SUMMARY
Differentiation of memory cells involves DNA-sequence changes in B lymphocytes but is less clearly defined in T cells. RNA rearrangement is identified here as a key event in memory T cell differentiation by analysis of a mouse mutation that altered the proportions of naive and memory T cells and crippled the process of Ptprc exon silencing needed to generate CD45RO in memory T cells. A single substitution in a memory-induced RNA-binding protein, hnRNPLL, destabilized an RNA-recognition domain that bound with micromolar affinity to RNA containing the Ptprc exon-silencing sequence. Hnrpll mutation selectively diminished T cell accumulation in peripheral lymphoid tissues but not proliferation. Exon-array analysis of Hnrpll mutant naive and memory T cells revealed an extensive program of alternative mRNA splicing in memory T cells, coordinated by hnRNPLL. A remarkable overlap with alternative splicing in neural tissues may reflect a co-opted strategy for diversifying memory T cells.
INTRODUCTION
Re-encounter with a foreign microbe or vaccine typically elicits a recall immunological response that is heightened in speed and magnitude. The simplest explanation for immunological memory is clonal expansion of antigen-specific cells (Burnet, 1959). Although most clonally expanded lymphocytes differentiate exhaustively into effector cells or undergo apoptosis, a small proportion persist for long periods after the immunological response has ceased (Intlekofer et al., 2006; Stockinger et al., 2006; Surh et al., 2006; Robertson et al., 2006; Williams and Bevan, 2007). Immunological memory may equally depend upon qualitative changes. “Memory” B cells are qualitatively distinct because somatic mutations in their antigen-receptor variable regions confer higher affinity (Neuberger et al., 2000), and DNA rearrangements in their antibody constant-region gene segments greatly enhance receptor signaling for plasma cell formation (Kaisho et al., 1997; Martin and Goodnow, 2002). These somatic DNA-sequence changes are not a feature of memory T cells, and most studies of memory T cell differentiation have focused instead on changes in transcription of cytokine- and homing-receptor genes (Kaech et al., 2002; Ansel et al., 2006; Intlekofer et al., 2006; Lee et al., 2006; Sallusto et al., 2004; Stockinger et al., 2006; Szabo et al., 2003; Vinuesa et al., 2005).
Memory T cells appear not to revert to a naive T cell state for the most part, an observation made on the basis of studies involving the most widely used marker for distinguishing naive T cells from memory T cells in man and many other mammals, CD45RO (Beverley, 2004). CD45RO is a variant of the CD45 major leukocyte transmembrane protein-tyrosine phosphatase, generated by alternative mRNA splicing (Hermiston et al., 2003). Naive T cells express distinct isoforms, typically CD45RB and CD45RAB or CD45RBC, due to inclusion in the N-terminal extracellular domain of O-glycan-rich A, B, or C segments encoded in the Ptprc mRNA by exons 4, 5, and 6, respectively. In memory T cells, as well as in recently activated T cells, exons 4–6 are silenced and excluded from Ptprc mRNA, resulting in the CD45RO isoform detected by specific monoclonal antibodies in man. Molecular biology studies of T cell lines and human polymorphisms have delineated a specific nucleotide sequence motif in Ptprc exons 4, 5, and 6—the activation-responsive sequence (ARS)—that is necessary and sufficient for silencing these exons in activated and memory T cells (Lynch and Weiss, 2000; Rothrock et al., 2003; Tong et al., 2005). Both T cell fusion with B lymphocytes (Rothstein et al., 1992), which do not silence these exons, and studies with inhibitors of protein-synthesis and protein kinase C (PKC) signaling (Konig et al., 1998; Lynch and Weiss, 2000) point to the existence of an inducible exon-silencing protein specifically expressed in T cells. Although regulated silencing of these exons in T cells is highly conserved across species, the nature of the trans-acting splicing factor(s) that reprogram CD45 splicing remains obscure. Here, we have identified a gene that was upregulated in memory T cells, encoded a protein with an RNA-recognition domain that bound directly to the Ptprc ARS element in vitro, and, in mice, played an essential, specific, and gene dosage-dependent role in alternative splicing of Ptprc and many other mRNAs differentiating memory T cells from their naive counterparts.
RESULTS
The thunder ENU Mouse Mutation Disrupts Peripheral T Cell Subsets
Pedigrees of ethylnitrosourea (ENU)-mutagenized C57BL/6 mice segregating thousands of random single-nucleotide substitutions (Nelms and Goodnow, 2001) were screened by flow cytometry for heritable mutations affecting the circulating proportions of memory and naive T cells. Pedigree ENU126 segregated a Mendelian recessive increase in the proportion of blood CD4+ and CD8+ cells with high CD44, a marker of activated and memory T cells (Figure 1A). Given the fewer overall CD4+ cells (see below), the strain was named thunder for Th cells under normal. Staining with antibodies to the RB isoform of CD45, which is present on naive CD4+ T cells but replaced by the CD45RO isoform on memory T cells, revealed no CD45RBlo T cells in peripheral lymphoid tissues (Figure 1B). Staining with antibodies to CD45RA and CD45RC isoforms, which are normally low on most CD4+ T cells and absent on memory T cells, showed that these were highly expressed on all T cells from the mutant strain (Figure 1B) and on dendritic cells, macrophages, and granulocytes (Figure S1, available online). Homozygous mutant mice were, nevertheless, born at the expected Mendelian frequency and were indistinguishable from WT (WT) littermates in appearance and behavior by visual inspection.
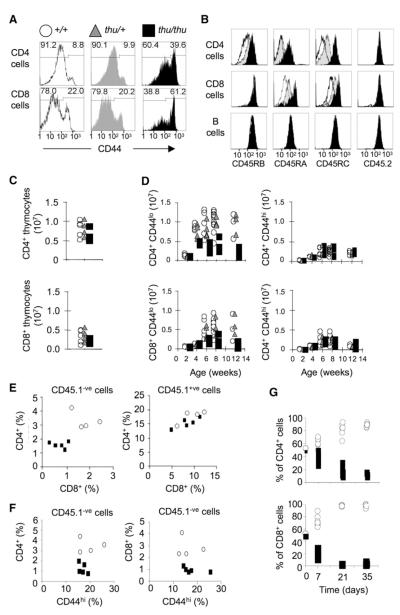
Figure 1. ENU Mouse Mutation that Disrupts Accumulation of Naive and Memory T Cells and Expression of CD45RO.
Flow-cytometric analysis of cells from WT +/+ (unfilled histograms and symbols), thu/+ (gray) and thu/thu (filled black) mice. This scheme of symbols and colors applies to all panels.
(A) CD44 on CD4+ or CD8+ blood T cells. The percentage of memory (CD44hi) and naive (CD44lo) subsets is shown.
(B) Staining for specific CD45 isoforms or an epitope common to all isoforms (CD45.2), gated on splenic CD4+ or CD8+ T cells or B cells.
(C) Number of CD4+ or CD8+ SP T cells in the thymus at 6 weeks.
(D) Number of naive or memory CD4+ and CD8+ T cells in the spleen at different ages.
(E) T cells in irradiated mice reconstituted with an equal mixture of +/+ CD45.1 and thu/thu CD45.2 (CD45.1-negative) bone marrow. Control mixed chimeras were reconstituted +/+ CD45.1 and +/+ CD45.2 bone marrow. Left panel shows percentage of CD4+ or CD8+ T cells that are CD45.1-negative, representing thu/thu T cells in the test mixed chimeras (filled squares) or +/+ T cells in the control mixed chimeras (open circles). Right panel shows percentage of CD4+ or CD8+ T cells that are CD45.1-positive, representing +/+ T cells either in the test mixed chimeras (filled squares) or in the control chimeras (open circles).
