7.9 | 7.6 | 7.3 | 7.6 | 7.0 | 7.7 | 7.5 | 6.8 | |
| 10 | 7.5 | 7.3 | 7.8 | 7.6 | 7.0 | 7.7 | 7.4 | 8.2 | 7.9 | 7.4 | |
| 14 | 7.8 | 7.8 | 8.6 | 8.2 | 7.9 | 8.3 | 7.7 | 8.5 | 8.2 | 7.6 | |
| Iquitos | 2 | 3.9 | 3.9 | 4.7 | 4.4 | 4.0 | 3.3 | 4.3 | 4.3 | 4.2 | 3.8 |
| 4 | 7.5 | 7.0 | 6.9 | 7.2 | 7.0 | 7.2 | 6.3 | 7.1 | 7.0 | 6.8 | |
| 6 | 7.7 | 7.5 | 7.9 | 7.7 | 7.1 | 6.7 | 6.9 | 8.0 | 7.7 | 7.4 | |
| 8 | 7.1 | 7.3 | 8.4 | 8.1 | 7.7 | 7.7 | 6.5 | 8.0 | 7.7 | 7.3 | |
| 10 | 7.8 | 7.7 | 8.7 | 8.1 | 7.7 | 8.0 | 7.3 | 7.8 | 7.8 | 7.3 | |
| 14 | 7.9 | 8.0 | 9.0 | 8.5 | 8.0 | 8.2 | 8.0 | 8.3 | 8.2 | 7.5 | |
Data are log10 viral RNA copies/mosquito. Six to 10 mosquitoes were tested at each time point, and titers represent the mean values of PCR-positive mosquitoes.
DISCUSSION
We found that dengue variants of the SE Asian genotype tended to infect mosquitoes more efficiently than did viruses of the American genotype; however, we also observed variation among viral strains within each genotype. Future studies will focus on increasing the number of virus strains and mosquito populations to determine whether this trend will hold. Genotype-dependent differences were most striking in field-derived colonies of Ae. aegypti, whereas disseminated infection rates were virtually identical in the laboratory-adapted Rockefeller colony. The Rockefeller colony has been maintained under laboratory conditions since the 1940s and, therefore, vector competence traits may have been affected by random genetic drift and selection over the course of many generations in the laboratory. Our findings underscore the importance of using recently colonized mosquitoes, which are more representative of original field populations.
Viral variants of particular genotype clearly varied in their ability to infect vector mosquitoes. Strain K0049 tended to infect fewer vector mosquitoes than did other members of the SE Asian genotype. It may be that mosquitoes were underinfected with infectious virus when strain K0049 was administered on the basis of viral RNA concentration. The discrepancy between PFU concentration and viral RNA concentration was greatest for strain K0049. When mosquitoes were fed an equivalent concentration of pfu, strain K0049 infected a greater proportion of mosquitoes relative to its American counterpart, strain 131. Dengue strain Ven2 infected more mosquitoes than did other members of the American genotype. This strain had undergone five passages in mosquito cell culture, more passages than any of the other viruses tested, and, therefore, phenotype could have changed as result of its passage history. It may be that serial passage of virus in mosquito cell lines selects for mosquito-adapted genotypes of virus. This possibility requires further study.
An array of techniques, including mosquito inoculation, plaque assay, and quantitative RT-PCR, are available to estimate the concentration of dengue virus. Each technique presents a different set of problems when attempting to compare titers between different virus strains. Viral infectivity assays measure a restricted phenotype and, therefore, may not accurately reflect the number of infectious particles if viral strains differ in their ability to form infectious plaques or to establish infection in the mosquito by intrathoracic inoculation. Moreover, results may be difficult to reproduce because of variation among lots of cell lines or populations of colonized mosquitoes. Quantitative RT-PCR may circumvent many of these limitations by directly measuring the concentration of viral RNA; however, it will also measure viral RNA that is not fully transcribed or packaged while replication is occurring in the cell. When preparing our viral stocks, we avoided this problem by estimating viral RNA concentration in the cell supernatant fraction, reflective of fully-formed virions that escaped the host cell. To compare viral replication rates in the vector mosquito, we extracted total RNA from the whole mosquito, thus incorporating viral RNA intermediates into our quantitative RT-PCR estimates. In this instance, estimate of viral RNA copy number serves as a surrogate for the concentration of infectious virus. This is a valid assumption unless dengue virus variants systematically differ in the ratio of infectious particles to viral RNA templates.
