Abstract
Protein ubiquitination is an essential post-translational modification (PTM) involved in the regulation of a variety of cellular functions, including transcription and protein degradation. Proteins can be both mono- or poly-ubiquitinated. Poly-ubiquitin chains vary in the manner by which the ubiquitin proteins are linked and their total length. Different poly-ubiquitin structures are thought to specify different fates for the target protein but the correlation between poly-ubiquitin structures and their specific cellular function(s) is not well understood. We have developed a set of specific and quantitative targeted mass spectrometry assays to determine the frequency of different types of inter-ubiquitin linkages in poly-ubiquitin chains relative to the total ubiquitin concentration. We chemically synthesized heavy isotope labeled reference peptides that represent the products generated by tryptic digestion of the known forms of inter-ubiquitin links for the yeast Saccharomyces cerevisiae and human, in addition to all peptides from tryptic digestion of a single ubiquitin molecule for these two species. We used these peptides to develop optimized Selected Reaction Monitoring (SRM) assays for their unambiguous detection in biological samples. We used these assays to profile the frequency of the different types of inter-ubiquitin linkages in a mixture of in vitro assembled human poly-ubiquitin chains and 15 isolated poly-ubiquitinated proteins from S. cerevisiae. We then applied the method to detect toxin induced changes in the poly-ubiquitination profile in complex and enriched protein samples.
Introduction
Ubiquitin (Ub) is a highly conserved protein found throughout the eukaryotic kingdom.1,2 The covalent conjugation of Ub to protein substrates plays an essential role in various cellular processes.3–8 Ubiquitinated proteins can be either mono- or poly-ubiquitinated, or both, depending on their fate and function.7,9–11 Substrate–Ub conjugation occurs via an amide bond between the ε-amino group of a lysine residue from the substrate and the carboxylic group of the C-terminal glycine (G76) of a Ub. Ubiquitin conjugation is catalyzed by ubiquitination enzymes (E1, E2, and E3) in a multi-step process.12–17 Poly-ubiquitination of a substrate happens when multiple Ub proteins are conjugated to a substrate-linked Ub via an inter-ubiquitin linkage. Inter-ubiquitin conjugation is the result of the same chemical reaction that catalyzes the attachment of Ub to the substrate protein and is catalyzed by the same enzymes (E1, E2 and E3). The only difference is that the lysine involved in the amide bond formation is from a Ub instead of a substrate protein.18 There are seven lysine residues in the Ub sequence (K6, K11, K27, K29, K33, K48, and K63) and all are known to participate in the formation of poly-Ub chains. Poly-Ub chains are mostly formed via K48 and K63 and poly-Ub chains formed via other lysines constitute a small portion of all linkages.8 Poly-Ub chains can be formed linearly where only one lysine in each Ub is involved in linkage formation or branched where multiple lysines from a Ub chain are involved in linkage formation. Linkages in the poly-Ub chains can be via the same lysines (homogeneous) in all chains or different lysines (heterogeneous). Even though the relationship between the type of linkages in a poly-Ub chain and their role in determining the modified protein’s fate is poorly understood, there are some hints that K63 is involved in stress response (non-degradative ubiquitination) while all other linkages play a role in protein degradation (degradative ubiquitination).8
The ability to analyze poly-Ub chains and their attachment sites to substrate proteins has significantly advanced in recent years. Large scale proteomic studies have been conducted successfully to identify ubiquitinated substrates and the respective Ub attachment sites.11,9–24 In most of these studies, the focus has been on identifying the ubiquitinated proteins and the site(s) of ubiquitination, while there has been less emphasis on determining the composition and topology of the poly-Ub chains. Determining the mono- or poly-ubiquitination status of the substrate as well as density (# of inter-ubiquitin linkages per unit of poly-Ub) and structure (type of inter-ubiquitin linkages) of the Ub moiety is as important as the identity of the substrate itself. That is because often the structure of the Ub moiety determines the substrate’s fate.8 Mono- and poly-ubiquitinated forms of a substrate can be distinguished by immunoblotting due to the significant contribution of the Ub moiety to the total mass of the protein conjugate. However, the linkages in poly-Ub chains cannot easily be characterized by this approach unless linkage-specific antibodies are used. The first limitation of characterizing poly-Ub chains by immunomethods relates to the development of suitable antibodies which so far have only been generated for K48 and K63 linkages.25 The second limitation relates to the binding mechanism of such antibodies which is more based on structural recognition rather than sequence recognition. This means that linkage specific antibodies recognize the linkages based on the structural features that they induce in poly-Ub chain topography, so the linkage can only be characterized on folded proteins. Even though these antibodies show a high level of affinity (Kd(K48) = 1.2 nM, Kd(K63) = 92 nM) and specificity for their targets, higher amounts of poly-ubiquitinated proteins are often required for detection because most proteins are fully or partially denatured during SDS-PAGE electrophoresis which eliminates structural epitopes. The third limitation arises from the fact that a significant fraction of ubiquitinated substrates exhibit heterogeneous poly-Ub chain linkages.25 The poly-Ub chain heterogeneity and lack of linkage specific antibodies for all linkages make the task of determining linkage frequency by immunoblotting even more complicated. The fourth limitation of linkage specific antibodies is in relative linkage quantification which requires comparing the western blot signals of different linkage specific antibodies.
Selected Reaction Monitoring (SRM), frequently referred to as multiple reaction monitoring (MRM), is a highly sensitive and quantitative mass spectrometric technique which is increasingly being used for proteomic applications. The method uses a series of quadrupole mass analyzers to detect and quantify specific, predetermined peptides in complex mixtures accurately and sensitively. SRM has been used before for the characterization of poly-Ub chain connectivity. Specifically, the Gygi group used 10 heavy isotope-labeled Ub peptides to develop an SRM assay that was used to characterize the ubiquitinated cyclin B1.26,27 However, a complete assay for comprehensive characterization and quantification of Ub and its linkages has not been described. Ubiquitination is a complex PTM with various manifestations and a systematic investigation of this PTM by SRM requires a comprehensive assay that targets all ubiquitin forms in their entirety. An ideal assay should include all peptides derived from ubiquitin to deal with issues such as: (1) variations in Ub peptide concentrations as a result of digestion inconstancies,26,28 (2) the fact that none of the tryptic peptides derived from a poly-Ub chain can be used individually to measure Ub concentration even when the digestion is complete and all peptides exist in equimolar ratios relative to intact Ub, and (3) PTMs of Ub itself. Previous studies have mainly focused on linked peptides derived from human poly-Ub digestion and a few unmodified peptides as mono-Ub representatives while neglecting the PTM-bearing Ub peptides altogether. In addition, previously targeted peptides have been assayed based on single transitions without extensive optimizations for maximum sensitivity.26 Such transitions may be sufficient for detection of peptides extracted from in-gel digestion of intact proteins but would certainly not be specific and sensitive enough for unambiguous detection of Ub peptides in complex mixtures. Previous assays have also not been well-characterized in terms of limit of detection (LOD) and linear dynamic range (LDR) which are necessary for high throughput analysis of substrates varying widely in concentration.
Here we introduce a complete set of SRM assays for the detection and quantification of ubiquitin and all inter-Ub linkages in yeast and human. Collectively, these assays represent a comprehensive, highly sensitive and specific method to probe the connectivity of poly-Ub chains. We show that the method is sensitive enough to measure the frequency of inter-ubiquitin linkages at the enriched protein level and specific enough to detect them in complex mixtures such as whole cell lysates. We used these assays to determine the frequency and density of different inter-ubiquitin linkages in synthetic human poly-Ub chains as well as in fifteen poly-ubiquitinated proteins enriched from yeast lysates to determine the level of degradative and non-degradative ubiquitination for each. We also used the assays to detect changes in the linkage density and frequency in an enriched sample of histone protein Htb2 and whole cell lysate when cells were treated with the DNA modifying agent methyl methanesulfonate (MMS). We demonstrate for the first time that Htb2 is poly-ubiquitinated. We then studied the effect of cadmium poisoning on Ub chain linkage density and frequency in a sample enriched for the transcription activator Met4, or whole cell lysate.