(F) Percentage of memory T cells among the CD45.1-negative T cells in (E), representing thu/thu T cells (filled squares) or +/+ T cells (open circles).
(G) A 50:50 mixture of +/+ CD45.1 and thu/thu CD45.2 thymocytes was injected into Rag1−/−° mice. The percentage of T cells of each donor type is shown at the time of injection and in the blood at different times after transfer.
The increased percentage of CD44hi T cells was secondary to a loss of CD44lo naive T cells from peripheral lymphoid tissues. Thymic T cell development and numbers of CD4+ and CD8+ single positive (SP) thymic T cells were indistinguishable from the WT (Figure 1C). Peripheral CD4+ and CD8+ T cells were also present in normal numbers in two-week-old mutant mice, consistent with normal production and emigration from the thymus, but the CD44lo subset failed to accumulate normally in the peripheral blood, spleen, and lymph node as the animals matured (Figure 1D). Transplantation of thunder bone marrow demonstrated that the mutation acted within T cells and not within radioresistant stromal elements of lymphoid tissues (Figure S2A). Cell-autonomous functions were delineated by reconstitution of irradiated mice with equal mixtures of WT and thunder bone marrow, distinguished by allelic variants in the constant extracellular segment of CD45: CD45.1 on WT and CD45.2 on mutant-derived leukocytes. The mutation acted cell autonomously to decrease accumulation of peripheral naive and memory T cells bearing the thunder mutation (CD45.1 negative cells, Figures 1E and 1F) but had no effect on thymic T cell differentiation, even under these competitive conditions (not shown). In contrast to animals in which all T cells bear the mutation, in mixed chimeras there was no increase in the percentage of CD44hi T cells among the peripheral T cell subset bearing the mutation (Figure 1F). From this result, we infer that CD44hi T cells compensate for peripheral T cell lymphopenia by homeostatic proliferation in animals in which all T cells bear the mutation, whereas in mixed chimeras, compensation occurs through replacement by WT T cells.
The evidence that the thunder mutation acts within peripheral T cells to diminish their persistence but not their proliferation was extended by cell-transfer, cell-culture, and viral-challenge experiments. Transfer of equal mixtures of WT and thu/thu thymocytes, distinguished by CD45.1 and CD45.2 allotypes, respectively, into Rag1−/− or irradiated mice lacking peripheral T cells showed that the mutant T cells (CD45.1-negative) selectively failed to compete with WT T cells for repopulating the recipient mice (Figure 1G). When the transferred cells were first labeled with CFSE dye for measuring lymphopenia-induced homeostatic cell division, thunder T cells nevertheless divided indistinguishably from controls (Figure S2B). Similarly, thunder T cells proliferated indistinguishably from WT T cells when stimulated with antibodies to CD3 and CD28 in vitro (Figure S3A) and differentiated into Th1 or Th17 effector cell subsets (Figure S3B). Upon challenge with influenza virus, thunder mice were capable of generating large populations of nucleoprotein-specific CD8 cells (Figure S3C).
thunder Mutation Cripples a Gene-Dosage-Sensitive Silencer of Ptprc Exons
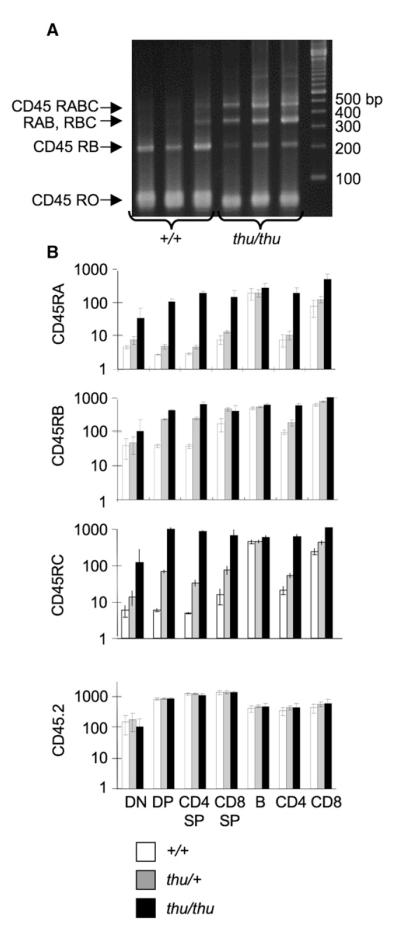
To understand the dramatic shift in CD45 isoforms expressed by thunder T cells, we analyzed Ptprc mRNA splicing by RT-PCR using primers spanning exons 3–7 (Figure 2A). WT thymocytes predominantly carried Ptprc mRNA lacking exons 4, 5, and 6 (encoding CD45R0) or containing only exon 5 (encoding CD45RB). By contrast, a greater proportion of Ptprc mRNA in thu/thu thymocytes contained exons 4, 5, and 6, encoding CD45 isoforms of higher molecular weight. The relative use of exons 4, 5, and 6 was quantified by flow-cytometric staining with monoclonal antibodies to the corresponding CD45RA, CD45RB, and CD45RC segments (histograms, see Figure 1B; quantitation, see Figure 2B). These were 10–100 times higher on thu/thu thymic and peripheral T cells than on WT counterparts and were comparable to the amounts on B cells, which do not silence these exons, whereas there was no difference in pan-CD45 staining intensity. In thu/+ heterozygotes, the mutation caused an intermediate silencing defect correlated with the number of ARS motifs in the corresponding exons: in CD4+8+ double positive (DP) or CD4 SP thymocytes, thu/+ heterozygosity caused only a subtle defect in CD45RA silencing (three motifs), an intermediate defect in CD45RC silencing (two motifs), and a marked failure to silence CD45RB (single motif) (Figure 2B). Thus, the thunder mutation disrupts an essential and gene-dosage-sensitive step in the silencing of each exon of Ptprc containing the ARS splicing-silencer motif.
Figure 2. Thunder T Cells Fail to Silence Ptprc Exons 4–6.
(A) Thymus cDNA from three different +/+ or thu/thu mice, PCR amplified with primers in Ptprc exons 3 and 7. Specific RNA splice products were deduced on the basis of their sequence-predicted sizes: RO, 71 bp; RB, 218 bp; RAB, 347 bp; RBC, 359 bp; RABC, 488 bp.
(B) Ptprc exon 4, 5, and 6 inclusion quantified in different cell subsets by geometric-mean fluorescence staining with antibodies against the corresponding CD45RA, CD45RB, and CD45RC segments. Relative CD45 expression was measured in parallel with antibody to the CD45.2 epitope in a membrane-proximal domain unaffected by alternative splicing. Abbreviations are as follows: DN, CD4−8− double-negative thymocytes; DP, CD4+8+ double-positive thymocytes; CD4 SP, CD4+ single-positive thymocytes; CD8 SP, CD8+ single-positive thymocytes; B, spleen B cells; CD4, spleen CD4 T cells; CD8, spleen CD8 T cells. Data are from six animals of each genotype. Columns indicate means, and error bars indicate standard deviations from n = 6 mice.