The ability of a mosquito population to transmit a pathogen is determined by a series of physiological and ecological variables. The relevant properties of the mosquito population include: vector competence, vector density, human biting rate, extrinsic incubation period, and vector longevity (Spielman and James 1990). Because both the American and SE Asian genotypes share the same vector mosquito, we reasoned that the selective advantage of one virus genotype over another may derive from their physiological interaction with the mosquito host. Viral genetics may directly affect vector competence, which measures the proportion of mosquitoes that can acquire, maintain, and transmit a pathogen. Prior studies indicate that vector strains and species may differ in their receptivity to midgut infection by dengue virus, whereas the rate of transmission is similar once virus has disseminated out of the midgut into the hemolymph (Gulber et al. 1979, Bosio et al. 1998). We, therefore, focused on the ability of viral genotypes to infect and disseminate in the mosquito by testing head squashes for the dengue antigen.
Any incremental decrease in vector competence could affect the capacity of dengue virus to establish and maintain transmission in a given locale. The basic reproductive number (Ro) refers to the average number of new infections generated from each case of current infection (Anderson and May 1991). The estimated Ro for dengue virus ranged from 1.3 to 2.5 during epidemics in Brazil and Mexico (Koopman et al. 1991 Marques et al. 1994). If Ro falls below 1.0, the virus will fail to perpetuate in a given host population. Because Ro appears to be relatively low for dengue virus, the threshold for epidemics will be sensitive to reductions in vector competence. This assumes, however, that host-related factors are equivalent for the two viral genotypes. It may be that human genetic traits and/or herd immunity select for one viral genotype over another. Or perhaps, some viral strains replicate more efficiently and sustain higher viremias within the human host, thereby infecting more mosquitoes. This last hypothesis is currently being tested in an effort to define the epidemic potential of dengue genotypes and strains.
Acknowledgments
We thank Dr. Richard Kinney for titrating virus stocks by plaque assay, Dr. Amy Morrison for providing mosquito eggs from Iquitos, Peru, Dr. Jack Hayes for his help with mosquito collections in Texas, and Dr. Mark Sharp for helping with statistical analyses. This work was supported by grants from NIH (AI 10427) and the Quillin Foundation.
ABBREVIATIONS
- ANCOVA
analysis of covariance
- DHF
dengue hemorrhagic fever
- FBS
fetal bovine serum
- PCR
polymerase chain reaction
- pfu
plaque-forming units
- Ro
basic reproductive number
- RT
reverse transcription
- SE
Southeast
References
- Anderson RM, May RM. Infectious Diseases of Humans. Oxford: Oxford University Press; 1991. A framework for discussing the population biology of infectious diseases; pp. 13–23. [Google Scholar]
- Bosio CF, Beaty BJ, Black WC. Quantitative genetics of vector competence for dengue-2 virus in Ae. aegypti. Am J Trop Med Hyg. 1998;59:965–970. doi: 10.4269/ajtmh.1998.59.965. [DOI] [PubMed] [Google Scholar]
- Freeman MF, Tukey WJ. Transformations related to the angular and the square root. Ann Math Stat. 1950;21:607–611. [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever: its history and resurgence as a global public health problem. In: Gubler DJ, Kuno G, editors. Dengue and Dengue Hemorrhagic Fever. New York: CAB International; 1997. pp. 1–22. [Google Scholar]
- Gubler DJ, Meltzer M. Impact of dengue/dengue hemorrhagic fever of the developing world. Adv Virus Res. 1999;53:35–70. doi: 10.1016/s0065-3527(08)60342-5. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Nalim S, Tan R, Saipan H, et al. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Ae. aegypti. Am J Trop Med. 1979;28:1045–1052. doi: 10.4269/ajtmh.1979.28.1045. [DOI] [PubMed] [Google Scholar]