Results and discussion
Building an array of validated SRM assays to monitor poly-Ub chains
First we generated an array of definitive and quantitative SRM assays to unambiguously detect and quantify the known inter-ubiquitin linkages in complex samples. To accomplish this task we chemically synthesized a set of reference peptides that correspond to the peptides expected after tryptic digestion of yeast and human inter-linked poly-Ub chains. Yeast and human Ubs differ by three amino acids: [S19P], [D24E], and [S28A]. These peptides were used to determine the optimal set of SRM transitions for each peptide. Specifically, the reference peptide list contained all linear peptides from a tryptic digestion of Ub and all linked peptides which carried a lysine residue from one Ub entity covalently attached to a diglycine moiety remnant of another. Phosphorylated versions of linear and linked peptides were also generated to cover known and potential sites of phosphorylation in Ub.22 A full list of the generated reference peptides is shown in Table 1. All targeted peptides were successfully synthesized with a heavy stable isotope labeled amino acid incorporated at the C-terminus (lysine or arginine), except for the tryptic peptide spanning residues 12 to 27 of yeast Ub which is unstable. To cover this region of yeast Ub we synthesized instead a commonly observed miscleaved tryptic peptide spanning residues 12 to 29. All synthetic peptides were solubilized and combined to generate a master mix which was used to generate a set of optimized SRM transitions. For each peptide the five most intense fragment ions were selected using SRM driven MS/MS scans (except for the peptide IQDK which only consists of four amino acids), whereby the collision energies were chosen to maximize the signal intensity for each transition (for details see Materials and Methods section). The optimized transitions are listed in Table 1. We will refer to this set of assays as the Ub-SRM assay.
Table 1.
The complete list of heavy isotope labeled peptides that were synthesized to detect and quantify yeast and human poly-Ub chains and the linkages they contain. Linear, branched (K-GG) and phosphorylated peptides were synthesized to cover Ub and poly-Ub chain as well as their post translationally modified forms. The five most intense transitions (Q1/Q3) for each peptide (except for peptide IQDK) in heavy and light form, respectively, are listed for a complete assay. The bold letters in the sequence of peptides (# 4–11) show sequence variability between human and yeast ubiquitin. Signal intensities of the specific transitions were normalized to the most intense signal set to 100%.
| # | Sequence | Location | Modification | Organism | Q1(H)_Q1(L) | Q3(H)_Q3(L) | Relative intensity (%) |
CE |
|---|---|---|---|---|---|---|---|---|
| 1 | MQIFVK | 1_6 | — | Yeast | 387.2_383.2 | 260.1_260.1 | 100 | 23 |
| Human | 387.2_383.2 | 514.3_506.3 | 91 | 23 | ||||
| 387.2_383.2 | 642.4_634.4 | 49 | 23 | |||||
| 387.2_383.2 | 254.2_246.2 | 45 | 30 | |||||
| 387.2_383.2 | 520.3_520.3 | 4 | 23 | |||||
| 2 | MQIFV[K_GG]TLTGK | 1_11 | Branched | Yeast | 463.3_460.6 | 313.2_305.2 | 100 | 27 |
| Human | 463.3_460.6 | 527.3_519.3 | 72 | 24 | ||||
| 463.3_460.6 | 385.2_381.2 | 56 | 22 | |||||
| 463.3_460.6 | 373.2_373.2 | 46 | 27 | |||||
| 463.3_460.6 | 564.8_560.8 | 38 | 22 | |||||
| 3 | TLTGK | 7_11 | — | Yeast | 264.2_260.2 | 313.2_305.2 | 100 | 17 |
| Human | 264.2_260.2 | 426.3_418.3 | 12 | 17 | ||||
| 176.4_173.8 | 212.1_204.1 | 4 | 17 | |||||
| 264.2_260.2 | 373.2_373.2 | 1 | 17 | |||||
| 264.2_260.2 | 316.2316.2 | 1 | 17 | |||||
| 4 | TLTG[K_GG]TITLEVESSDTIDNVK | 7_27 | Branched | Yeast | 796.1_793.4 | 986.5_978.5 | 100 | 40 |
| 796.1_793.4 | 1115.5_1107.5 | 84 | 40 | |||||
| 796.1_793.4 | 899.5_891.4 | 61 | 40 | |||||
| 796.1_793.4 | 1214.6_1206.6 | 52 | 39 | |||||
| 796.1_793.4 | 697.4_689.4 | 52 | 42 | |||||
| 5 | TLTG[K_GG]TITLEVEPSDTIENVK | 7_27 | Branched | Human | 804.1_801.4 | 1010.5_1002.5 | 100 | 38 |
| 804.1_801.4 | 1139.6_1131.6 | 18 | 38 | |||||
| 804.1 801.4 | 1098.5_1094.6 | 13 | 30 | |||||
| 804.1_801.4 | 913.5_905.5 | 12 | 46 | |||||
| 804.1_801.4 | 1172.6_1172.7 | 12 | 38 | |||||
| 6 | TITLEVESSDTIDNVKSK | 12_29 | — | Yeast | 663_660.3 | 887_882.9 | 100 | 27 |
| 663_660.3 | 779.9_775.9 | 17 | 27 | |||||
| 663_660.3 | 836.4_832.4 | 16 | 27 | |||||
| 663_660.3 | 1330.7_1322.6 | 12 | 34 | |||||
| 663_660.3 | 1201.6_1193.6 | 11 | 34 | |||||
| 7 | TITLEVEPSDTIENVK | 12_27 | — | Human | 599.3_596.6 | 505.8_501.8 | 100 | 27 |
| 898.5_894.5 | 1010.5_1002.5 | 84 | 41 | |||||
| 898.5_894.5 | 1139.6_1131.6 | 39 | 41 | |||||
| 898.5_894.5 | 1238.6_1230.6 | 27 | 41 | |||||
| 599.3_596.6 | 497.3_489.3 | 23 | 27 | |||||
| 8 | TITLEVESSDTIDNV[K_GG]SK | 12_29 | Branched | Yeast | 701_698.4 | 944_940 | 100 | 29 |
| 701_698.4 | 812.4_804.41 | 28 | 36 | |||||
| 701_698.4 | 315.7_1307.6 | 21 | 36 | |||||
| 701_698.4 | 893.5_889.4 | 14 | 29 | |||||
| 701_698.4 | 836.9_832.9 | 8 | 29 | |||||
| 9 | TITLEVEPSDTIENV[K_GG]AK | 12_29 | Branched | Human | 703.7_701 | 948_944 | 100 | 28 |
| 703.7_701 | 1323.7_1315.7 | 59 | 32 | |||||
| 703.7_701 | 810.5_802.4 | 51 | 36 | |||||
| 703.7_701 | 840.9_836.9 | 31 | 32 | |||||
| 703.7_701 | 776.4_772.4 | 20 | 32 | |||||
| 10 | S[K_GG]IQDK | 28_33 | Branched | Yeast | 420.7_416.7 | 511.3_503.3 | 100 | 25 |
| 280.8_278.2 | 398.2_390.2 | 65 | 16 | |||||
| 280.8_278.2 | 330.2_330.2 | 59 | 12 | |||||
| 280.8_278.2 | 511.3_503.3 | 51 | 16 | |||||
| 420.7_416.7 | 270.2_262.1 | 33 | 25 | |||||
| 11 | A[K_GG]IQDK | 28_33 | Branched | Human | 412.7_408.7 | 511.3_503.3 | 100 | 28 |
| 275.5_272.8 | 398.2_390.2 | 53 | 15 | |||||
| 412.8_408.7 | 270.2_262.1 | 39 | 32 | |||||
| 275.5_272.8 | 511.3_503.3 | 34 | 15 | |||||
| 275.5_272.8 | 314.2_314.2 | 28 | 15 | |||||
| 12 | IQDK | 30_33 | — | Yeast | 256.2_252.1 | 398.2_390.2 | 100 | 15 |
| Human | 171.1_168.4 | 270.2_262.1 | 13 | 13 | ||||
| 256.2_252.1 | 357.2_357.2 | 4 | 16 | |||||
| 171.1_168.4 | 242.2_242.2 | 3 | 12 | |||||