Missense Mutation in a Memory T Cell-Induced Gene, Hnrpll
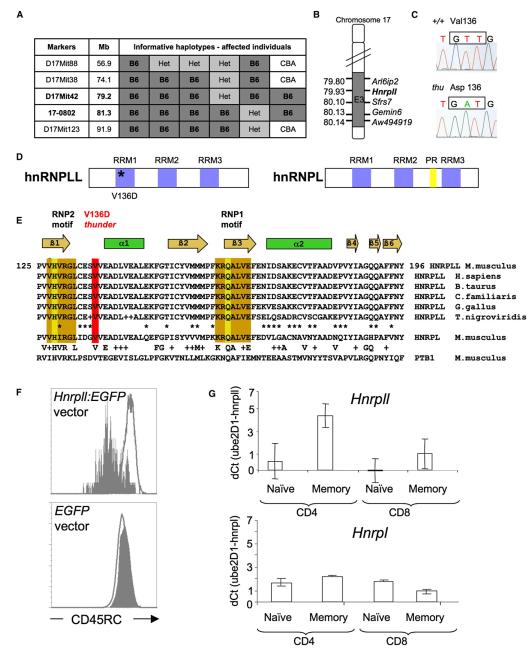
The causative thunder mutation was identified by standard meiotic mapping, sequencing, and gene complementation. The thunder T cell abnormalities—both the failure to silence CD45 variable exons and the deficiency of naive T cells—mapped to a single 2.1 Mb locus on distal chromosome 17, containing 21 transcription units (Figures 3A and 3B, Figure S4). This result excludes the possibility that mutations in genes outside this interval account for the T cell abnormalities defined in Figure 1. All 21 transcripts within the interval were sequenced and compared with parental B6 controls, but only Hnrpll contained a mutation: an exonic T-to-A substitution resulting in a Val136-to-Asp codon change (Figure 3C). The mutated valine was in the first of three RNA recognition motif (RRM) domains in the hnRNPLL protein (Figure 3D) and was absolutely conserved in vertebrates and in similar domains of other hnRNPs (Figure 3E). Strikingly, the most closely related protein, hnRNPL (Hnrpl), has previously been shown to bind the Ptprc ARS motif (Rothrock et al., 2005; Tong et al., 2005). We confirmed that the Hnrpll mutation was responsible for the thunder splicing defect by restoring CD45RC silencing in thu/thu T cells transduced with WT Hnrpll cDNA expressed in a bicistronic retroviral vector (Figure 3F).
Figure 3. thunder Is a Missense Mutation in the RNA-Recognition Motif of hnRNPLL.
(A) Meiotic mapping of thunder to a 2.1 Mb interval derived from the B6-thunder strain.
(B) Candidate RNA-regulating genes within the interval.
(C) T→A substitution in thunder mRNA nucleotide 684, codon 136.
(D) Predicted domains of the hnRNPLL and hnRNPL proteins. Abbreviations are as follows: RRM, RNA recognition motif domain; PR, proline rich domain.
(E) Amino acid sequence alignment of the hnRNPLL RRM1 domain from different vertebrate species and with corresponding RRM domains from hnRNPL and PTB1. Gold boxes denote two RNP motifs that form a central binding site for RNA, with key RNA contacts on H and Q residues. The regular secondary-structure elements in the RRM1 domain of hnRNPLL are indicated at the top.
(F) thu/thu T lymphoblasts were transduced with a bicistronic retroviral vector containing WT Hnrpll cDNA and EGFP or containing WT EGFP alone. Flow-cytometric staining for CD45RC is shown gated on GFP+ cells (gray histograms) compared with nontransduced GFP° cells in the same culture (open histograms), showing correction of CD45RC silencing by the WT cDNA.
(G) Relative mRNA abundance of Hnrpll (upper panel) and Hnrpl (lower panel) in WT naive and memory CD4+ and CD8+ T cells measured by qRT-PCR, normalized to Ube2D1. Columns indicate means, and error bars indicate standard deviations from n = 3 mice.
The data above established that hnRNPLL is an essential trans-acting factor for silencing Ptprc exons in vivo but did not explain how silencing is developmentally regulated between naive and memory T cells. Measurement of Hnrpll mRNA in naive and memory T cells by real-time PCR or by microarray showed that it was increased ~300% in memory T cells, whereas Hnpl was not (Figure 3G and Figure S5A). B cells expressed much lower Hnrpll mRNA than did T cells (Figures S5B and S5E), consistent with the absence of a Ptprc exon-silencing factor in B cells (Rothstein et al., 1992). Analysis of GEO-deposited microarray data showed that Hnrpll mRNA was induced ~400% by T cell receptor (TCR) stimulation through PKC-dependent signaling (Figure S5C; Safford et al., 2005), consistent with the characteristics for ARS-dependent silencing (Lynch and Weiss, 2000). Foxp3+ CD25+ T cells are also CD45RBlo and had elevated Hnrpll mRNA compared to CD4+25− T cells (Figure S5D; Fontenot et al., 2005). Given the marked effects on CD45 silencing when the WT Hnrpll gene dose was halved (Figure 2B), increased expression of Hnrpll is sufficient for explaining the regulated silencing of CD45 in memory T cells.
thunder Mutation Destabilizes an ARS RNA-Binding Domain
Structural and binding studies were carried out for understanding how the mutation affected the RRM1 domain of hnRNPLL (Figure 4). The domain adopted the canonical RRM fold (Maris et al., 2005) and was structurally very similar to the RRM1 domain of PTB1 (Simpson et al., 2004), with which it shared 52% sequence similarity and an average pairwise backbone rmsd of 1.9Å (Figure S6G). The V136 side chain was buried, suggesting that the V136D mutation could disrupt the structure of the domain. Comparison of NMR spectra of bacterially expressed WT and mutant hnRNPLL RRM1 domains (Figures S6A and S6B) revealed that the spectral changes caused by the V136D mutation were limited to the vicinity of helix α1, which lay beneath the β sheet surface containing the conserved residues responsible for RNA binding in other RRM domains (Figures 4A and 4C; Figure S7A).
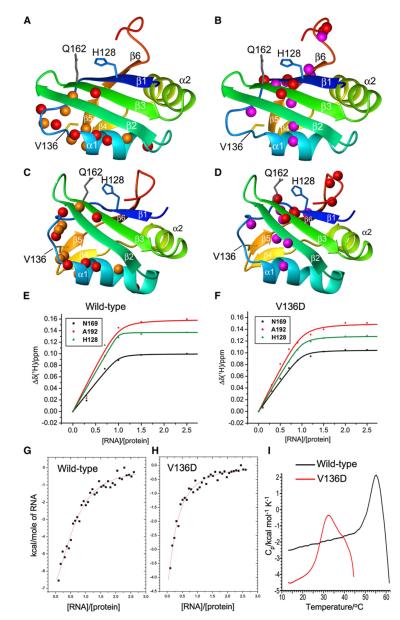
Figure 4. hnRNPLL RRM1 Domain Structure, ARS RNA Binding, and Consequences of the thunder V136D Mutation.
(A) Ribbon representation of the hnRNPLL RRM1 domain (residues 124–202), including side chains at the site of mutation (V136, yellow) and RNA contact residues conserved in other RRM domains (His128, Gln162). Red spheres: backbone amides broadened beyond detection in the NMR spectra of the V136D mutant. Orange spheres: backbone amides for which the mutation results in different chemical shifts.
(B) Same as (A) except that spheres identify backbone amides with different chemical shifts in the presence of the 15-mer RNA 5′-CCUUACCUGCACGCA-3′, with red colors indicating amide protons broadened beyond detection in the presence of the 15-mer RNA.
(C and D) Same as (A) and (B), but with ribbon representation of the PTB1 RRM1 domain (PDB: 1SJQ).
(E) Binding affinity of RRM1 domain to 15-mer RNA, measured by NMR spectroscopy at 25°C. Kd values obtained from the chemical-shift changes of His128, Asn169 and Ala192 were 0.1 ± 0.5 μM, 0.7 ± 0.7 μM, and 1.5 ± 1.3 μM, respectively, indicating an upper limit of about 1.5 μM.
(F) Same as (E), but for V136D mutant. Corresponding Kd values were 1.1 ± 0.4 μM, 1.0 ± 0.4 μM, and 2.0 ± 0.7 μM, indicating an upper limit of about 2 μM.