- Halstead SB. Immunological parameters of togavirus disease syndromes. In: Schlesinger RW, editor. The Togaviruses:Biology, Structure, Replication. New York: Academic Press; 1980. pp. 107–173. [Google Scholar]
- Halstead SB. The XXth century dengue pandemic: need for surveillance and research. World Health Stat Q. 1992;45:2395–2400. [PubMed] [Google Scholar]
- Halstead SB, Nimmannitya S, Cohen SN. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J Biol Med. 1970;42:311–328. [PMC free article] [PubMed] [Google Scholar]
- Huang CYH, Butrapet S, Pierro DJ, Chang GJ, et al. Chimeric dengue type 2 (vaccine strain PDK-53)/dengue type 1 virus as a potential candidate dengue type 1 virus vaccine. J Virol. 2000;74:3020–3028. doi: 10.1128/jvi.74.7.3020-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliks SC, Nimmannitya S, Nisalak A, Burke DS. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am J Trop Med Hyg. 1988;38:411–419. doi: 10.4269/ajtmh.1988.38.411. [DOI] [PubMed] [Google Scholar]
- Kliks SC, Nisalak A, Brandt WE, Wahl L, et al. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am J Trop Med Hyg. 1989;40:444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- Koopman JS, Prevots DR, Vaca Marin MA, Gomez Dantes H, et al. Determinants and predictors of dengue infection in Mexico. Am J Epidemiol. 1991;133:1168–1177. doi: 10.1093/oxfordjournals.aje.a115829. [DOI] [PubMed] [Google Scholar]
- Kuberski TT, Rosen L. A simple technique for the detection of dengue antigen in mosquitoes by immunofluorescence. Am J Trop Med Hyg. 1977;26:533–537. doi: 10.4269/ajtmh.1977.26.533. [DOI] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, et al. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999;73:4738–4747. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangada MN, Igarashi A. Molecular and in vitro analysis of eight dengue type 2 viruses isolated from patients exhibiting different disease severities. Virology. 1998;244:458–466. doi: 10.1006/viro.1998.9093. [DOI] [PubMed] [Google Scholar]
- Marques CA, Forattini OP, Massad E. The basic reproductive number for dengue fever in Sao Paulo State, Brazil: 1990–1991 epidemic. Trans R Soc Trop Med Hyg. 1994;88:58–59. doi: 10.1016/0035-9203(94)90498-7. [DOI] [PubMed] [Google Scholar]
- Pan American Health Organization. Dengue and Dengue Hemorrhagic Fever in the Americas: Guidelines for Prevention and Control. Washington DC: PAHO publications; 1994. Historical overview of the Americas; pp. 3–8. [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479–493. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R, Harrison LM, Salas RA, Tovar D, et al. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230:244–251. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- Rothman AL, Kurane I, Ennis FA. Multiple specificities in the murine CD4+ and CD8+ T-cell response to dengue virus infection. J Virol. 1996;70:6540–6546. doi: 10.1128/jvi.70.10.6540-6546.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, et al. Risk factors in dengue shock syndrome: a prospective study in Rayong, Thailand. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- Spielman A, James AA. Transmission of vector-borne disease. In: Warren KS, Mahmoud AAF, editors. Tropical and Geographic Medicine. 2. New York: McGraw-Hill; 1990. pp. 146–159. [Google Scholar]
- Tesh RB. A method for the isolation and identification of dengue viruses, using mosquito cell cultures. Am J Trop Med Hyg. 1979;28:1053–1059. doi: 10.4269/ajtmh.1979.28.1053. [DOI] [PubMed] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, et al. Dengue in the early febrile phase: viremia and antibody responses. J Infect Dis. 1997;176:322–330. doi: 10.1086/514048. [DOI] [PubMed] [Google Scholar]
- Vaughn DW, Green S, Kalayanarooj S, Innis BL, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- Watts DM, Porter K, Putvatana R, Vasquez B, et al. Failure of secondary infection with American genotype dengue 2 virus to cause dengue hemorrhagic fever. Lancet. 1999;354:1431–1434. doi: 10.1016/S0140-6736(99)04015-5. [DOI] [PubMed] [Google Scholar]