| 13 | IQD[K_GG]EGIPPDQQR | 30_42 | Branched | Yeast | 549.9_546.6 | 653.3_643.3 | 100 | 36 |
| Human | 549.9_546.6 | 750.4_740.4 | 100 | 29 | ||||
| 549.9_546.6 | 441.2_431.2 | 78 | 43 | |||||
| 549.9_546.6 | 498.3_493.3 | 19 | 22 | |||||
| 549.9_546.6 | 785.4_785.4 | 13 | 29 | |||||
| 14 | EGIPPDQQR | 34_43 | — | Yeast | 525.3_520.3 | 750.4_740.4 | 100 | 29 |
| Human | 525.3_520.3 | 653.3_643.3 | 52 | 30 | ||||
| 350.5_347.2 | 375.7_370.7 | 37 | 17 | |||||
| 525.3_520.3 | 441.2_431.2 | 30 | 31 | |||||
| 525.3_520.3 | 556.3_546.3 | 7 | 31 | |||||
| 15 | LIFAGK | 43_48 | — | Yeast | 328.7_324.7 | 430.3_422.2 | 100 | 20 |
| Human | 328.7_324.7 | 543.3_535.3 | 29 | 20 | ||||
| 219.5_216.8 | 283.2_275.2 | 12 | 12 | |||||
| 328.7_324.7 | 272.2_268.2 | 3 | 20 | |||||
| 328.7_324.7 | 374.2_374.2 | 2 | 20 | |||||
| 16 | LIFAG[K_GG]QLEDGR | 43_54 | Branched | Yeast | 490.9_487.6 | 622.8_617.8 | 100 | 19 |
| Human | 490.9_487.6 | 357.2_347.2 | 26 | 33 | ||||
| 490.9_487.6 | 599.3_589.3 | 24 | 33 | |||||
| 490.9_487.6 | 549.3_544.3 | 19 | 26 | |||||
| 490.9_487.6 | 727.4_717.4 | 16 | 33 | |||||
| 17 | QLEDGR | 49_54 | — | Yeast | 364.2_359.2 | 486.2_476.2 | 100 | 25 |
| Human | 364.2_359.2 | 486.2_486.2 | 100 | 25 | ||||
| 364.2_359.2 | 599.3_589.3 | 13 | 25 | |||||
| 364.2_359.2 | 300.2_295.2 | 5 | 21 | |||||
| 364.2_359.2 | 543.2_543.2 | 2 | 26 | |||||
| 18 | TLSDYNIQK | 55_63 | — | Yeast | 545.3_541.3 | 875.4_867.4 | 100 | 29 |
| Human | 545.3_541.3 | 673.4_665.4 | 20 | 31 | ||||
| 545.3_541.3 | 396.3_388.3 | 12 | 32 | |||||
| 545.3_541.3 | 788.4_780.4 | 9 | 30 | |||||
| 545.3_541.3 | 494.8_490.8 | 5 | 30 | |||||
| 19 | TL[S_Phos]DYNIQK | 55_63 | Phosphorylated | Yeast | 585.3_581.3 | 955.4_947.4 | 100 | 24 |
| Human | 585.3_581.3 | 510.3_502.3 | 30 | 31 | ||||
| 585.3_581.3 | 673.4_665.4 | 28 | 38 | |||||
| 585.3_581.3 | 534.7_530.7 | 14 | 31 | |||||
| 585.3_581.3 | 788.4_780.4 | 13 | 31 | |||||
| 20 | TLSDYNIQ[K_GG]ESTLHLVLR | 55_72 | Branched | Yeast | 752.1_748.7 | 1020.5_1015.5 | 100 | 32 |
| Human | 752.1_748.7 | 1077.6_1067.6 | 24 | 46 | ||||
| 752.1_748.7 | 647.4_637.4 | 21 | 46 | |||||
| 752.1_748.7 | 1319.81309.8 | 13 | 46 | |||||
| 752.1_748.7 | 948.6_938.6 | 12 | 46 | |||||
| 21 | TLSDYNIQ[K_GG]E[S_Phos]TLHLVLR | 55_72 | Branched | Yeast | 778.7_775.4 | 1060.51055.5 | 100 | 37 |
| Phosphorylated | Human | 778.7_775.4 | 647.4_637.4 | 45 | 43 | |||
| 778.7_775.4 | 10171012 | 33 | 33 | |||||
| 778.7_775.4 | 959.5_954.5 | 31 | 37 | |||||
| 778.7_775.4 | 700.4_695.4 | 9 | 37 | |||||
| 22 | TL[S_Phos]DYNIQ[K_GG]ESTLHLVLR | 55_72 | Branched | Yeast | 778.7_775.4 | 1060.51055.5 | 100 | 37 |
| Phosphorylated | Human | 778.7_775.4 | 1077.6_1067.6 | 53 | 43 | |||
| 778.7_775.4 | 647.4_637.4 | 45 | 43 | |||||
| 778.7_775.4 | 948.6_938.6 | 44 | 43 | |||||
| 778.7_775.4 | 838_833 | 36 | 38 | |||||
| 23 | ESTLHLVLR | 64_72 | — | Yeast | 359.9_356.5 | 431.2_431.2 | 100 | 17 |
| Human | 359.9_356.5 | 298.2_288.2 | 78 | 18 | ||||
| 359.9_356.5 | 431.3_426.3 | 77 | 17 | |||||
| 359.9_356.5 | 510.4_500.4 | 60 | 23 | |||||
| 359.9_356.5 | 397.3_387.3 | 43 | 23 | |||||
| 24 | E[S_Phos]TLHLVLR | 64_72 | Phosphorylated | Yeast | 579.3_574.3 | 510.4_500.4 | 100 | 38 |
| Human | 386.5_383.2 | 510.4_500.4 | 83 | 27 | ||||
| 579.3_574.3 | 647.4_637.4 | 76 | 38 | |||||
| 386.5_383.2 | 324.2_319.2 | 51 | 27 | |||||
| 386.5_383.2 | 324.6_324.6 | 50 | 27 |
Characterizing the Ub-SRM assays
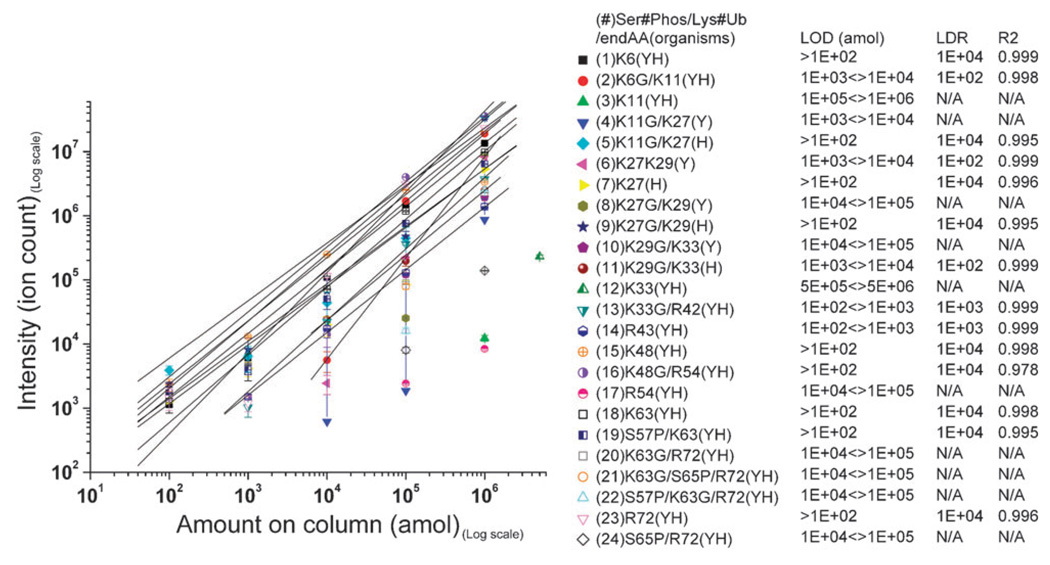
We next determined the LOD and LDR for each Ub-SRM assay. To determine the limit of detection we assayed a dilution series of an equimolar mixture of all 24 peptides prepared in such a manner that 2 µL injections to the chromatography column contained 100 amol, 1 fmol, 10 fmol, 100 fmol, and 1 pmol of each peptide, respectively. Each dilution was analyzed in triplicate by SRM in non-scheduled mode with a dwell time of 15 milliseconds. Quadrupole resolutions were set at 1 Da and all peptides were separated on a 60 minute gradient. The results of these measurements are shown in Fig. 1. Reported values are the means of the triplicate measurements and the errors are the standard error of the mean. The data indicate that Ub was detected at the 100 amol level with 6 peptides, at the 1 fmol level with 8 peptides, at the 10 fmol level with 11 peptides, and at the 100 fmol level with 18 peptides. Two very short peptides (peptides #3 and 12) were only detectable at the pmol level. K6 and K11 linkages were detectable at the 10 fmol level, K27 and K29 linkages were detectable at the 100 fmol level, K33 linkage was detectable at the 1 fmol level, K48 linkage was detectable at the 100 amol level, and K63 linkage was detectable at the 100 fmol level. Ub phosphorylation at serine 57 and 65 was detectable at the 100 amol and 100 fmol levels, respectively. K63 linkages with phosphorylation at serine 57 and 65 were detectable at the 100 fmol level. The signals for all peptides were linear with high correlations from the lowest detectable signals up to 1 pmol where the chromatography column capacity was reached. The precision of these measurements increased as a function of amount loaded into the column. At the 100 amol level the average relative standard deviation for all detected peptides was 34%, at the 1 fmol level this value dropped to 30%, at the 10 fmol level it was 21% and at the 100 fmol level it dropped to mere 8%. At the 1 pmol level this value was only 5%. All these values are reported assuming that the synthetic peptides were fully solubilized. We used established guidelines to minimize the effect of incomplete solubilization on quantitative measurements.29 These results indicate that by the SRM method, Ub can be detected easily at low amol levels and that inter-ubiquitin linkages (with the exception of K48 linkages) were detectable at higher concentrations. The linear dynamic range for all peptides was from the limit of detection up to at least 1 pmol (4 orders of magnitude for most peptides) which guarantees accurate quantification via isotope dilution analysis over a wide range of concentrations.