(G) Binding affinity measured by isothermal calorimetry at 15°C. The continuous line represents the least-squares fit of the data, yielding a Kd value of 1.9 ± 0.4 μM.
(H) Same as (G), but for the V136D mutant. The fitted Kd value is 3.1 ± 1.4 μM.
(I) DSC curves of mutant (red) or WT (black) RRM1 domains. Both proteins precipitated irreversibly at high temperature.
Binding of the RRM1 domain to RNA containing the Ptprc ARS motif was established by NMR spectroscopy (Figures 4B and 4D; Figures S6 and S7), monitoring the titration of the RRM1 domain with single-stranded 15-mer RNA comprising the potent ARS exon-silencing sequence from Ptprc exon 6 region 7, defined by Tong et al. (2005). Both NMR analysis (Figure 4E) and isothermal calorimetry (Figure 4G; Figure S8) yielded an affinity of the WT hnRNPLL RRM1 domain for ARS RNA of 1–2 μM at 25°C and 15°C. This value is comparable to the RNA affinity of other isolated RRM domains (Maris et al., 2005). In particular, 1–3 μM affinity for oligonucleotides has been reported for the isolated RRM1 and RRM2 domains of PTB1, which are closely related to the RRM1 domain of hnRNPLL. Cooperativity among the four RRMs in the PTB1 protein leads to nM affinity for RNA (Auweter et al., 2007), and a similar mechanism may operate among the three RRM domains to enhance the binding affinity of full-length hnRNPLL.
The NMR data showed that RNA interacts selectively with conserved RNA contact residues, such as His128 and Gln162, across the β1 and β3 sheet surface defined previously as the RNA binding site for PTB1 and other RRM domains (Maris et al., 2005). We confirmed specificity of binding by probing the interaction of the WT RRM1 domain with a mutant RNA oligonucleotide bearing two substitutions, from GCACGCA to GGACGUA, that are sufficient to inactivate exon 6 silencing activity in cells (Tong et al., 2005). The RNA sequence changes greatly reduced the number of shifted peaks in the NMR spectrum and the binding affinity, as expected for nonspecific binding (Figure S6E). Thus, the hnRNPLL RRM1 domain binds RNA with sequence specificity corresponding to the inducible activity defined previously by Tong et al. (2005).
The affinity of RRM1 for WT ARS RNA was barely altered by the thunder mutation (Figures 4F and 4H), consistent with conformational changes far from the RNA-binding surface. However, when stabilities were assessed by differential scanning calorimetry (DSC), the mutation resulted in a much lower melting temperature and a significant degree of unfolding above 25°C, whereas the structure of the WT domain was stable until at least 45°C(Figure 4I). We infer from the combined NMR and DSC data that the V136D mutation interferes with ARS RNA binding indirectly by producing a thermolabile structure that is mostly unfolded at 37°C.
Hnrpll Controls Reprogramming of Alternative Splicing in Memory T Cells
We then asked whether splicing of other genes was also dependent upon Hnrpll. RNA was prepared from flow-sorted spleen memory CD4 and memory CD8 cells, their naive CD44lo counterparts, and CD4+ and CD8+ SP thymocytes. Four pools of each cell type were prepared from more than four independent thu/thu and +/+ mice, and mRNA was converted to labeled cRNA and hybridized to Affymetrix Mouse Exon 1.0 ST Arrays. The complete data set comprised 24 biologically independent samples of mutant T cell RNA and 24 independent samples of WT T cell RNA, thereby providing a high degree of replication. These data have been deposited in the GEO database, and a web-enabled interface for reviewing the exon data has been established at http://www.systemsimmunology.org/Thunder. Data for 15,788 informative genes was first analyzed for changes in discrete exons between thu/thu and +/+ cells within each T cell subset by two-way (genotype × probe set) ANOVA. ANOVA values of p = 0.001 and p = 0.05 were calculated as having false discovery rates (FDRs) of 0.012–0.059 and 0.189–0.463, respectively, in individual T cell types (Table S1). Each gene was then ranked for reproducibility of two-way ANOVA evidence for altered splicing in thu/thu cells across the six T cell subsets. Ptprc ranked sixth overall, with an ANOVA p < 0.001 in five of the six T cell subsets, thereby validating the analysis. For distiguishing true alternative-splicing events from artifacts arising from unresponsive or crosshybridizing probes or confounding effects of changes in gene expression overall, the data for each high-ranked gene was manually inspected by plotting of the mean signal and standard error for each probe set and cell type in alignment with annotated mouse exons and UCSC reference transcripts (Figure 5). Of the 35 probe sets for Ptprc exons, probe set signals for exons 4, 5, and 6 were selectively elevated in thu/thu compared to +/+ for all six T cell subsets, providing striking validation of the exon-array data and our analysis approach (Figure 5A).
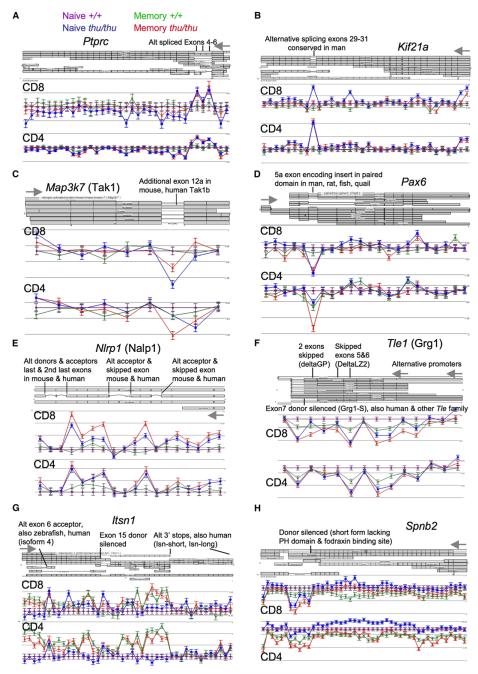
Figure 5. Exon-Array Probe-Set Data According to Hnrpll Genotype and T Cell Subset, Aligned with Corresponding Exons and Reference Transcripts.
Log2 mean and standard error of the signal for each probe set is shown for either CD4+ or CD8+ T cells (memory thu/thu in red, memory +/+ in green, naive thu/thu in blue, naive +/+ in purple), normalized against the mean signal on the same probe set in corresponding naive +/+ cells. Grey arrows indicate sites of transcript initiation.
(A and B) Examples of genes in Group A, in which signal is consistently increased in thu/thu cells for probe sets corresponding to known alternatively spliced internal exons.
(C and D) Examples of genes in Group B, in which signal is consistently decreased in thu/thu cells for probe sets corresponding to known alternatively spliced internal exons.
(E and F) Examples of genes in which multiple internal but noncontiguous exons, corresponding to known transcript isoforms, show consistent Hnrpll-dependent changes.
(G and H) Examples of genes in which variable silencing of an internal splice donor creates alternative 3′ mRNA and carboxy termini.