Fig. 1.
The signal (ion counts) recorded for all 24 synthetic peptides (see Table 1 for peptide reference) as a function of the amounts loaded on the chromatography column. 100 amol, 1 fmol, 10 fmol, 100 fmol and 1 pmol of each peptide was loaded onto the column for determination of the limit of detection (LOD) and the linear dynamic range (LDR). LDR is not reported for the peptides with less than three data points. R2 is used as a measure of linearity.
Assessment of the Ub-SRM assay
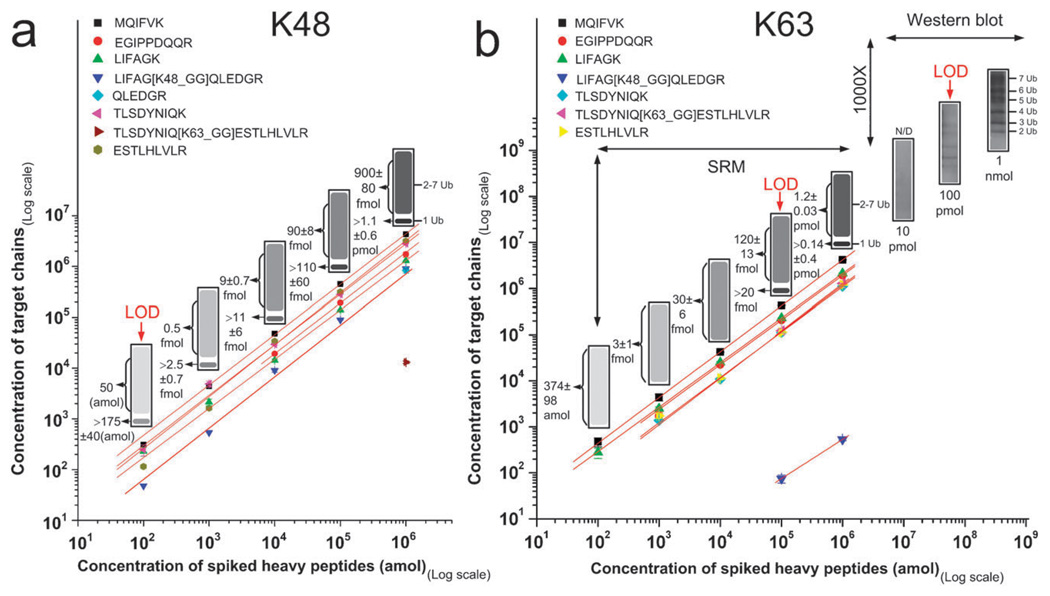
Next we used in vitro assembled human poly-Ub chain mixtures consisting entirely of pure K48 or K63 linkages to determine the quantification accuracy of the SRM assays at various concentrations.30–33 These mixtures contain mono- and poly-Ub chains of 2 to 7 inter-linked Ubs. The K48-linked poly-Ub mixture was first digested by trypsin then used to prepare a dilution series spanning four orders of magnitude from 100 amol to 1 pmol. These samples were spiked with equimolar mixtures of human heavy isotope labeled peptides with the same concentration as the digested samples. These mixtures were analyzed in triplicate via the Ub-SRM assay to determine the concentration of poly-Ub chains in the mixture. Even though the concentration of mono-Ub in the mixture cannot directly be determined from the SRM data, a minimum concentration for mono-Ub can be calculated based on the concentration of linear and linked peptides (see Methods for more details). The results, shown in Fig. 2a, demonstrate that K48 linkage can be detected at low amol level. Quantification of linkage specific peptides (peptides #16 and 20) allowed us to measure the concentration of poly-Ub chains in each sample. We also measured the linear peptides that are expected from both mono- and poly-Ub chains (peptides #1, 14, 15, 17, 18, and 23) in order to calculate a minimum concentration for mono-Ub. The relative error in calculating the minimum concentration of mono-Ub using multiple linear peptides on average was 25%. The average relative error for K48 links was 8%. Quantitative measurements at increasing concentrations were linear. Trace amounts of K63 linkage were detected at the pmol level in K48 chains digest (1.4% of all links were K63 links).
Fig. 2.
(a) The quantification of Ub peptides derived from tryptic digestion of human K48 multi-Ub chains. Eight peptides were detected including K48 linked peptides. K63 was detected in trace amounts. Mono and poly-Ub entities were detected at low amol levels. These quantifications were linear over 4 orders of magnitude. Quantification data are also shown using an SDS-gel simulation for better visualization. The lower band in the simulated SDS-gel represents mono-Ub and the smear on top represents unresolved poly-Ub chains; (b) same as (a), except that human K63 multi-Ub chains were analyzed. The SRM data were compared with linkage specific immunodetection of K63 linkages. K63 linkage was detectable at the 100 fmol level which is a 1000 fold lower than the limit of detection by immunoblotting. Ub was detected at the 100 amol level as shown in simulated SDS-gel images on top of SRM peptide measurements.
We next analyzed K63-linked poly-Ub chains. K63-linked poly-Ub chains were digested and analyzed via SRM in the exact same manner as the K48 chains. The results are shown in Fig. 2b. The detection limit for K63 linkages by SRM in this experiment was 100 fmol. The relative error in calculating the concentration of total Ub and the minimum concentration of mono-Ub using linear and linked peptides was 26% and 29%, respectively. We also found trace amounts of K48 linkages in K63 specific chains (0.04% of all links were K48 links). The average relative error for K63 and K48 links was 6.5% and 13%, respectively. Quantitative measurements at increasing concentrations were linear. For further analysis we utilized the widely used commercially available K63 linkage specific antibody, and compared the LOD between SRM and immunoblotting for K63 poly-Ub chains. Increasing amounts of K63-linked poly-Ub chains (10 pmol, 100 pmol, and 1 nmol) were separated via SDS-PAGE and immunoblotted using the K63-linkage-specific Ub antibody (BML-PW0600-0025, Enzo Life Sciences, PA, USA) to establish the LOD. We detected K63 linked poly-Ub chains of up to 7 chains and the LOD was found to be 100 pmol by this method. By comparing the SRM and immunoblotting methods we determined that SRM was at least a 1000 fold more sensitive for the detection of K63 linkage. While SRM can detect mono-Ub in the mixture, information that is not attainable by linkage specific antibodies, it cannot resolve the poly-Ub chains of various sizes which is possible by immunoblotting. Even though K48 linkage specific antibodies have been developed, they are not commercially available and there are no linkage specific antibodies for other linkages. Therefore, SRM provides the only available assay for the detection of such linkages at the sub-picomole level.
SRM analysis of enriched poly-ubiquitinated, TAP-tagged yeast proteins
Next we utilized the set of Ub-SRM assays to establish quantitative profiles of the inter-ubiquitin linkages in the poly-Ub chains of specific Tandem Affinity Purification (TAP) tagged proteins enriched from yeast lysate. The purpose of this experiment was to test the specificity and detection limit of the Ub-SRM assay when used to characterize the Ub substructure of individual, enriched proteins of different concentrations (copy per cell), diverse poly-Ub chain sizes, and various enrichment efficiencies. We selected fifteen Ub substrates previously validated to be poly-ubiquitinated with high confidence under normal cell growth conditions.34 TAP-tagged target proteins were enriched from the lyaste of cells grown to mid-log phase via affinity purification. The enrichment efficiency for target proteins was evaluated using western blots (ESI†, Fig. S1).
Digested and desalted poly-ubiquitinated protein samples were spiked with a mixture of all synthetic peptides. The concentration of each peptide was adjusted so that all five heavy transitions could be easily detected when spiked into poly-ubiquitinated protein digests. The SRM transitions were monitored for detection and quantification of all yeast Ub peptides from the digests. All experiments were done in triplicate and the mean and standard error of the mean were reported for each peptide. The results are shown in Fig. 3.
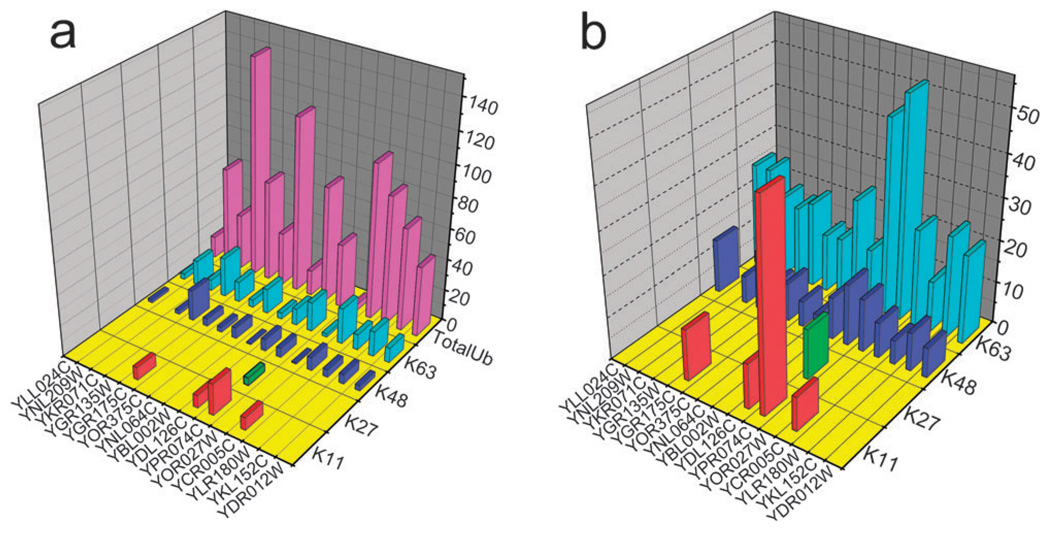
Fig. 3.
(a) The absolute concentration of total ubiquitin and inter-Ub links of affinity purified proteins. Four of 7 inter-ubiquitin links tested were detected. Total Ub concentration was calculated by averaging the summed concentrations of linked peptides and their preceding and following linear peptides. All concentrations are reported in fmol µg−1 of digest; (b) this plot shows the relative concentrations of inter-Ub links to the total Ub in enriched protein samples. In the case of YPR074C that the summation of linkage/total Ub% reaches 100% we can conclude that mono-ubiquitinated form of this protein was not present in the enriched sample.
Most of the linear peptides from Ub (peptides #1, 3, 14, 15, 17, 18, and 23) were detected and quantified for all proteins. The often undetected peptide #6 is a miscleaved tryptic peptide and peptide #12 is too hydrophilic to be retained on the column. Two linked peptides (peptides #16 and 20) containing lysines 48 and 63, respectively, were consistently detected from the digests. Branched peptides #4 and 8 which represent K11 and K27 linkages, respectively, were also detected for several proteins. Data for a complete SRM assay (TAP-tagged protein YPR074C) are shown in ESI†, Fig. S2. The quantification is based on the average ratio between the area under the curves for transitions from heavy peptides with known concentrations and their light counterparts. In summary: (1) we found all target proteins to be poly-ubiquitinated, (2) the presence of mono-ubiquitinated target proteins was confirmed for thirteen out of fifteen targets, (3) the fraction of ubiquitin that is involved in poly-ubiquitination varied amongst the substrates.
The concentration of total Ub cannot be determined using the linear peptides alone because none of the Ub peptides are independent of Ub linkages. In other words, all Ub peptides are either preceded by a lysine or have one at their C-terminus so their concentration varies if one of these lysines participates in linkage formation. For example, the concentrations of peptides #15 and 17 depend on the concentration of K48 linkage and the concentrations of peptides #18 and 23 depend on the concentration of K63 linkage. The concentration of linear peptides preceding and following a linkage is expected to be the same which is the case for peptides #15 and 17 surrounding K48 linkage (26% variations) and peptides #18 and 23 surrounding K63 linkage (8.5%). This concentration dependency among Ub peptides may explain variation in measured concentrations. Another explanation for variation in peptide concentrations is the possibility of multiple linkages on the same chain. Peptides carrying adjacent linkages cannot be detected by our assay and cause variation in concentration of preceding and following peptides. We are aware that there might be other ubiquitinated proteins in these samples but we expect their contribution to the total Ub content to be minimal, due to the effective TAP enrichment. These data also suggest that there are considerable differences among the target substrates in terms of ubiquitination quantity and linkages.
Detecting local and global quantitative changes in ubiquitination induced by DNA damage
Another important aspect of Ub analysis is the ability to quantify changes in the ubiquitination level and in the distribution of poly-Ub inter-links of an enriched protein or the entire proteome, as a result of a biological stimulus. Therefore, we next attempted to quantify changes in the ubiquitination level and inter-link profiles of affinity enriched histone protein Htb2 (YBL002W) as well as the whole cell lysate when yeast cells were treated with MMS, a chemical known to induce DNA damage.35 It is known that Htb2 ubiquitination is critical for transcription and the cell cycle response, but the dynamics of Htb2 ubiquitination are not well understood.36,37 We used the Ub-SRM assay to answer the following questions regarding Htb2 ubiquitination: (1) does a certain level of ubiquitinated Htb2 exist in the cell at all times and if it does what is its Ub status (mono- or poly, link types, etc.) and (2) if ubiquitinated Htb2 exists in normal conditions, are there changes in its Ub status as a result of DNA damage.
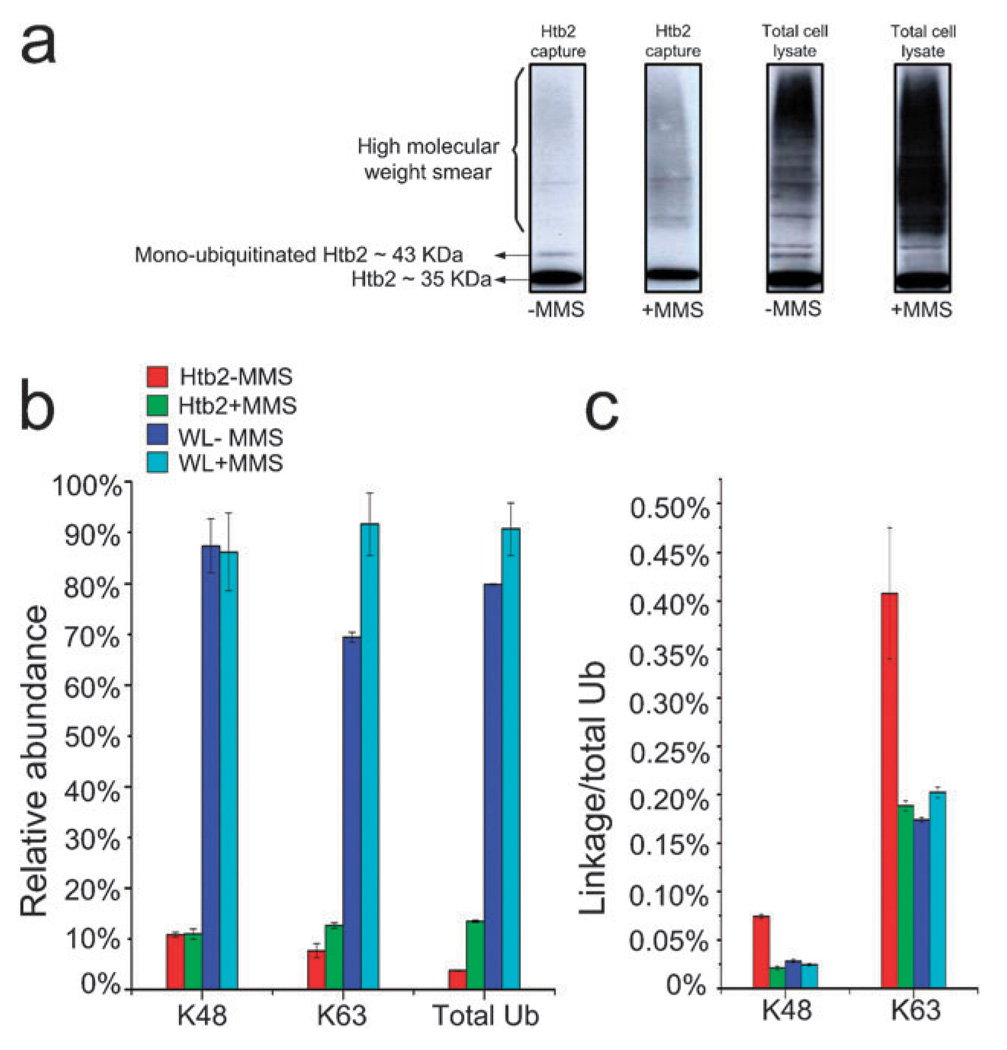
To examine regulation of ubiquitination in the Htb2 enriched sample, the Htb2 TAP-tagged strain was treated with 0.1% MMS, known to induce DNA damage. Mock treated cells were used as a control. Htb2 protein was enriched using the TAP tag from both the control and treated lysates. These samples and corresponding total cell lysates were separated on SDS-PAGE, immunoblotted, and probed with rabbit polyclonal antibody against Ub protein conjugates (Fig. 4a). We then digested and analyzed equal amounts of each capture and the corresponding total cell lysate by SRM in the same manner as described in the previous section. The results are summarized in Fig. 4b and c. The western blot data indicate that mono-ubiquitinated Htb2 existed under normal conditions and was significantly reduced when cells were treated with MMS. The unmodified form of Htb2 was also detected by western blot due to the protein A tag cross-reactivity with the blotting antibody. The Ub western blots of the control and treated total lysates indicate a general increase in ubiquitination after MMS treatment (Fig. 4a).