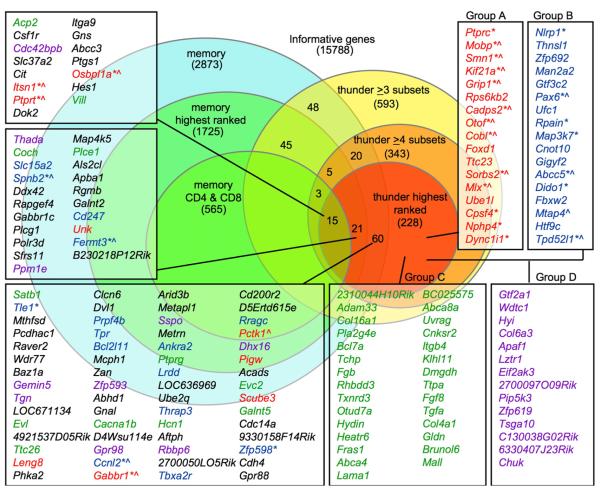
The validated analysis strategy outlined above was then applied to the full data set. We found that 343 genes exhibited a genotype × probe set ANOVA p < 0.05 in four or more cell types. Data for the 228 most highly ranked genes were manually reviewed against UCSC mouse reference transcripts (representative examples, see Figures 5B–5H; all annotations, see Figures S9–S14 and Tables S2–S7), placing them into one of six categories. Categories A–D comprised 125 genes with probe sets that reproducibly changed in Hnrpll mutant T cells corresponding to internal exons in UCSC reference mouse mRNAs (listed in Figure 6). Of these, 59 genes (47%) had Hnrpll-dependent probe sets corresponding to alternatively spliced exons already identified in UCSC reference transcripts. The fact that such a high proportion correspond to sequence-validated alternative-splicing events represents further validation of the exon-array data and analysis, because the current set of reference mRNAs appear to cover less than half of the alternative splice events (Blencowe, 2006; Clark et al., 2007; Einstein et al., 2008; Yeo et al., 2007). The 59 transcript-validated genes were subdivided into Groups A and B. Group A comprised 27 genes, including Ptprc, in which the signal increased in thu/thu cells, indicating a failure of exon silencing in the mutant T cells. In group B (32 genes), the signal decreased in thunder T cells, implying decreased exon inclusion resulting from Hnrpll deficiency. Group C (42 genes) and Group D (24 genes) mirrored Groups A and B, respectively, except that the Hnrpll-dependent probe sets were not currently identified as an alternatively spliced exon by a reference mouse mRNA. Genes with Hnrpll-dependent probe sets corresponding to the first exon of UCSC reference transcripts were assigned to Group E (64 genes). These potentially represent alternative promoters and mRNA start sites, and this was clearly the case for a number that were manually compared to transcript, EST, and published evidence (Figure S13 and Table S6). Genes assigned to Group F (38 genes) exhibited complex patterns of changes in Hnrpll-dependent probe sets, such as changes in overall mRNA expression from single-transcript or multiple-transcript gene complexes.
Figure 6. hnRNPLL Controls Alternative Splicing of Numerous mRNAs.
Summary of exon-array analysis of mRNA from naive and memory CD4+ and CD8+ T cells and SP thymocytes sorted from thu/thu and +/+ mice (four biological replicates of each cell and genotype). Of the 228 most highly ranked Hnrpll-regulated genes, those assigned to Group A (red font) and Group B (blue font) contained Hnrpll-dependent probe sets corresponding to known alternatively spliced exons in reference mouse transcripts. Asterisks denote genes with transcript or published data for alternative splicing of the same exons in man. Chevrons denote genes with published data for alternative splicing of the same exons in neural tissue. Groups A and C (green font): genes with Hnrpll-dependent exons exhibiting consistently increased signal in thu/thu. Groups B and D (purple font): genes with Hnrpll-dependent exons exhibiting consistently decreased signal in thu/thu. Black font denotes genes in Group E, with Hnrpll-dependent probe sets that appear to reflect alternative promoter use, and in Group F, with a complex pattern of changes in Hnrpll-dependent probe sets.
Additional manual analysis was performed for the 59 transcript-validated Hnrpll-dependent genes in Groups A and B. First, the probe set and transcript-gene views were manually compared to all mouse transcripts, mouse ESTs, and transcripts of other species in the UCSC Genome Browser (July 2007 assembly) for detection of conserved alternative-splicing events in man and other species. Second, published information on alternative splicing for these 59 genes was identified by a search of the gene symbols and names of the encoded proteins in NCBI Homologene, OMIM, and PubMed databases and then a reading of the abstracts and papers for the identification of information about alternative splicing. Two important points emerged. First, the Hnrpll-regulated alternative-splicing events identified in 63% of genes in Group A and 38% in Group B were conserved between mouse and man, ascertained on the basis of sequenced mRNA variants in the two species (asterisks in Figure 6). This level of human-mouse conservation provided additional validation of the Hnrpll-dependent splicing events identified by the array analysis, because only 10%–20% of alternatively spliced exons appear to be conserved between man and mouse overall (Blencowe, 2006).
Many of the evolutionarily conserved, Hnrpll-dependent alternatively spliced forms alter the encoded proteins in important ways. For example, thu/thu T cells had a marked increase in mRNA signal selectively for alternatively spliced Kif21a exons encoding part of the cargo-binding segment of this microtubular transport kinesin (Figure 5B), which may alter polarized interactions with antigen-presenting cells. The thu-diminished signal for Map3k7 exon 12a (Figure 5C) encodes a 27 aa insert in the carboxy-terminal interaction domain of TGF-β-activated kinase 1 (TAK1), a key enzyme for inflammatory signaling to NF-kB (Kondo et al., 1998; Sakurai et al., 1998). Pax6 exon 5a (Figure 5D) encodes a conserved insert within the DNA-binding paired domain that alters the specificity of this transcriptional regulator of neuronal cell development (Dominguez et al., 2003). In thunder T cells, there was increased expression of a spliced form of Nlrp1 mRNA (Figure 5E) encoding a shorter form of the NALP1 inflammasome sensor for bacterial muramyl dipeptide, lacking two of the leucine rich repeats and other segments between the caspase recruitment domain and the nucleotide binding domains (Bruey et al., 2007; Hlaing et al., 2001). The multiple alternatively spliced exons in Tle1 (also called Grg1; Figure 5F) encode glycine-proline rich (GP) and leucine zipper (LZ) protein-interaction domains and an alternative stop codon and 3′UTR that are variably excluded from the long forms and delta GP or delta LZ forms of this transcriptional repressor (Lepourcelet and Shivdasani, 2002).
The second finding from manual analysis was that for 15 of the 27 genes (56%) in Group A, the exons showing Hnrpll-dependent alternative splicing have been published as undergoing alternative splicing in the nervous system (chevrons in Figure 6, details in Supplemental Data). This remarkable overlap could reflect a shared strategy for cellular diversification, consistent with the observation that alternative splicing appears to be more common in genes expressed in functionally complex tissues with diverse cell types, such as the brain and the immune system (Blencowe, 2006). Indeed, many of the alternative-splicing events regulated by Hnrpll in T cells have important functions in the nervous system (see Supplemental Data). For example, the Hnrpll-dependent exons in Kif21a (Figure 5B) are also alternatively spliced in neurons and this gene is mutated in congenital fibrosis of the extraocular muscles, types 1 and 3 (CFEOM; Yamada et al., 2003). The internal exons of Itsn1 that are enhanced in thu/thu T cells, as well as the short mRNA form enhanced in memory T cells (Figure 5G), encode different isoforms of Intersectin 1, a multidomain protein functioning in clathrin-mediated endocytosis, which through variable silencing of 3′ splice donors produces a neuronal-specific long form and two shorter forms (Tsyba et al., 2004). The enhanced alternative Grip1 exon in thunder cells (Figure S9) encodes a palmitoylated, membrane-associated Grip1b form of Glutamate Receptor Interacting Protein 1, a multi-PDZ-domain intracellular scaffolding protein that plays a key role in cell-matrix interactions during development, neuronal dendrite morphogenesis, and synaptic reorganization of AMPA receptors (Charych et al., 2006). Although thunder mutant mice display no abnormalities in gross behavioral or motor skills, these observations, coupled with the wide expression of Hnrpll in the brain (Figure S5E), make it logical for future studies to test thunder mutant animals for specific neural defects.