Fig. 4.
(a) Anti-Ub protein conjugate western blots for Htb2 protein (tagged with protein A which causes cross-reactivity with the Anti-Ub antibody) enriched from control and 0.1% MMS treated cells and their corresponding total cell lysates. The Htb2 blots suggest that the mono-ubiquitinated form of Htb2 disappears after the MMS treatment, while a high molecular weight smear appears after the MMS treatment. The total cell lysate immunoblots show a general increase in ubiquitination as a result of MMS treatment; (b) shows the average abundance of K48, K63 links and total Ub normalized to the highest value in each category. The data indicate that overall ubiquitination increases in both enriched Htb2 and total lysate after MMS treatment, whereas poly-ubiquitination via lysine 48 remains the same. An increase in the concentration of the K63 links suggests that additional poly-Ub conjugates in the Htb2 enriched sample are formed via lysine 63 as a result of MMS treatment; (c) the K48 and K63/total Ub ratio (representing poly-ubiquitination via K63) both show a decrease as a result of MMS treatment.
SRM analysis of the same samples provided detailed quantitative information regarding the Ub status of Htb2 and the whole cell lysate before and after MMS treatment (Fig. 4b and c). By monitoring for K48 and K63 linkages we observed poly-Ub forms of Htb2 in the untreated culture, which had not been detected before this study. To validate this finding we digested and extracted the peptides in the high molecular weight smear (MW > 50 kDa) of a Coomassiestained SDS-PAGE gel for mass spectrometric analysis. We detected Htb2 in the high molecular weight smear confirming that Htb2 is poly-ubiquitinated (data not shown). We are examining this observation further. The K48 links did not appear to change as a result of MMS treatment in the Htb2 enriched sample, while K63 links showed a 1.6 fold increase in MMS treated samples. The total Ub content of the enriched Htb2 showed a 3.6 fold increase following MMS treatment (Fig. 4b). We also investigated the linkage to Ub ratio for each sample (Fig. 4c). These data show that the linkage/total Ub ratio for both K48 and K63 links in enriched Htb2 samples decreased as a result of MMS treatment. There are two explanations for this observation. One explanation is that the majority of MMS-induced ubiquitination of Htb2 is mono-ubiquitination. The other explanation is that MMS treatment induces higher density poly-ubiquitination with Ub chains linked via adjacent lysines.19 These links are not detectable by Ub-SRM assay but they contribute to the total Ub content. The first explanation is in direct contrast with our western blot that suggests a decrease in mono-ubiquitinated form of Htb2. But the second explanation is more compatible with our western blot (Fig. 4a) that shows a smear appearing in the higher molecular weight regions of the gel after MMS treatment.
The Ub-SRM assay was also used to detect and quantify Ub and Ub links in control and treated whole cell lysates without any fractionations or enrichment. We were able to detect linear ubiquitin peptides and branched ubiquitin peptides for K48 and K63 links directly from the whole cell lysate but we were not able to observe the other links. K6, K11, K27, K29, and K33 are rare and have not ever been detected without prior enrichment. As before we found the level of K48 links to remain unchanged by MMS treatment, and the K63 links showed a slight increase (1.3 fold) following MMS treatment. The total Ub content of the cells also showed a slight increase (Fig. 4b). The K48 linkage/ total Ub ratio showed a slight decrease while the ratio for K63 links showed an increase (Fig. 4c). These results indicate that formation of K63 linked Ub chains is the prominent detectable change in the Ub content of the cell as a result of MMS treatment, which is compatible with other findings connecting K63 links to DNA damage response.33
Detecting local and global quantitative changes in ubiquitination induced by heavy metal toxicity
The Met4 (YNL103W) transcription activator regulates the MET gene network responsible for the biosynthesis of the sulfur-containing amino acids. In yeast, Cd2+ activates the MET gene network in a Met4-dependent fashion by preventing both degradative and non-degradative ubiquitination of Met4.38,39 To further investigate these previously reported findings at the molecular level, we treated yeast cells containing a TAP-tagged version of Met4 with 0.1 mM Cd2+ for 30 minutes (mock treated cells were used as a control). The Met4 protein was then enriched using the TAP tag. Control and treated total cell lysates and Met4 captures were separated on SDS-PAGE, immunoblotted, and probed with rabbit polyclonal antibody against Ub protein conjugates. We digested and analyzed equal amounts of each capture and the corresponding total lysate by SRM in the same manner as explained in the previous sections (Fig. 5).
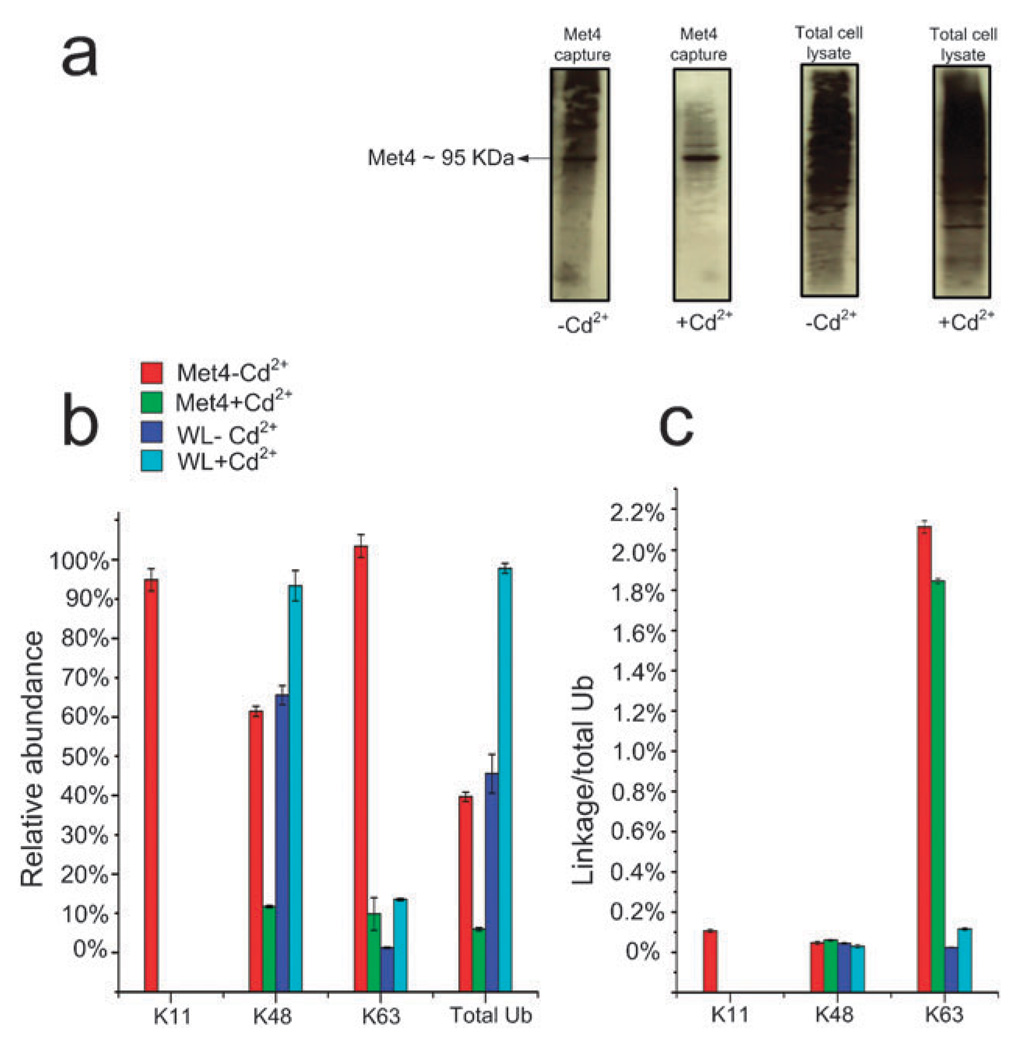
Fig. 5.
(a) The anti-Ub protein conjugate western blots for the Met4 enriched samples from control and 0.1 mM Cd2+ treated cells and their corresponding total cell lysates are shown. The western blots of the Met4 captures show a decrease in overall ubiquitination of Met4 as a result of Cd2+ treatment. The western blots of the whole cell lysates on the other hand do not show a significant change in ubiquitination as a result of Cd2+ treatment; (b) the SRM assay shows that all ubiquitinated forms of Met4 in enriched samples including poly-ubiquitinated form via lysine 11, 48, and 63 decrease as a result of Cd2+ treatment while in the total cell lysates of the Cd2+ treated cells this trend is the opposite; (c) shows the link/total Ub ratio for all detected inter-Ub links. K11/total Ub ratio (representing poly-ubiquitination via K11) is only detectable in Met4 enriched sample before Cd2+ treatment. K48/total Ub ratio (representing poly-ubiquitination via K48) remains unchanged for the Met4 enriched samples and whole cell lysate after the Cd2+ treatment. In contrast, K63/total Ub ratio (representing poly-ubiquitination via K63) shows a decrease in Met4 enriched sample after the treatment and an increase in the whole cell lysate.