In parallel, we queried the array data for alternative splicing occurring in WT T cells between naive and memory subsets (Figure 6). This revealed 2873 genes (18%) with two-way ANOVA evidence at p < 0.05 (FDR 0.607–0.693) for alternative splicing between naive and memory CD4 or CD8 cells. We found that 1725 genes (10.9%) exhibited ANOVA p < 0.001 (FDR 0.026–0.036) in either CD4 or CD8 memory cells, with 565 (3.6%) scoring p < 0.001 in both memory cell types (Table S7). Genes with altered splicing in thunder T cells were significantly enriched for genes with evidence of altered splicing between WT naive and memory cells: 96 of the 228 most highly ranked Hnrpll-dependent genes (42%) also had evidence (p < 0.05) for altered splicing in normal memory cell differentiation, contrasting with 18% expected by chance (chi-square test, p < 0.0001). Manual inspection of the probe set data showed the congruence between Hnrpll dependency and altered splicing in WT memory T cell differentiation also applied at the level of the individual probe sets and exons (Figures S9–S14).
DISCUSSION
The findings above revealed Hnrpll to be a regulated gene in lymphocytes, most highly expressed in CD45RO memory T cells and Treg cells that fully silence the Ptprc variable exons, expressed at lower amounts in naive T cells that fail to silence exon 5 encoding CD45RB, and expressed at the lowest amounts in B cells that do not silence any of these exons and express CD45RABC. Genetic and structural studies delineated a critical RRM1 domain in the hnRNPLL protein that bound RNA containing the consensus ARS exon-silencing sequence but not a mutant ARS sequence lacking exon-silencing activity. A point mutation that destabilized the RRM1 domain inactivated silencing of all three exons when homozygous, and in the heterozygous state, the mutation partially compromised silencing in concordance with the known relative strength of the ARS sequence in each exon. The hnRNPLL protein thus fulfils the criteria for the inducible silencing factor in T cells and explains the regulated changes in CD45 splicing that serve as a key marker for memory T cells in many species.
Oberdoerffer et al. (2008) recently performed a complementary study using an shRNA screen of potential splicing factors in human Jurkat T lymphoma cells to find that hnRNPLL is essential for silencing CD45 exons 4, 5, and 6. The fact that two different screens identify the same gene and protein underscores the specific requirement for hnRNPLL in CD45 exon silencing. Nevertheless, the two studies differ in their conclusions about the global function of hnRNPLL in T cells. Oberdoerffer et al. observe that during puromycin selection for a vector encoding an shRNA against hnRNPLL, human T lymphoblasts expressing this vector proliferate poorly compared to those expressing a control vector. This result contrasts with the identical proliferation of hnRNPLL mutant and WT T cells observed here. Oberdoerffer et al. suggest other targets of hnRNPLL by exon-array analysis, but the study does not appear to employ biological replicates or cell sorting of matched subsets to control for the confounding effect of reduced proliferation in cells expressing Hnrpll shRNA. Few genes listed by Oberdoerffer et al. correspond to Hnrpll-dependent alternative-splicing events identified in the 48 independent samples of sorted T cells analyzed here. In particular, STAT5 and CD44 exhibit alternative splicing in T lymphoblasts expressing hnRNPLL shRNA, whereas we found no evidence for this in any subset of hnRNPLL mutant T cells (Figure S15). Because STAT5, CD44, and IL2RA are correlated with T cell proliferation, the differences described by Oberdoerffer may be secondary to reduced proliferation of the shRNA-expressing cells compared to the controls. Nevertheless, it is also possible that the discrepant findings reflect differences between mouse and human T cells or between knockdown of hnRNPLL compared to point mutation of the RRM1 domain.
A key finding from our study is that changes in Ptprc splicing represent the tip of a large network of alternative splicing in memory T cells—with hundreds of other genes undergoing Hnrpll-dependent splicing. The set of genes identified here thus provides a unique resource for discovering new markers andmechanisms of memory T cell differentiation. Although many genes exhibited failure of exon silencing like Ptprc (Groups A and C), an equal number showed the opposite effect (Groups B and C). hnRNPLL may act directly as an exon-splicing enhancer in the latter instances, consistent with evidence for other hnRNPs (Hung et al., 2008). Alternatively, hnRNPLL may promote exon inclusion indirectly by affecting splicing-enhancer factors. It is indeed likely that the direct effects of hnRNPLL are amplified by a cascade of secondary changes, given that alternative splicing in thunder T cells affected a number of RNA-processing regulators, including Smn1, Cpsf4, Prpf4b, Ccnl2, Thrap3, and Sfrs11. There were also many instances of genes (Group E) with Hnrpll-dependent changes in the relative use of alternative first exons and apparent mRNA start sites. This, again, may reflect a cascade, given that many thunder-altered genes encode transcriptional regulators, including Arid3b, Smarca, Nfatc1, Mlx, Foxd1, Tle1, Grg1, Tcf7l2, and Pax6.
The intrinsic failure of Hnrpll mutant T cells to persist in the periphery may reflect altered splicing of many genes controlling cell signaling and survival among the Hnrpll-regulated set. These include regulators of apoptosis Nlrp1 (encoding NALP1), Lrdd (encoding PIDD), Madd (encoding DENN), Apaf1, Cyc1 (encoding Cytochrome-c), and Bcl2l11 (encoding Bim). Multiple components of the TCR and other receptor signaling pathways are alternatively spliced by Hnrpll, including Ptprc, other transmembrane protein tyrosine phosphatases Ptprt and Ptprk; kinases Map3k7 (TAK1), Rps6kb2 (S6-kinase), Pctk1, and Abelson tyrosine kinase; the TCR CD3-zeta subunit Cd247; phospholipase c gamma 1 Plcg1; adaptors Dok2, Sorbs2, Fam48a; and regulators of posttranslational modification by ubiquitin, NEDD8, and ISG15: Ube1l, Ufc1, and Fbxw2. Other Hnrpll-dependent genes are involved in the intracellular calcium response (Cadps2, Kcnip2, Ankra2, Otof, and NFATc1), cell adhesion and cytoskeletal reorganization (Grip1, Cobl, Sorbs2, Spnb2, Fermt3, Mtap4, Myo7a, and Nisch), or vesicle and intracellular transport and targeting (Kif21a, Mtap4, Grip1, Itsn1, Dync1i1, Tpd52l1, Pctk1, Osbpl1a, Nphp4, Myo7a, and Nisch). The shortened longevity of T cells when Hnrpll function is crippled in thu/thu animals is likely to reflect an accumulation of changes, rather than simply being ascribed to an individual hnRNPLL target, such as Ptprc. A similar conclusion has been reached for the shortened lifespan of neurons brought about by mutations in the splicing regulator Smn1 (Zhang et al., 2008), the splicing of which is itself regulated by Hnrpll.
It is remarkable that more than half of the genes with known internal alternatively spliced exons that are found here to be silenced by hnRNPLL (Group A) are well known as a result of studies of alternative splicing in the brain and other neural tissues. Like the immune system, the nervous system must adapt to ever-changing environmental stimuli, and to do this, it must generate an enormous diversity of cell types and plasticity of firing patterns within these cell types (Muotri and Gage, 2006). Unlike B lymphocytes, in which DNA rearrangement and mutation play a central role in diversifying memory B cells from their naive precursors for adaptaion to new environmental agents, neuronal differentiation and memory does not appear to involve DNA recombination or mutation. Instead, RNA rearrangement by alternative splicing has emerged as a major element of neuronal differentiation, diversity, plasticity, and memory (Li et al., 2007; Muotri and Gage, 2006; Ule and Darnell, 2006; Zipursky et al., 2006). The extensive RNA rearrangements revealed here in memory T cells, as well as their overlap with neuronal alternative splicing, suggests that memory T cells diversify their synapse formation and response patterns by co-opting solutions evolved originally for the brain.