The western blots of the captured Met4 samples, using the rabbit polyclonal antibody against Ub protein conjugates before and after the Cd2+ treatment, are not significantly different but show a general decrease in the high molecular weight smears as a result of the treatment. There was no significant change in Ub profile for the whole cell lysates when visualized by western blot (Fig. 5a). We then applied the Ub-SRM assay to investigate changes in Ub profile of the Met4 protein in detail. For Met4 we were able to detect K11, K48 and K63 links which were all significantly reduced as a result of Cd2+ treatment. Total Ub content of the enriched samples was also reduced after the Cd2+ treatment. All these results are consistent with previous findings that indicate that the exposure of yeast cells to heavy metals leads to activation of Met4 by a complete loss of ubiquitinated Met4 species. At the proteome level this trend was reversed. K48 and K63 links as well as total Ub showed an increase as a result of Cd2+ treatment. Heavy metal toxicity leads to oxidative stress which causes both protein and DNA damage resulting in an increase in both K48 and K63 linked Ub chains (Fig. 5b). We also investigated linkage/Ub ratio for all samples. Only a very small fraction of Met4 Ub was linked via K11 (0.1%) which was not detected after treatment. The K48/total Ub ratio (representing poly-ubiquitination via K48) did not show a significant change before or after treatment with Cd2+ in the Met4-enriched or whole cell lysate samples. The K63/total Ub ratio (representing poly-ubiquitination via K63) showed a slight decrease in the Met4-enriched sample after the treatment and a slight increase after the treatment in the whole cell lysate. These data indicate that Cd2+ exposure does not significantly affect the link density in poly-Ub chains in Met4 or at the whole cell lysate level. However, Cd2+ treatment does induce a significant rearrangement within the poly-Ub chains.
Experimental procedures
Materials
Heavy isotope labeled reference peptides (see Table 1 for a complete list) were synthesized by Sigma-Aldrich USA (St. Louis, MO, USA). Multi-Ub chains (Ub2–7, K48-linked product # UW8860 and Ub2–7, K63-linked product # UW9570) and K63-linkage-specific Ub Antibody were purchased from BIOMOL (Enzo Life Sciences, PA, USA). Dynabeads® M-280 tosylactivated were purchased from Invitrogen (Carlsbad, Ca, USA). Rabbit affinity purified antibody to mouse IgG (whole molecule) was purchased from MP Biomedicals (Irvine, CA, USA). Yeast TAP-tagged strain (endogenous promoter) was purchased from Thermo Scientific (Huntsville, AL, USA). Trifluoroacetic acid was purchased from Pierce Co. (Rockford, IL, USA). HPLC-grade acetonitrile (ACN) and HPLC grade water with 0.1% formic acid were purchased from Mallinkrodt (St. Louis, MO, USA). C18 PepMap 100 reversed-phase C18 columns (0.075 × 150 mm) were purchased from Dionex, Inc. (Sunnyvale, CA, USA). Mass Spectrometry Grade Trypsin Gold was purchased from Promega (Madison, WI, USA). Mass spectra were acquired using a Tempo 2D-NanoLC-4000QTrap (Applied Biosystems, CA, USA).
Methods
Yeast growth, harvest and lysis
Selected strains of Saccharomyces cerevisiae (see ESI†, Table S1 for a complete list) were transferred from a TAP-tagged library (with protein A as the tag) to YPD plates which were incubated at 30 °C until yeast colonies were detected. A single colony was then grown overnight (16–20 h) in 5 mL of media at 30 °C with agitation. The cultured cells were used to seed 50 mL of media to a starting OD600 of 0.3 and the cells were allowed to grow for 4 h under the conditions same as the seeding culture. The entire 50 mL culture was transferred to 2 L cultures, and the strain grown to a final OD600 of 1.0–1.5 before harvesting. Cells were pelleted by centrifugation for 5 min at 4000 rpm and washed twice with cold PBS, pH 7.4. The cells were flash frozen in liquid nitrogen and stored at −80 °C. Frozen cells were then mechanically disrupted by grinding using a Retsch PM100 Planetary Ball Mill (Retsch, Newtown, PA).40
Yeast cells treatment
Yeast cells were grown in YPD to an OD600 = 0.5 and were exposed to 0.1 mM cadmium for 30 min. Alternatively, yeast cells were treated with 0.1% MMS in similar fashion for 1 h. Mock treated cells were used as controls for both experiments. Cell harvest and lysis were handled as described.
Conjugation of tosylactivated Dynabeads to Cappel rabbit affinity purified antibody to mouse IgG
M-280 tosylactivated Dynabeads (Invitrogen, Carlsbad, CA) were used to capture proteins of interest. Dynabeads were coupled to Cappel rabbit affinity purified antibody to mouse IgG (whole molecule, MP Biomedicals, Irvine, CA). For coupling, 2 mL of Dynabeads (2 × 109 beads per mL) were first washed twice with 2 mL of PBS, pH 7.4. Beads were resuspended in 2 mL of Cappel antibody (5 µg antibody per 107 beads) and incubated overnight at 37 °C with end over end rotation. Unbound antibody was removed by washing the beads with 2 mL of 0.2 M Tris, pH 8.3, at 4 °C, then performing a second wash for 4 h at 37 °C with end over end rotation. After incubation, beads were washed once with 2 mL of PBS at 4 °C. Beads were resuspended in 2 mL PBS with 0.02% NaN3 and 0.05% Tween 20. The slurry stored at 4 °C.
Affinity capture of the TAP-tagged proteins
Frozen cells were first mechanically disrupted by grinding using a Retsch PM100 Planetary Ball Mill (Retsch, Newtown, PA).40 IgG-coupled Dynabeads were prepared for conjugation by washing 300 µL of the bead slurry three times with 1 mL cold lysis buffer. (Lysis buffer—50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% NP-40, 10% glycerol, 400 µM Na3VO4, 10 mM NaF and 10 mM Na4P2O7 passed through a 0.2 µM filter and supplemented with one Complete Protease Inhibitor cocktail tablet per 50 mL (Roche Diagnostics, Indianapolis, IN) and 10 mM N-ethylmaleimide just before use. The washed beads were resuspended in 150 µL of lysis buffer, divided into 6 equal aliquots into 2 mL microcentrifuge tubes, and placed on ice. Cells were prepared by suspending 1 g of the ground cell powder in 900 µL of cold lysis buffer. The suspension was added to the prepared beads in equal aliquots and incubated at 4 °C for one hour with end over end rotation. After incubation, 1 mL of the unbound fraction was removed from each tube and replaced with 1 mL of cold lysis buffer for washing. One aliquot was saved for comparison analyses. The beads were washed a total of three times with 1 mL of cold lysis buffer for each wash. Bound proteins were eluted by adding an equal volume of 0.2% SDS (25 µL). All six aliquots were combined into one tube, which was placed on magnetic stand to remove any remaining beads. The eluate was transferred to a new tube and stored at −80 °C until further processing.
Tryptic digestion of proteins
Ammonium bicarbonate powder was added to the protein sample (whole cell lysate, captured proteins, or multi-Ub chains) in 0.2% SDS to the final concentration of 100 mM. Proteins were reduced by adding DTT to a final concentration of 5 mM and incubated for 30 min at 37 °C. Proteins were then alkylated by adding chloroacetamide to a final concentration of 10 mM and incubated for 30 min at room temperature in the dark.41 Samples were then diluted to 0.05% SDS by adding 100 mM ammonium bicarbonate. Trypsin was added next using a 1 : 50 ratio of trypsin to protein. Samples were incubated overnight at 37 °C with end over end rotation. 5 µL of each sample was separated by SDS-PAGE to verify completeness of digestion (data not shown). Residual SDS was removed from all samples using Mixed-Mode Ion-Exchange Cartridges from Waters (Milford, MA, USA).