EXPERIMENTAL PROCEDURES
Mice and Flow Cytometry
C57BL/6 male mice were mutagenized with four i.p. injections of 85 mg/kg ENU, each 1 week apart (Nelms and Goodnow, 2001). All animal procedures were approved by the Australian National University Animal Experimentation Ethics Committee. Antibody conjugates were from BD PharMingen or Caltag. For CFSE labeling, 5.0 × 107 cells/ml in RPMI 1640 were incubated for 10 min at 37°C with 5 μM CFSE (Molecular Probes) and washed three times in ice-cold RPMI/10% FCS. Data were acquired on a FACS Calibur and analyzed with FlowJo software. For cell transfers, thymocytes from WT CD45.1 and thu/thu (CD45.2) mice were mixed at a 50:50 ratio and CFSE labeled, and 2 × 107 cells were injected into the tail vein of RAG.1−/−° or irradiated C57BL6J mice.
Mapping, cDNA Amplification, and Retroviral Complementation
DNA from (B6 × CBA/H)F2 mice was analyzed by PCR for single-sequence-length-polymorphism and single-nucleotide-polymorphism markers. cDNA from +/+ and thu/thu spleens was PCR amplified and sequenced with primers complementary for each Ensembl transcript in the linked interval. For retroviral complementation, hnRNPLL cDNA was subcloned into the polylinker of bicistronic MSCV-IRES-EGFP vector, retrovirus-containing supernatant collected from transfected Phoenix cells, spinoculated into magnetic-bead-enriched splenic CD4+ T cells that had been activated with anti-CD3 and anti-CD28 for 24 hr, and analyzed by FACS 24 hr after spinoculation. CD45 exon 3–7 cDNA was amplified with 10 uM each of 5′-GGCAAACACCTACACCCA-3′ and 5′-GCTTGCAAGGCCCAGAGTGGATGGTGTAAG-3′ with the Elongase polymerase kit (Invitrogen). Products were electrophoresed in 1.5% agarose gel, stained with EtBr, and imaged with GeneSnap software.
Exon-Array Analysis
CD4-, CD8-, and CD44-stained spleen and lymph-node cells or thymocytes were sorted into ice-cold RPMI 10% FCS on a FACS Vantage DIVA or FACS Aria (Becton Dickinson). For each of the six cell types, multiple independent +/+ or thu/thu donor mice were used for isolating four independent replicate pools of mRNA. For each pool, total RNA was extracted from 2–5 million sorted cells with the RNeasy Plus Mini Kit (QIAGEN), purity was analyzed with an Agilent Bioanalyzer Chip, 1 ug RNA was in vitro transcribed to cRNA, and after the first round of amplification, 10 ug cRNA was used in a second round of amplification. A hybridization cocktail that included 5.5 ug fragmented end-labeled ssDNA was applied to Affymetrix GeneChip Mouse Exon 1.0 ST Array chips. Hybridization was performed with F450-001 fluidics wash and stain script with the use of the Affymetrics Gene Chip Fluidics Station 450. Arrays were scanned with the Affymetrix GCS3000 7G and Gene Chip operating software for the production of CEL files. Array data were deposited in NCBI’s Gene Expression Omnibus (GEO) database (see Accession Numbers section).
Probe-set intensities were calculated from the CEL files of the 48 samples in Partek Genomics Suite 6.3 with the use of the core probe sets defined by Affymetrix. We included 15,788 informative genes in the analysis, defining an informative gene with a unique transcript ID, with mean signals for individual probe sets greater than 3 in the T cell samples analyzed. Alternative splicing was analyzed by two-way (genotype × probe-set ID) ANOVA with the use of Partek algorithms with no normalization. FDRs for the two-way ANOVA comparison of each T cell subset were calculated by the Benjamini and Hochberg step-up method in Partek. Gene views were created with Partek, with the use of known transcripts in the UCSC Mouse Genome database (July 2007 version).
RRM1-Protein Production
RRM1 domains were PCR amplified from plasmids carrying WT and thunder Hnrpll cDNA and cloned into pETMCS III for the encoding of constructs of 111 residues including an N-terminal His tag. Residues 15–106 were identical to residues 7–98 in the NMR structure 1WEX. Residues 1–14 were MHHHHHHMLSTEGG, and residues 107–112 were TDDPSG. The proteins were expressed in E. coli BL21(DE3) by induction with 1 mM IPTG and purified by Ni-NTA affinity chromatography (QIAGEN) with a step gradient of imidazole (25–500 mM) used for elution. 15N/13C double-labeled samples of both proteins were produced in M9 medium containing 1 g/l of 15NH4Cl and 4 g/l of 13C-glucose.
NMR Measurements and Fold Determination
We recorded NMR spectra of 0.8 mM (WT) and 0.4 mM (mutant) solutions in NMR buffer (90% H2O/10% D2O, 50 mM Tris$HCl [pH 7.2], 50 mM NaCl, 1 mM NaN3, 3 mM DTT). All data were acquired at 25°C, with Bruker Avance 800 or 600 MHz NMR spectrometers equipped with cryoprobes. The resonance assignments of the WT domain were performed with HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HNHA, NOESY-15N-HSQC, H(C)CH-TOCSY, and (H)CCH-TOCSY spectra, supplemented by recorded 15N-HSQC spectra of five samples produced cell free with combinatorial 15N-labeling (Wu et al., 2006). The 15N-HSQC spectrum of the mutant was assigned by a NOESY-15N-HSQC spectrum and comparison to the assignments of the WT protein. Interactions with RNA were probed by titration of a 67.5 μM solution of 15N-labeled WT RRM1 domain in NMR buffer with a 1.5 mM solution of the WT oligonucleotide 5′-CCUUACCUGCACGCA-3′ or the mutant 5′-CCUUACCUGGACGUA-3′ (Biospring, Germany) and monitoring of the chemical-shift changes of the protein by 15N-HSQC spectra. The NMR resonance assignments have been deposited at the Biological Magnetic Resonance Bank (BMRB) (see Accession Numbers). The chemical shifts of the Cα, Cβ, C’, HN, Hα, and N spins were used for calculating the fold of the domain (residues 8–112, or 109-213 in the numbering of Figure 3E), with the use of CS-Rosetta with the standard protocol of Shen et al. (2008) without rescoring of the final structures. The lowest all-atom energy structure from 10,000 calculations (Figure S6F) is shown in Figures 4A and 4B. The coordinates can be downloaded from http://rsc.anu.edu.au/~go/coordinates and will be deposited in the Protein Data Bank once the procedure for accepting Rosetta outputs has been established.
Differential Scanning and Isothermal Calorimetry
Thermal stability was probed in NMR buffer with a differential scanning calorimeter (VP-DSC, MicroCal). Thermograms were measured between 10°C and 62°C for the WT RRM1 domain and between 10°C and 45°C for the V136D mutant, with a scan rate of 1.5°C/min and protein concentrations below 200 μM. For isothermal calorimetry measurement of RNA binding, a 5 μM solution of RRM1 domain was titrated with a 60 μM solution of RNA oligomer and the heat absorbed per mole of titrant was monitored. Figures 4E and 4F display the data obtained after subtraction of a control experiment, where the RNA oligomer was titrated into buffer without protein (see Figure S8). A constant of 0.6 kcal/mole (WT RRM1) or 2.5 kcal/mole (mutant RRM1) was subtracted for elimination of a spurious sign change in the data after correction by the data from the control titration into buffer.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the Australian Research Council Federation Fellowships (C.C.G. and G.O.) by an Australian Partnership for Advanced Computing supercomputer grant, by National Health and Medical Research Council Program grant 316956 (C.C.G.), and by National Institutes of Health (NIH) grant RO1-AI52127 (C.C.G.) and NIH contract BAA NIAID-DAIT-07-35 (A.A., E.B., and C.C.G.).