Heavy isotope labeled peptide preparation and mass spectral analysis
A list of ten or more SRM transitions was generated for each heavy isotope labeled peptide, labeled at the C-terminus with a heavy isotope-labeled lysine (+8.0 Da) or arginine (+10 Da), using the TIQAM algorithm.42 Heavy isotope labeled peptides (provided by Sigma in vials each containing 1 nmol pure peptide) were resuspended in 50 µ 20% ACN, 1% formic acid and mixed in a 1 : 1 ratio (500 fmol final concentration of each peptide). All 24 synthetic peptides were then separated on a LC PACKINGS PepMap 100 reversed-phase C18 column (0.075 × 150 mm) using a Tempo 2D nano-LC system from Eksigent (Eksigent Technologies, LLC, Dublin, CA, USA) at 200 nL min−1. Flow from the column was directed to a 4000QTrap workstation (Applied Biosystems, Framingham, MA) equipped with a nano-ESI source. The five most intense fragment ions for each peptide were determined by monitoring all singly and doubly charged y and b ions of all doubly and triply charged peptide ions. Collision energies for each transition (m/z values of a peptide and one of its fragment ions usually monitored in SRM for peptide detection) were optimized to ensure maximum sensitivity and lowest possible detection limit (Table 1). A list of SRM transitions for light peptides was generated from the list of SRM transitions for heavy isotope labeled peptides by subtracting 8 or 10 mass units from the peptide and transition ions mass depending on the C-terminal amino acid (lysine or arginine). The lists of light and heavy transitions were then combined to generate the final SRM transition list. All samples were spiked with an equimolar mixture of the heavy isotope labeled peptides and analyzed using our Ub-SRM assay.
Data processing
All SRM data were manually validated using ABI’s Multiquant software. The concentrations of light peptides were calculated using the formula below:
All measurements were done in triplicate. The values reported in this manuscript are all means of the triplicate measurements and errors are reported as the standard error of the mean.
Calculating the minimum concentration for mono-Ub
The exact concentration of mono-Ub in a mixture of mono- and poly-Ub chains cannot be calculated directly, because the most distal ubiquitin of a poly-Ub chain does not bear a linkage so produces the same peptides as a single Ub directly linked to a substrate. However a minimum concentration for mono-Ub can be calculated based on the concentration of linear and linked peptides in the mixture, and this can be used to confirm the presence of mono-ubiquitinated species in the mixture. The minimum mono-Ub concentration is calculated based on the fact that the concentration of linear peptides generated by digestion of a poly-Ub chain cannot exceed double the total concentration of Ub links in the mixture (see ESI†, Table S3). Therefore, if the concentration of linear peptides is more than twice the concentration of all Ub linkages, some of the linear peptides must have originated from mono-Ub species in the mixture. Hence, the minimum mono-Ub concentration is calculated by subtracting double the concentration of linked peptides from the average concentration of linear peptides.
Conclusion
Ubiquitination, degradative or non-degradative, is an essential PTM that is involved in a plethora of regulatory mechanisms and signaling pathways. Quantitative analysis of poly-Ub chains is crucial for the mechanistic understanding of numerous cellular functions. Traditionally, poly-ubiquitinated proteins have been studied using protein immunoblotting and other antibody-based detection methods. Mass spectrometry has also been used to study ubiquitination but to a lesser extent, due to detection and specificity limits. Most mass spectrometric analyses have focused on the identification of ubiquitinated proteins and their respective sites of Ub modification. Here we introduce a comprehensive targeted mass spectrometry approach based on SRM that is capable of detecting and quantifying various inter-ubiquitin links in complex mixtures in a single analysis. We demonstrate that this method is sensitive enough to quantify and characterize the poly-Ub chain of enriched proteins and specific enough to quantify and characterize the entire Ub population present in a whole cell lysate without any fractionation or enrichment. We demonstrate that Ub and all its possible linkages and phosphorylation sites can be detected in quantities as low as 100 amol. We show that the targeted MS assay is at least a 1000 fold more sensitive than the only commercially available Ub linkage-specific antibody. We used the targeted assay to characterize the poly-Ub chains of 15 enriched proteins. We also used this assay to detect changes in the ubiquitination level of an enriched histone protein Htb2 sample and the corresponding total lysate treated with MMS as well as an enriched sample for the transcription activator Met4 and the corresponding total yeast lysate treated with Cd2+. This study shows that the Ub-SRM assay is capable of characterizing poly-Ub chains of single proteins or whole cell lysates in a fast, reproducible, sensitive, and quantitative manner—a task that was not possible before.
Supplementary Material
Acknowledgements
This work was funded with federal (US) funds from the National Heart, Lung, and Blood Institute via the Seattle Proteome Center (contract No. N01-HV-8179 to RA).
Footnotes
Electronic supplementary information (ESI) available: Western blot evidence for purification of poly-Ubiquitinated proteins and SRM traces used to detect and quantify poly-Ub chains.
References
- 1.Ciechanover A, Elias S, Heller H, Ferber S, Hershko A. J. Biol. Chem. 1980;255:7525–7528. [PubMed] [Google Scholar]
- 2.Ciechanover A. Biochem. Biophys. Res. Commun. 2009;387:1–10. doi: 10.1016/j.bbrc.2009.06.065. [DOI] [PubMed] [Google Scholar]
- 3.Bernassola F, Ciechanover A, Melino G. Cell Death Differ. 2010;17:1–3. doi: 10.1038/cdd.2009.189. [DOI] [PubMed] [Google Scholar]
- 4.Finley D. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravid T, Hochstrasser M. Nat. Rev. Mol. Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W, Ye Y. Cell. Mol. Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Cell. Vol. 137. Cambridge, Mass: 2009. pp. 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 10.Pickart CM, Fushman D. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Bianchi K, Meier P. Mol. Cell. 2009;36:736–742. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 12.Hershko A, Heller H, Elias S, Ciechanover A. J. Biol. Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 13.Hershko A. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 15.King RW, Deshaies RJ, Peters JM, Kirschner MW. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 16.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Cell. Vol. 139. Cambridge Mass.: 2009. pp. 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshaies RJ, Joazeiro CA. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 18.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. J. Biol. Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser P, Wohlschlegel J. Methods Enzymol. 2005;399:266–277. doi: 10.1016/S0076-6879(05)99018-6. [DOI] [PubMed] [Google Scholar]
- 20.Hatakeyama S, Matsumoto M, Nakayama KI. Methods Enzymol. 2005;399:277–286. doi: 10.1016/S0076-6879(05)99019-8. [DOI] [PubMed] [Google Scholar]
- 21.Peng J, Cheng D. Methods Enzymol. 2005;399:367–381. doi: 10.1016/S0076-6879(05)99025-3. [DOI] [PubMed] [Google Scholar]
- 22.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 23.Hitchcock AL, Auld K, Gygi SP, Silver PA. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12735–12740. doi: 10.1073/pnas.2135500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denison C, Kirkpatrick DS, Gygi SP. Curr. Opin. Chem. Biol. 2005;9:69–75. doi: 10.1016/j.cbpa.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Cell. Vol. 134. Cambridge, Mass.: 2008. pp. 668–678. [DOI] [PubMed] [Google Scholar]
- 26.Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 27.Mollah S, Wertz IE, Phung Q, Arnott D, Dixit VM, Lill JR. Rapid Commun. Mass Spectrom. 2007;21:3357–3364. doi: 10.1002/rcm.3227. [DOI] [PubMed] [Google Scholar]
- 28.Denis NJ, Vasilescu J, Lambert J-P, Smith JC, Figeys D. Proteomics. 2007;7:868–874. doi: 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
- 29.Mirzaei H, McBee JK, Watts J, Aebersold R. Mol. Cell. Proteomics. 2008;7:813–823. doi: 10.1074/mcp.M700495-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piotrowski J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. J. Biol. Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson KD, Tashayev VL, O’Connor LB, Larsen CN, Kasperek E, Pickart CM. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 32.Hofmann RM, Pickart CM. Cell. Vol. 96. Cambridge, Mass.: 1999. pp. 645–653. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann RM, Pickart CM. J. Biol. Chem. 2001;276:27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 34.Seyfried NT, Xu P, Duong DM, Cheng D, Hanfelt J, Peng J. Anal. Chem. 2008;80:4161–4169. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lis ET, Romesberg FE. Mol. Cell. Biol. 2006;26:4122–4133. doi: 10.1128/MCB.01640-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogiso Y, Sugiura R, Kamo T, Yanagiya S, Lu Y, Okazaki K, Shuntoh H, Kuno T. Mol. Cell. Biol. 2004;24:2324–2331. doi: 10.1128/MCB.24.6.2324-2331.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 38.Yen JL, Su NY, Kaiser P. Mol. Biol. Cell. 2005;16:1872–1882. doi: 10.1091/mbc.E04-12-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaiser P, Su NY, Yen JL, Ouni I, Flick K. Cell Division. 2006;1:16. doi: 10.1186/1747-1028-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Nat. Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Mann M. Nat. Methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 42.Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, Buhlmann P, Bjorck L, Domon B, Aebersold R. Mol. Cell. Proteomics. 2008;7:1489–1500. doi: 10.1074/mcp.M800032-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.