Footnotes
SUPPLEMENTAL DATA Supplemental Data include supplemental notes on group A neural spliced genes, 15 figures, and eight tables and can be found with this paper online at http://www.immunity.com/supplemental/S1074-7613(08)00508-6.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS The GEO accession number for the microarray data reported in this paper is GSE13416.
The BMRB accession number for the NMR resonance assignments is 15820.
REFERENCES
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FH. Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein-RNA complexes of weak affinities. J. Mol. Biol. 2007;367:174–186. doi: 10.1016/j.jmb.2006.12.053. [DOI] [PubMed] [Google Scholar]
- Beverley PC. Kinetics and clonality of immunological memory in humans. Semin. Immunol. 2004;16:315–321. doi: 10.1016/j.smim.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Bruey JM, Bruey-Sedano N, Luciano F, Zhai D, Balpai R, Xu C, Kress CL, Bailly-Maitre B, Li X, Osterman A, et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129:45–56. doi: 10.1016/j.cell.2007.01.045. [DOI] [PubMed] [Google Scholar]
- Burnet F. The clonal selection theory of acquired immunity. Cambridge University Press; London: 1959. [Google Scholar]
- Charych EI, Li R, Serwanski DR, Li X, Miralles CP, Pinal N, De Blas AL. Identification and characterization of two novel splice forms of GRIP1 in the rat brain. J. Neurochem. 2006;97:884–898. doi: 10.1111/j.1471-4159.2006.03795.x. [DOI] [PubMed] [Google Scholar]
- Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino F, Speicher S, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat. Genet. 2003;36:31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- Einstein R, Jordan H, Zhou W, Brenner M, Moses EG, Liggett SB. Alternative splicing of the G protein-coupled receptor superfamily in human airway smooth muscle diversifies the complement of receptors. Proc. Natl. Acad. Sci. USA. 2008;105:5230–5235. doi: 10.1073/pnas.0801319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Weiss A. CD45: A critical regulator of signalling thresholds in immune cells. Annu. Rev. Immunol. 2003;21:107–137. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- Hlaing T, Guo RF, Dilley KA, Loussia JM, Morrish TA, Shi MM, Vincenz C, Ward PA. Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of the mammalian Ced-4 family of apoptosis proteins. J. Biol. Chem. 2001;276:9230–9238. doi: 10.1074/jbc.M009853200. [DOI] [PubMed] [Google Scholar]
- Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Wherry EJ, Reiner SL. Not-so-great expectations: re-assessing the essence of T-cell memory. Immunol. Rev. 2006;211:203–213. doi: 10.1111/j.0105-2896.2006.00396.x. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- Kaisho T, Schwenk F, Rajewsky K. The roles of gamma 1 heavy chain membrane expression and cytoplasmic tail in IgG1 responses. Science. 1997;276:412–415. doi: 10.1126/science.276.5311.412. [DOI] [PubMed] [Google Scholar]
- Kondo M, Osada H, Uchida K, Yanagisawa K, Masuda A, Takagi K, Takahashi T, Takahashi T. Molecular cloning of human TAK1 and its mutational analysis in human lung cancer. Int. J. Cancer. 1998;75:559–563. doi: 10.1002/(sici)1097-0215(19980209)75:4<559::aid-ijc11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Konig H, Ponta H, Herrlich P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998;17:2904–2913. doi: 10.1093/emboj/17.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lepourcelet M, Shivdasani RA. Characterization of a novel mammlian groucho isoform and its role in transcriptional regulation. J. Biol. Chem. 2002;277:47732–47740. doi: 10.1074/jbc.M208154200. [DOI] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Lynch KW, Weiss A. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol. Cell. Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat. Immunol. 2002;3:182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Nelms KA, Goodnow CC. Genome-wide ENU mutagenesis to reveal immune regulators. Immunity. 2001;15:409–418. doi: 10.1016/s1074-7613(01)00199-6. [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Ehrenstein MR, Rada C, Sale J, Batista FD, Williams G, Milstein C. Memory in the B-cell compartment: antibody affinity maturation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:357–360. doi: 10.1098/rstb.2000.0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson JM, Macleod M, Marsden VS, Kappler JW, Marrack P. Not all CD4+ memory T cells are long lived. Immunol. Rev. 2006;211:49–57. doi: 10.1111/j.0105-2896.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- Rothrock C, Cannon B, Hahm B, Lynch KW. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Rothrock CR, House AE, Lynch KW. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein DM, Saito H, Streuli M, Schlossman SF, Morimoto C. The alternative splicing of the CD45 tyrosine phosphatase is controlled by negative regulatory trans-acting splicing factors. J. Biol. Chem. 1992;267:7139–7147. [PubMed] [Google Scholar]
- Safford M, Collins S, Lutz MA, Allen A, Huang CT, Kowalski J, Blackford A, Horton MR, Drake C, Schwartz RH, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 2005;6:472–480. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- Sakurai H, Shigemori N, Hasegawa K, Sugita T. TGF-beta-activated kinase 1 stimulates NF-kappa B activation by an NF-kappa-B inducing kinase-independent mechanism. Biochem. Biophys. Res. Commun. 1998;13:545–549. doi: 10.1006/bbrc.1998.8124. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapy KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson PJ, Monie TP, Szendroi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S, Matthews S. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure. 2004;12:1631–1643. doi: 10.1016/j.str.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Stockinger B, Bourgeois C, Kassiotis G. CD4+ memory T cells: functional differentiation and homeostasis. Immunol. Rev. 2006;211:39–48. doi: 10.1111/j.0105-2896.2006.00381.x. [DOI] [PubMed] [Google Scholar]
- Surh CD, Boyman O, Purton JF, Sprent J. Homeostasis of memory T cells. Immunol. Rev. 2006;211:154–163. doi: 10.1111/j.0105-2896.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- Tong A, Nguyen J, Lynch KW. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J. Biol. Chem. 2005;280:38297–38304. doi: 10.1074/jbc.M508123200. [DOI] [PubMed] [Google Scholar]
- Tsyba L, Skrypkina I, Rynditch A, Nikolaienko O, Ferenets G, Fortna A, Gardiner K. Alternative splicing of mammalian Intersectin 1: domain associations and tissue specificities. Genomics. 2004;84:106–113. doi: 10.1016/j.ygeno.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr. Opin. Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Vinuesa CG, Tangye SG, Moser B, Mackay CR. Follicular B helper T cells in antibody responses and autoimmunity. Nat. Rev. Immunol. 2005;5:853–865. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu. Rev. Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- Wu PS, Ozawa K, Jergic S, Su XC, Dixon NE, Otting G. Amino-acid type identification in 15N-HSQC spectra by combinatorial selective 15N-labelling. J. Biomol. NMR. 2006;34:13–21. doi: 10.1007/s10858-005-5021-9. [DOI] [PubMed] [Google Scholar]
- Yamada K, Andrews C, Chan WM, McKeown CA, Magli A, de Berardinis T, Loewenstein A, Lazar M, O’Keefe M, Letson R, et al. Heterozygous mutations of the kinesin KIF21A in congenital fibrosis of the extraocular muscles type 1 (CFEOM1) Nat. Genet. 2003;35:318–321. doi: 10.1038/ng1261. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky SL, Wojtowicz WM, Hattori D. Got diversity? Wiring the fly brain with Dscam. Trends Biochem. Sci. 2006;31:581–588. doi: 10.1016/j.tibs.2006.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.